Introduction
Hypertrophic cardiomyopathy (HCM) is one of the most
common genetic heart disorders with a prevalence of 1 in 200
worldwide (0.6% of the general population) (1). HCM is characterized by increased
thickness of the ventricular wall. Over 1,500 mutations in at least
11 genes, encoding components of the cardiac sarcomere-associated
proteins, have been identified as potential cause for HCM (2-4).
MYH7 and MYBPC3, encoding β-myosin heavy chain and
myosin-binding protein C, respectively, are the 2 most common genes
involved, accounting for 50-60% of the HCM families. However, in
about 40% of HCM patients, the causal genes remain to be
identified, highlighting the need for precision medicine as genetic
diversity of HCM-associated individuals (3,4). The
clinical outcomes of HCM are diverse, ranging from no symptoms to
cardiac ischemia, cardiac arrhythmia, congestive heart failure and
other organ system dysfunctions (5).
HCM is also associated with an increased risk of sudden death,
heart failure and thromboembolic events, which results in an annual
mortality rate of 1% worldwide (6).
A challenging characteristic of diagnosing HCM is
the lack of association between genotype and phenotype, as family
members carrying the same mutation develop different symptoms
(7). Therefore, HCM is regarded as a
highly complex disease due to its diverse clinical presentations,
heterogeneous phenotypes, large number of mutations and broad
spectrum of complications (4,7). The
development of HCM likely results from a combination of endogenous
genetic mutations with an exogenous decline in protein-protective
mechanisms, and environmental factors such as lifestyle, blood
pressure and physical exercise (8).
In this regard, it is crucial to identify transcriptional profiles,
epigenetic modifications and post-translational modifications which
may initiate the onset of HCM.
In the last decade, accumulating evidence has
indicated that rather than being transcriptional noise, diverse
noncoding RNAs (ncRNAs) act as important regulators of HCM
initiation and progression at the post-transcriptional level
(9-15).
To date, studies have primarily focused on small, noncoding
endogenous RNAs (9,10). MicroRNAs (miRNAs) are 22-25
nucleotides in length and regulate gene expression by directly
targeting mRNAs for degradation or translational repression
(10). Several miRNAs, such as
miR-21, miR-1, miR-29a, miR-133a and miR-130b have been described
as important regulators of HCM in murine and human hearts (9,11-14).
Furthermore, these miRNAs can be detected in the bloodstream,
highlighting the possibility of their use as circulating biomarkers
of HCM (11,14,15).
However, the role of miRNAs in the progression of HCM remains to be
elucidated.
In addition to miRNAs, long noncoding RNAs (lncRNAs)
are known as important regulators of cardiac pathology (16,17).
lncRNAs are transcripts >200 nucleotides in length with no
protein-coding capacity. Based on their diverse biochemical roles,
lncRNAs can perform their functions via RNA-DNA, RNA-RNA or
RNA-protein interactions (18).
Notably, lncRNAs have been reported to competitively interact with
miRNAs and thus inhibit target mRNA degradation by a competitive
endogenous RNA (ceRNA) regulatory mechanism (19,20).
However, little is known regarding the function of lncRNAs in HCM,
particularly in human hearts. Current studies of lncRNAs in HCM
primarily describe expression profiling by RNA sequencing or
microarray technology (21,22). However, the underlying mechanisms
involving miRNAs or mRNAs are still unclear. Therefore, considering
the large number lncRNAs expressed in the heart and limited
knowledge of their function physiologically and
pathophysiologically, it is hypothesized that functionally-related
lncRNAs are functionally associated with mRNAs or miRNAs. Similar
associations have been detected in several diseases such as
ischemic cardiomyopathy (23),
diabetic cardiomyopathy (24), and
heart failure (25), but have not
yet been revealed in HCM.
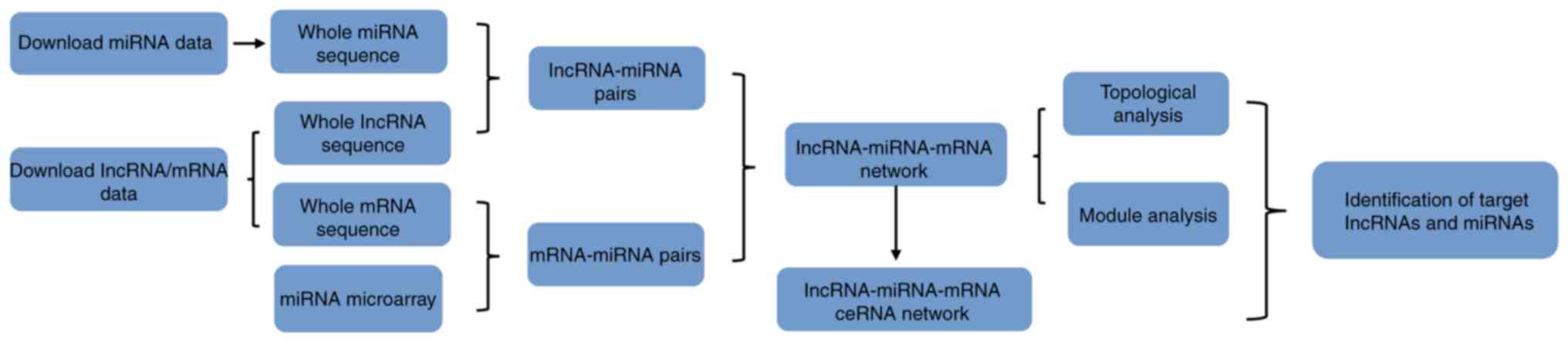
In the present study, a triple network was
constructed using human data from the National Center for
Biotechnology Information Gene Expression Omnibus (NCBI GEO). Based
on the ceRNA mechanism in which lncRNA, miRNA and mRNA form a
triplet, where the lncRNA and mRNA share the same miRNA. The
present study constructed a lncRNA-miRNA-mRNA network with high
reliability, providing a potentially novel understanding of the
mechanisms of the development of HCM and potential therapeutic
targets.
Materials and methods
Raw data
NCBI GEO (ncbi.nlm.nih.gov/geo/) is a publicly available
genomics database containing data obtained from array- and
sequence-based analysis. Users can query and download experimental
data and curated gene expression profiles from NCBI-GEO. In the
present study, human lncRNA and mRNA expression data were
downloaded from dataset GSE68316 in myocardial tissues (21), and human miRNA expression data were
downloaded from GSE36946 of surgical myectomy tissues. The age of
patients corresponding to the heart samples in GSE68316 ranged from
31-60 years, whereas the age range of the patients in GSE36946 was
9-78 years; therefore, data for heart samples corresponding to
patients aged from 31-60 years in GSE36946 were selected.
Subsequently, the two datasets were merged to construct a
lncRNA-miRNA-mRNA network based on the ceRNA theory. The
experimental design and flowchart of the steps performed in the
present study are presented in Fig.
1.
Screening of differentially expressed
lncRNAs, miRNAs and mRNAs
To detect differentially expressed lncRNAs, miRNAs
and mRNAs between patients with HCM and healthy donors, two-class
differential analyses was used. The threshold of dysregulated
lncRNAs/mRNAs/miRNAs was a fold change of >2. Student's t-test
was performed using SPSS 23.0 (IBM Corp.). P<0.05 was considered
to indicate a statistically significant difference.
Prediction of target lncRNAs and mRNAs
of miRNAs
miRNA sequences were downloaded from miRBase
(mirbase.org/) and lncRNA sequences were
downloaded from the NCBI nucleotide database (ncbi.nlm.nih.gov/nucleotide/). The miRNA targets
of lncRNAs were predicted and the minimum free energy of
lncRNA-miRNA duplexes was calculated using RNAhybrid (bibiserv.cebitec.uni-bielefeld.de/rnahybrid)
(26). miRNA-mRNA interactions were
predicted using miRanda (http://www.microrna.org/) and TargetScan (targetscan.org/vert_72/).
Construction of a lncRNA-miRNA-mRNA
network
Pearson's correlation coefficient was used to
calculate the correlation between lncRNA and mRNA expression. The
pairs of lncRNA-mRNA with r score >0.99 and P score <0.05
were regarded as target pairs. Among all the selected lncRNA-mRNA
pairs, if the lncRNA and mRNA were both targeted and were sharing a
common miRNA, the lncRNA-miRNA-mRNA ceRNA network was identified as
a co-expression competing triplet. Cytoscape software (3.7.1)
(27) was used to visualize the
lncRNA-miRNA-mRNA network.
Functional enrichment analysis
To study functional enrichment, Gene Ontology (GO;
geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; genome.jp/kegg/) pathway analyses of mRNAs in the
lncRNA-miRNA-mRNA network were performed using the Cytoscape
plugins BiNGO and Database (http://apps.cytoscape.org). The GO database provides
gene ontologies, annotations of genes and gene products based on
terms. KEGG is a relational database comprising searchable
molecular interaction pathways and reaction networks in metabolism,
various cellular processes and multiple human diseases.
Reconstruction of a key
lncRNA-miRNA-mRNA subnetwork
Cytoscape software was used to construct a new
subnetwork extracted from all lncRNAs and their related miRNAs and
mRNAs in the triple network. The target lncRNAs were identified by
calculating the number and node of related lncRNA-miRNA-mRNA
triplets,
Results
Screening for differentially expressed
lncRNAs, miRNAs and mRNAs
Human lncRNA/mRNA expression data were obtained from
GEO dataset GSE68316 and human miRNA expression data were
downloaded from GEO dataset GSE36946. lncRNAs, mRNAs and miRNAs
were considered as significantly differentially expressed when the
fold change was >2 with P<0.05. A total of 8 miRNAs (Table I), 431 lncRNAs and 609 mRNAs were
selected for subsequent analyses. Considering the specific
description of miRNAs in different microarrays, the names of
selected miRNAs were redefined according to miRBase (Table I). Subsequently, the two datasets
were merged to construct a ceRNA based lncRNA-miRNA-mRNA
network.
 | Table ISelected differently expressed miRNAs
in samples of hypertrophic cardiomyopathy patients and healthy
controls. |
Table I
Selected differently expressed miRNAs
in samples of hypertrophic cardiomyopathy patients and healthy
controls.
| miRNA | Redefined name | P-value | Fold-change | Expression |
|---|
| hsa-miR-514 |
hsa-miR-514a-3p |
1.72x10-6 | 3.069 | Upregulated |
| hsa-miR-373 | hsa-miR-373-3p |
2.33x10-6 | 2.577 | Upregulated |
|
hsa-miR-30c-1a | hsa-miR-30c-3p |
2.49x10-3 | 2.451 | Downregulated |
|
hsa-miR-144a | hsa-miR-144-5p |
3.38x10-6 | 2.398 | Downregulated |
| hsa-miR-1247 |
hsa-miR-1247-5p |
1.80x10-3 | 2.290 | Downregulated |
|
hsa-miR-10aa | hsa-miR-10a-3p |
6.63x10-6 | 2.286 | Downregulated |
| hsa-miR-10a | hsa-miR-10a-5p |
3.18x10-6 | 2.101 | Downregulated |
| hsa-miR-1268 | hsa-miR-1268a |
2.30x10-6 | 2.096 | Downregulated |
ceRNA based lncRNA-miRNA-mRNA
network
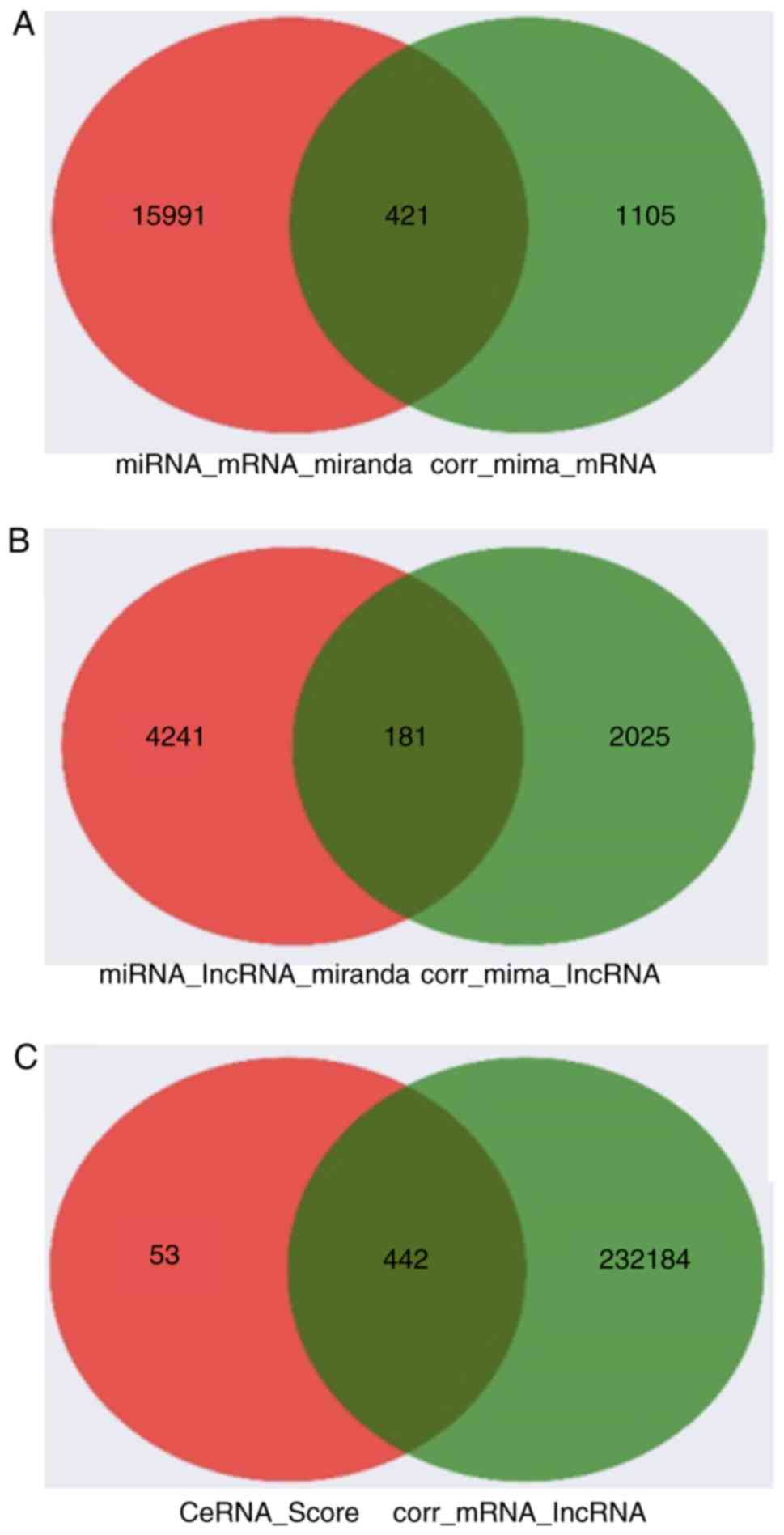
lncRNA-miRNA and miRNA-mRNA pairs were identified
according to both base sequence and expression levels. A total of
181 miRNA-lncRNA pairs and 421 miRNA-mRNA pairs were predicted
based on the intersecting elements (Fig.
2A and B). In addition, 442
lncRNA-mRNA pairs were further selected according to expression
levels and ceRNA score (Fig. 2C).
Finally, a network composed of 30 lncRNAs and 94 mRNAs (Fig. 2D), and a ceRNA network composed of 30
lncRNAs, 94 mRNAs and 8 miRNAs were constructed (Fig. 2E).
Topological analysis of the
HCM-related lncRNA-miRNA-mRNA network
Hub nodes play important roles in biological
networks. The topological properties of the HCM-related ceRNA
network composed of lncRNA-miRNA-mRNA were first analyzed. The
degree (the level of a gene related to its surroundings, where
genes with a higher degree is more likely to be the core
information exchange of the network), closeness (the length of
information transferred from one node to the other) and betweenness
(the function and influence of the corresponding gene in the whole
network, where the larger the value is, the more capable of the
gene is to participate in the communication between other genes) of
the network were calculated and the topological features of all the
nodes in the network were ranked. The top 20 genes in the network
were listed in Table II.
Interestingly, 5 lncRNAs appeared in the list. The number of
primary lncRNA-miRNA pairs and secondary miRNA-mRNA pairs were
calculated as presented in Table
III. Among the top 10 lncRNA-miRNA pairs, the 5 lncRNAs
identified in the ceRNA network were determined. Therefore, these 5
lncRNAs (ENST00000597346.1, ENST00000458178.1, ENST00000544461.1,
ENST00000567093.1 and ENST00000571219.1) not only had higher
betweenness and node degree but were also involved in a higher
number of lncRNA-miRNA and miRNA-mRNA pairs, which suggested that
these 5 lncRNAs may serve a role in the initiation and/or
development of HCM.
 | Table IITop 20 genes in degree, betweenness
and closeness. |
Table II
Top 20 genes in degree, betweenness
and closeness.
| Betweenness | Closeness | Degree | Pagerank | Name | Gene type |
|---|
| 2950 | 0.439597 | 60 | 0.107238 | hsa-miR-514 | miRNA |
| 2950 | 0.439597 | 60 | 0.107238 | hsa-miR-373 | miRNA |
| 1462.053 | 0.314904 | 46 | 0.06756 |
hsa-miR-30c-1a | miRNA |
| 2786.915 | 0.41195 | 34 | 0.049831 | hsa-miR-1268 | miRNA |
| 568.3588 | 0.293722 | 31 | 0.04458 | hsa-miR-10a | miRNA |
| 2332.149 | 0.40184 | 30 | 0.0444 |
hsa-miR-144a | miRNA |
| 184.1859 | 0.274059 | 15 | 0.022549 |
hsa-miR-10aa | miRNA |
| 147.3383 | 0.272917 | 14 | 0.020977 | hsa-miR-1247 | miRNA |
| 132.3279 | 0.348404 | 6 | 0.008662 |
ENST00000597346.1 | lncRNA |
| 92.107 | 0.346561 | 5 | 0.007389 | AAK1 | mRNA |
| 92.107 | 0.346561 | 5 | 0.007389 |
ENST00000458178.1 | lncRNA |
| 54.59979 | 0.337629 | 4 | 0.006136 |
ENST00000544461.1 | lncRNA |
| 4273.936 | 0.458042 | 4 | 0.006679 | RELL1 | mRNA |
| 58.5175 | 0.344737 | 4 | 0.006111 | SCN5A | mRNA |
| 49.93945 | 0.344737 | 4 | 0.006131 | FNDC1 | mRNA |
| 49.93945 | 0.344737 | 4 | 0.006131 | FAM123C | mRNA |
| 48.78154 | 0.341146 | 4 | 0.006126 |
ENST00000567093.1 | lncRNA |
| 48.78154 | 0.341146 | 4 | 0.006126 | TNS1 | mRNA |
| 35.3917 | 0.332487 | 3 | 0.004916 |
ENST00000571219.1 | lncRNA |
| 28.90726 | 0.339378 | 3 | 0.004853 | ARGFX | mRNA |
 | Table IIINumber of lncRNA-miRNA and miRNA-mRNA
pairs. |
Table III
Number of lncRNA-miRNA and miRNA-mRNA
pairs.
| lncRNA | lncRNA-miRNA
pairs | miRNA-mRNA
pairs | Total number |
|---|
|
ENST00000597346.1 | 6 | 58 | 64 |
|
ENST00000458178.1 | 5 | 58 | 63 |
|
ENST00000567093.1 | 4 | 53 | 57 |
|
ENST00000544461.1 | 4 | 44 | 48 |
|
ENST00000570167.1 | 3 | 51 | 54 |
|
ENST00000506222.2 | 3 | 50 | 53 |
|
ENST00000571219.1 | 3 | 44 | 47 |
|
ENST00000499521.2 | 2 | 238 | 240 |
|
ENST00000602172.1 | 2 | 238 | 240 |
Key lncRNA-miRNA-mRNA subnetworks
The 5 key lncRNAs in the ceRNA network were further
investigated. The 5 lncRNAs were found to primarily target
miR-10a-5p, miR-30c-3p, miR-1247-5p, miR-1268a and miR-144-5p
(Table IV). The mRNAs and miRNAs
associated with these 5 lncRNAs were identified in the global
triple network and new subnetworks were constructed (Table IV). Among the 5 lncRNAs, sequence
Basic Local Alignment Search Tool (BLAST) analysis showed that the
sequence of lncRNA ENST00000597346.1 is part of ENST00000567093.1,
and the sequence of ENST00000544461.1 is also part of
ENST00000458178.1. ENST00000567093.1 and ENST00000458178.1 were
conserved among human, mouse and rat genomes, whereas
ENST00000571219.1 was only detected in the human genome. GO
function and KEGG pathway analyses for the lncRNA-related mRNAs
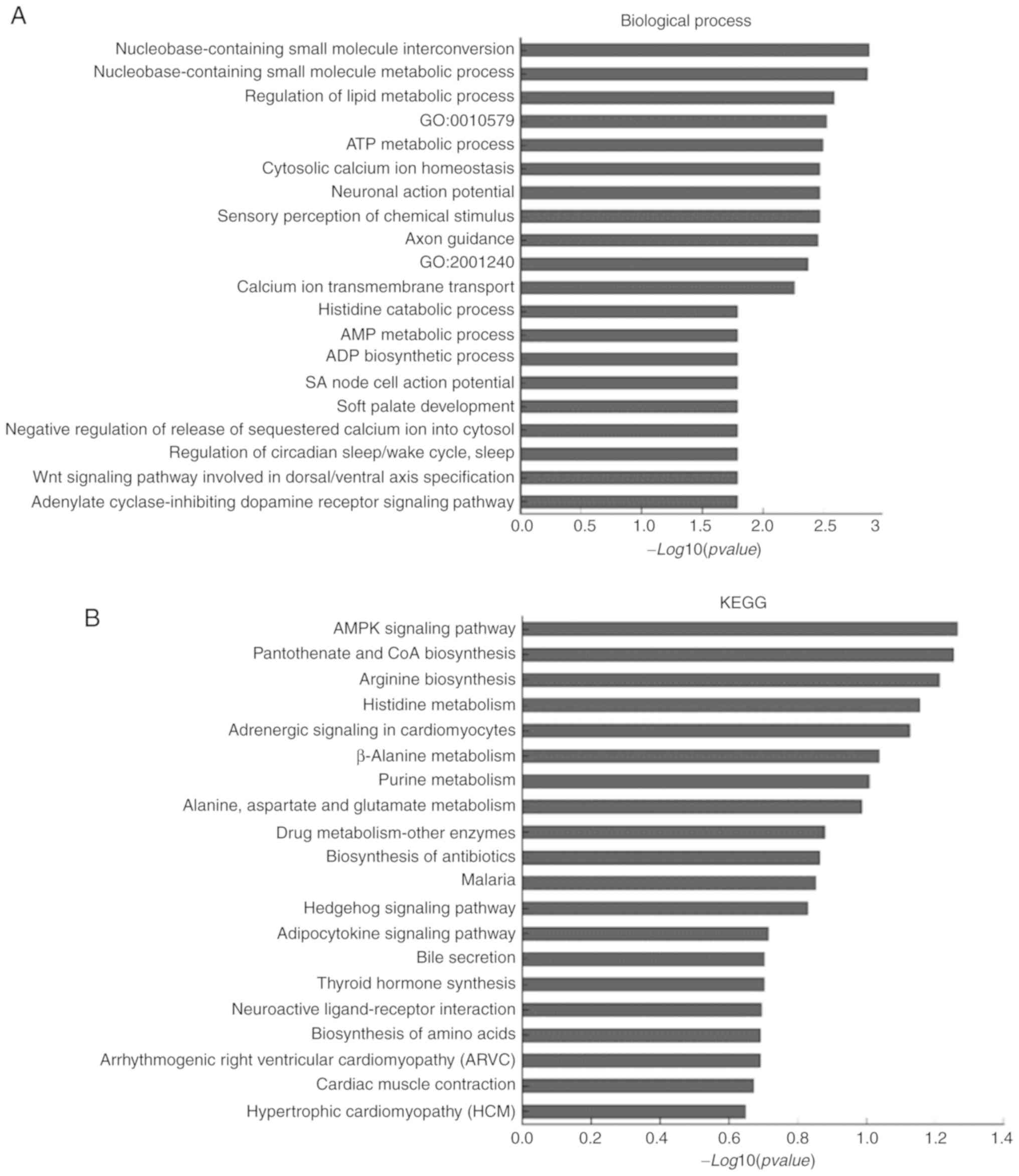
were then performed. For ENST00000458178.1, differentially
expressed mRNAs were primarily enriched in ‘Nucleobase-containing
small molecule interconversion’, ‘Nucleobase-containing small
molecule metabolic process’ and ‘Regulation of lipid metabolic
process’ (Fig. 3A). Enriched KEGG
pathways included the ‘AMPK signaling pathway’, ‘Pantothenate and
CoA biosynthesis’ and ‘Arginine biosynthesis’ (Fig. 3B), which shared a similar trend with
ENST00000567093.1 (Fig. 3D and
E). The subnetworks of
ENST00000458178.1 and ENST00000567093.1 are presented in Fig. 3C and F, respectively. The biological processes,
KEGG pathways and subnetwork of ENST00000571219.1-related mRNAs are
presented in Fig. S1A-C. The HCM
pathway was enriched in all 5 lncRNA-related KEGG pathways,
indicating the potential roles of these 5 lncRNAs in HCM (Fig. 3B and E; Fig.
S1B).
 | Table IVlncRNA-miRNA-mRNA networks of the 5
selected lncRNAs. |
Table IV
lncRNA-miRNA-mRNA networks of the 5
selected lncRNAs.
| lncRNA | ceRNA score | miRNA | mRNA |
|---|
|
ENST00000567093.1 | 1 | hsa-miR-10a;
hsa-miR-30c-1a;
hsa-miR-1268; hsa-miR-1247 | TNS1 |
|
ENST00000458178.1 | 0.687 | hsa-miR-10a;
hsa-miR-30c-1a;
hsa-miR-1268 | ANK1 |
|
ENST00000544461.1 | 0.666 | hsa-miR-10a;
hsa-miR-144a;
hsa-miR-1268 | KCNN2, SCN5A |
| | | hsa-miR-10a;
hsa-miR-1268; hsa-miR-1247 | TNS1 |
|
ENST00000597346.1 | 0.666 | hsa-miR-10a;
hsa-miR-30c-1a;
hsa-miR-1268 | ANK1 |
|
ENST00000571219.1 | 0.666 |
hsa-miR-30c-1a; hsa-miR-1247 | FAM22F, UROC1, AK1,
CRYGN, PPP1R26, TNS1 |
| | |
hsa-miR-144a; hsa-miR-30c-1a | RFT1, FAM196B,
CACNB2, EPB49, RGPD1, GRIK2, SCN5A, AAK1 |
Module analysis of a HCM-related
lncRNA-miRNA network
To further investigate the crosstalk between mRNAs
and lncRNAs, a bidirectional hierarchical clustering analysis was
performed using gplot in R software (version 3.5.1; http://www/r-project.org/). The heat map showed two
modules screened by cluster analysis that were closely related to
HCM (Fig. 4A-C). The two modules
included two different gene populations. GO enrichment analysis and
KEGG analysis of genes were performed in the modules (Fig. 4D-G). In module 1, ‘Positive
regulation of adenylate cyclase activity involved in
G-protein-coupled receptor signaling pathways (GO: 0010579)’ was
significantly and closely related to HCM. KEGG pathway analysis
further demonstrated that the ‘AMPC signaling pathway’ was the most
significant signaling pathway in HCM. In module 2, ‘Translation’
had the most notable relationship with HCM, and KEGG pathway
analysis showed that ‘Ribosome’ served a predominant role in HCM
(Fig. 4D-G). Among all the lncRNAs
in the two modules, 8 lncRNAs were included in the ceRNA network
(ENST00000499521.2, ENST00000458178.1, ENST00000567093.1,
ENST00000591866.1, ENST00000367207.3, ENST00000425185.1,
ENST00000596350.1 and ENST00000416301.1). According to the results
of the current study, ENST00000458178.1 and ENST00000567093.1 not
only had higher betweenness and node degree (Table II) but were also involved in a
higher number of lncRNA-miRNA and miRNA-mRNA pairs (Table III), which were regarded as
potential targets for HCM and were confirmed by module
analysis.
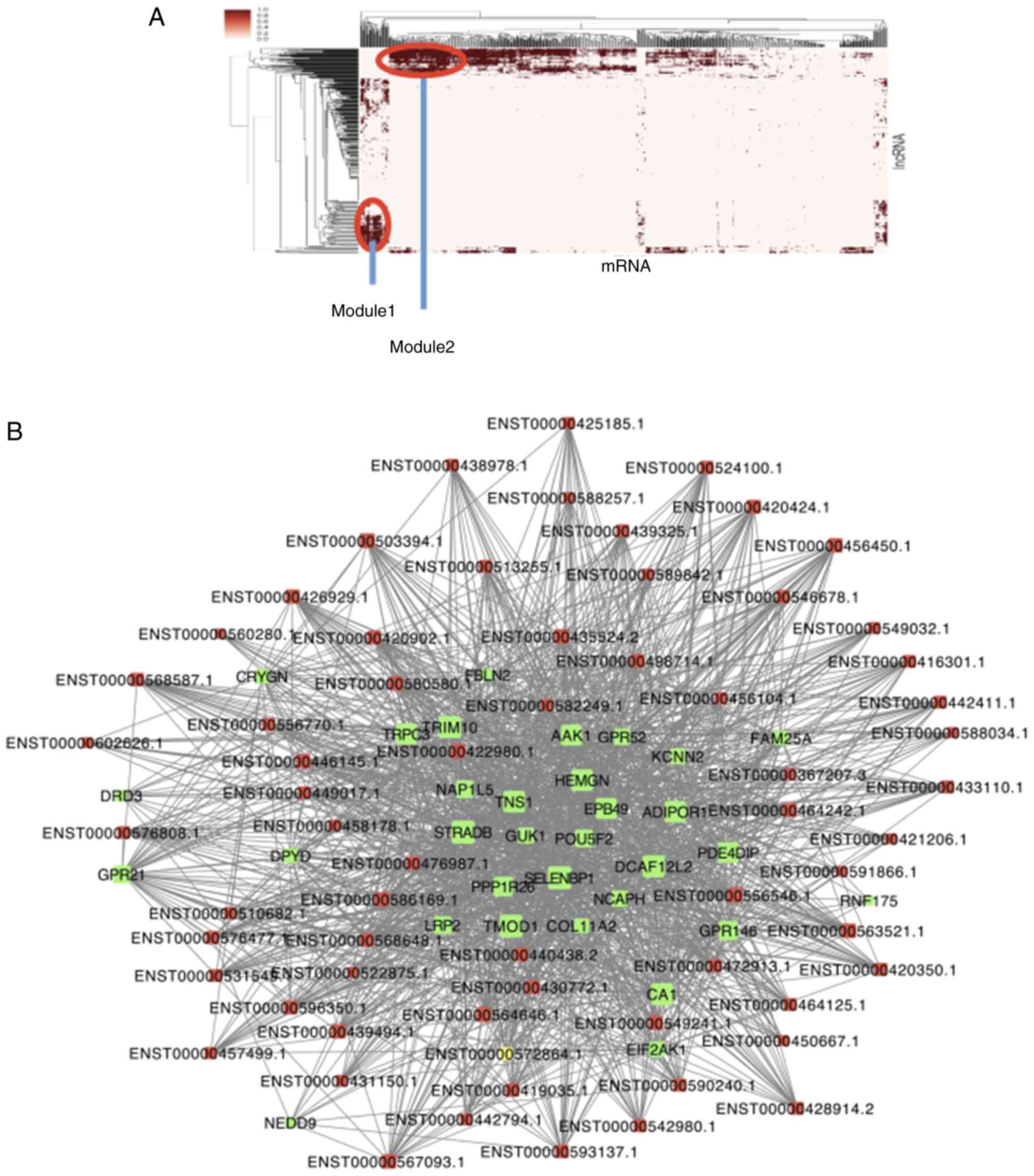 | Figure 4Module analysis of the HCM-related
lncRNA-miRNA network. (A) Two modules were determined to be highly
related to HCM. lncRNA-mRNA networks in (B) module 1. HCM,
hypertrophic cardiomyopathy; lncRNA, long non-coding RNA; miRNA,
microRNA; KEGG, Kyoto Encyclopedia of Genes and Genomes. Module
analysis of the HCM-related lncRNA-miRNA network. lncRNA-mRNA
networks in (C) module 2. Red nodes represent lncRNAs and green
nodes represent mRNAs. (D) Biological processes and (E) KEGG
analysis of differentially expressed mRNAs related to module 1.
HCM, hypertrophic cardiomyopathy; lncRNA, long non-coding RNA;
miRNA, microRNA; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Module analysis of the HCM-related lncRNA-miRNA network. (F)
Biological processes and (G) KEGG analysis of differentially
expressed mRNAs related to module 2. HCM, hypertrophic
cardiomyopathy; lncRNA, long non-coding RNA; miRNA, microRNA; KEGG,
Kyoto Encyclopedia of Genes and Genomes. |
Discussion
HCM is a commonly inherited cardiovascular disease
that is the most frequent cause of sudden death in younger
individuals and functional disability resulting from heart failure
and stroke (28). Although
therapeutic advancements have been made in HCM, the treatment of
HCM remains limited. Currently, major efforts have been made to
detect the underlying mechanisms of HCM (29-31).
ncRNAs including miRNAs and lncRNAs, have been identified as novel
regulators underlying HCM (32-35).
However, current studies of lncRNAs are focused on RNA sequencing
or microarray technology (21,22), the
functional studies are relative few and complicated compared to
miRNAs. Therefore, it is necessary to identify an efficient and
accurate way to infer the potential function of lncRNAs with miRNAs
and/or mRNAs whose functions have already been annotated. In the
present study, interaction data from NCBI GEO (lncRNA-mRNA dataset:
GSE68316, miRNA dataset: GSE36946) were used to generate a triple
network based on ceRNA theory, which suggested that lncRNAs and
mRNAs share the common miRNA in one triplet. The ceRNA network was
composed of 30 lncRNA nodes, 94 mRNA nodes and 8 miRNA nodes.
Subsequently, hub nodes and the number of relationship pairs were
used to perform topological and subnetwork analysis. 2 lncRNAs
(ENST00000458178.1 and ENST00000567093.1) were observed to be key
topological nodes, whose node degrees and number of lncRNA-miRNA
and miRNA-mRNA interaction pairs were higher compared with other
lncRNAs, and these 2 lncRNAs were highly conserved in human, mouse
and rat genomes. According to the ceRNA network, the 2 lncRNAs
primarily targeted miR-10a-5p, miR-30c-3p, miR-1247-5p and
miR-1268a.
A number of transcriptomic and proteomic profiling
analyses of HCM have been previously performed and several
dysregulated miRNAs have been identified. For example, Song et
al (36) identified 13
dysregulated miRNAs involved in myocardial tissues in patients with
HCM. The list of miRNAs in GSE36946 and previous reports on HCM or
cardiac hypertrophy were compared. Several miRNAs, including
miR-10a, miR-30c and miR-373, were common between the present study
and previous studies (37-41).
Additionally, studies have shown that miR-10a was downregulated in
a transverse abdominal aortic constriction-induced cardiac
hypertrophy model and angiotensin II (Ang II)-stimulated
cardiomyocytes. Overexpression of miR-10a in Ang II-treated
cardiomyocytes ameliorated cell hypertrophy and decreased the
expression of natriuretic peptides A and B by directly inhibiting
T-box transcription factor TBX5 (37,38).
Diabetic cardiomyopathy (DCM) is characterized by endothelial
dysfunction, myocyte hypertrophy, necrosis, apoptosis and increased
fibrosis deposition (39). A
previous study demonstrated that cardiac miR-30c expression was
decreased in rats and patients with DCM and in high HG-treated
cardiomyocytes (40). Overexpression
of miR-30c attenuated HG-induced cardiomyocyte hypertrophy by
inhibiting cell division control protein 42 homolog and
serine/threonine-protein kinase PAK 1(40). Furthermore, a study reported that
concurrent overexpression of miR-30c and miR-181a resulted in a
greater decrease in cardiomyocyte hypertrophy and apoptosis via the
p53-p21 pathway compared with the overexpression of miR-30c or
miR-181a alone, suggesting a synergistic effect of these two miRNAs
in DCM-induced cardiac hypertrophy (41). Contrary to our results, miR-373
levels were decreased in the plasma of patients with HCM, and
overexpression of miR-373 improved cardiac hypertrophy induced by
DCM. The discrepancy in the expression profiles of miRNAs may have
arose from differences in tissue samples and models of cardiac
hypertrophy. However, although studies have shown that these miRNAs
serve important roles in cardiac hypertrophy, the functions and
underlying mechanisms of the miRNAs in HCM remains to be
elucidated.
In the present study, GO and KEGG pathway analyses
were used to assess biological functions that are enriched among
differentially expressed coding genes. Owing to similar miRNA
targets (miR-10a-5p, hsa-miR-30c-3p and miR-1268a), the significant
GO terms of ENST00000567093.1 and ENST00000458178.1 shared common
trends involving ‘Nucleobase-containing small molecule
interconversion’, ‘Nucleobase-containing small molecule metabolic
process’ and ‘Regulation of lipid metabolic process’, and the
results were consistent with previous studies on HCM (42,43).
Pathway analysis of lncRNA-related mRNAs showed that the HCM
pathway was highly enriched. The other top pathways based on KEGG
pathway analyses, including ‘AMPK signaling pathway’, ‘Pantothenate
and CoA biosynthesis’, ‘Arginine biosynthesis’, ‘Adrenergic
signaling in cardiomyocytes’ and ‘Histidine metabolism’ were
primarily metabolic pathways that have been shown to serve
important roles in HCM (44-47).
Bidirectional hierarchical clustering analysis was
performed to investigate the crosstalk between mRNAs and lncRNAs.
GO and KEGG pathway analyses of these 2 modules indicated that in
module 1, the dysregulated genes were primarily involved in
metabolic regulation, including lipid regulation,
nucleobase-containing small molecule metabolic process and CoA
biosynthesis, which has been discussed earlier. In module 2, the
mRNAs were primarily enriched in translational regulation,
including translation initiation, termination and elongation,
ribosome and RNA degradation. Once the transcriptional process is
impaired, the expression levels of numerous proteins are
dysregulated, and this may alter normal physiological processes. To
the best of our knowledge, there are no previous studies which have
addressed translational regulation in HCM, and thus, it would be
meaningful to investigate transcriptional dysfunction in HCM.
Nevertheless, additional studies are required to fully understand
the molecular mechanisms underlying HCM.
Currently, identifying ncRNA-disease associations is
playing an increasingly vital role in diagnostic and therapeutic
tools for diseases including HCM. However, since the fact that it
is expensive and time-consuming via experimental studies to uncover
associations between ncRNAs and disease, novel and effective
computational models for the identification of ncRNAs associated
with HCM or other diseases are being developed. Several novel
computational methods have been used to calculate potential
ncRNAs-disease association scores (48,49).
lncRNAs have emerged as one of the largest and significantly
diverse type of RNA family (50,51). The
biological role and functions of lncRNAs are diverse and still
mostly uncharacterized. Their target-mimetic and sponge/decoy
function on miRNAs have been identified. Previous studies
demonstrated that lncRNAs can act as miRNA sponges, reducing their
regulatory effect on mRNAs (52,53).
This function introduces an extra layer of complexity in the
miRNA-mRNA interaction network. In addition, while several studies
examining. lncRNAs or miRNAs in cardiac hypertrophy have been
performed (54-56),
to the best of our knowledge, there are relatively fewer studies
focusing on a ceRNA network between lncRNAs and miRNAs of HCM
(57,58), particularly in human models. In the
present study, a ceRNA based lncRNA-miRNA-mRNA network in human HCM
models was constructed. A total of 2 lncRNAs (ENST00000458178.1 and
ENST00000567093.1) were determined to be key topological nodes and
highly conserved in human, mouse and rat genomes. According to the
ceRNA network, the 2 lncRNAs primarily targeted miR-10a-5p,
miR-30c-3p, miR-1247-5p and miR-1268a, which were highly related to
the development of HCM. Therefore, the study provides a framework
for constructing powerful computational methods in human HCM models
to predict potential lncRNA-miRNA-disease associations and select
the most promising lncRNAs/miRNAs related to HCM or other diseases
for experimental validation.
However, there are several disadvantages to using
bioinformatics analysis. Firstly, the changes and potential
functions of lncRNAs/miRNAs/mRNAs are hypothetical, and need to be
further verified in animal or human models of hypertrophic
cardiomyopathy. Secondly, a lncRNA-miRNA-mRNA ceRNA network was
constructed by combining two different GSE datasets (GSE68313 and
GSE36946); there may be missing and overlapping data between
different databases. HCM myocardial samples will be collected and a
complete lncRNA/miRNA/mRNA profile will be performed in future
studies. The functional role of the 2 lncRNAs on HCM and the
relationship between lncRNAs and miRNAs detected in the present
study will be further explored. The underlying mechanisms can be
investigated by western blotting and proteomics studies. A
luciferase reporter assay will also be performed to confirm the
target genes of miRNAs in future work.
The present study has some limitation. Due to the
lack of samples, the changes in lncRNAs, miRNAs and mRNAs were not
validated in myocardial tissues from patients with HCM, and thus,
there may have been false positives. Additionally, during the
process of converting gene IDs from different databases, a number
of genes may have been lost, which may have decreased the accuracy
of the present results. Finally, the present study was primarily
focused on alterations in lncRNAs/miRNAs in HCM samples, whereas
the specific functions of ceRNAs in HCM remain unknown, therefore
the underlying biological functions and mechanisms warrants further
exploration.
In conclusion, a ceRNA based lncRNA-miRNA-mRNA
network was constructed in the present study, providing a new
strategy for studying HCM and other diseases. Furthermore,
lncRNA-miRNA pairs may be regarded as candidate diagnostic
biomarkers or potential therapeutic targets for treatment of
HCM.
Supplementary Material
lncRNA-miRNA-mRNA subnetwork of lncRNA
ENST00000544461. (A) Biological function and (B) pathway analysis
of differentially expressed mRNAs related to lncRNA
ENST00000544461.1. (C) lncRNA ENST00000544461.1 related miRNA-mRNA
subnetwork. Green nodes represent miRNAs and red nodes represent
mRNAs. miRNA, microRNA; lncRNA, long non-coding RNA.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National
Natural Science Foundation of China (grant no. 81700343), Natural
Science Foundation of Jiangsu Province (grant no. BK20170296),
China Postdoctoral Science Foundation (grant no. 2018M642317),
Post-Doctoral Foundation of Jiangsu Province (grant no. 2018K095B),
Six Talent Peaks Project of Jiangsu Province (grant nos. WSN-202
and WSW-183), Maternal and Child Health Research Project of Jiangsu
Province (grant no. F201803), and Changzhou High-Level Medical
Talents Training Project (grant no. 2016ZCL J020).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GEO repository. GSE36949:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36949GSE68316:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68316
Authors' contributions
FH and JS analyzed, interpreted and ensured the
quality of the data. LT conceived and designed the study. LY and XH
developed the methodology. LT wrote and reviewed the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Maron BJ, Casey SA, Olivotto I, Sherrid
MV, Semsarian C, Autore C, Ahmed A, Boriani G, Francia P, Winters
SL, et al: Clinical course and quality of life in high-risk
patients with hypertrophic cardiomyopathy and implantable
cardioverter-defibrillators. Circ Arrhythm Electrophysiol.
11(e005820)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Maron BJ: Recognition of hypertrophic
cardiomyopathy as a contemporary, relatively common, and treatable
disease (from the international summit V). Am J Cardiol.
113:739–744. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Spudich JA: Hypertrophic and dilated
cardiomyopathy: Four decades of basic research on muscle lead to
potential therapeutic approaches to these devastating genetic
diseases. Biophys J. 106:1236–1249. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Roma-Rodrigues C and Fernandes AR:
Genetics of hypertrophic cardiomyopathy: Advances and pitfalls in
molecular diagnosis and therapy. Appl Clin Genet. 7:195–208.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
de Gregorio C and Andò G: Risk of sudden
death and outcome in patients with hypertrophic cardiomyopathy with
benign presentation and without risk factors: A word of comfort to
younger patients? Am J Cardiol. 114:500–501. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Spirito P, Autore C, Formisano F, Assenza
GE, Biagini E, Haas TS, Bongioanni S, Semsarian C, Devoto E,
Musumeci B, et al: Risk of sudden death and outcome in patients
with hypertrophic cardiomyopathy with benign presentation and
without risk factors. Am J Cardiol. 113:1550–1555. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Towe EC, Bos JM, Ommen SR, Gersh BJ and
Ackerman MJ: Genotype-phenotype correlations in apical variant
hypertrophic cardiomyopathy. Congenit Heart Dis. 10:E139–E145.
2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cirino AL, Harris S, Lakdawala NK, Michels
M, Olivotto I, Day SM, Abrams DJ, Charron P, Caleshu C, Semsarian
C, et al: Role of genetic testing in inherited cardiovascular
disease: A review. JAMA Cardiol. 2:1153–1160. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Roma-Rodrigues C, Raposo LR and Fernandes
AR: MicroRNAs based therapy of hypertrophic cardiomyopathy: The
road traveled so far. BioMed Res Int. 2015(983290)2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sartorio CL, Lazzeroni D, Bertoli G and
Camici PG: Theranostic biomarkers in hypertrophic cardiomyopathy:
Insights in a long road ahead. Front Biosci (Landmark Ed).
22:1724–1749. 2017.PubMed/NCBI
|
|
11
|
Liebetrau C, Mollmann H, Dörr O, Szardien
S, Troidl C, Willmer M, Voss S, Gaede L, Rixe J, Rolf A, et al:
Release kinetics of circulating muscle-enriched microRNAs in
patients undergoing transcoronary ablation of septal hypertrophy. J
Am Coll Cardiol. 62:992–998. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bagnall RD, Tsoutsman T, Shephard RE,
Ritchie W and Semsarian C: Global microRNA profiling of the mouse
ventricles during development of severe hypertrophic cardiomyopathy
and heart failure. PLoS One. 7(e44744)2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sayed D, Hong C, Chen IY, Lypowy J and
Abdellatif M: MicroRNAs play an essential role in the development
of cardiac hypertrophy. Circ Res. 100:416–424. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ntelios D, Meditskou S, Efthimiadis G,
Pitsis A, Nikolakaki E, Girtovitis F, Parcharidou D, Zegkos T,
Kouidou S, Karvounis H and Tzimagiorgis G: Elevated plasma levels
of miR-29a are associated with hemolysis in patients with
hypertrophic cardiomyopathy. Clin Chim Acta. 471:321–326.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fang L, Ellims AH, Moore XL, White DA,
Taylor AJ, Chin-Dusting J and Dart AM: Circulating microRNAs as
biomarkers for diffuse myocardial fibrosis in patients with
hypertrophic cardiomyopathy. J Transl Med. 13(314)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li X, He X, Wang H, Li M, Huang S, Chen G,
Jing Y, Wang S, Chen Y, Liao W, et al: Loss of AZIN2 splice variant
facilitates endogenous cardiac regeneration. Cardiovasc Res.
114:1642–1655. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ono K, Kuwabara Y, Horie T and Kimura T:
Long non-coding RNAs as key regulators of cardiovascular diseases.
Circ J. 82:1231–1236. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kaur H, Sarmah D, Saraf J, Vats K, Kalia
K, Borah A, Yavagal DR, Dave KR, Ghosh Z and Bhattacharya P:
Noncoding RNAs in ischemic stroke: Time to translate. Ann N Y Acad
Sci. 1421:19–36. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liang H, Pan Z, Zhao X, Liu L, Sun J, Su
X, Xu C, Zhou Y, Zhao D, Xu B, et al: LncRNA PFL contributes to
cardiac fibrosis by acting as a competing endogenous RNA of let-7d.
Theranostics. 8:1180–1194. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
He L, Chen Y, Hao S and Qian J: Uncovering
novel landscape of cardiovascular diseases and therapeutic targets
for cardioprotection via long noncoding RNA-miRNA-mRNA axes.
Epigenomics. 10:661–671. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu X, Ma Y, Yin K, Li W, Chen W, Zhang Y,
Zhu C, Li T, Han B, Liu X, et al: Long non-coding and coding RNA
profiling using strand-specific RNA-seq in human hypertrophic
cardiomyopathy. Sci Data. 6(90)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yang W, Li Y, He F and Wu H: Microarray
profiling of long non-coding RNA (lncRNA) associated with
hypertrophic cardiomyopathy. BMC Cardiovasc Disord.
15(62)2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liang H, Su X, Wu Q, Shan H, Lv L, Yu T,
Zhao X, Sun J, Yang R, Zhang L, et al: LncRNA 2810403D21Rik/Mirf
promotes ischemic myocardial injury by regulating autophagy through
targeting Mir26a. Autophagy. 12:1–15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Feng Y, Xu W, Zhang W, Wang W, Liu T and
Zhou X: LncRNA DCRF regulates cardiomyocyte autophagy by targeting
miR-551b-5p in diabetic cardiomyopathy. Theranostics. 9:4558–4566.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang M, Jiang Y, Guo X, Zhang B, Wu J,
Sun J, Liang H, Shan H, Zhang Y, Liu J, et al: Long non-coding RNA
cardiac hypertrophy-associated regulator governs cardiac
hypertrophy via regulating miR-20b and the downstream PTEN/AKT
pathway. J Cell Mol Med. 23:7685–7698. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Maron BJ and Maron MS: Hypertrophic
cardiomyopathy. Lancet. 381:242–255. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Rakowski H: Determining hypertrophic
cardiomyopathy mortality: Gaining wisdom from knowledge. JAMA
Cardiol 2019.
|
|
30
|
Liu F, Fu J, His D, Sun C, He G, Hu R,
Zhang J and Liu L: Percutaneous intramyocardial septal
radiofrequency ablation for interventricular septal reduction: An
ovine model with 1-year outcomes. Cardiology. 20:1–10.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Boll G, Rowin EJ, Maron BJ, Wang W,
Rastegar H and Maron MS: Efficacy of combined cox-maze IV and
ventricular septal myectomy for treatment of atrial fibrillation in
patients with obstructive hypertrophic cardiomyopathy. Am J
Cardiol. 125:120–126. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kitow J, Derda AA, Beermann J, Kumarswarmy
R, Pfanne A, Fendrich J, Lorenzen JM, Xiao K, Bavendiek U,
Bauersachs J and Thum T: Mitochondrial long noncoding RNAs as blood
based biomarkers for cardiac remodeling in patients with
hypertrophic cardiomyopathy. Am J Physiol Heart Circ Physiol.
311:H707–H712. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gómez J, Lorca R, Reguero JR, Martin M,
Moris C, Alonso B, Iglesias S, Diaz-Molina B, Avanzas P and Coto E:
Genetic variation at the long noncoding RNA H19 gene is associated
with the risk of hypertrophic cardiomyopathy. Epigenomics.
10:865–873. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Shi H, Li J, Song Q, Cheng L, Sun H, Fan
W, Li J, Wang Z and Zhang G: Systematic identification and analysis
of dysregulated miRNA and transcription factor feed-forward loops
in hypertrophic cardiomyopathy. J Cell Mol Med. 23:306–316.
2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li M, Chen X, Chen L, Chen K, Zhou J and
Song J: MiR-1-3p that correlates with left ventricular function of
HCM can serve as a potential target and differentiate HCM from DCM.
J Trans Med. 16(161)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Song L, Su M, Wang S, Zou Y, Wang X, Wang
Y, Cui H, Zhao P, Hui R and Wang J: MiR-451 is decreased in
hypertrophic cardiomyopathy and regulates autophagy by targeting
TSC1. J Cell Mol Med. 18:2266–2274. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang D, Zhai G, Ji Y and Jing H:
microRNA-10a targets T-box 5 to inhibit the development of cardiac
hypertrophy. Int Heart J. 58:100–106. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang F, Yang XY, Zhao JY, Yu LW, Zhang P,
Duan WY, Chong M and Gui YH: miR-10a and miR-10b target the
3'-untranslated region of TBX5 to repress its expression. Pediatr
Cardiol. 35:1072–1079. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jia G, DeMarco VG and Sowers JR: Insulin
resistance and hyperinsulinaemia in diabetic cardiomyopathy. Nat
Rev Endocrinol. 12:144–153. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Raut SK, Kumar A, Singh GB, Nahar U,
Sharma V, Mittal A, Sharma R and Khullar M: miR-30c mediates
upregulation of Cdc42 and Pak1 in diabetic cardiomyopathy.
Cardiovasc Ther. 33:89–97. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Raut SK, Singh GB, Rastogi B, Saikia UN,
Mittal A, Dogra N, Singh S, Prasad R and Khullar M: miR-30c and
miR-181a synergistically modulate p53-p21 pathway in diabetes
induced cardiac hypertrophy. Mol Cell Biochem. 417:191–203.
2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu X, Ye B, Miller S, Yuan H, Zhang H,
Tian L, Nie J, Imae R, Arai H, Li Y, et al: Ablation of ALCAT1
mitigates hypertrophic cardiomyopathy through effects on oxidative
stress and mitophagy. Mol Cell Biol. 32:4493–4504. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Krishnan J, Suter M, Windak R, Krebs T,
Felley A, Montessuit C, Tokarska-Schlattner M, Aasum E, Bogdanova
A, Perriard E, et al: Activation of a HIF1alpha-PPARgamma axis
underlies the integration of glycolytic and lipid anabolic pathways
in pathologic cardiac hypertrophy. Cell Metab. 9:512–524.
2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Murphy RT, Mogensen J, McGarry K, Bahl A,
Evans A, Osman E, Syrris P, Gorman G, Farrell M, Holton JL, et al:
Adenosine monophosphate-activated protein kinase disease mimicks
hypertrophic cardiomyopathy and wolff-parkinson-white syndrome:
Natural history. J Am Coll Cardiol. 45:922–930. 2005.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Bauersachs J, Störk S, Kung M, Waller C,
Fidler F, Hoyer C, Frantz S, Weidemann F, Ertl G and Angermann CE:
HMG CoA reductase inhibition and left ventricular mass in
hypertrophic cardiomyopathy: A randomized placebo-controlled pilot
study. Eur J Clin Invest. 37:852–859. 2007.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Rani DS, Nallari P, Priyamvada S,
Narasimhan C, Singh L and Thangaraj K: High prevalence of arginine
to glutamine substitution at 98, 141 and 162 positions in troponin
I (TNNI3) associated with hypertrophic cardiomyopathy among
Indians. BMC Med Genet. 13(69)2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Jimenez J and Tardiff JC: Abnormal heart
rate regulation in murine hearts with familial hypertrophic
cardiomyopathy-related cardiac troponin T mutations. Am J Physiol
Heart Circ Physiol. 300:H627–H635. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chen X, Xie D, Zhao Q and You ZH:
MicroRNAs and complex diseases: From experimental results to
computational models. Brief Bioinform. 20:515–539. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chen X, Wang L, Qu J, Guan NN and Li JQ:
Predicting miRNA-disease association based on inductive matrix
completion. Bioinformatics. 34:4256–4265. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li Y, Zhang J, Huo C, Ding N, Li J, Xiao
J, Lin X, Cai B, Zhang Y and Xu J: Dynamic organization of lncRNA
and circular RNA regulators collectively controlled cardiac
differentiation in humans. EBioMedicine. 24:137–146.
2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Li Z, Zhang Y, Ding N, Zhao Y, Ye Z, Shen
L, Yi H and Zhu Y: Inhibition of lncRNA XIST improves myocardial
I/R injury by targeting miR-133a through inhibition of autophagy
and regulation of SOCS2. Mol Ther Nucleic Acids. 18:764–773.
2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Dykes IM and Emanueli C: Transcriptional
and post-transcriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Oh JG, Watanabe S, Lee A, Gorski PA, Lee
P, Jeong D, Liang L, Liang Y, Baccarini A, Sahoo S, et al: miR-146a
suppresses SUMO1 expression and induces cardiac dysfunction in
maladaptive hypertrophy. Circ Res. 123:673–685. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wang K, Long B, Liu F, Wang JX, Liu CY,
Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al: A circular RNA
protects the heart from pathological hypertrophy and heart failure
by targeting miR-223. Eur Heart J. 37:2602–2611. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Bang C, Batkai S, Dangwal S, Gupta SK,
Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, et al:
Cardiac fibroblast-derived microRNA passenger strand-enriched
exosomes mediate cardiomyocyte hypertrophy. J Clin Invest.
124:2136–2146. 2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Yuan Y, Wang J, Chen Q, Wu Q, Deng W, Zhou
H and Shen D: Long non-coding RNA cytoskeleton regulator RNA
(CYTOR) modulates pathological cardiac hypertrophy through
miR-155-mediated IKKi signaling. Biochim Biophys Acta Mol Basis
Dis. 1865:1421–1427. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Li J, Wu Z, Zheng D, Sun Y, Wang S and Yan
Y: Bioinformatics analysis of the regulatory lncRNAmiRNAmRNA
network and drug prediction in patients with hypertrophic
cardiomyopathy. Mol Med Rep. 20:549–558. 2019.PubMed/NCBI View Article : Google Scholar
|