Introduction
Renal cell carcinoma (RCC), the most common
urogenital neoplasm, accounts for ~5% of human adult malignancies
worldwide (1). It is estimated that
64,000 new cases and 14,000 deaths occurred as a result of RCC in
the United States in 2017(1).
Additionally, the worldwide incidence and mortality rates of RCC
are steadily increasing (1). To
date, metastatic lesions are present in ~30% of patients at initial
presentation due to an absence of symptoms at the early stages
(2). Despite advances in therapeutic
approaches, the prognosis of patients with RCC remains poor and
<40% of patients survive ≥5 years post diagnosis, as therapeutic
options are limited (3). Moreover,
RCC recurrence is primarily due to the lack of routine adjuvant
therapy in the clinic, as RCC is both chemotherapy and radiotherapy
resistant (4,5). Therefore, it is necessary to screen new
compounds and develop novel targeted therapies for RCC.
The leaves and seeds of Ginkgo biloba L., an
ancient gymnosperm species, have a long history of use in
traditional Chinese medicine (6,7). GA,
which is characterized by a mixture of components containing 13-19
side chain carbon atoms at site 6 and 0-3 side chain double bonds,
can be extracted from ginkgo fruit, exotesta and leaves (8-10).
GA has been reported to display a wide range of bioactive
properties, including antimicrobial, antivirus and molluscicidal
activities (11). Accumulating
evidence has demonstrated that several monomer structures of GA
(C13:0, C15:0 and C17:1) exert antitumor activity in a number of
human malignancies, including hepatocellular carcinoma as well as
colon, breast and lung cancer (12-15).
However, to the best of our knowledge, no previous study has been
conducted to investigate the pharmacological effect of GA in
RCC.
Network pharmacology, a novel systems pharmacology
model, was established to explore the therapeutic mechanism of
individual drugs on the basis of pharmacokinetics synthesis
screening, target identification and network analysis (16,17). A
network pharmacology-based approach has displayed potential for
investigating the therapeutic mechanism of natural agents in a
number of malignancies, including lung cancer, breast cancer and
hepatocellular carcinoma (18,19). The
identification of the therapeutic mechanism of GA could facilitate
more precise GA application in malignant cancer treatment.
Therefore, the present study utilized a network pharmacology-based
approach to explore the key target of GA. Subsequently, molecular
docking, a theoretical simulation method, was used to analyze the
interaction between molecules and to predict the binding site and
affinity of GA to ensure the accuracy and effectiveness of the
network analysis.
Materials and methods
Cell culture and chemicals
The human RCC cell lines 786-O and A498 were
purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Cells were grown in RPMI 1640 medium
(Sigma-Aldrich; Merck KGaA) supplemented with 10% fetal bovine
serum (HyClone; GE Healthcare Life Sciences) and 100 µg/ml
penicillin/streptomycin at 37˚C in a humidified incubator with 5%
CO2. GA (C17:1;
C24H38O3; Tauto Biotech Co., Ltd.;
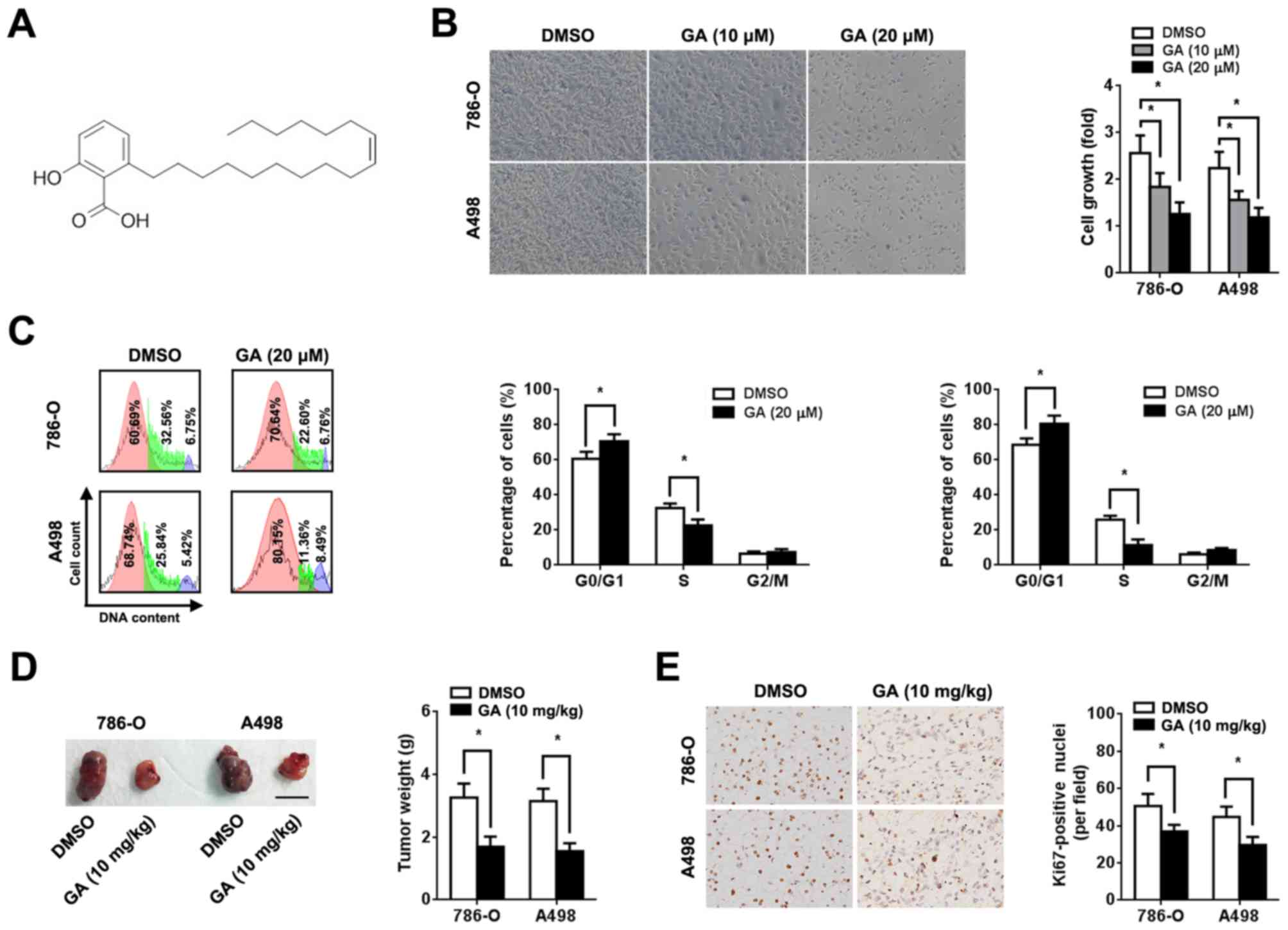
Fig. 1A) was dissolved in DMSO
(Sigma-Aldrich; Merck KGaA) to make a 20 mM stock solution. For
cell treatment, 10 and 20 µM GA (diluted in RPMI 1640 medium) were
used.
Cell viability assay
786-O and A498 cells were seeded at a density of
1x103 cells per well in 96-well plates. After culture
for 12 h at 37˚C, the medium was replaced with maintenance medium
containing the desired concentrations (10 and 20 µM) of GA or DMSO.
Cell viability was assessed after 72 h incubation at 37˚C using an
MTT assay kit (Nanjing KeyGen Biotech Co., Ltd.) according to the
manufacturer's protocols. DMSO was used to dissolve formazan
crystals and absorbance was measured in each well at 450 nm using a
318C microplate reader (Shanghai Peiou Analytical Instruments Co.,
Ltd.).
Transwell invasion assay
To determine c ell invasion, invasion assays were
performed using 24-well Matrigel-coated invasion chambers (8-µm
pore size; Corning Inc.). RCC cells were pre-treated with GA for 24
h, seeded into the upper chamber of the Transwell insert and
incubated for 24 h at 37˚C. Subsequently, cells on the upper side
of the membrane were wiped off and invading cells were fixed in
100% cold methanol at 4˚C for 20 min and stained with 0.2% crystal
violet at room temperature for 30 min (Sigma-Aldrich; Merck KGaA).
The stained cells were quantified under a light microscope, from
five randomly selected fields of view (magnification, x400).
Wound healing assay
RCC cells were pre-treated with 10 and 20 µM GA for
24 h at 37˚C and allowed to grow to 90-95% confluence without
serum. The cell monolayers were scratched with a sterile 200-µl
pipette tip. Subsequently, the cells were washed with RPMI 1640
medium to remove cell debris and fresh cell culture medium (RPMI
1640 medium supplemented with 10% FBS) was added. Images of the
scratches were obtained at 0 and 24 h using a light microscope
(magnification, x200). Migratory ability was measured as the
percentage of wound closure with the initial wound width defined as
1.
Cell cycle and early apoptosis
assay
Cell cycle distribution and apoptosis were analyzed
using the Annexin V-FITC Apoptosis kit (Solarbio Life Sciences)
according to the manufacturer's protocols. Briefly, RCC cells were
cultured in RPMI 1640 medium in 6-well plates at a density of
1x106 cells for 12 h at 37˚C. Then, the medium was
replaced with RPMI 1640 medium containing 20 µM GA. Cells were
stained with Annexin V-FITC and propidium iodide buffer at room
temperature for 15 min followed by the addition of Annexin V
binding buffer. Cell cycle and apoptosis assays were performed on a
BD Accuri C6 (BD Biosciences) flow cytometer and BD Accuri C6
software Version 1.0 (Becton, Dickinson and Company) was used to
analyze the data.
Xenograft implantation
Animal experiments were approved by The Animal Care
and Use Committee of the Second Military Medical University
(approval no. NMU20181032). Animals had free access to autoclaved
chow diet and water in a pathogen-free state, at 22±1˚C with 45±10%
humidity and 12-h light/dark cycles. The health and behaviour of
the mice were monitored every 12 h. Moribund mice (exhibiting
anorexia, immobility or an unkempt coat) would be euthanized by
CO2 at the rate of 3.5 l/min displacing 20% of the cage
volume per minute. Death was verified by monitoring cessation of
breathing. To detect the effect of GA on tumorigenicity in nude
mice, 6-8-week-old male BALB/c mice (n=10; weight, 20-30 g; Second
Military Medical University) were injected subcutaneously with
786-O or A498 cells (1x107 dissolved in PBS per site;
one tumor burden per mouse) on the right flank. Mice were randomly
divided into two groups (n=5 per group) that either received an
intragastric administration of GA (10 mg/kg) or 0.9% saline twice
per week (13). At 6 weeks after GA
injection, mice were euthanized by decapitation. The tumor volumes
and weight were measured. Prior to 6 weeks after GA injection, no
mice were euthanized or found dead.
Protein-protein interaction (PPI)
network construction and analysis
The Online Mendelian Inheritance in Man (OMIM)
Database (version 1.0; www.ncbi.nlm.nih.gov/omim), from which the original
data used in the present study was obtained, is a central-level
database covering relevant information and literature on human
genetic diseases and gene loci. Data were input into the Search
Tool for the Retrieval of Interacting Genes/Proteins (version 11.0;
string-db.org) database for further network
construction. The obtained PPI networks were then visualized using
Cytoscape software (version 3.6.1; https://cytoscape.org). Topology analysis (CentiScaPe
plug-in; version 2.2; http://apps.cytoscape.org/download/stats/centiscape/)
was used to identify key targets in the network. Degrees of
betweenness were assessed using the key hubs of centrality, namely
betweenness (the number between nodes) and closeness (the distance
between nodes).
Molecular docking
The docking between GA and key targets of RCC was
implemented using Sybyl X software (version 2.1.1; https://en.freedownloadmanager.org/Windows-PC/SYBYL-X.html).
The conformation of GA was downloaded from the Traditional Chinese
Medicine Systems Pharmacology Database (version 1.0; https://tcmspw.com/tcmsp.php). The mol2 format file
was further transformed and extracted by Openbabel software
(release date, 2017-02-22; https://sourceforge.net/projects/openbabel/). The
X-ray crystal structures of key targets were obtained from the RCSB
Protein Data Bank (version 1.0; www.rcsb.org).
Prior to docking, targets were structurally optimized by adding
hydrogen atoms and removing water molecules, and the representative
protein activity docking pockets were subsequently generated.
Molecular docking was implemented based on the Docking Suite module
(release date, 2013-04-08; https://sourceforge.net/projects/wpfdocking/) and
hydrogen bond stability and domain structure similarity were used
to evaluate the reliability of results. The criteria of binding
scoring: 0-3, Poor; 4-6, moderate; and 7-10, good.
Immunohistochemistry and
immunoblotting assay
Cells were pre-treated with 10 ng/ml epidermal
growth factor (EGF; PeproTech, Inc.) for 12 h at 37˚C.
Immunohistochemical assays of xenografts were conducted using Ki-67
(cat. no. ab15580; 1:50; Abcam) and EGF receptor (EGFR; cat. no.
ab52894; 1:50; Abcam) monoclonal antibodies, as previously
described (20). The expression
level was analyzed from images obtained using a light microscope
(magnification, x200) from five random fields of view. Immunoblot
assays were performed as previously described (20). Primary antibodies included those
targeted against phosphorylated (p)-EGFR (cat. no. 4370; 1:1,000;
Cell Signaling Technology, Inc.), EGFR (cat. no. 2085; 1:1,000;
Cell Signaling Technology, Inc.), p-Akt (Ser473; cat. no. 4060;
1:1,000; Cell Signaling Technology, Inc.), Akt (cat. no. 4691;
1:1,000; Cell Signaling Technology, Inc.), p-ERK (Thr202/Tyr204;
cat. no. 4370; 1:1,000; Cell Signaling Technology, Inc.) and ERK
(cat. no. 4370; 1:1,000; Cell Signaling Technology, Inc.).
Horseradish peroxidase-conjugated secondary antibody (1:2,000; cat.
no. ab6112; Abcam) was used. β-actin antibody (cat. no. ab8226;
1:5,000; Abcam) was used as a control. Relative protein expression
and staining intensity was calculated based on the densitometric
analysis by Image Lab software Version 3.0 (Bio-Rad Laboratories,
Inc.).
Statistical analysis
All data are presented as the mean ± SD. SPSS
software (version 18.0; SPSS Inc.) was used for statistical
analysis. The statistical significance between two groups was
analyzed using an unpaired Student's t-test. One-way ANOVA followed
by a Dunnett's post hoc test was performed to compare data among ≥3
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
GA inhibits RCC cell proliferation in
vitro and in vivo
To investigate the role of GA in RCC, human RCC
cells (786-O and A498) were incubated with different concentrations
of GA. An MTT assay was performed 72 h after treatment and a
dose-dependent decrease in cell viability was observed (Fig. 1B). In addition, GA increased the
proportion of cells in the G0/G1 phase and
decreased the proportion of cells in the S phase (Fig. 1C). However, GA treatment did not
significantly affect RCC cell apoptosis (Fig. S1). Based on in vitro data,
xenograft implantation in BALB/c nude mice was performed to clarify
the effect of GA on RCC in vivo. RCC cell suspensions
(1x107) were subcutaneously injected into nude mice. The
tumor volumes and weights of the group treated with GA were
significantly lower compared with those of the group treated with
0.9% saline (P<0.05; Figs. 1D and
S2A). In the present study, the maximum diameter of a single tumor
was 1.8 cm. However, no significant difference in body weight loss
was observed between the two groups (Fig. S2C). Additionally,
immunohistochemical staining revealed that GA significantly reduced
Ki-67 expression in the xenograft (P<0.05; Fig. 1E).
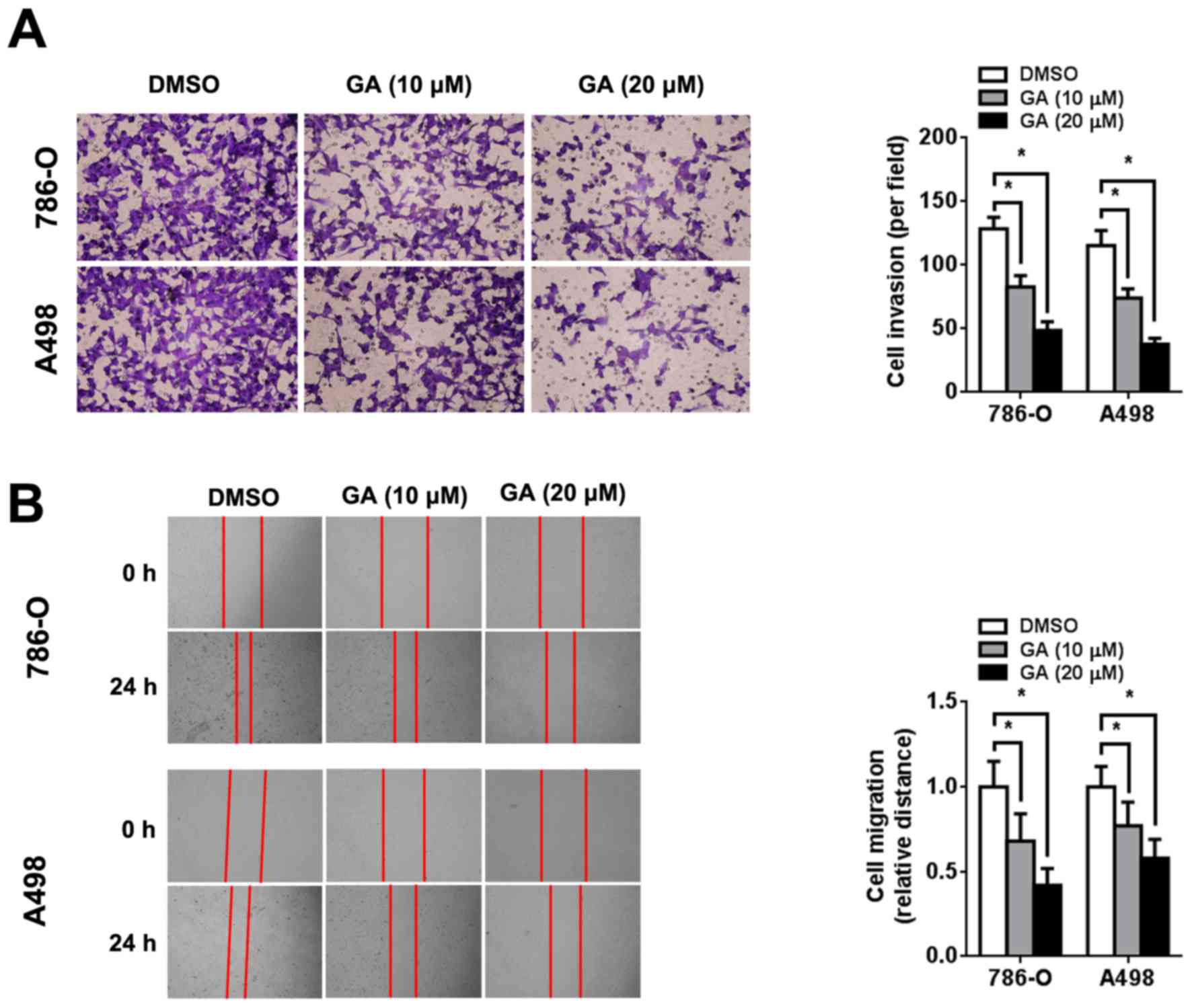
GA inhibits RCC cell invasion and
migration
The effect of GA on the invasion and migration of
RCC cells was evaluated using Transwell invasion and wound healing
assays, respectively. Transwell invasion assays showed that GA
reduced the number of invading RCC cells in a dose-dependent manner
(Fig. 2A). Consistent with this
result, wound healing assays also showed that GA inhibited RCC cell
migration in a dose-dependent manner (Fig. 2B).
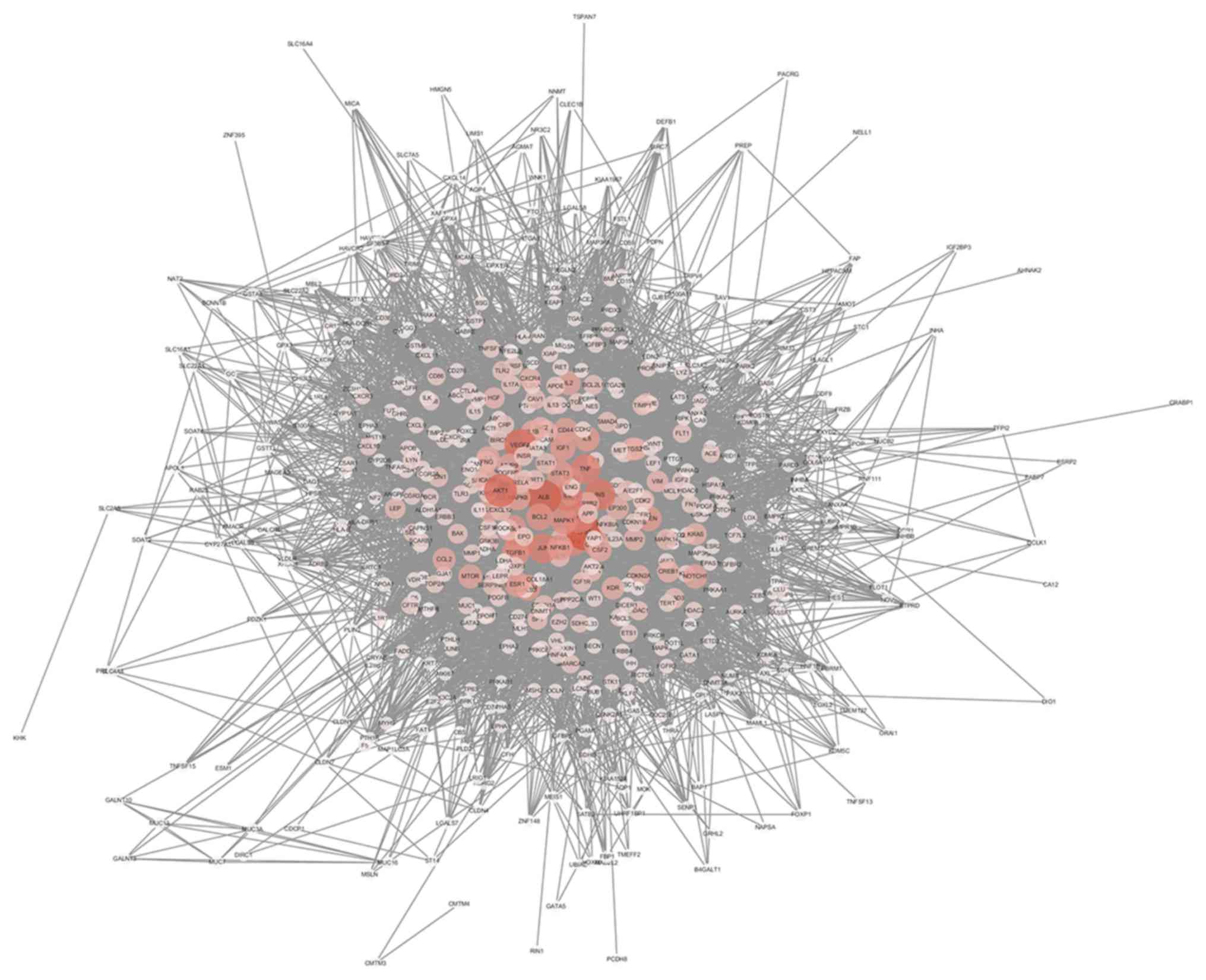
PPI network and topology analysis
The interaction network was constructed based on
OMIM original data and is depicted in Fig. 3, and included 549 nodes and 11,836
edges. Larger areas and deeper colours represent higher node
connectivity, which likely indicates greater importance of the node
in the network. The results displayed the scale-free property of
the network, meaning the number of nodes decreases as the number of
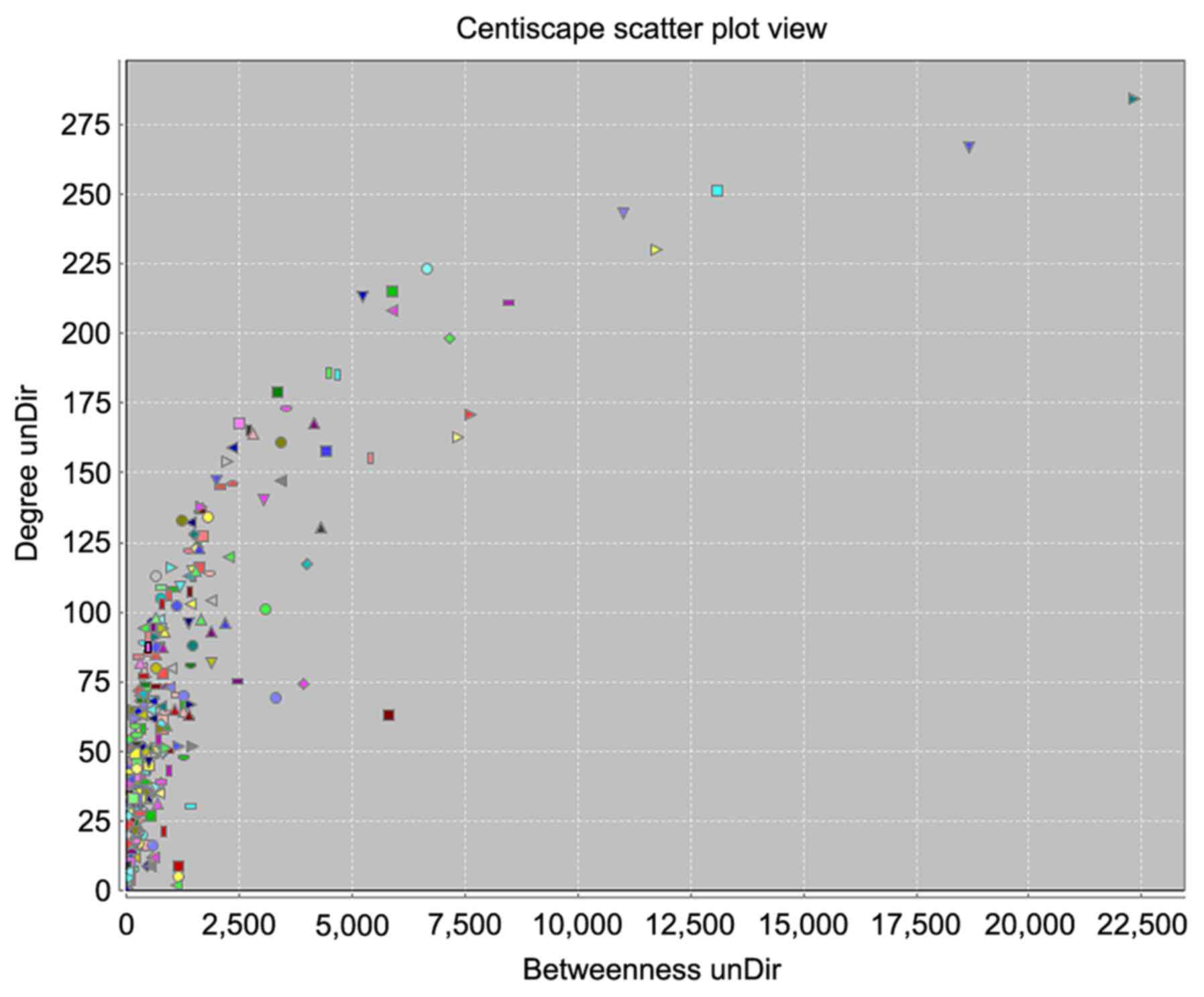
edges in the network increases. Betweenness degree was used to
distinguish these highly connected key nodes from other nodes in
the network (Fig. 4). Finally, 15
key targets were selected: Tumor protein p53, 284 points; albumin,
267 points; AKT1, 251 points; vascular endothelial growth factor A,
243 points; insulin, 230 points; interleukin 6, 223 points; JUN,
215 points; tumor necrosis factor, 213 points; MYC, 211 points;
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit-α,
208 points; EGFR, 198 points; mitogen-activated protein kinase 1,
186 points; transforming growth factor β1, 185 points; BCL2, 179
points; and STAT3, 173 points.
GA inactivates the EGFR pathway in
RCC
Molecular docking, a tool for studying the
interaction strength between molecules, was used to explore the
accuracy and effectiveness of the PPI network. To assess the
reliability of the aforementioned drug-target interactions and to
further investigate binding mode and affinity, molecular docking
models were constructed for GA and the 15 key targets. Analysis of
the binding data indicated stable conformational binding and good
scoring (>7 points) of EGFR to GA (9.4558 points; Fig. 5A). Immunohistochemical staining
displayed lower EGFR expression in the xenograft after GA treatment
compared to before treatment (Fig.
5B). The immunoblotting assay demonstrated that EGFR, p-Akt and
p-Erk (downstream of the EGFR signaling pathway) were significantly
downregulated after GA treatment in xenografts, as well as in
vitro (P<0.05; Fig. 5C).
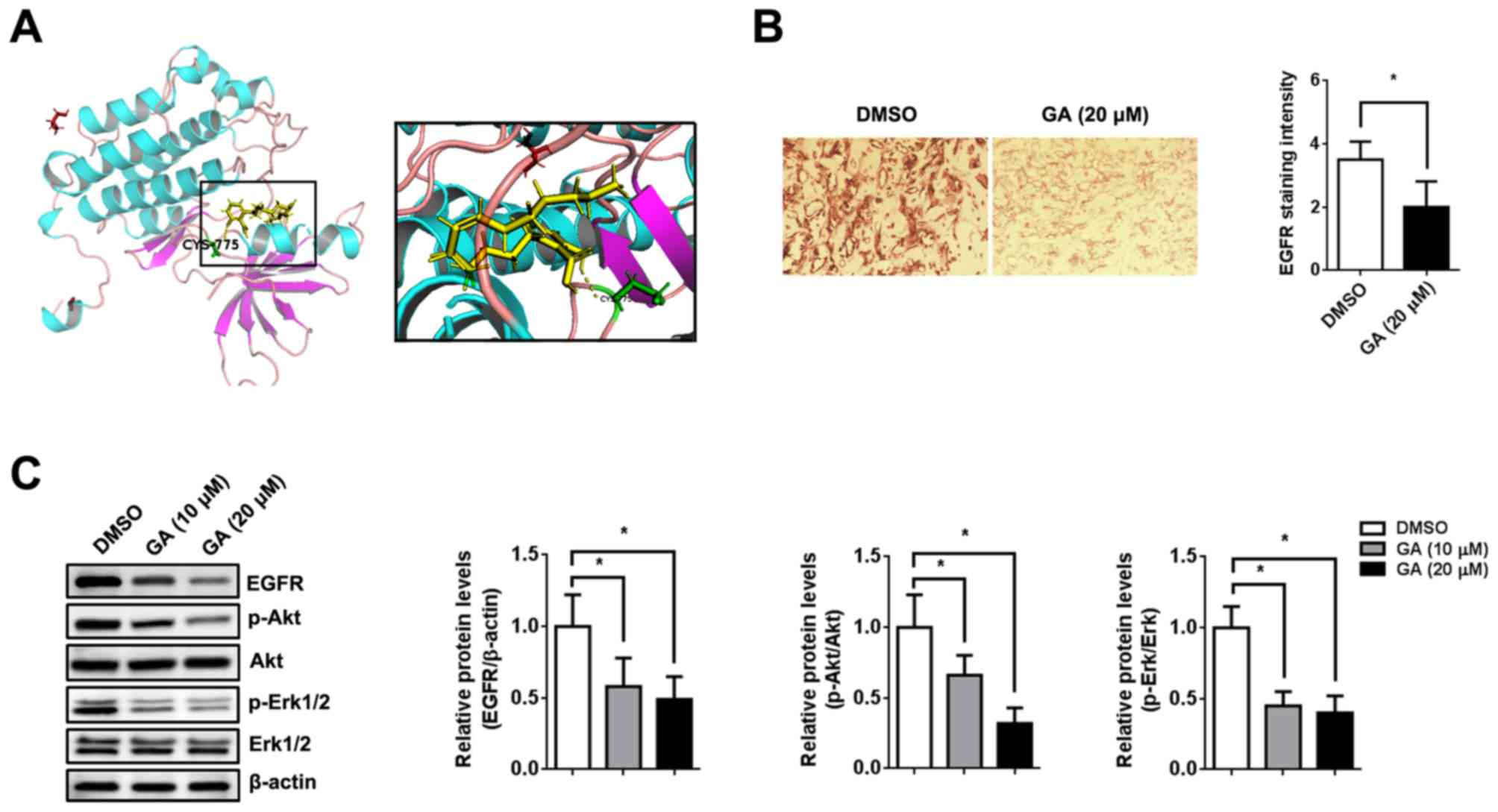 | Figure 5GA exerts its function via
suppressing the EGFR pathway. (A) Binding interaction model of EGFR
and GA. (B) Mice were injected subcutaneously with 786-O cells
(1x107 cells) followed by intragastric administration of
GA (10 mg/kg) or 0.9% saline twice per week. Representative image
and quantification of immunohistochemical staining for EGFR in the
xenograft (magnification, x200). (C) 786-O cells were pre-treated
with 10 ng/ml EGF for 12 h at 37˚C followed by DMSO or GA for 48 h
at 37˚C. The expression of p-EGFR, EGFR, p-Akt, Akt, p-Erk1/2 and
Erk1/2 was analyzed by immunoblotting. β-actin was used as an
internal control. n=5. *P<0.05, as indicated. GA,
ginkgolic acid; EGFR, epidermal growth factor receptor; EGF,
epidermal growth factor; p, phosphorylated. |
Discussion
Despite great advances in cancer therapy, major
limitations still exist in the management of RCC (21). Moreover, patients with metastatic RCC
have a short life expectancy (median overall survival, 22.5 months)
(22). Extracts from natural plants
are becoming an increasingly popular strategy to identify novel
antitumor compounds (23,24). Recently, several studies have
reported that GA displays promising antitumor activities (12-15).
Therefore, identification of the regulatory mechanism of GA in
tumorigenesis would provide valuable insight for the treatment of
malignancies. Collectively, the results of the present study
indicated that GA suppressed the development of RCC. These results
were consistent with previous studies demonstrating that GA plays
an inhibitory biological role in cancer (12-15,25).
However, the biological effect of GA on RCC was observed at high
concentrations of GA in vitro and it is uncertain whether
this concentration would be possible in vivo. Therefore,
further in vivo investigation (pharmacokinetics and
toxicology analysis) is required.
GA suppresses tumor growth and invasion via various
signaling pathways, including the hepatocyte growth factor/c-Met
signaling pathway in hepatocellular carcinoma (12), the protein kinase AMP-activated
catalytic subunit α2 signaling pathway in colon cancer (13), the NF-κB signaling pathway in breast
cancer (14) and the PI3K/Akt/mTOR
signaling pathway in lung cancer (15). A number of studies have reported
that, in addition to inducing cell cycle arrest, GA also increases
apoptosis in colon cancer and hepatocellular carcinoma (26,27).
Therefore, it is difficult to clarify the antitumor mechanism of GA
in RCC. Network pharmacology provides a strategy to investigate
complex mechanisms of drug action and to identify potential drug
targets (28). In a network
pharmacology system, therapy response can be taken into account
based on the robustness of complex disease networks in dealing with
node attacks (node linking degree) due to inherent diversity and
redundant compensation signaling pathways (12,19). The
result of network pharmacology is a highly resilient network system
with topological interaction (19,29).
Therefore, computational prediction with a PPI network and topology
analyses reveals the potential interactions among compounds and
multiple signaling transduction molecules underlying a disease
phenotype (30). According to the
algorithm value, 15 key targets associated with RCC were identified
in the current study. Furthermore, analysis of the molecular
docking model demonstrated a stable conformational affinity between
EGFR and GA. EGFR was validated as a direct target for GA both
in vitro and in vivo. Furthermore, GA negatively
modulated p-Akt and p-Erk expression via the EGFR/Akt/Erk signaling
pathway in RCC cells. However, the mechanism of EGFR protein
degradation (autophagy and ubiquitination) requires further
investigation.
EGFR belongs to the ErbB family of receptor tyrosine
kinases, which can be activated by several ligands, including EGF,
transforming growth factor-α, amphiregulin, heparin-binding EGF,
β-cellulin and epiregulin (31-34).
In humans, EGFR is upregulated and/or activated in various
malignancies and suppression of EGFR activation is considered a
valid strategy for tumor treatment (34,35). Akt
and Erk are downstream signaling molecules of EGFR, and the
aberrant activation of either triggers a cascade of proliferative
responses (36). Akt and Erk
phosphorylation are increased when EGFR is activated in RCC
(37,38). Previous studies have provided direct
evidence that the EGFR signaling pathway is highly activated in RCC
progression and EGFR expression is associated with prognosis in
patients with clear cell RCC (38-40).
Furthermore, inactivation of the EGFR signaling pathway inhibited
RCC cell migration and invasion in vitro (41). These data imply that activation of
the EGFR signaling pathway may be an important event that occurs
during RCC development. The present study demonstrated that GA,
which acts as a suppressor of EGFR, exhibited a biological effect
in RCC cells via the EGFR/Akt/Erk signaling pathway.
The present study investigated the molecular
mechanism by which GA exerts its negative effects on proliferation,
invasion and migration in human RCC cells. GA significantly
inhibited the EGFR signaling pathway by directly binding to EGFR.
The downstream targets of EGFR, including p-Akt and p-Erk, were
also downregulated following GA treatment. EGFR, as a novel target
of GA, may be a promising therapeutic agent for human RCC.
Supplementary Material
Effect of GA on RCC cell apoptosis.
786-O and A498 cells were pre-treated with 20 μM GA or left
untreated. Representative cell cycle plots 2 days after GA
treatment. Cell apoptosis assay was performed using flow cytometry.
n=5. GA, ginkgolic acid; RCC, renal cell carcinoma; ns, not
significant.
Effect of GA on tumor growth in
vivo. Mice were injected subcutaneously with 786-O or A498
cells (1x107 cells) followed by intragastric administration of GA
(10 mg/kg) or 0.9% saline twice per week. (A) Tumor volumes, (B)
tumor weight as a percentage of body weight and (C) the percentage
of body weight loss were measured 6 weeks after GA administration.
n=5. *P<0.05, as indicated. GA, ginkgolic acid; ns, not
significant.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Medical
Research Project of Hongkou District of Shanghai (grant no.
1702-11) and The Natural Science Foundation of Fujian Province
(grant no. 2016J05196).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZD and BF contributed to the conception and design
of the study. CZ, NN, HW and MZ performed the experiments. PZ and
WZ analyzed the data. HS participated in the design of the study.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of the Second Military Medical University (approval no.
NMU20181032; Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cindolo L, Patard JJ, Chiodini P, Schips
L, Ficarra V, Tostain J, de La Taille A, Altieri V, Lobel B,
Zigeuner RE, et al: Comparison of predictive accuracy of four
prognostic models for nonmetastatic renal cell carcinoma after
nephrectomy: A multicenter European study. Cancer. 104:1362–1371.
2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Patil S, Ishill N, Deluca J and Motzer RJ:
Stage migration and increasing proportion of favorable-prognosis
metastatic renal cell carcinoma patients: Implications for clinical
trial design and interpretation. Cancer. 116:347–354.
2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Redova M, Svoboda M and Slaby O: MicroRNAs
and their target gene networks in renal cell carcinoma. Biochem
Biophys Res Commun. 405:153–156. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mangolini A, Bonon A, Volinia S, Lanza G,
Gambari R, Pinton P, Russo GR, Del Senno L, Dell'Atti L and Aguiari
G: Differential expression of microRNA501-5p affects the
aggressiveness of clear cell renal carcinoma. FEBS Open Bio.
4:952–965. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
van Beek TA and Wintermans MS: Preparative
isolation and dual column high-performance liquid chromatography of
ginkgolic acids from Ginkgo biloba. J Chromatogr A.
930:109–117. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Major RT: The ginkgo, the most ancient
living tree. The resistance of Ginkgo biloba L. to pests
accounts in part for the longevity of this species. Science.
157:1270–1273. 1967.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Siegers CP: Cytotoxicity of alkylphenols
from Ginkgo biloba. Phytomedicine. 6:281–283.
1999.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hecker H, Johannisson R, Koch E and
Siegers CP: In vitro evaluation of the cytotoxic potential of
alkylphenols from Ginkgo biloba L. Toxicology. 177:167–177.
2002.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhou C, Li X, Du W, Feng Y, Kong X, Li Y,
Xiao L and Zhang P: Antitumor effects of ginkgolic acid in human
cancer cell occur via cell cycle arrest and decrease the Bcl-2/Bax
ratio to induce apoptosis. Chemotherapy. 56:393–402.
2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lü JM, Yan S, Jamaluddin S, Weakley SM,
Liang Z, Siwak EB, Yao Q and Chen C: Ginkgolic acid inhibits HIV
protease activity and HIV infection in vitro. Med Sci Monit.
18:BR293–BR298. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li H, Meng X, Zhang D, Xu X, Li S and Li
Y: Ginkgolic acid suppresses the invasion of HepG2 cells via
downregulation of HGF/cMet signaling. Oncol Rep. 41:369–376.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Qiao L, Zheng J, Jin X, Wei G, Wang G, Sun
X and Li X: Ginkgolic acid inhibits the invasiveness of colon
cancer cells through AMPK activation. Oncol Lett. 14:5831–5838.
2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hamdoun S and Efferth T: Ginkgolic acids
inhibit migration in breast cancer cells by inhibition of NEMO
sumoylation and NF-κB activity. Oncotarget. 8:35103–35115.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam
D, Lee J, Lee SG, Yang WM, Um JY, et al: Ginkgolic acid inhibits
invasion and migration and TGF-β-induced EMT of lung cancer cells
through PI3K/Akt/mTOR inactivation. J Cell Physiol. 232:346–354.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ye HZ, Zheng CS, Xu XJ, Wu MX and Liu XX:
Potential synergistic and multitarget effect of herbal pair
Chuanxiong Rhizome-Paeonia Albifora Pall on osteoarthritis disease:
A computational pharmacology approach. Chin J Integr Med.
17:698–703. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xenarios I, Salwínski L, Duan XJ, Higney
P, Kim SM and Eisenberg D: DIP, the database of interacting
proteins: A research tool for studying cellular networks of protein
interactions. Nucleic Acids Res. 30:303–305. 2002.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xu R and Wunsch DC II: Clustering
algorithms in biomedical research: A review. IEEE Rev Biomed Eng.
3:120–154. 2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ramadan E, Naef A and Ahmed M: Protein
complexes predictions within protein interaction networks using
genetic algorithms. BMC Bioinformatics. 17:(Suppl 7): S269.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang D, Zhu C, Zhang Y, Zheng Y, Ma F, Su
L and Shao G: MicroRNA-30e-3p inhibits cell invasion and migration
in clear cell renal cell carcinoma by targeting Snail1. Oncol Lett.
13:2053–2058. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Renal cell cancer treatment (PDQ(R)):
Patient version, in PDQ cancer information summaries. 2002:
Bethesda (MD).
|
|
22
|
Shingarev R and Jaimes EA: Renal cell
carcinoma: New insights and challenges for a clinician scientist.
Am J Physiol Renal Physiol. 313:F145–F154. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang C, Ren Q, Chen XT, Song ZQ, Ning ZC,
Gan JH, Ma XL, Liang DR, Guan DG, Liu ZL and Lu AP: System
pharmacology-based strategy to decode the synergistic mechanism of
Zhi-zhu Wan for functional dyspepsia. Front Pharmacol.
9(841)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang J, Li Y, Chen X, Pan Y, Zhang S and
Wang Y: Systems pharmacology dissection of multi-scale mechanisms
of action for herbal medicines in stroke treatment and prevention.
PLoS One. 9(e102506)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ma J, Duan W, Han S, Lei J, Xu Q, Chen X,
Jiang Z, Nan L, Li J, Chen K, et al: Ginkgolic acid suppresses the
development of pancreatic cancer by inhibiting pathways driving
lipogenesis. Oncotarget. 6:20993–21003. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu Y, Yang B, Zhang L, Cong X, Liu Z, Hu
Y, Zhang J and Hu H: Ginkgolic acid induces interplay between
apoptosis and autophagy regulated by ROS generation in colon
cancer. Biochem Biophys Res Commun. 498:246–253. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhou CC, Du W, Wen Z, Li JY and Zhang P:
Effects of natural plant ginkgolic acids on the apoptosis of human
Hep-2 cancer cells. Sichuan Da Xue Xue Bao Yi Xue Ban. 40:459–461.
2009.PubMed/NCBI(In Chinese).
|
|
28
|
Yuan H, Ma Q, Cui H, Liu G, Zhao X, Li W
and Piao G: How can synergism of traditional medicines benefit from
network pharmacology? Molecules. 22(E1135)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Stelzl U, Worm U, Lalowski M, Haenig C,
Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A,
Koeppen S, et al: A human protein-protein interaction network: A
resource for annotating the proteome. Cell. 122:957–968.
2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Boezio B, Audouze K, Ducrot P and
Taboureau O: Network-based approaches in pharmacology. Mol Inform.
36:2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Harris RC, Chung E and Coffey RJ: EGF
receptor ligands. Exp Cell Res. 284:2–13. 2003.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Freed DM, Bessman NJ, Kiyatkin A,
Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM,
Leahy DJ, Lidke DS and Lemmon MA: EGFR ligands differentially
stabilize receptor dimers to specify signaling kinetics. Cell.
171:683–695.e18. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sigismund S, Avanzato D and Lanzetti L:
Emerging functions of the EGFR in cancer. Mol Oncol. 12:3–20.
2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang Z: ErbB receptors and cancer. Methods
Mol Biol. 1652:3–35. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Liu Q, Yu S, Zhao W, Qin S, Chu Q and Wu
K: EGFR-TKIs resistance via EGFR-independent signaling pathways.
Mol Cancer. 17(53)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cosmai L, Gallieni M and Porta C: Renal
toxicity of anticancer agents targeting HER2 and EGFR. J Nephrol.
28:647–657. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu F, Shangli Z and Hu Z: CAV2 promotes
the growth of renal cell carcinoma through the EGFR/PI3K/Akt
pathway. Onco Targets Ther. 11:6209–6216. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Liu L, Miao L, Liu Y, Qi A, Xie P, Chen J
and Zhu H: S100A11 regulates renal carcinoma cell proliferation,
invasion, and migration via the EGFR/Akt signaling pathway and
E-cadherin. Tumour Biol. 39(1010428317705337)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Feng ZH, Fang Y, Zhao LY, Lu J, Wang YQ,
Chen ZH, Huang Y, Wei JH, Liang YP, Cen JJ, et al: RIN1 promotes
renal cell carcinoma malignancy by activating EGFR signaling
through Rab25. Cancer Sci. 108:1620–1627. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen F, Deng J, Liu X, Li W and Zheng J:
HCRP-1 regulates cell migration and invasion via EGFR-ERK mediated
up-regulation of MMP-2 with prognostic significance in human renal
cell carcinoma. Sci Rep. 5(13470)2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liang L, Li L, Zeng J, Gao Y, Chen YL,
Wang ZQ, Wang XY, Chang LS and He D: Inhibitory effect of silibinin
on EGFR signal-induced renal cell carcinoma progression via
suppression of the EGFR/MMP-9 signaling pathway. Oncol Rep.
28:999–1005. 2012.PubMed/NCBI View Article : Google Scholar
|