Colorectal cancer (CRC) is one of the most common
cancers, causing the third highest cancer-associated morbidity and
the second highest of cancer-associated mortality worldwide
(1). According to a previous
estimation, >1.8 million new cases of CRC and 881,000 cases of
mortality occurred in 2018. According to current trends, in 2030,
the incidence of colon and rectal cancers will increase by 90.0%
and 124.2%, respectively (1). The
5-year survival rate for patients with CRC ranges from 12.5–70.4%
and the prognosis for this disease is poor (2). Due to the fact clear clinical symptoms
are not commonly seen in most CRC cases and diagnostic methods with
high sensitivity and accuracy are lacking, the early detection of
CRC is difficult. Currently, the main treatments for CRC surgery,
radiotherapy and chemotherapy. However, one-third of patients
exhibit recurrent issues following radical surgery and distant
metastasis (3,4). Furthermore, the radiation dose is
difficult to control during radiotherapy and the identification of
the location of lesions in patients with CRC is required for an
accurate dose to be administered (5). Additionally, resistance to chemotherapy
can be easily ignored due to lack of monitoring indicators during
treatment. The aforementioned issues highlight the requirement for
the development of novel therapeutic treatments and the
identification of predictive markers in CRC. The pathogenesis of
CRC has been studied extensively, yet the mechanism governing the
disease remains to be determined and this inhibits the research and
development progress (6).
MicroRNA (miRNA/miR) are small noncoding RNAs that
are 18–24 nucleotides (nt) in length and have been indicated to be
associated with the post-transcriptional regulation of genes in
CRC. Since October 2018, 38,589 miRNAs have been identified
(miRBase Sequence Database). Studies have demonstrated that changes
in the expression of miRNAs are associated with the majority of
cellular processes within tumor pathology (7). miRNAs are regulatory RNA molecules in
the genome and serve an important role in a number of physiological
conditions, including cell differentiation, development, apoptosis,
immune response, hematopoiesis, cell death and proliferation
(8,9). miRNAs function by binding to
complementary sequences on the 3′-untranslated regions, or the open
reading frames, of target genes to regulate gene expression at the
post-transcriptional level, leading to the degradation of target
mRNAs or the inhibition of mRNA translation (10).
Studies have indicated that numerous miRNAs are
aberrantly expressed in CRC and regulate multiple targets (Table I) (11–35). It
has been revealed that all members of the miR-17-92 cluster
participate in the process of CRC. The miR-17-92 family includes
miR-17, miR-18, miR-19a, miR-19b, miR-20a and miR-92a. Currently,
the miR-17-92 cluster is one of the most researched miRNA clusters
and is an example of a multi-sequence miRNA gene that is associated
with the development of a variety of malignancies (36). miR-17 has been indicated to be
overexpressed in CRC and promote the invasion and metastasis of
tumors by targeting PTEN (11). High
levels of miR-18a in CRC have been revealed to attenuate the repair
function of DNA and induce carcinogenesis by targeting ATM to
suppress ATM expression (12). The
overexpression of miR-19a in CRC cells has been indicated to
promote cell invasion and the epithelial-mesenchymal transition
(EMT), and has been revealed to be associated with lymph node
metastasis (37). In a previous
study, miR-92a upregulated β-catenin and vimentin, and
downregulated epithelial-cadherin, targeting PTEN induced EMT in
CRC through the PTEN/phosphoinositide 3 kinase/protein kinase B
pathway (22). Using the Integrating
Gene Expression Omnibus DataSets portal (GEO DataSets) of 3 cohorts
from different regions (GSE41655, GSE35834 and GSE48267),
bioinformatics analysis has identified common changes in 14
differentially expressed miRNAs in CRC, including miR-145, miR-497,
miR-30a, miR-31 and miR-20a. Target prediction of differentially
expressed miRNAs has previously demonstrated that tumorigenesis is
associated with a series of transcription factors. A previous study
indicated that miR-20a may serve an important role in CRC,
according to the network constructed by these microRNAs (38). Furthermore, miR-20a was revealed to
be upregulated in the serum, plasma, tissue and fecal samples of
patients with CRC, and was indicated to serve a role in the
development of CRC and the mechanisms regulating chemotherapy
resistance (39–43). The aforementioned results provide
novel information regarding CRC and can aid in the development of
novel target drugs for use in the treatment of this disease. In the
present review, miR-20a is introduced, the mechanisms of miR-20a in
CRC are elaborated on and then the potential of miR-20a as a
diagnostic indicator of CRC is described. Additionally, the
expression of miR-20a in CRC during chemotherapy is described,
explaining why miR-20a may be used as a biomarker for predicting
drug resistance to CRC. Finally, the research prospects of miR-20a
in CRC are discussed.
Human miR-20a, which is located at 13q31.3, is also
known as MIR20, MIRH1, MIRHG1, MIRN20, miR-20, MIR17HG, MIRN20A,
mir-20a, C13orf25, miRNA20A, hsa-mir-20 or hsa-mir-20a.
miR-20a is an important molecule in a variety of
biological processes and in cancer progression. Previous studies
have shown that the serum exhibits high miR-20a expression and has
been indicated to be associated with poor prognosis in patients
with nasopharyngeal carcinoma and gastric cancer (44,45). A
study has shown that miR-20a can directly target run-related
transcription factor 1 to attenuate cell-death in cervical cancer
cells by preventing natural killer (NK) cells from releasing
interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α), which
promote tumor growth (46). miR-20a
can affect RB1CC1/FIP200 cellular levels, which are associated with
autophagosome formation and the regulation of the autophagy pathway
in breast cancer cells (47).
miR-20a can also inhibit proliferation and induce the apoptosis of
hepatocellular carcinoma cells in vitro by targeting the
anti-apoptotic member myeloid cell leukemia sequence 1 protein of
the Bcl-2 family (48). The
expression of miR-20a in glioma, cervical cancer, gastric cancer,
lung carcinoma, neuroblastoma and prostate cancer, and its
corresponding target genes and their functions, are presented in
Table II (46,49–68).
However, the upregulation or downregulation of miR-20a expression
in a number of tumors is not consistent in numerous previous
studies due to the differences in cell lines and limited sample
sizes (47,69–79). The
current review article outlines the role of miR-20a in CRC and
provides information that can be used in future research on
miR-20a.
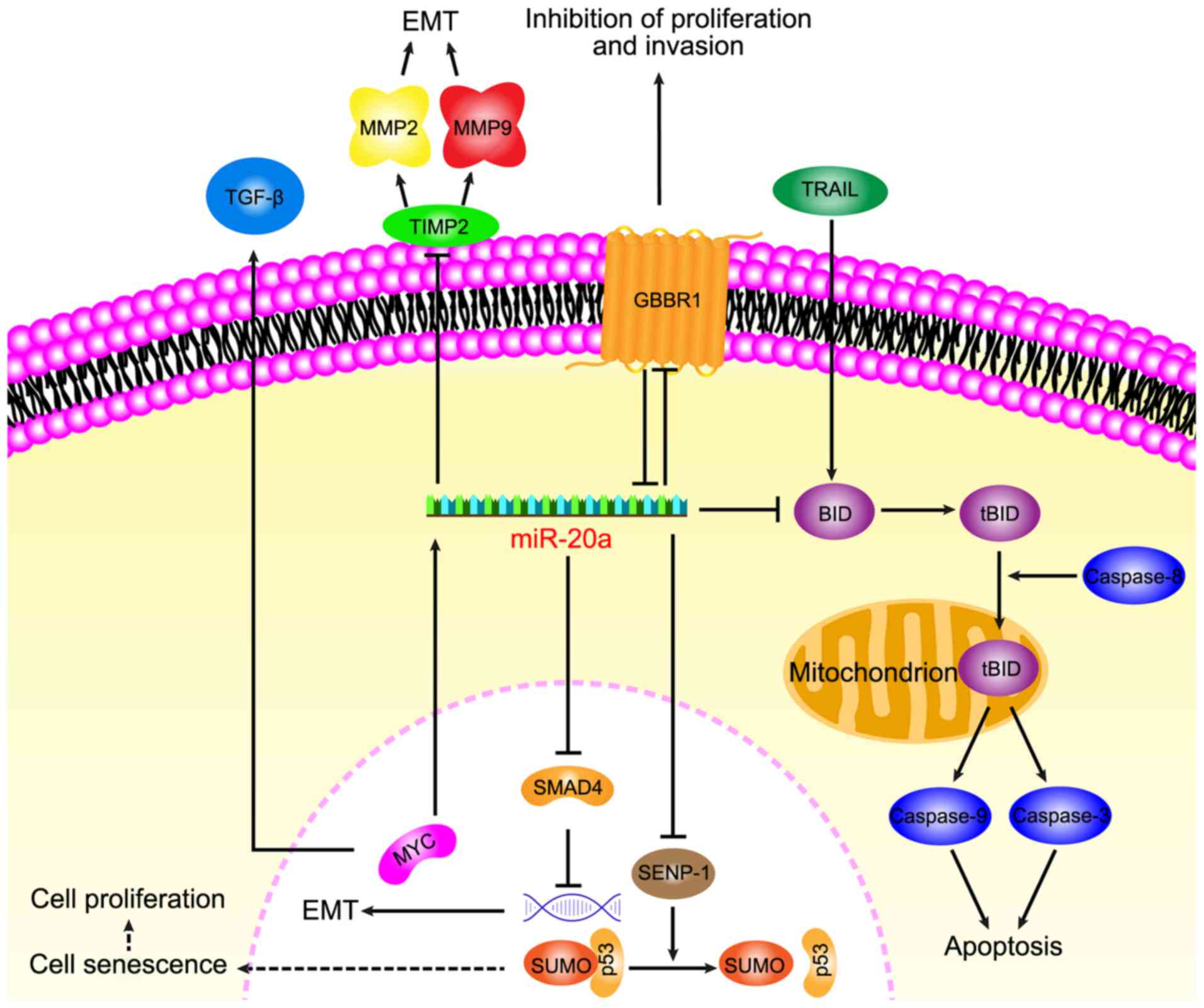
Increasing data has suggested that miR-20a serves a
key role in CRC and its diverse functions in relation to
carcinogenesis of CRC, including anti-apoptotic, EMT, migration,
invasion and senescence, have recently been focused on (Fig. 1).
A previous study demonstrated that TNF-related
apoptosis-inducing ligand (TRAIL) -induced apoptosis is associated
with a higher expression of miR-20a in CRC SW480 cells compared
with the normal colorectal epithelial cell line FHC (15). Mitochondrial apoptosis, that is
induced by a combination of the knockdown miR-20a and TRAIL,
depends on the upregulation of BH3 interacting domain death agonist
(BID). The knockdown of miR-20a was indicated to inhibit the
translocation of truncated BID (tBID) into the mitochondria, which
induced the mitochondrial pathway of apoptosis. TRAIL is a TNF
superfamily member that can selectively induce apoptosis in cancer
cells and induce exogenous apoptosis, however TRAIL is associated
with the activation of caspase-8 (80). BID, which is a pro-apoptotic member
of the Bcl-2 family, serves as a bridge between death receptor
signaling and mitochondrial apoptosis (81). Following the interaction between the
death receptor and the ligand, BID is cleaved by cystatin-8 into an
activated form, tBID (82). The
translocation of tBID to the mitochondria is followed by
mitochondrial outer membrane permeabilization and depolarization
(83), leading to the activation of
caspase-9, caspase-3 and apoptotic effects (80). Furthermore, the activation of
caspase-9 and caspase-3 has been demonstrated to be initiated by
caspase-8 activation. The knockdown of miR-20a in SW480 cells has
been revealed to increase the antitumor effect of TRAIL through the
caspase-8 dependent pathway (15).
miR-20a regulates BID apoptotic genes that are associated with
TRAIL sensitivity of CRC and therefore anti-miR-20a is a promising
target to promote apoptosis.
During EMT, cancer cells lose epithelial
characteristics to form migratory and invasive mesenchymal cell
phenotypes, leading to the consideration that EMT is a prologue to
cancer cell invasion and migration. By promoting EMT, miR-20a aids
the detachment of CRC cells from the tissue parenchyma and their
entry into systemic circulation during cancer metastasis. Studies
have indicated that miR-20a induced EMT by inhibiting mothers
against decapentaplegic homolog 4 (Smad4) expression via direct
targeting of Drosophila Smad4 3′-untranslated region (UTR),
whereas the overexpression of Smad4 abolished EMT that was mediated
by miR-20a overexpression. Furthermore, among a variety of miRNAs,
miR-20a has been indicated to exhibit the highest potential for
binding to 3′-UTR of Smad4, which is a central signal transduction
element of the transforming growth factor (TGF) superfamily and its
mutation leads to a functional transition of TGF-β into a tumor
promoter (84,85). Early carcinogenesis is caused by WNT
signaling activation and TGF-β inactivation. Additionally,
experiments have indicated that miR-20a interfered with the colonic
epithelium homeostasis by disrupting the regulation of Myc/p21 via
TGF-β. Research has also revealed that miR-20a enhances EMT by
modulating the expression of tissue inhibitor of
metalloproteinases-2 (TIMP2) and matrix metalloproteinase 9 (MMP9),
and that the overexpression of miR-20a inhibits the expression of
TIMP2 and induces the expression of MMP2 and MMP 9 to promote EMT
(16).
However, EMT can also lead to decreased cell
adhesion, cytoskeletal dynamics, morphological changes and
increased invasion and migration ability (86). The overexpression of miR-20a has been
revealed to promote migration and invasion in CRC cells and to be
inversely correlated with Smad4 levels (85). Longqiu et al (87) demonstrated that miR-20a was
upregulated in HCT116 and HT-29 cells (CRC lines). Through
specifically binding to the 3′-UTR of γ-amino-butyric acid type B
receptor 1 (GABBR1), miR-20a downregulated the expression of GABBR1
and promoted proliferation and invasion. GABBR1 is a
7-transmembrane receptor and its expression is indicated to be
decreased in CRC tissues (88,89). A
study has shown that the overexpression or activation of GABBR1
inhibited the proliferation and invasion of CRC HCT116 cells,
indicating GABBR1 may be a target for use in CRC treatment. These
results indicated that miR-20a may function through the
downregulation of GABBR1 to promote cell proliferation and
invasion, leading to CRC. However, the validity of this mechanism
requires verification in several other CRC cell lines.
In conclusion, miR-20a has been revealed to
participate in EMT by regulating Smad4 and TIMP2. During the EMT
process, miR-20a can also regulate GABBR1 to increase CRC invasion
and metastasis. Further study of miR-20a in CRC may provide new
research prospects for the mechanisms regulating EMT.
To reduce the high morbidity and mortality observed
in patients with CRC, the identification of novel biomarkers is
urgently required. Observing the expression of miRNAs in CRC and
the development of diagnostic markers has become a priority in
current research (93). Slattery
et al (94) analyzed large
sample data and demonstrated that >86% of the differentially
expressed miRNAs between CRC tissues and normal mucosa are present
in 80% of the population. Tan et al (95) indicated that although the difference
in miRNA levels in ulcerative colitis was not significant in mouse
experiments, in CRC significant changes were observed in miRNAs and
the upregulation of miR-20a in CRC tissues was higher compared with
normal tissues. Analysis of the expression profiles of miRNAs
between tissues from patients with CRC and normal mucosa revealed
that miR-20a was an important miRNA with a high predictive value
for cancer (94,96,97).
Analysis of 20 pairs of CRC tissues and adjacent tissues also
indicated that miR-20a was differentially expressed between tissues
(98). The level of miR-20a is
enhanced in CRC tissues and its expression level has been indicated
to be positively correlated with histological markers (including
Ki-67 and cluster of differentiation 34) (99). High levels of miR-20a predict poor
prognosis in patients with CRC and particularly in patients with
tumor recurrence (85).
Previously, studies have indicated that the level of
miR-20a in the serum and plasma of patients with CRC increased and
was associated with late stage CRC (100,101).
The results of custom miScript miRNA PCR array analysis of 100 CRC
cases, performed by Zekri et al (102), demonstrated that miR-20a was
significantly increased in the serum of female patients with CRC.
The selective detection of small RNA expression in blood samples
from 74 patients with stage II–IV CRC and 32 healthy controls
indicated that miR-20a increased significantly in patients with
CRC, and plasma levels increased in accordance with the extent of
malignancy (103). Liu et al
(98) demonstrated that miR-20a
exhibits a diagnostic value for CRC following the comprehensive
analysis of GEOs and The Cancer Genome Atlas databases. miR-20a in
the serum of patients with CRC is increased compared with healthy
patient serum. Furthermore, reverse transcription quantitative
(RT-q)-PCR analysis of samples from 15 patients with CRC indicated
that the levels of circulating miR-20a in plasma samples were
significantly reduced following surgical resection. Despite these
outcomes not supporting the results of a previous study (104), the staging and grading of tumors
should be taken into consideration to determine whether miR-20 is a
novel marker for CRC. A larger sample size is required for future
research. These aforementioned studies provided new insight for use
in the diagnosis and monitoring of CRC, and indicated that miR-20a
is a promising serum and plasma marker in CRC.
Chemotherapy is the widely used treatment for
patients with CRC, especially in patients with metastatic CRC.
Adjuvant chemoradiotherapy following surgery is also a widely used
treatment, leading to a high cure rate and increased positive
patient prognosis. The association between miRNA and chemotherapy
has been reported in a variety of diseases, for example miR-21
antisense oligonucleotide has been indicated to increase the
sensitivity of cholangiocarcinoma cells to gemcitabine (107), miR-15b and miR-16 have been
revealed to regulate multidrug resistance by targeting Bcl2 in
gastric cancer cells (108) and
miR-451 has been demonstrated to downregulates multidrug resistance
1 and induces breast cancer cell resistance to doxorubicin
(109). Furthermore, miRNAs are
associated with the expression of a number of target genes that are
related to chemosensitivity or the sensitivity of drugs to cancer
cells (110). Therefore,
identifying an indicator to monitor CRC chemotherapy is important
and recent studies have indicated that miR-20a can be used as a
monitoring indicator for chemotherapy (111,112).
Due to the difference in the location of colon
cancer and rectal cancer, the corresponding treatment will often
differ. A study has demonstrated that miR-20a and a number of
miRNAs are reliable prognostic predictors for patients with stage
II colon cancer (101). For
advanced rectal cancer, preoperative chemoradiotherapy (CRT) can
improve overall survival and reduce the local recurrence rate. The
reduced expression of circulating miR-20a is inversely associated
with negative postoperative lymph nodes, which may contribute to
the stratification of the entire mesorectal surgery. Preoperative
CRT blood circulating miR-20a can be used as a marker to indicate
lymph node status of patients with locally advanced rectal cancer
following neoadjuvant CRT. Additionally, the differential
regulation of miR-20a before and after radiotherapy and
chemotherapy, in patients with rectal cancer, may be used as a
marker for monitoring tumor response before and after radiotherapy
and chemotherapy (113–115).
Chemotherapy resistance is the main cause of
treatment failure in patients with cancer, especially in patients
with advanced cancer. miRNAs can serve as potential biomarkers for
predicting therapeutic responses in CRC and are major regulators of
CRC drug resistance (116). miR-20a
is associated with resistance to chemotherapy in a variety of
cancer types, including in cancers of the digestive system
(113). A previous study
demonstrated that the expression of miR-20a in patients exhibiting
chemoresistant CRC was significantly upregulated compared with
patients that exhibited chemosensitive CRC (117). Therefore, miR-20a may be used as a
biomarker for predicting the chemical sensitivity of CRC.
Platinum-based chemotherapy is one of the most
important strategies for the treatment of CRC. Cisplatin is widely
used in the treatment of CRC and exhibits a number of anticancer
activities (118). However, the
chemotherapy resistance of CRC cells is challenge in the treatment
of patients with CRC (119).
Studies have indicated that miR-20a induces cisplatin resistance in
numerous cancers, potentially including in CRC (55,57,111,112).
The miR-20a inhibitor can sensitize cisplatin-induced CRC
cytotoxicity via the reactive oxygen species (ROS) pathway. ROS
induce apoptosis during drug treatments and can also exhibit an
effect on the chemosensitivity of cancer cells (111). In combination therapy with an
miR-20a inhibitor and cisplatin, ROS production is important for
the death of CRC cells. ASK1 is a key mediator in the ROS-dependent
cell death pathway and the knockdown of miR-20a upregulates ASK1
expression and promotes cisplatin-dependent phosphorylation of
ASK1. Treatment to knockdown miR-20a can increase the expression of
ASK1 to enhance ROS-dependent cell death. A combination of
cisplatin and anti-miR-20a have been revealed to significantly
increased JNK activation and subsequent mitochondrial apoptosis in
CRC cells. Therefore, cisplatin-induced mitochondrial apoptosis is
promoted by the ROS/ASK1/JNK pathway through combination with
anti-miR-20a in CRC. Furthermore, the introduction of an miR-20a
inhibitor eliminates cisplatin resistance and enhances the
anti-tumor effect of cisplatin in vivo (112).
Fluorouracil, oxaliplatin and teniposide are
additional chemotherapeutic agents that are used for CRC therapy
and miR-20a has been indicated to regulate the sensitivity of these
drugs. The overexpression of miR-20a exhibits resistance to these
drugs in CRC. Furthermore, miR-20a directly downregulates BNIP2
mRNA and BNIP2 protein levels by binding to BNIP2 3′ UTR to
increase the resistance of CRC cells. BNIP2 is a proapoptosis
factor, which is a member of the BCL-2 protein family (120). Therefore, miR-20a serves a role in
multidrug resistance in CRC cells and may be a therapeutic target
for anti-chemotherapeutic drug resistance in CRC (121).
The incidence of CRC has been increasing in recent
years and many patients are in advanced stages of the disease. The
early detection and treatment of CRC is important for controlling
the progression of the disease. A number of studies have
demonstrated that miRNA can be used as a biomarker for the
diagnosis and prognosis of CRC, and this can provide information on
tumor initiation, development, invasion, metastasis and response to
chemotherapy (16,39,111,112,122,123).
A number of studies have also confirmed that miR-20a is
significantly upregulated and serves an important role in the
development, progression and therapeutic response of CRC, and it
can be used as a biomarker for CRC in tissues, serum and fecal
samples (124). miR-20a has
indicated efficacy in predicting risk of CRC recurrence and is
associated with low survival rates (125,126).
Furthermore, miR-20a has been revealed to promote CRC through an
association with anti-apoptosis mechanisms, EMT, migration,
invasion and senescence, indicating it to be a promising
therapeutic target for use in the treatment of CRC. Chemotherapy is
a widely used treatment in patients with CRC, however resistance
can occur and the mechanisms behind this are undetermined. miR-20a
has been indicated to be an effective indicator in monitoring
response to treatment (112–115,121).
Furthermore, carcinoembryonic antigen (CEA) is a common tumor
marker in gastrointestinal cancer, but its specificity and
sensitivity are low. The combination of miR-20a with CEA and CA19-9
can improve the sensitivity and specificity of CEA, allowing
patients to obtain the most accurate prognosis (127).
Currently, the targeted therapeutic drugs for CRC,
which are used clinically, can be classified into two types. One is
cetuximab and panitumumab, which target epidermal growth factor
receptor and bevacizumab, regorafenib, aflibercept and ramucirumab,
which target vascular endothelial growth factor. Although research
into molecular targeted therapies is increasing, a large number of
genes have been associated with tumor development and it is
difficult to determine the genes that serve a leading role in a
number of different stages in development. Molecular targeted
therapy also exhibits the problem of multi-target combination
therapy. Currently, the types of drugs available for clinical use
are limited but exhibit great development potential. miR-20a serves
an important role in the treatment and drug resistance of CRC. In
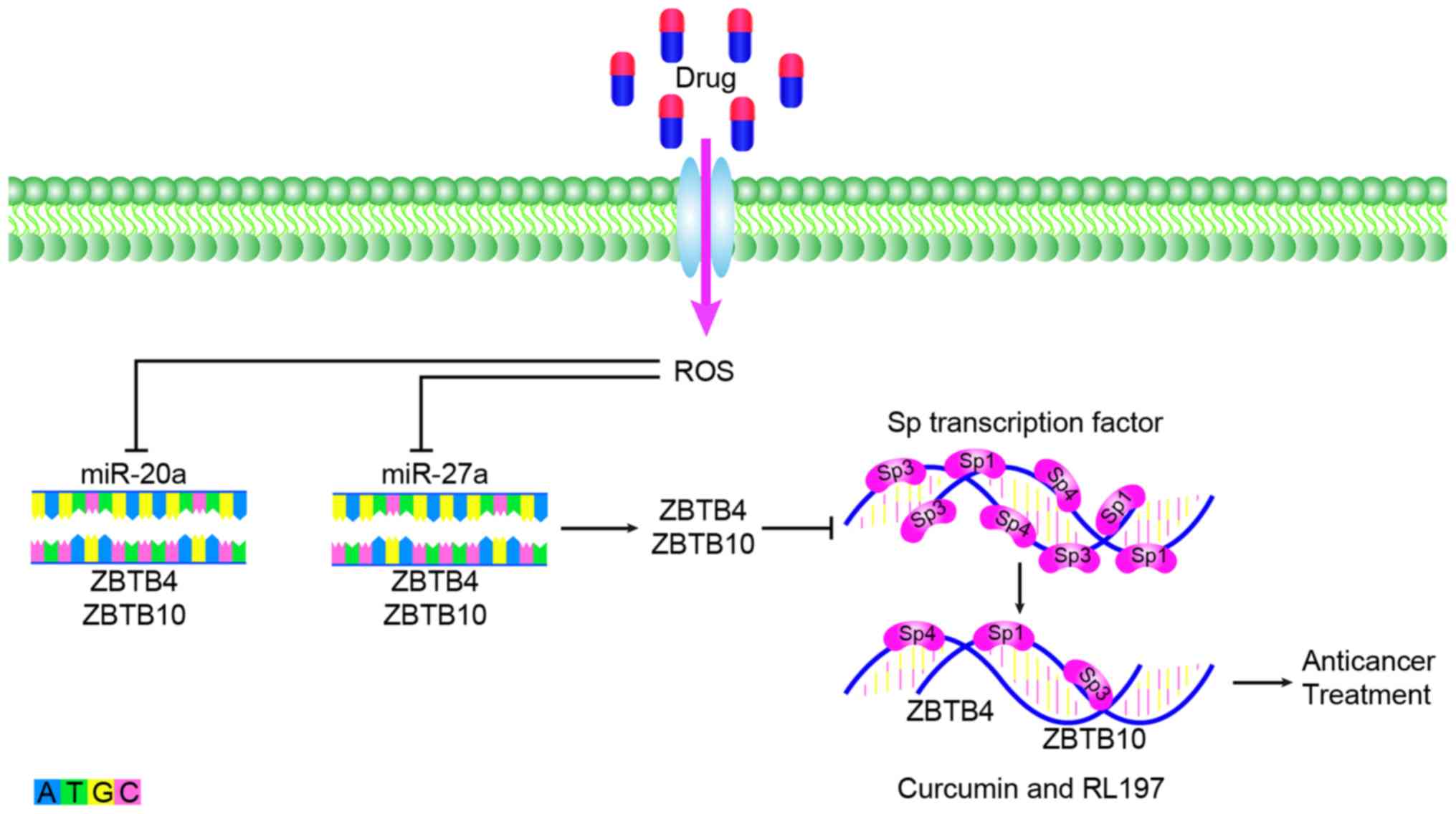
anticancer therapy, miR-20a can inhibit the Sp repressor ZBTB4 and
enhance the expression of Sp transcription factor. Anti-colon
cancer drugs have been revealed to eradicate the miR-20a-ZBTB4 and
miR-ZBTB10 axes within the colon by targeting SP regulators and SP
transcription factors. Subsequently, Sp transcription factors and
Sp-regulated genes competitively bind to GC-rich sites on their
promoter sequences and induce the downregulation of Sp proteins and
Sp-dependent genes. After disruption of the miR-20a-ZBTB4 axis,
ZBTB10 and ZBTB4 serve as transcriptional inhibitors of the SP
regulatory gene, which can reduce Sp1, Sp3, Sp4 and SP regulatory
proteins to inhibit the growth, proliferation, survival,
angiogenesis and metastasis of cancer (Fig. 2). These regulatory proteins include
curcumin and a synthetic analog of curcumin, (3E,5E)-3,5-bis
(2,5-dimethoxybenzylidene)-1-tert-butoxycarbonylpiperidin-4-one
(RL197) (128,129). Sorafenib is a multi-kinase
inhibitor that is currently approved for the treatment of liver,
kidney and thyroid cancer. Studies have indicated that miR-20ain
CRC cell lines exhibit significant changes before and after
treatment with sorafenib and may become an inhibitor of CRC
(130). miR-20a is associated with
chemoresistance, which provides a new direction for studying
anti-transformation resistance (121). Therefore, the development of drugs
for the treatment of CRC based on miR-20a is promising. In CRC,
over-expression and knocking down of miR-20a followed by
high-throughput screening could be applied to discover downstream
target genes. This method can identify molecules that regulate the
expression of miR-20a in CRC cells and thus find drug targeting
molecules for the CRC. Part of mechanisms of CRC indicated that
miR-20a induces high expression of certain proteins and low
expression of some proteins, making high expression protein
inhibitors a promising treatment for CRC. As mentioned above,
miR-20a can inhibit TIMP2 expression, induce expression of both
MMP2 and MMP9 to promote the occurrence of EMT (16). Whether CRC can be treated by
inhibiting MMP2 and MMP9 is also worth discussing. Since miR-20a is
a cancer-promoting factor in CRC, specific inhibition could be
considered as a potential method for the treatment of CRC. However,
specifically targeting miR-20a is technically difficult. Recent
studies have found that circular (circ) RNA can selectively bind to
miRNA as a sponge (131,132). circRNA binding to miRNA can be
predicted by bioinformatics analysis and the screened circRNA can
be used as a therapeutic strategy for miRNA (133). For example, circ_0009910 inhibits
cell proliferation and promotes apoptosis through negative
regulation of miR-20a in acute myeloid leukemia (AML) cells, which
may become a potential therapeutic target for future AML treatment
(134). In addition, long noncoding
(lnc) RNA can interact with miRNA as an competitive endogenous RNA
to participate in the regulation of target gene expression
(135). For example, lncRNA MEG3
can competently bind to miR-23a to regulate the expression of
apoptotic protease activating factor-1 and thus regulate the
progression of laryngeal cancer (136). Therefore, the establishment of
lncRNA-miRNA-mRNA or circRNA-miRNA-RNA pathway in CRC is expected
to provide a new direction for the treatment of CRC in the future
(137).
Therefore, further study into the molecular
mechanisms and functions of the miR-20a network is required for use
in clinical practice. Elucidating the mechanism of elevated miR-20a
expression in cancer, of miR-20a's association with chemotherapy
resistance, the mechanism network of miR-20a promoting cancer,
systemic effects and the side effects of anti-miRNA therapy,
require further investigation. Furthermore, the presence of miRNAs
in serum, plasma and other body fluids (including the saliva, urine
and amniotic fluid) encourages the study of miRNAs, and the
detection of miR-20a in these indicates that this miRNA may be used
as a non-invasive biomarker for early detection and monitoring of
disease progression. An increased amount of data is required to
identify miR-20a in fecal, blood and other screenings, and this
will provide evidence that will increase attention and lead to the
assessment of the potential utility of miRNAs in clinical
practice.
The authors would like to thank Prof. Qian Tao,
Department of Clinical Oncology, the Chinese University of Hong
Kong, for their comments on the manuscript.
The present study was funded by the National Natural
Science Foundation of China (grant no. 81101643), the Natural
Science Foundation of Hunan Province of China (grant no.
2019JJ50521), the Hunan Province Cooperative Innovation Center for
Molecular Target New Drug Study (2014–405) and the Foundation of
the Construct Program of the Key Discipline in Hunan Province of
China (2011–76).
Not applicable.
XZ and YZ designed the present study. ZX and SC
prepared and wrote the manuscript. SF and YL revised the
manuscript. JZ and HL performed a literature search and selected
the studies to be included. All authors approved the final version
of the article.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal Cancer Statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lan YT, Chang SC, Yang SH, Lin CC, Wang
HS, Jiang JK, Chen WS, Lin TC, Chiou SH and Lin JK: Comparison of
clinicopathological characteristics and prognosis between early and
late recurrence after curative surgery for colorectal cancer. Am J
Surg. 207:922–930. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guraya SY: Pattern, stage, and time of
recurrent colorectal cancer after curative surgery. Clin Colorectal
Cancer. 18:e223–e228. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Makishima H, Yasuda S, Isozaki Y, Kasuya
G, Okada N, Miyazaki M, Mohamad O, Matsufuji N, Yamada S, Tsuji H,
et al Liver Cancer Working Group, : Single fraction carbon ion
radiotherapy for colorectal cancer liver metastasis: A dose
escalation study. Cancer Sci. 110:303–309. 2019.PubMed/NCBI
|
|
6
|
Chen H, Xu Z and Liu D: Small non-coding
RNA and colorectal cancer. J Cell Mol Med. 23:3050–3057. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xi XP, Zhuang J, Teng MJ, Xia LJ, Yang MY,
Liu QG and Chen JB: MicroRNA-17 induces epithelial-mesenchymal
transition consistent with the cancer stem cell phenotype by
regulating CYP7B1 expression in colon cancer. Int J Mol Med.
38:499–506. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17-5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu CW, Dong YJ, Liang QY, He XQ, Ng SS,
Chan FK, Sung JJ and Yu J: MicroRNA-18a attenuates DNA damage
repair through suppressing the expression of ataxia telangiectasia
mutated in colorectal cancer. PLoS One. 8:e570362013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui
S, Gu Y, Sun W, You C, Liu Z, et al: miR-19a promotes colorectal
cancer proliferation and migration by targeting TIA1. Mol Cancer.
16:532017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang L, Cai JL, Huang PZ, Kang L, Huang
MJ, Wang L and Wang JP: miR19b-3p promotes the growth and
metastasis of colorectal cancer via directly targeting ITGB8. Am J
Cancer Res. 7:1996–2008. 2017.PubMed/NCBI
|
|
15
|
Huang G, Chen X, Cai Y, Wang X and Xing C:
miR-20a-directed regulation of BID is associated with the TRAIL
sensitivity in colorectal cancer. Oncol Rep. 37:571–578. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu T, Jing C, Shi Y, Miao R, Peng L, Kong
S, Ma Y and Li L: microRNA-20a enhances the
epithelial-to-mesenchymal transition of colorectal cancer cells by
modulating matrix metalloproteinases. Exp Ther Med. 10:683–688.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dalmasso G, Cougnoux A, Delmas J,
Darfeuille-Michaud A and Bonnet R: The bacterial genotoxin
colibactin promotes colon tumor growth by modifying the tumor
microenvironment. Gut Microbes. 5:675–680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin H, Shi X, Zhao Y, Peng M, Kong Y, Qin
D and Lv X: MicroRNA-30a mediates cell migration and invasion by
targeting metadherin in colorectal cancer. Technol Cancer Res
Treat. 17:15330338187581082018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J,
Zhang CY, Chen J and Zhang J: MicroRNA-31 activates the RAS pathway
and functions as an oncogenic microRNA in human colorectal cancer
by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol
Chem. 288:9508–9518. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Akao Y, Noguchi S, Iio A, Kojima K, Takagi
T and Naoe T: Dysregulation of microRNA-34a expression causes
drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer
Lett. 300:197–204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang G, Zhou H, Xiao H, Liu Z, Tian H and
Zhou T: MicroRNA-92a functions as an oncogene in colorectal cancer
by targeting PTEN. Dig Dis Sci. 59:98–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cappuzzo F, Sacconi A, Landi L, Ludovini
V, Biagioni F, D'Incecco A, Capodanno A, Salvini J, Corgna E,
Cupini S, et al: MicroRNA signature in metastatic colorectal cancer
patients treated with anti-EGFR monoclonal antibodies. Clin
Colorectal Cancer. 13:37–45.e4. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lian B, Yang D, Liu Y, Shi G, Li J, Yan X,
Jin K, Liu X, Zhao J, Shang W, et al: miR-128 Targets the
SIRT1/ROS/DR5 Pathway to Sensitize Colorectal Cancer to
TRAIL-Induced Apoptosis. Cell Physiol Biochem. 49:2151–2162. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Salem SM, Hamed AR, Fayez AG and Nour
Eldeen G: Non-target genes regulate miRNAs-mediated migration
steering of colorectal carcinoma. Pathol Oncol Res. 25:559–566.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu K, Liu X, Mao X, Xue L, Wang R, Chen L
and Chu X: MicroRNA-149 suppresses colorectal cancer cell migration
and invasion by directly targeting forkhead box transcription
factor FOXM1. Cell Physiol Biochem. 35:499–515. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li LX, Lam IH, Liang FF, Yi SP, Ye LF,
Wang JT, Guo WW and Xu M: MiR-198 affects the proliferation and
apoptosis of colorectal cancer through regulation of
ADAM28/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci.
23:1487–1493. 2019.PubMed/NCBI
|
|
28
|
Zhang Z, Zhong X, Xiao Y and Chen C:
MicroRNA-296 inhibits colorectal cancer cell growth and enhances
apoptosis by targeting ARRB1-mediated AKT activation. Oncol Rep.
41:619–629. 2019.PubMed/NCBI
|
|
29
|
Yan F, Tu Z, Duan L, Wang D and Lin F:
MicroRNA-383 suppresses cell proliferation and invasion in
colorectal cancer by directly targeting paired box 6. Mol Med Rep.
17:6893–6901. 2018.PubMed/NCBI
|
|
30
|
Wang W, He Y, Rui J and Xu MQ: miR-410
acts as an oncogene in colorectal cancer cells by targeting
dickkopf-related protein 1 via the Wnt/β-catenin signaling pathway.
Oncol Lett. 17:807–814. 2019.PubMed/NCBI
|
|
31
|
Li T, Jian X, He H, Lai Q, Li X, Deng D,
Liu T, Zhu J, Jiao H, Ye Y, et al: MiR-452 promotes an aggressive
colorectal cancer phenotype by regulating a Wnt/β-catenin positive
feedback loop. J Exp Clin Cancer Res. 37:2382018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li KP, Fang YP, Liao JQ, Duan JD, Feng LG,
Luo XZ and Liang ZJ: Upregulation of miR-598 promotes cell
proliferation and cell cycle progression in human colorectal
carcinoma by suppressing INPP5E expression. Mol Med Rep.
17:2991–2997. 2018.PubMed/NCBI
|
|
33
|
Wang L, Xu M, Lu P and Zhou F:
microRNA-769 is downregulated in colorectal cancer and inhibits
cancer progression by directly targeting cyclin-dependent kinase 1.
OncoTargets Ther. 11:9013–9025. 2018. View Article : Google Scholar
|
|
34
|
Yang D, Li R, Xia J, Li W and Zhou H:
miR-3666 suppresses cellular proliferation and invasion in
colorectal cancer by targeting SATB2. Mol Med Rep. 18:4847–4854.
2018.PubMed/NCBI
|
|
35
|
Liu H, Li D, Fang H and Ning J:
Species-specific function of microRNA-7702 in human colorectal
cancer cells via targeting TADA1. Am J Transl Res. 10:2579–2589.
2018.PubMed/NCBI
|
|
36
|
Mogilyansky E and Rigoutsos I: The
miR-17/92 cluster: A comprehensive update on its genomics,
genetics, functions and increasingly important and numerous roles
in health and disease. Cell Death Differ. 20:1603–1614. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang L, Wang X, Wen C, Yang X, Song M,
Chen J, Wang C, Zhang B, Wang L, Iwamoto A, et al: Hsa-miR-19a is
associated with lymph metastasis and mediates the TNF-α induced
epithelial-to-mesenchymal transition in colorectal cancer. Sci Rep.
5:133502015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hao S, Huo S, Du Z, Yang Q, Ren M, Liu S,
Liu T and Zhang G: MicroRNA-related transcription factor regulatory
networks in human colorectal cancer. Medicine (Baltimore).
98:e151582019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shirafkan N, Mansoori B, Mohammadi A,
Shomali N, Ghasbi M and Baradaran B: MicroRNAs as novel biomarkers
for colorectal cancer: New outlooks. Biomed Pharmacother.
97:1319–1330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo Y, Bao Y and Yang W: Regulatory miRNAs
in colorectal carcinogenesis and metastasis. Int J Mol Sci.
18:182017. View Article : Google Scholar
|
|
41
|
Motoyama K, Inoue H, Takatsuno Y, Tanaka
F, Mimori K, Uetake H, Sugihara K and Mori M: Over- and
under-expressed microRNAs in human colorectal cancer. Int J Oncol.
34:1069–1075. 2009.PubMed/NCBI
|
|
42
|
Yau TO, Wu CW, Tang CM, Chen Y, Fang J,
Dong Y, Liang Q, Ng SS, Chan FK, Sung JJ, et al: MicroRNA-20a in
human faeces as a non-invasive biomarker for colorectal cancer.
Oncotarget. 7:1559–1568. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Moradi-Marjaneh R, Hassanian SM, Mehramiz
M, Rezayi M, Ferns GA, Khazaei M and Avan A: Reactive oxygen
species in colorectal cancer: The therapeutic impact and its
potential roles in tumor progression via perturbation of cellular
and physiological dysregulated pathways. J Cell Physiol.
234:10072–10079. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zeng X, Xiang J, Wu M, Xiong W, Tang H,
Deng M, Li X, Liao Q, Su B, Luo Z, et al: Circulating miR-17,
miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers
in nasopharyngeal carcinoma. PLoS One. 7:e463672012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li
L, Xu H, Zhong J and Zeng X: Serum miR-20a is a promising biomarker
for gastric cancer. Biomed Rep. 6:429–434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhu SY, Wu QY, Zhang CX, Wang Q, Ling J,
Huang XT, Sun X, Yuan M, Wu D and Yin HF: miR-20a inhibits the
killing effect of natural killer cells to cervical cancer cells by
downregulating RUNX1. Biochem Biophys Res Commun. 505:309–316.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li S, Qiang Q, Shan H, Shi M, Gan G, Ma F
and Chen B: miR-20a and miR-20b negatively regulate autophagy by
targeting RB1CC1/FIP200 in breast cancer cells. Life Sci.
147:143–152. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX
and Zhong L: Decrease expression of microRNA-20a promotes cancer
cell proliferation and predicts poor survival of hepatocellular
carcinoma. J Exp Clin Cancer Res. 32:212013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liao C, Chen W and Wang J: MicroRNA-20a
regulates glioma cell proliferation, invasion, and apoptosis by
targeting CUGBP elav-like family member 2. World Neurosurg.
121:e519–e527. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu X: Up-regulation of miR-20a by HPV16
E6 exerts growth-promoting effects by targeting PDCD6 in cervical
carcinoma cells. Biomed Pharmacother. 102:996–1002. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhou L, Li X, Zhou F, Jin Z, Chen D, Wang
P, Zhang S, Zhuge Y, Shang Y and Zou X: Downregulation of
leucine-rich repeats and immunoglobulin-like domains 1 by
microRNA-20a modulates gastric cancer multidrug resistance. Cancer
Sci. 109:1044–1054. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wei L and Ran F: MicroRNA-20a promotes
proliferation and invasion by directly targeting early growth
response 2 in non-small cell lung carcinoma. Oncol Lett.
15:271–277. 2018.PubMed/NCBI
|
|
53
|
Yu Y, Zhang J, Jin Y, Yang Y, Shi J, Chen
F, Han S, Chu P, Lu J, Wang H, et al: MiR-20a-5p suppresses tumor
proliferation by targeting autophagy-related gene 7 in
neuroblastoma. Cancer Cell Int. 18:52018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhao F, Pu Y, Qian L, Zang C, Tao Z and
Gao J: MiR-20a-5p promotes radio-resistance by targeting NPAS2 in
nasopharyngeal cancer cells. Oncotarget. 8:105873–105881. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xiong Y, Sun F, Dong P, Watari H, Yue J,
Yu MF, Lan CY, Wang Y and Ma ZB: iASPP induces EMT and cisplatin
resistance in human cervical cancer through miR-20a-FBXL5/BTG3
signaling. J Exp Clin Cancer Res. 36:482017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang D, Bian G, Pan Y, Han X, Sun Y, Wang
Y, Shen G, Cheng M, Fang X and Hu S: MiR-20a-5p promotes
radio-resistance by targeting Rab27B in nasopharyngeal cancer
cells. Cancer Cell Int. 17:322017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhu M, Zhou X, Du Y, Huang Z, Zhu J, Xu J,
Cheng G, Shu Y, Liu P, Zhu W, et al: miR-20a induces cisplatin
resistance of a human gastric cancer cell line via targeting CYLD.
Mol Med Rep. 14:1742–1750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dhar S, Kumar A, Rimando AM, Zhang X and
Levenson AS: Resveratrol and pterostilbene epigenetically restore
PTEN expression by targeting oncomiRs of the miR-17 family in
prostate cancer. Oncotarget. 6:27214–27226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Du Y, Zhu M, Zhou X, Huang Z, Zhu J, Xu J,
Cheng G, Shu Y, Liu P, Zhu W, et al: miR-20a enhances cisplatin
resistance of human gastric cancer cell line by targeting NFKBIB.
Tumour Biol. 37:1261–1269. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wei J, Qi X, Zhan Q, Zhou D, Yan Q, Wang
Y, Mo L, Wan Y, Xie D, Xie J, et al: miR-20a mediates
temozolomide-resistance in glioblastoma cells via negatively
regulating LRIG1 expression. Biomed Pharmacother. 71:112–118. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhao S, Yao D, Chen J, Ding N and Ren F:
MiR-20a promotes cervical cancer proliferation and metastasis in
vitro and in vivo. PLoS One. 10:e01209052015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang Y, Han T, Wei G and Wang Y:
Inhibition of microRNA-17/20a suppresses cell proliferation in
gastric cancer by modulating UBE2C expression. Oncol Rep.
33:2529–2536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang X, Kong Y, Xu X, Xing H, Zhang Y,
Han F, Li W, Yang Q, Zeng J, Jia J, et al: F-box protein FBXO31 is
down-regulated in gastric cancer and negatively regulated by miR-17
and miR-20a. Oncotarget. 5:6178–6190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou J, Liu R, Luo C, Zhou X, Xia K, Chen
X, Zhou M, Zou Q, Cao P and Cao K: MiR-20a inhibits cutaneous
squamous cell carcinoma metastasis and proliferation by directly
targeting LIMK1. Cancer Biol Ther. 15:1340–1349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xiong Y, Zhang L and Kebebew E: MiR-20a is
upregulated in anaplastic thyroid cancer and targets LIMK1. PLoS
One. 9:e961032014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xie J, Liu M, Li Y, Nie Y, Mi Q and Zhao
S: Ovarian tumor-associated microRNA-20a decreases natural killer
cell cytotoxicity by downregulating MICA/B expression. Cell Mol
Immunol. 11:495–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Qiang XF, Zhang ZW, Liu Q, Sun N, Pan LL,
Shen J, Li T, Yun C, Li H and Shi LH: miR-20a promotes prostate
cancer invasion and migration through targeting ABL2. J Cell
Biochem. 115:1269–1276. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chang Y, Liu C, Yang J, Liu G, Feng F,
Tang J, Hu L, Li L, Jiang F, Chen C, et al: MiR-20a triggers
metastasis of gallbladder carcinoma. J Hepatol. 59:518–527. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bai X, Han G, Liu Y, Jiang H and He Q:
MiRNA-20a-5p promotes the growth of triple-negative breast cancer
cells through targeting RUNX3. Biomed Pharmacother. 103:1482–1489.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhao W, Geng D, Li S, Chen Z and Sun M:
LncRNA HOTAIR influences cell growth, migration, invasion, and
apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer
Med. 7:842–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yuan G, Zhao Y, Wu D, Gao C and Jiao Z:
miRNA-20a upregulates TAK1 and increases proliferation in
osteosarcoma cells. Future Oncol. 14:461–469. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Si W, Shen J, Du C, Chen D, Gu X, Li C,
Yao M, Pan J, Cheng J, Jiang D, et al: A miR-20a/MAPK1/c-Myc
regulatory feedback loop regulates breast carcinogenesis and
chemoresistance. Cell Death Differ. 25:406–420. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhao F, Pu Y, Cui M, Wang H and Cai S:
MiR-20a-5p represses the multi-drug resistance of osteosarcoma by
targeting the SDC2 gene. Cancer Cell Int. 17:1002017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu L, He J, Wei X, Wan G, Lao Y, Xu W, Li
Z, Hu H, Hu Z, Luo X, et al: MicroRNA-20a-mediated loss of
autophagy contributes to breast tumorigenesis by promoting genomic
damage and instability. Oncogene. 36:5874–5884. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Karimkhanloo H, Mohammadi-Yeganeh S,
Ahsani Z and Paryan M: Bioinformatics prediction and experimental
validation of microRNA-20a targeting Cyclin D1 in hepatocellular
carcinoma. Tumour Biol. 39:10104283176983612017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Shen J, Pan J, Du C, Si W, Yao M, Xu L,
Zheng H, Xu M, Chen D, Wang S, et al: Silencing NKG2D
ligand-targeting miRNAs enhances natural killer cell-mediated
cytotoxicity in breast cancer. Cell Death Dis. 8:e27402017.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen Y, Wang X, Cheng J, Wang Z, Jiang T,
Hou N, Liu N, Song T and Huang C: MicroRNA-20a-5p targets RUNX3 to
regulate proliferation and migration of human hepatocellular cancer
cells. Oncol Rep. 36:3379–3386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Pu Y, Yi Q, Zhao F, Wang H, Cai W and Cai
S: MiR-20a-5p represses multi-drug resistance in osteosarcoma by
targeting the KIF26B gene. Cancer Cell Int. 16:642016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang Y, Zheng L, Ding Y, Li Q, Wang R,
Liu T, Sun Q, Yang H, Peng S, Wang W, et al: MiR-20a Induces Cell
Radioresistance by Activating the PTEN/PI3K/Akt Signaling Pathway
in Hepatocellular Carcinoma. Int J Radiat Oncol Biol Phys.
92:1132–1140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Laussmann MA, Passante E, Hellwig CT,
Tomiczek B, Flanagan L, Prehn JH, Huber HJ and Rehm M: Proteasome
inhibition can impair caspase-8 activation upon submaximal
stimulation of apoptotic tumor necrosis factor-related apoptosis
inducing ligand (TRAIL) signaling. J Biol Chem. 287:14402–14411.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li H, Zhu H, Xu CJ and Yuan J: Cleavage of
BID by caspase 8 mediates the mitochondrial damage in the Fas
pathway of apoptosis. Cell. 94:491–501. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Orzechowska EJ, Girstun A, Staron K and
Trzcinska-Danielewicz J: Synergy of BID with doxorubicin in the
killing of cancer cells. Oncol Rep. 33:2143–2150. 2015.PubMed/NCBI
|
|
83
|
Eskes R, Desagher S, Antonsson B and
Martinou JC: Bid induces the oligomerization and insertion of Bax
into the outer mitochondrial membrane. Mol Cell Biol. 20:929–935.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang GJ, Li Y, Zhou H, Xiao HX and Zhou
T: miR-20a is an independent prognostic factor in colorectal cancer
and is involved in cell metastasis. Mol Med Rep. 10:283–291. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cheng D, Zhao S, Tang H, Zhang D, Sun H,
Yu F, Jiang W, Yue B, Wang J, Zhang M, et al: MicroRNA-20a-5p
promotes colorectal cancer invasion and metastasis by
downregulating Smad4. Oncotarget. 7:45199–45213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Longqiu Y, Pengcheng L, Xuejie F and Peng
Z: A miRNAs panel promotes the proliferation and invasion of
colorectal cancer cells by targeting GABBR1. Cancer Med.
5:2022–2031. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Jiang X, Su L, Zhang Q, He C, Zhang Z, Yi
P and Liu J: GABAB receptor complex as a potential target for tumor
therapy. J Histochem Cytochem. 60:269–279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Peters HC, Kämmer G, Volz A, Kaupmann K,
Ziegler A, Bettler B, Epplen JT, Sander T and Riess O: Mapping,
genomic structure, and polymorphisms of the human GABABR1 receptor
gene: Evaluation of its involvement in idiopathic generalized
epilepsy. Neurogenetics. 2:47–54. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yates KE, Korbel GA, Shtutman M, Roninson
IB and DiMaio D: Repression of the SUMO-specific protease Senp1
induces p53-dependent premature senescence in normal human
fibroblasts. Aging Cell. 7:609–621. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Melchior F and Hengst L: SUMO-1 and p53.
Cell Cycle. 1:245–249. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Agostini M, Pucciarelli S, Calore F, Bedin
C, Enzo M and Nitti D: miRNAs in colon and rectal cancer: A
consensus for their true clinical value. Clin Chim Acta.
411:1181–1186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Slattery ML, Herrick JS, Pellatt DF,
Stevens JR, Mullany LE, Wolff E, Hoffman MD, Samowitz WS and Wolff
RK: MicroRNA profiles in colorectal carcinomas, adenomas and normal
colonic mucosa: Variations in miRNA expression and disease
progression. Carcinogenesis. 37:245–261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tan YG, Zhang YF, Guo CJ, Yang M and Chen
MY: Screening of differentially expressed microRNA in ulcerative
colitis related colorectal cancer. Asian Pac J Trop Med. 6:972–976.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Bovell L, Shanmugam C, Katkoori VR, Zhang
B, Vogtmann E, Grizzle WE and Manne U: miRNAs are stable in
colorectal cancer archival tissue blocks. Front Biosci (Elite Ed).
4:1937–1940. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
97
|
Pellatt DF, Stevens JR, Wolff RK, Mullany
LE, Herrick JS, Samowitz W and Slattery ML: Expression profiles of
miRNA subsets distinguish human colorectal carcinoma and normal
colonic mucosa. Clin Transl Gastroenterol. 7:e1522016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu X, Xu T, Hu X, Chen X, Zeng K, Sun L
and Wang S: Elevated circulating miR-182 acts as a diagnostic
biomarker for early colorectal cancer. Cancer Manag Res.
10:857–865. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Emami SS, Akbari A, Zare AA, Agah S,
Masoodi M, Talebi A, Minaeian S, Fattahi A and Moghadamnia F:
MicroRNA expression levels and histopathological features of
colorectal cancer. J Gastrointest Cancer. 50:276–284. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Brunet Vega A, Pericay C, Moya I, Ferrer
A, Dotor E, Pisa A, Casalots À, Serra-Aracil X, Oliva JC, Ruiz A,
et al: microRNA expression profile in stage III colorectal cancer:
Circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep.
30:320–326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang JX, Song W, Chen ZH, Wei JH, Liao
YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, et al: Prognostic and
predictive value of a microRNA signature in stage II colon cancer:
A microRNA expression analysis. Lancet Oncol. 14:1295–1306. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zekri AR, Youssef AS, Lotfy MM, Gabr R,
Ahmed OS, Nassar A, Hussein N, Omran D, Medhat E, Eid S, et al:
Circulating serum miRNAs as diagnostic markers for colorectal
cancer. PLoS One. 11:e01541302016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Eslamizadeh S, Heidari M, Agah S,
Faghihloo E, Ghazi H, Mirzaei A and Akbari A: The role of microRNA
signature as diagnostic biomarkers in different clinical stages of
colorectal cancer. Cell J. 20:220–230. 2018.PubMed/NCBI
|
|
104
|
Yang Q, Wang S, Huang J, Xia C, Jin H and
Fan Y: Serum miR-20a and miR-486 are potential biomarkers for
discriminating colorectal neoplasia: A pilot study. J Cancer Res
Ther. 14:1572–1577. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Yamazaki N, Koga Y, Yamamoto S, Kakugawa
Y, Otake Y, Hayashi R, Saito N and Matsumura Y: Application of the
fecal microRNA test to the residuum from the fecal occult blood
test. Jpn J Clin Oncol. 43:726–733. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Rotelli MT, Di Lena M, Cavallini A,
Lippolis C, Bonfrate L, Chetta N, Portincasa P and Altomare DF:
Fecal microRNA profile in patients with colorectal carcinoma before
and after curative surgery. Int J Colorectal Dis. 30:891–898. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Meng F, Henson R, Lang M, Wehbe H,
Maheshwari S, Mendell JT, Jiang J, Schmittgen TD and Patel T:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: miR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Kovalchuk O, Filkowski J, Meservy J,
Ilnytskyy Y, Tryndyak VP, Chekhun VF and Pogribny IP: Involvement
of microRNA-451 in resistance of the MCF-7 breast cancer cells to
chemotherapeutic drug doxorubicin. Mol Cancer Ther. 7:2152–2159.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ma J, Dong C and Ji C: MicroRNA and drug
resistance. Cancer Gene Ther. 17:523–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Li X, Wang H, Wang J, Chen Y, Yin X, Shi
G, Li H, Hu Z and Liang X: Emodin enhances cisplatin-induced
cytotoxicity in human bladder cancer cells through ROS elevation
and MRP1 downregulation. BMC Cancer. 16:5782016. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhang L, He L, Zhang H and Chen Y:
Knockdown of miR-20a enhances sensitivity of colorectal cancer
cells to cisplatin by increasing ASK1 expression. Cell Physiol
Biochem. 47:1432–1441. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Molinari C, Salvi S, Foca F, Teodorani N,
Saragoni L, Puccetti M, Passardi A, Tamberi S, Avanzolini A, Lucci
E, et al: miR-17-92a-1 cluster host gene (MIR17HG) evaluation and
response to neoadjuvant chemoradiotherapy in rectal cancer. Onco
Targets Ther. 9:2735–2742. 2016.PubMed/NCBI
|
|
114
|
Azizian A, Kramer F, Jo P, Wolff HA,
Beißbarth T, Skarupke R, Bernhardt M, Grade M, Ghadimi BM and
Gaedcke J: Preoperative prediction of lymph node status by
circulating mir-18b and mir-20a during chemoradiotherapy in
patients with rectal cancer. World J Surg. 39:2329–2335. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Jo P, Azizian A, Salendo J, Kramer F,
Bernhardt M, Wolff HA, Gruber J, Grade M, Beißbarth T, Ghadimi BM,
et al: Changes of microrna levels in plasma of patients with rectal
cancer during chemoradiotherapy. Int J Mol Sci. 18:182017.
View Article : Google Scholar
|
|
116
|
Okugawa Y, Toiyama Y and Goel A: An update
on microRNAs as colorectal cancer biomarkers: Where are we and
what's next? Expert Rev Mol Diagn. 14:999–1021. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhang J, Zhang K, Bi M, Jiao X, Zhang D
and Dong Q: Circulating microRNA expressions in colorectal cancer
as predictors of response to chemotherapy. Anticancer Drugs.
25:346–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Xie T, Li Y, Li SL and Luo HF:
Astragaloside IV enhances cisplatin chemosensitivity in human
colorectal cancer via regulating NOTCH3. Oncol Res. 24:447–453.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Ma MZ, Chen G, Wang P, Lu WH, Zhu CF, Song
M, Yang J, Wen S, Xu RH, Hu Y, et al: Xc- inhibitor sulfasalazine
sensitizes colorectal cancer to cisplatin by a GSH-dependent
mechanism. Cancer Lett. 368:88–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Aouacheria A, Brunet F and Gouy M:
Phylogenomics of life-or-death switches in multicellular animals:
Bcl-2, BH3-Only, and BNip families of apoptotic regulators. Mol
Biol Evol. 22:2395–2416. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Chai H, Liu M, Tian R, Li X and Tang H:
miR-20a targets BNIP2 and contributes chemotherapeutic resistance
in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta
Biochim Biophys Sin (Shanghai). 43:217–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ashrafizadeh M, Ezzati H, Ahmadi Z,
Farkhondeh T and Samarghandian S: Anti-tumor activity of propofol:
A focus on microRNAs. Curr Cancer Drug Targets. 19:2019, https://doi.org/10.2174/1568009619666191023100046
View Article : Google Scholar
|
|
123
|
Ji R, Zhang X, Gu H, Ma J, Wen X, Zhou J,
Qian H, Xu W, Qian J and Lin J: miR-374a-5p: A New Target for
Diagnosis and Drug Resistance Therapy in Gastric Cancer. Mol Ther
Nucleic Acids. 18:320–331. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Luo X, Burwinkel B, Tao S and Brenner H:
MicroRNA signatures: Novel biomarker for colorectal cancer? Cancer
Epidemiol Biomarkers Prev. 20:1272–1286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Caritg O, Navarro A, Moreno I,
Martínez-Rodenas F, Cordeiro A, Muñoz C, Ruiz-Martinez M,
Santasusagna S, Castellano JJ and Monzó M: Identifying high-risk
stage II colon cancer patients: A three-microRNA-based score as a
prognostic biomarker. Clin Colorectal Cancer. 15:e175–e182. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Pesta M, Kucera R, Topolcan O, Karlikova
M, Houfkova K, Polivka J, Macanova T, Machova I, Slouka D and Kulda
V: Plasma microRNA levels combined with CEA and CA19-9 in the
follow-up of colorectal cancer patients. Cancers (Basel).
11:112019. View Article : Google Scholar
|
|
128
|
Gandhy SU, Kim K, Larsen L, Rosengren RJ
and Safe S: Curcumin and synthetic analogs induce reactive oxygen
species and decreases specificity protein (Sp) transcription
factors by targeting microRNAs. BMC Cancer. 12:5642012. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Choi JB, Kim JH, Lee H, Pak JN, Shim BS
and Kim SH: Reactive Oxygen Species and p53 Mediated Activation of
p38 and Caspases is Critically Involved in Kaempferol Induced
Apoptosis in Colorectal Cancer Cells. J Agric Food Chem.
66:9960–9967. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Pehserl AM, Ress AL, Stanzer S, Resel M,
Karbiener M, Stadelmeyer E, Stiegelbauer V, Gerger A, Mayr C,
Scheideler M, et al: Comprehensive Analysis of miRNome Alterations
in Response to Sorafenib Treatment in Colorectal Cancer Cells. Int
J Mol Sci. 17:172016. View Article : Google Scholar
|
|
131
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
2019:https://doi.org/10.1007/s00018-019-03345-5
|
|
132
|
Su Q and Lv X: Revealing new landscape of
cardiovascular disease through circular RNA-miRNA-mRNA axis.
Genomics. S0888-7543(19)30565-8. 2019. View Article : Google Scholar
|
|
133
|
Xiu Y, Jiang G, Zhou S, Diao J, Liu H, Su
B and Li C: Identification of potential immune-related
circRNA-miRNA-mRNA regulatory network in intestine of paralichthys
olivaceus during Edwardsiella tarda infection. Front Genet.
10:7312019. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L
and Ming Z: Silencing of circ_0009910 inhibits acute myeloid
leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol
Dis. 75:41–47. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang X, Wu N, Wang J and Li Z: LncRNA
MEG3 inhibits cell proliferation and induces apoptosis in laryngeal
cancer via miR-23a/APAF-1 axis. J Cell Mol Med. 23:6708–6719. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Huang QR and Pan XB: Prognostic lncRNAs,
miRNAs, and mRNAs form a competing endogenous RNA network in colon
cancer. Front Oncol. 9:7122019. View Article : Google Scholar : PubMed/NCBI
|