Introduction
Vitiligo is a common skin disorder characterized by
melanocyte dysfunction in the epidermis of the skin, which results
in the appearance of white spots on the skin of affected patients
(1). The prevalence of vitiligo is
~1% in the USA (2) and ~0.56% in
China (3). Although vitiligo is not
life-threatening, it is associated with a significant psychological
burden for patients (4) and it may
result in large financial costs for public health systems. In fact,
it is estimated that the cost of vitiligo is ~175 million dollars
per year in the USA (5). The
pathogenesis of vitiligo remains unclear and currently, there is no
gold standard treatment for the disease (6). Therefore, there is an urgent
requirement for the identification of novel therapeutic targets and
the development of new methods to treat vitiligo with an improved
therapeutic efficacy.
Previously, clinical and epidemiological
investigations have demonstrated that vitiligo is a complex disease
that can be affected by various environmental and genetic factors,
such as heredity and gene mutation (7-9). In
addition, it was reported that NLR family pyrin domain containing 1
(NLRP1) and the histocompatibility complex are associated with an
increased risk of developing vitiligo (10-12).
Notably, genome-wide analysis previously suggested that zinc finger
MIZ-type containing 1 (ZMIZ1) is significantly associated with the
occurrence and development of vitiligo (13). ZMIZ1, also termed hZiMP10, is located
in the 10q22.3 region of the chromosome (14). ZMIZ1 belongs to the protein inhibitor
of activated STAT (PIAS) family and it encodes a PIAS-like protein
containing 1,067 amino acid residues (15). Similar to other PIAS proteins, ZMIZ1
contains a ring finger region termed Miz, which serves an important
role in protein-protein interactions (16). However, the exact role of ZMIZ1 in
vitiligo remains unknown and requires further investigation.
Therefore, the present study aimed to investigate
the role of ZMZ1 in vitiligo, in addition to the related mechanism.
ZMIZ1 overexpression and knockdown melanocyte cell lines were
established, and the effects of ZMIZ1 on the proliferation,
apoptosis, migration and invasion of melanocytes were investigated.
The results of the present study may provide a theoretical basis
for the clinical application of ZMIZ1 as a potential therapeutic
target for the treatment of vitiligo.
Materials and methods
Cell culture
The human normal melanocyte cell line PIG1 and human
vitiligo melanocyte cell line PIG3V were purchased from ScienCell
Research Laboratories, Inc. Cells were cultured in Medium 254
(Gibco; Thermo Fisher Scientific, Inc.), supplemented with 0.5%
FBS, and 100 U/ml penicillin and 100 U/ml streptomycin Cells were
maintained in a humidified incubator at 37˚C and 5% CO2.
The medium was changed every other day.
Lentiviral infection
The gene fragment of ZMIZ1 and three short hairpin
RNA (shRNA/sh) interference fragments, sh-ZMIZ1-1, sh-ZMIZ1-2 and
sh-ZMIZ1-3 (purchased from Shanghai GenePharma Co., Ltd.),
targeting ZMIZ1 were subcloned into lentiviral vectors
(pGLVH1/GFP+Puro; Shanghai GenePharma Co., Ltd.), alongside the
overexpression negative control (OE NC; empty vector) and the
sh-NC. Packaging plasmid (pAX2), envelope plasmid (pMD2.G) and
pGLVH1/GFP+Puro, 293T cells (seeded onto the cell culture plates
with 5x106 cells/well) (ScienCell Research Laboratories,
Inc.) were infected with these lentiviral vectors as previously
described (17). The sequences of
the shRNAs were as follows: Sh-ZMIZ1-1,
5'-GGAGAGCCCAACTATGGAAAC-3'; sh-ZMIZ1-2,
5'-GCCCATCAAGTCGGACTTAC-3'; sh-ZMIZ1-3,
5'-GCCAGATGATCATGCCCAATG-3', sh-NC, 5'-TAATGCTATCCGTCTAATC-3'. 293T
cells (Cell Bank of the Chinese Academy of Sciences;
3x106 cells/10 cm dish) were transfected using
Lipofectamine™ 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The following ratio was
used for vector transfection: Lentiviral vectors: packaging
plasmid: envelope plasmid; 10 µg: 7.5 µg: 3 µg for 72 h at 37˚C in
an incubator before the 293T lentiviral supernatant was harvested
by centrifugation at 200 x g for 5 min at room temperature. PIG1
and PIG3V cells (2x105 cells/ml) in six-well plates were
infected with the 293T lentiviral supernatant in the presence of 5
µg/ml polybrene (Beyotime Institute of Biotechnology). Cells were
collected 48 h after infection for further analysis, and the
infection efficiency was determined by fluorescence microscopy
(magnification, x400).
Cells were divided into 7 groups: i) Control
(Con) group (uninfected cells); ii) OE NC group; iii) OE ZMIZ1
group; iv) sh-NC group; v) sh-ZMIZ1-1 group; vi) sh-ZMIZ1-2 group;
and vii) sh-ZMIZ1-3 group.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
To determine the mRNA expression levels of ZMIZ1 in
PIG1 and PIG3V cells, total RNA was extracted from the cells using
the RNeasy Mini kit (Qiagen GmbH), according to the manufacturer's
protocol. Total RNA was reverse transcribed into cDNA using the
PrimeScript RT Master mix (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. The expression levels of
ZMIZ1 were quantified by qPCR using the SYBR-Greem Premix Ex Taq
kit (Takara Biotechnology, Co., Ltd.) according to the
manufacturer's protocol and an ABI 7500 Biosystems thermocycler.
The following thermocycling conditions were used: Initial
denaturation at 94˚C for 5 min, 36 cycles at 94˚C for 20 sec, 54˚C
for 20 sec and 72˚C for 20 sec. ZMIZ1 mRNA levels were quantified
using the 2-ΔΔCq method (18) and normalized to the internal
reference gene GAPDH. The following primers sequences were used:
ZMIZ1 forward, 5'-TGTTTGACGGTGGTCAGTCG-3' and reverse,
5'-CTTGTCTCGGTTTGCAGCAC-3'; and GAPDH forward,
5'-TGTGGGCATCAATGGATTTGG-3' and reverse,
5'-ACACCATGTATTCCGGGTCAAT-3'.
Western blotting
Total protein was extracted from PIG1 and PIG3V
cells using RIPA lysis buffer (Beyotime Institute of
Biotechnolohy). Total protein was quantified using a bicinchoninic
acid assay kit (Beyotime Institute of Biotechnology) and 20 µg
protein/lane was separated by 8% SDS-PAGE. The separated proteins
were subsequently transferred onto PVDF membranes and blocked with
5% non-fat dry milk in Tris-buffered saline (TBS; pH 7.4)
containing 0.05% Tween-20 at room temperature for 1 h. The
membranes were incubated with the following primary antibodies
overnight at 4˚C: anti-ZMIZ1 (1:1,000; cat. no. abs126964a; Absin,
Biotechnology Co., Ltd.), anti-Bcl-2 (1:1,000; cat. no. ab196495;
Abcam), anti-caspase-3 (1:1,000; cat. no. 9662; Cell Signaling
Technology, Inc.), anti-cleaved caspase-3 antibody (1:1,000; cat.
no. 9661; Cell Signaling Technology, Inc.) anti-GAPDH (1:2,000;
cat. no. 60004-1-1; ProteinTech Group, Inc.) and anti-α-tubulin
(1:2,000; cat. no. 66031-1-lg; ProteinTech Group, Inc.). Following
the primary antibody incubation, the membranes were incubated with
horseradish peroxidase-conjugated secondary antibodies (anti-rabbit
antibody; 1:2,000; cat. no. A0208 and anti-mouse antibody; 1:2,000;
cat. no. A0216; both from Beyotime Institute of Biotechnology) at
room temperature for 1 h and then washed using TBS-Tween-20.
Protein bands were visualized using a SuperSignal West Pico
Chemiluminescent substrate (Pierce; Thermo Fisher Scientific, Inc.)
and the chemiluminescence signals were detected using a Tanon-5200
Imaging system (Tanon Science and Technology, Co., Ltd.). α-tubulin
or GAPDH served as the loading control. Densitometric analysis was
performed using ImageJ (v1.46; National Institutes of Health).
Cell proliferation assay
The MTT assay was performed 48 h post-lentiviral
infection to determine the effect of ZMIZ1 on the proliferation of
PIG1 and PIG3V cells. Briefly, cells were harvested by
centrifugation (300 x g) at 4˚C for 5 min, resuspended, seeded into
96-well plates (5x103 cells/well). Next, 5 mg/ml MTT
solution was added into each well and the cells were incubated for
an additional 4 h in a humidified incubator at 37˚C and 5%
CO2. Finally, the absorbance at a wavelength of 490 nm
was measured using a microplate reader.
Flow cytometric analysis of
apoptosis
The effect of ZMIZ1 on the apoptosis of PIG1 and
PIG3V cells was determined using the Annexin V apoptosis detection
kit (Nanjing KeyGen Biotech Co., Ltd.), according to the
manufacturer's protocol. Briefly, cells (5x105 cells/ml)
were harvested and double-stained with 7-aminoactinomycin D and
Annexin V/allophycocyanin at room temperature for 15 min. Apoptotic
cells were subsequently analyzed in the different groups using a BD
FACS Calibur flow cytometer (BD Biosciences) and FlowJo software
(v10.4; FlowJo LLC).
Transwell assay
The effect of ZMIZ1 on the migration of PIG1 and
PIG3V cells was determined using a Transwell assay. PIG1 and PIG3V
cells were harvested and plated into the upper chambers of 24-well
Transwell plates (Corning, Inc.) at a density of 5x104
cells/well serum-free Medium 254. The lower chamber was filled with
Medium 254 containing 10% FBS. Following the incubation at 37˚C for
24 h, the migratory cells in the lower chamber were fixed in 4%
paraformaldehyde for 20 min at room temperature and stained with
crystal violet for 10 min at room temperature. Stained cells were
viewed under a light microscope (magnification, x200). The number
of migratory cells was counted and quantified using ImageJ (v1.46;
National Institutes of Health).
Immunocytochemistry
PIG1 and PIG3V cells (3x104
cells/coverslip) were seeded on glass coverslips and washed with
PBS, fixed in 4% paraformaldehyde for 30 min at room temperature
and permeabilized with 0.3% Triton X-100 for 20 min at room
temperature. Subsequently, the cells were blocked with 1% BSA
(Gibco; Thermo Fisher Scientific, Inc.) for 30 min at room
temperature. The Phalloidin-iFluor 488 reagent (cat. no. ab176753;
Abcam) was used to stain the cells, according to the manufacturer's
protocol, for 30 min at room temperature. Stained cells were imaged
under a confocal microscope (magnification, x63).
Statistical analysis
Statistical analysis was performed using SPSS
software (v22.0; IBM Corp.) and data are presented as the mean ± SD
of ≥3 independent experimental repeats. Statistical differences
among the groups were determined using one-way ANOVA followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Establishment of ZMIZ1 knockdown and
overexpression in vitro cell models
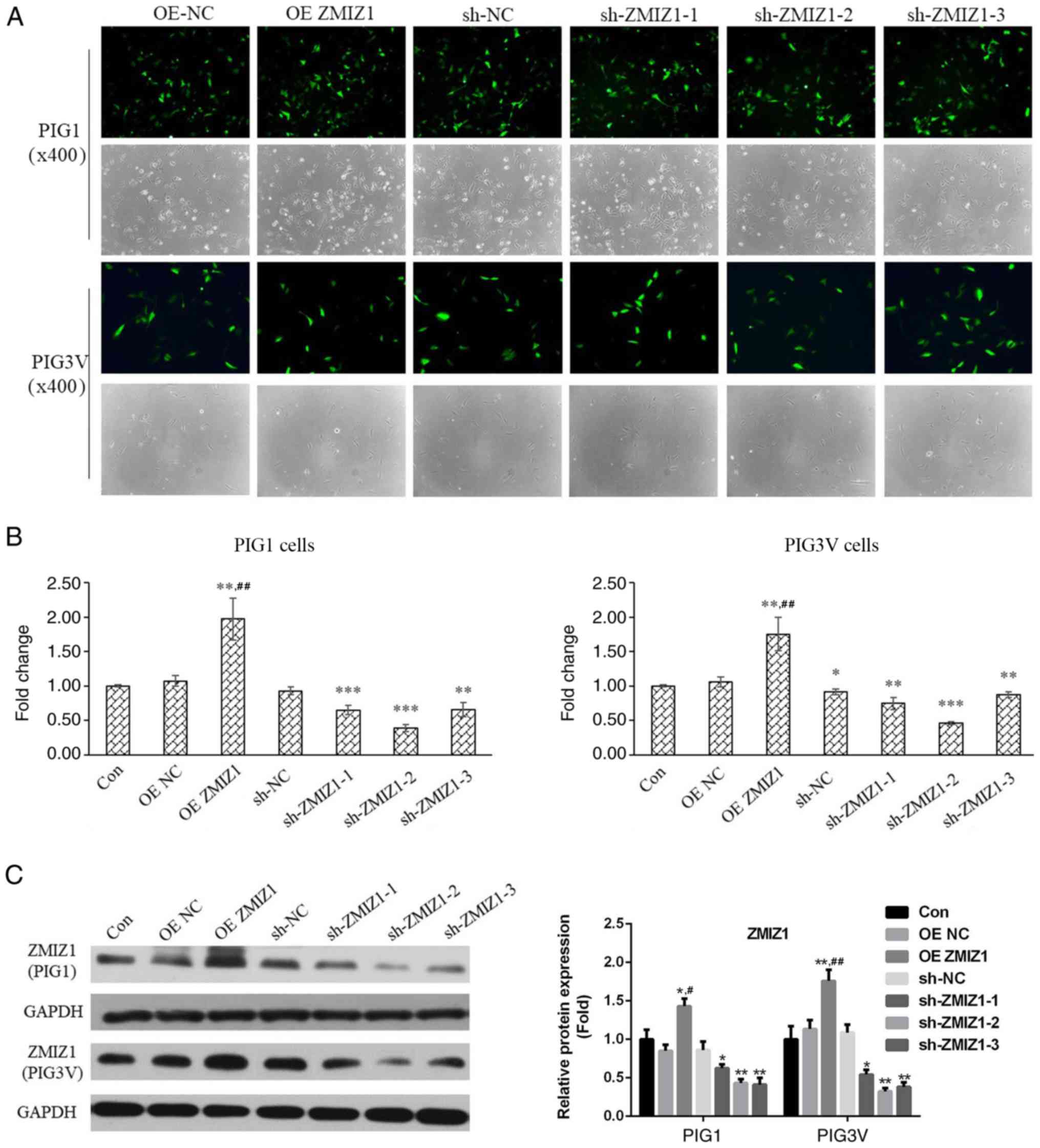
PIG1 and PIG3V cells were infected with lentiviral
vectors to establish ZMIZ1 knockdown and overexpression cell
models. Fluorescence was observed in each infected group,
suggesting that the lentivirus infection had been successful
(Fig. 1A). Moreover, RT-qPCR and
western blotting analysis were performed to evaluate the efficiency
of the infection. ZMIZ1 expression levels were significantly
increased in the OE ZMIZ1 group at both the mRNA Fig. 1B) and protein (Fig. 1C) levels. Conversely, sh-ZMIZ1-1,
sh-ZMIZ1-2 and sh-ZMIZ1-3 significantly decreased the mRNA and
protein expression levels of ZMIZ1 in both PIG1 and PIG3V cells
compared with the sh-NC (Fig. 1B and
C). Based on these results,
sh-ZMIZ1-1 and sh-ZMIZ1-2 were used for ZMIZ1 knockdown in
subsequent experiments.
Effects of ZMIZ1 on the proliferation
and apoptosis of PIG1 and PIG3V cells in vitro
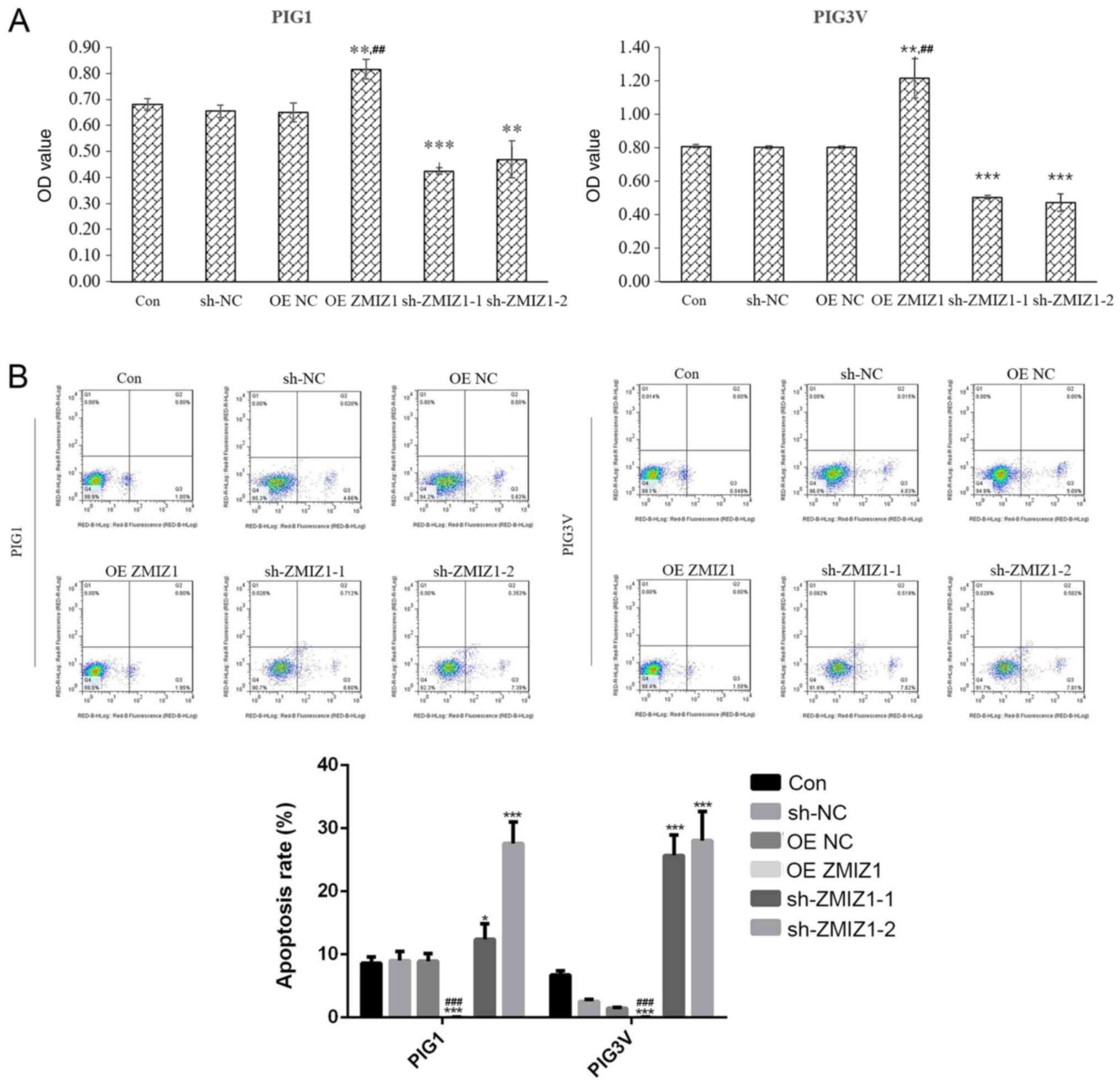
The effect of ZMIZ1 on cell proliferation was
analyzed using a MTT assay and flow cytometry 48 h post-lentiviral
infection. The overexpression of ZMIZ1 significantly increased the
proliferation (P<0.01; Fig. 2A)
and decreased the apoptotic rate (Fig.
2B) compared with the Con group. Conversely, the genetic
knockdown of ZMIZ1 significantly inhibited the proliferation
(Fig. 2A) and induced the apoptosis
of PIG1 and PIG3V cells in vivo (Fig. 2B) compared with the Con group.
ZMIZ1 regulates the expression levels
of Bcl-2 and caspase-3 in PIG1 and PIG3V cells in vitro
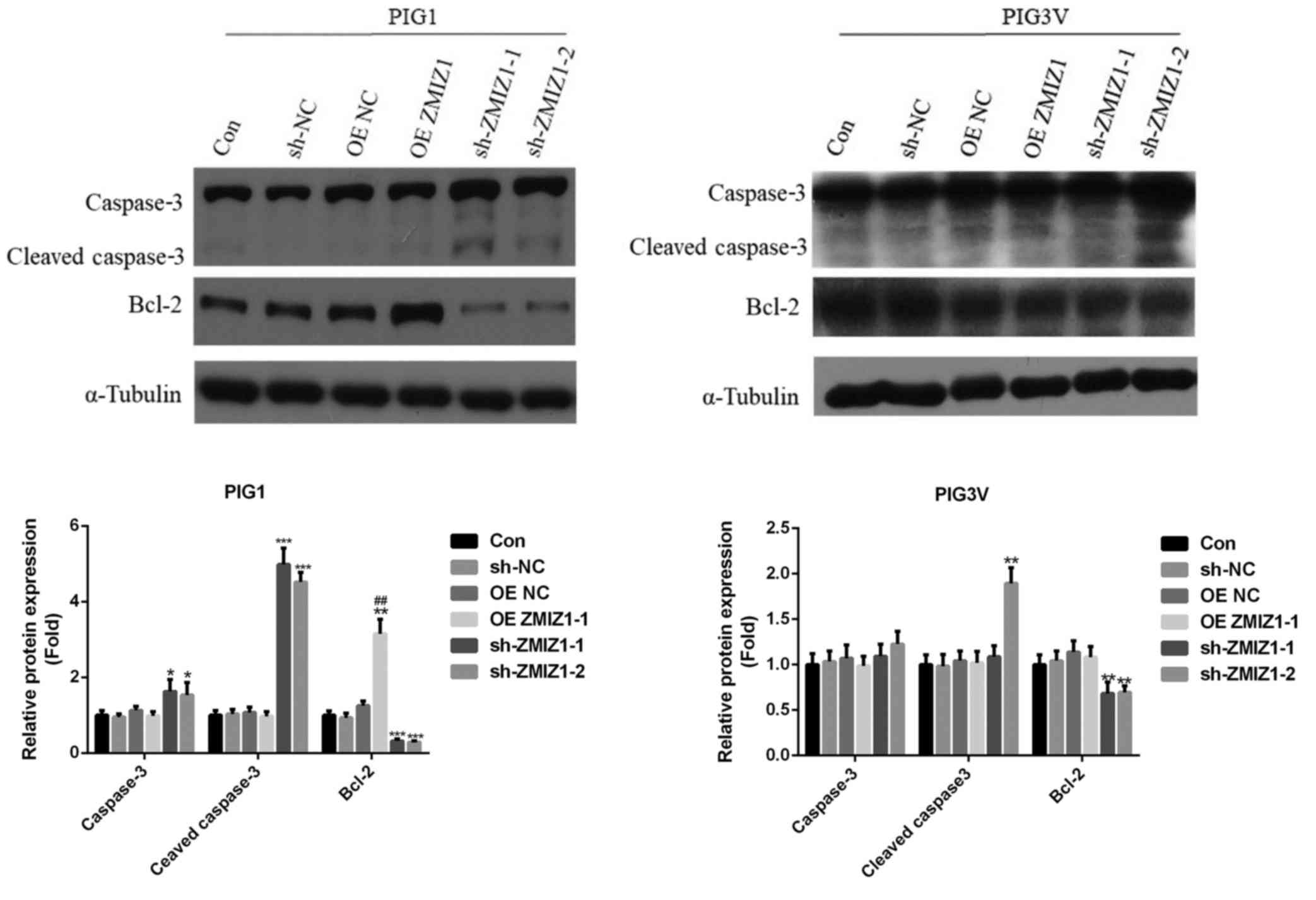
The effects of ZMIZ1 on the expression levels of
proliferation- and apoptosis-associated proteins were analyzed
using western blotting. The overexpression of ZMIZ1 significantly
increased the expression levels of the anti-apoptotic protein Bcl-2
in PIG1 cells compared with the Con group (Fig. 3). Conversely, the genetic knockdown
of ZMIZ1 significantly decreased the expression levels of Bcl-2,
and increased the expression levels of the proapoptotic protein
caspase-3 and its activated form, cleaved caspase-3, in PIG1 cells
(Fig. 3). Moreover, the genetic
knockdown of ZMIZ1 significantly decreased the expression levels of
Bcl-2 in PIG3V cells compared with the Con group and sh-ZMIZ1-2
significantly increased the expression levels of cleaved caspase-3
in PIG3V cells compared with the Con group. On the other hand, the
overexpression of ZMIZ1 had no significant effects on the
expression levels of caspase-3 and cleaved caspase-3 in PIG1 and
PIG3V cells, and the genetic knockdown of ZMIZ1 had no significant
effects on the expression levels of caspase-3 in PIG3V cells
(Fig. 3).
Effect of ZMIZ1 overexpression and
knockdown on the migration of PIG1 and PIG3V melanocytes
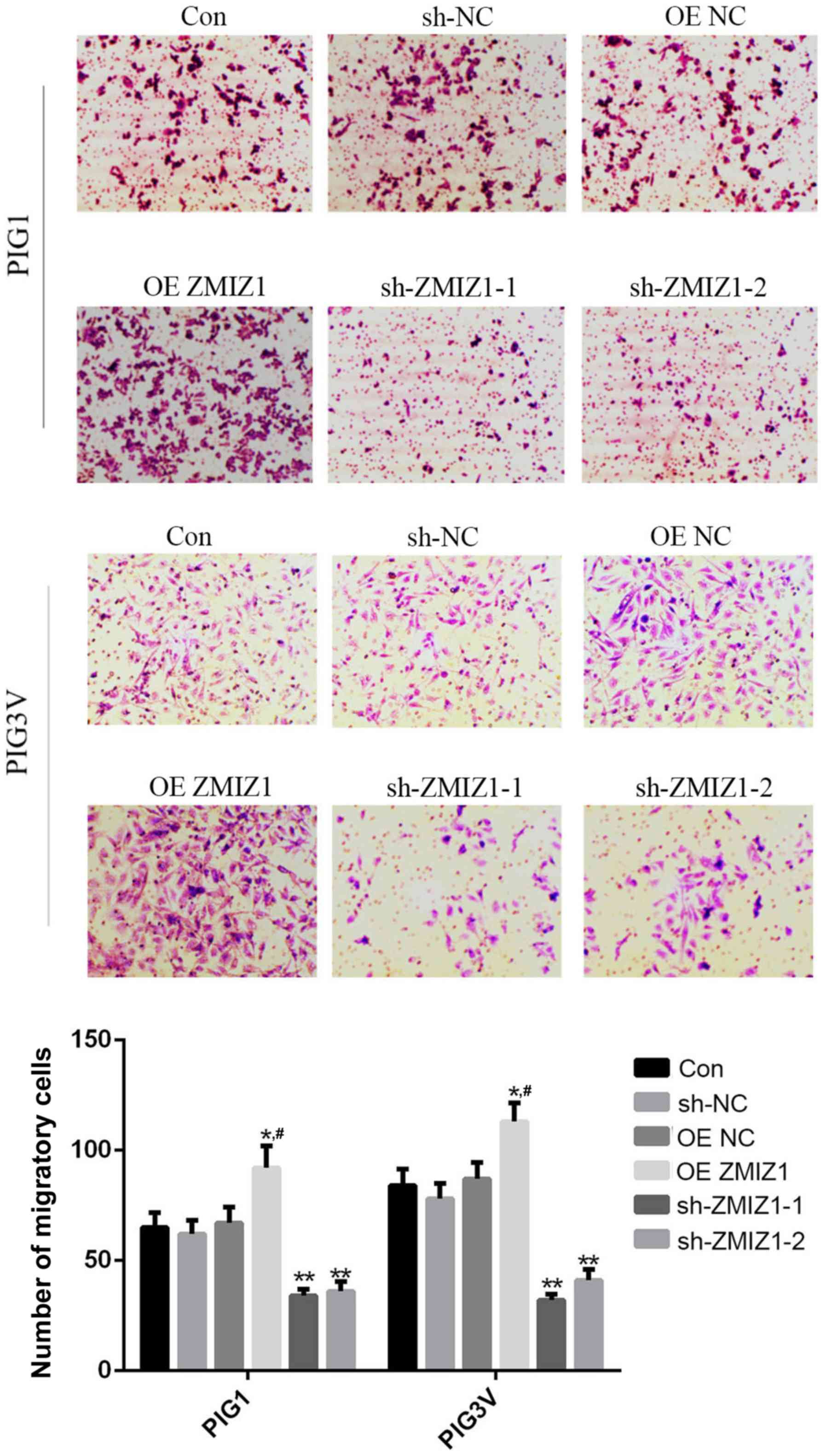
The effects of ZMIZ1 on the migration of PIG1 and
PIG3V cells were investigated using a Transwell assay. The
overexpression of ZMIZ1 significantly increased the migratory
ability of PIG1 and PIG3V cells compared with the Con group
(Fig. 4), whereas the genetic
knockdown of ZMIZ1 demonstrated the opposite effects (Fig. 4).
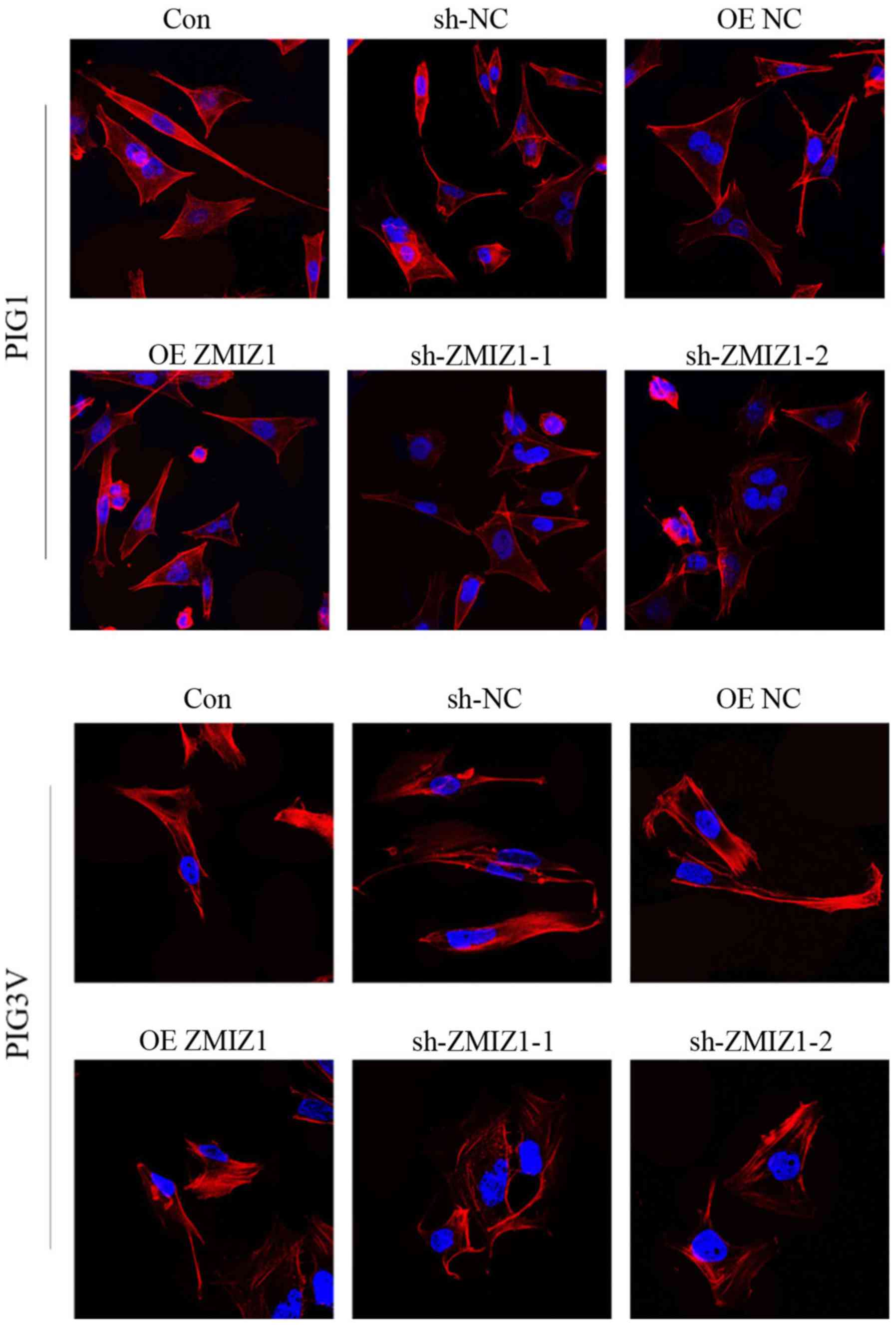
ZMIZ1 affects the remodeling of actin
cytoskeleton in PIG1 and PIG3V melanocytes in vitro
The remodeling of the actin cytoskeleton has been
considered as a necessary process for the migration of different
cell types, including melanocytes (19,20).
Therefore, the effects of ZMIZ1 on actin remodeling in the
cytoskeleton were investigated using immunocytochemistry. Genetic
knockdown of ZMIZ1 resulted in morphology of the cells to become
irregular, and the cell shape tend to be more rounded, indicating
the degradation of the cytoskeleton and apoptosis of the cell
(Fig. 5).
Discussion
The present study successfully established zinc
finger MIZ-type containing 1 (ZMIZ1) overexpression and knockdown
PIG1 and PIG3V model cell lines, which were subsequently used to
investigate the effects of ZMIZ1 on the biological behaviors of
melanocytes. ZMIZ1 promoted proliferation and migration and
inhibited apoptosis of PIG1 and PIG3V cells, suggesting that ZMIZ1
may be a potential therapeutic target for the treatment of
vitiligo. A previous report discovered that PIAS3, a member of the
PIAS family, inhibited the transcriptional activity of melanocyte
inducing transcription factor and STAT3 in vivo (21), and it has been further reported that
both of these transcription factors may serve important roles in
the growth and maintenance of melanocytes (21). Similar to other PIAS family proteins,
ZMIZ1 has been found to enhance the transcriptional activity of the
SMAD3/4 complex; this subsequently activated the TGF-β/SMAD
signaling pathway, which serves an important role in the regulation
of melanocyte proliferation, differentiation and regulation
(22,23). Moreover, it was also suggested that
ZMIZ1 may induce autoreactive T cells and the loss of tolerance to
melanocyte antigens during vitiligo (24). The aforementioned studies indicated
that ZMIZ1, as well as other PIAS members, may regulate the growth
and function of melanocytes, and that the aberrant expression
levels of ZMIZ1 may cause melanocyte dysfunction, which may lead to
the development of vitiligo. However, reports on the effects of
ZMIZ1 on melanocytes are limited.
Increasing evidence has suggested that the impaired
proliferation and increased apoptosis of melanocytes may lead to
the occurrence and progression of vitiligo (25,26). In
the present study, the effect of ZMIZ1 on the proliferation and
apoptosis of the human melanocyte cell lines PIG1 and PIG3V cells
was investigated. It was discovered that the overexpression of
ZMIZ1 significantly increased the proliferation and significantly
decreased the apoptotic rate of the cells, whereas the genetic
knockdown of ZMIZ1 exhibited the opposite effects. Bcl-2 is a
well-known anti-apoptotic protein and caspase-3 is considered as a
proapoptotic protein (27,28). The results obtained in the present
study discovered that the overexpression of ZMIZ1 increased the
expression levels of Bcl-2, while the genetic knockdown of ZMIZ1
decreased the expression levels of Bcl-2, in addition to increasing
the expression levels of caspase-3 and its activated form cleaved
caspase-3 in PIG1 and PIG3V cells. Taken together, these results
suggested that ZMIZ1 may affect the proliferation and apoptosis of
melanocytes in vivo.
Human melanocytes originate from the neural crest
and following proliferation, migrate to the nearby epidermis or the
hair matrices to produce melanin (29). Therefore, migration is an important
step in the function of melanocytes. In the case of vitiligo,
functional melanocytes are destroyed in skin lesions and
consequently, promoting the migration of functional melanocytes to
the depigmented area is the main aim of current anti-vitiligo
strategies (26,30). In the present study, the
overexpression of ZMIZ1 increased the migratory ability and the
remodeling of the actin cytoskeleton in PIG1 and PIG3V cells, while
the genetic knockdown of ZMIZ1 resulted in the opposite effects.
These results indicated that ZMIZ1 may facilitate the migration and
cytoskeleton rearrangement of melanocytes, suggesting that the
overexpression of ZMIZ1 may be a potential strategy to recruit
functional melanocytes to the site of the vitiliginous skin.
Nevertheless, the present study only performed in
vivo cellular studies, therefore the role of ZMIZ1 in the
pathogenesis of vitiligo requires further investigation using in
vivo animal studies and clinical analysis in the future.
In conclusion, to the best of our knowledge, the
present study was the first to suggest that ZMIZ1 may regulate the
proliferation, apoptosis and migration of human melanocytes in
vitro. The results obtained in the present study provided novel
evidence to indicate that ZMIZ1 may serve as a novel therapeutic
target in vitiligo.
Acknowledgements
Not applicable.
Funding
The present study was supported by funds from the
Initial Funding for Outstanding Young People in Zhejiang Provincial
People's Hospital (grant no. ZRY2018C004) and the Shanghai Natural
Science Foundation (grant no. 16ZR1419200).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ML performed the experiments and wrote the
manuscript; YF and YW performed the experiments; JX and HX designed
the study and revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yadav AK, Singh P and Khunger N:
Clinicopathologic analysis of stable and unstable vitiligo: A study
of 66 cases. Am J Dermatopathol. 38:608–613. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Shi H, Lin B, Huang Y, Wu J, Zhang H, Lin
C, Wang Z, Zhu J, Zhao Y, Fu X, et al: Basic fibroblast growth
factor promotes melanocyte migration via activating
PI3K/Akt-Rac1-FAK-JNK and ERK signaling pathways. IUBMB Life.
68:735–747. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Wang X, Du J, Wang T, Zhou C, Shen Y, Ding
X, Tian S, Liu Y, Peng G, Xue S, et al: Prevalence and clinical
profile of vitiligo in China: A community-based study in six
cities. Acta Derm Venereol. 93:62–65. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Grimes PE and Miller MM: Vitiligo: Patient
stories, self-esteem, and the psychological burden of disease. Int
J Womens Dermatol. 4:32–37. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hadi A, Wang JF, Uppal P, Penn LA and
Elbuluk N: Comorbid diseases of vitiligo: A 10-year cross-sectional
retrospective study of an urban US population. J Am Acad Dermatol.
82:628–633. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yuan X, Meng D, Cao P, Sun L, Pang Y, Li
Y, Wang X, Luo Z, Zhang L and Liu G: Identification of pathogenic
genes and transcription factors in vitiligo. Dermatol Ther
(Heidelb). 32(e13025)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Galeone M, Colucci R, Dragoni F and
Moretti S: Can environmental factors contribute in triggering
vitiligo and associated autoimmune thyroid diseases? Clinical
series possibly related to the Chernobyl nuclear accident. G Ital
Dermatol Venereol. 153:729–730. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Laddha NC, Dwivedi M, Gani AR, Mansuri MS
and Begum R: Tumor necrosis factor B (TNFB) genetic variants and
its increased expression are associated with vitiligo
susceptibility. PLoS One. 8(e81736)2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Al-Harthi F, Zouman A, Arfin M, Tariq M
and Al-Asmari A: Tumor necrosis factor-α and -β genetic
polymorphisms as a risk factor in Saudi patients with vitiligo.
Genet Mol Res. 12:2196–2204. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Veiga-Castelli L, Oliveira ML, Pereira A,
Debortoli G, Marcorin L, Fracasso N, Silva G, Souza A, Massaro J,
Simões AL, et al: HLA-G polymorphisms are associated with
non-segmental vitiligo among Brazilians. Biomolecules.
9(9)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yi X, Cui T, Li S, Yang Y, Chen J, Guo S,
Jian Z, Li C, Gao T, Liu L, et al: Identification of the risk HLA-A
alleles and autoantigen in Han Chinese vitiligo patients and the
association of CD8+T cell reactivity with disease characteristics.
Med Sci Monit. 24:6489–6497. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li J, Yan M, Zhang Y, Feng C, Wang H, Wang
C and Sun L: Meta-analysis of the association between NLRP1
polymorphisms and the susceptibility to vitiligo and associated
autoimmune diseases. Oncotarget. 8:88179–88188. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sun Y, Zuo X, Zheng X, Zhou F, Liang B,
Liu H, Chang R, Gao J, Sheng Y, Cui H, et al: A comprehensive
association analysis confirms ZMIZ1 to be a susceptibility gene for
vitiligo in Chinese population. J Med Genet. 51:345–353.
2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang SK, Hong M, Choi H, Zhao W, Jung Y,
Haritunians T, Ye BD, Kim KJ, Park SH, Lee I, et al: Immunochip
analysis identification of 6 additional susceptibility loci for
Crohn's disease in Koreans. Inflamm Bowel Dis. 21:1–7.
2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Rakowski LA, Garagiola DD, Li CM, Decker
M, Caruso S, Jones M, Kuick R, Cierpicki T, Maillard I and Chiang
MY: Convergence of the ZMIZ1 and NOTCH1 pathways at C-MYC in acute
T lymphoblastic leukemias. Cancer Res. 73:930–941. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Peng Y, Lee J, Zhu C and Sun Z: A novel
role for protein inhibitor of activated STAT (PIAS) proteins in
modulating the activity of Zimp7, a novel PIAS-like protein, in
androgen receptor-mediated transcription. J Biol Chem.
285:11465–11475. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xu L, Zhang Z, Xie T, Zhang X and Dai T:
Inhibition of BDNF-AS Provides Neuroprotection for Retinal Ganglion
Cells against Ischemic Injury. PLoS One.
11(e0164941)2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Schnoor M: Endothelial actin-binding
proteins and actin dynamics in leukocyte transendothelial
migration. J Immunol. 194:3535–3541. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang DG, Xu XH, Ma HJ, Li CR, Yue XZ, Gao
J and Zhu WY: Stem cell factor combined with matrix proteins
regulates the attachment and migration of melanocyte precursors of
human hair follicles in vitro. Biol Pharm Bull. 36:1317–1325.
2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Thingnes J, Lavelle TJ, Gjuvsland AB,
Omholt SW and Hovig E: Towards a quantitative understanding of the
MITF-PIAS3-STAT3 connection. BMC Syst Biol. 6(11)2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kim HJ, Choi CP, Uhm YK, Kim YI, Lee JW,
Yoon SH, Chung JH and Lee MH: The association between endothelin-1
gene polymorphisms and susceptibility to vitiligo in a Korean
population. Exp Dermatol. 16:561–566. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jeong KH, Shin MK, Uhm YK, Kim HJ, Chung
JH and Lee MH: Association of TXNDC5 gene polymorphisms and
susceptibility to nonsegmental vitiligo in the Korean population.
Br J Dermatol. 162:759–764. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ben Ahmed M, Zaraa I, Rekik R,
Elbeldi-Ferchiou A, Kourda N, Belhadj Hmida N, Abdeladhim M, Karoui
O, Ben Osman A, Mokni M, et al: Functional defects of peripheral
regulatory T lymphocytes in patients with progressive vitiligo.
Pigment Cell Melanoma Res. 25:99–109. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lan CC, Wu CS, Chiou MH, Chiang TY and Yu
HS: Low-energy helium-neon laser induces melanocyte proliferation
via interaction with type IV collagen: Visible light as a
therapeutic option for vitiligo. Br J Dermatol. 161:273–280.
2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Goldstein NB, Koster MI, Hoaglin LG,
Spoelstra NS, Kechris KJ, Robinson SE, Robinson WA, Roop DR, Norris
DA and Birlea SA: Narrow band ultraviolet B treatment for human
vitiligo is associated with proliferation, migration, and
differentiation of melanocyte precursors. J Invest Dermatol.
135:2068–2076. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Song Y, Zhong M and Cai FC: Oxcarbazepine
causes neurocyte apoptosis and developing brain damage by
triggering Bax/Bcl-2 signaling pathway mediated caspase 3
activation in neonatal rats. Eur Rev Med Pharmacol Sci. 22:250–261.
2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hu PF, Chen WP, Bao JP and Wu LD:
Paeoniflorin inhibits IL-1β -induced chondrocyte apoptosis by
regulating the Bax/Bcl-2/caspase-3 signaling pathway. Mol Med Rep.
17:6194–6200. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Keswell D, Davids LM and Kidson SH:
Migration of human melanocytes into keratinocyte monolayers in
vitro. J Dermatol Sci. 66:160–163. 2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
AlGhamdi KM, Kumar A, Ashour AE and
AlGhamdi AA: A comparative study of the effects of different
low-level lasers on the proliferation, viability, and migration of
human melanocytes in vitro. Lasers Med Sci. 30:1541–1551.
2015.PubMed/NCBI View Article : Google Scholar
|