Introduction
The incidence of fractures is highest among the
young and older populations (1). Of
note, fractures in pediatric patients are common injuries that
result in pain, as well as short- and long-term morbidity; it has
been estimated that 27-50% of all individuals experience a fracture
prior to turning 18 years old (2,3).
Goulding et al (4) estimated
the risk of recurrent fracture to be 12-20% in pediatrics patients
aged <16 years. Furthermore, these figures may indicate that
pediatric patients who do not have any diseases affecting bone
integrity may still be at risk to suffer a bone fracture or even
recurrent fractures (2-4).
The causes for this may arise from the diet, preferred free-time
activities or sex, but may also be linked to genetic factors
(3).
The causes and etiopathogenetic pathways of
pediatric fractures are not well studied (5). It has been previously reported that
bone-associated and bone-independent factors contribute to the
fracture risk (2,6). Furthermore, bone mineral content (BMC)
and bone mineral density (BMD) have been indicated to be highly
heritable in genetic studies (7-11).
Mora and Gilsanz (12) revealed that
80% of the variance of the BMC is influenced by genetics.
Furthermore, BMD has been reported to be a risk factor for
fractures in adults (1), but there
is limited research on pediatric fractures. Previous studies have
linked a low BMD to increased fracture risk during childhood
(3,13); however, in other studies, this
association was not identified (6,14). In
adults, genome-wide association studies (GWAS) have identified
genes from ≥11 different chromosomes (chrs) that may affect BMD
either locally or throughout the body (15-21).
However, with regards to pediatrics, only few studies have reported
on genetic loci that influence BMD or cortical bone thickness
(7,8). Furthermore, the heritability of the
fractures and BMD differ, and thus, it has been suggested that
fracture risk should be analyzed independently of BMD (22).
BMD and BMC are useful when examining the factors
underlying fractures (23,24); however, their association with
fractures in childhood remains to be fully elucidated. Therefore,
the present study focused on fractures rather than BMD or BMC. To
the best of our knowledge, only few studies have examined the
association between genetics and childhood fracture risk.
Therefore, the aim of the present study was to identify whether any
genetic loci are associated with fractures prior to the age of 7
years using GWAS.
Patients and methods
Patient population
The study population was a part of the Northern
Finland Birth Cohort 1986 (NFBC1986) that originally included all
pregnant females (n=9,362) living in the geographic area of the two
northernmost provinces of Finland with expected date of delivery
between 1st July 1985 and 30th June 1986. These mothers and their
live-born infants (n=9,432) were followed up regularly since
pregnancy. Immigration during this period to northern Finland was
minimal, and therefore, the cohort was predominantly Finnish
(25). The detailed description of
the cohort is provided in the study published by Järvelin et
al (26) and on the NFBC website
(27).
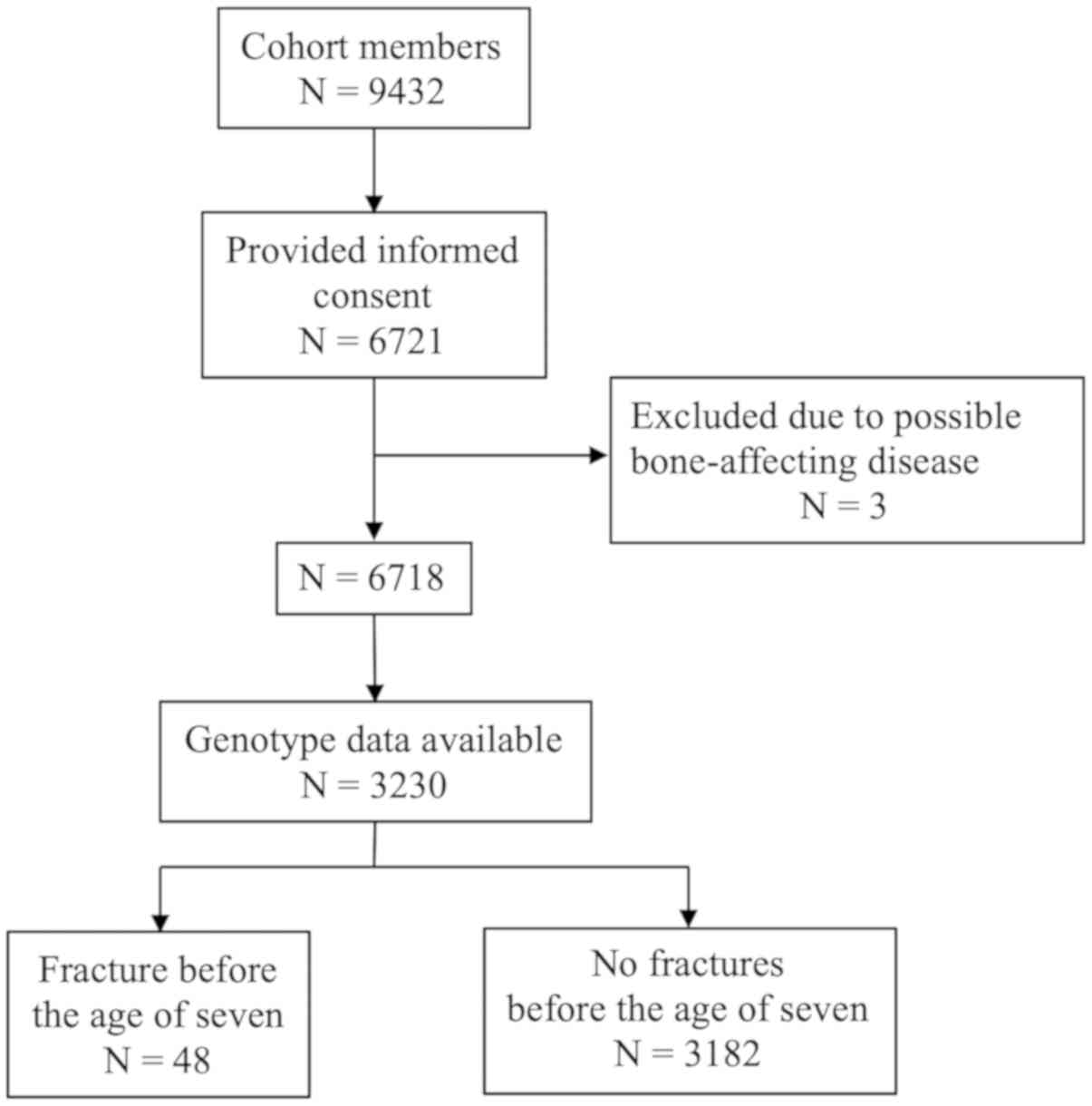
Of the cohort population, 6,721 consented to the use
of their information for research purposes. The information for the
individuals who did not provide consent was not available. Study
subjects with diseases of the bone, including osteogenesis
imperfecta, or possible bone-affecting malignancies, were excluded
from the original study population (n=3). Informed consent from the
parents was provided orally during antenatal clinic visits. At the
16-year follow-up (in 2001), the cohort participants and their
parents provided written informed consent for the use of all of
their collected data for scientific purposes. The participation was
voluntary.
Genotype data
Genotypes were determined for the cohort members who
had provided consent and their blood samples were used for this
purpose. The genotype data were available for 3,230 individuals,
including 48 cases who had a fracture and 3,182 controls without
fractures. Genotyping was performed using the Illumina
HumanOmniExpressExome v1.2 bead chip (Illumina, Inc.) and
imputation was performed using the 1000G phase 3 reference panel
(28). After quality control (QC),
the study included 3,515,000 single-nucleotide polymorphisms (SNPs)
(28). The QC process is described
in detail in a previous study (28).
The data was controlled based on genotype call rate, minor allele
frequency, multidimensional scaling outliers, relatedness,
heterozygosity rate and Hardy-Weinberg Equilibrium. The imputation
quality threshold was 0.4. The flow chart for inclusion is
presented in Fig. 1.
Fractures
The National Hospital Discharge Register (NHDR)
provided the data on the fractures (the name of the register has
changed and it is now known as Care Register for Health Care). The
register contains information on all in-hospital treated fractures
with diagnostic codes and discharge dates. The NHDR is coordinated
by the Finnish institute for health and welfare and it contains all
hospital admissions in Finland (29). This register is the oldest nationwide
discharge register worldwide and thoroughly covers acute injuries
of the population (annual coverage of injuries, ~100%) and
classifies these injuries accurately (annual accuracy, >95%)
(30).
In the study area, pediatric fractures are
exclusively treated in public hospitals and it is mandatory to send
information of these injuries to the NHDR. During the study period,
out-patient visits were not recorded in the NHDR and any patients
treated outside the hospital were not included. Simple fractures,
including non-displaced fracture of wrist or foot, were not
included in the NHDR, as these are treated by general practitioners
in primary healthcare centers. All the in hospital treated
fractures were recognized for the analysis and the patients were
classified according to whether they had a fracture or not. The
International Classification of Diseases, version-9 (ICD-9) was
used to identify the fractures (31). If the patient had several
hospitalizations with the same ICD code, the fractures were
determined to be caused by ≥2 separate injuries if there was an
interval of ≥6 months between the hospitalization dates. The ICD-9
codes reported for the fractures are presented in Table I.
 | Table IReported ICD-9 codes. |
Table I
Reported ICD-9 codes.
| ICD-9 code | Explanation |
|---|
| 800 | Fracture of vault
of skull |
| 802 | Fracture of facial
bones |
| 810 | Fracture of
clavicle |
| 812 | Fracture of
humerus |
| 813 | Fracture of radius
or ulna |
| 816 | Fracture of one or
more phalanges of the hand |
| 821 | Fracture of other
or unspecified parts of femur |
| 823 | Fracture of tibia
or fibula |
The cause of the fracture may also be included in
the NHDR data. Out of the 48 individuals who had a fracture in the
population of the present study, 37 had a reported mechanism.
Furthermore, 15 patients had low-energy accidents, including
falling onto a plane, tripping, slipping or falling from a height
of <1 m. For the other fractures, it was not possible to
reliably determine the energy involved in the injury.
Data analyses
The association between specific genomic regions and
fractures was assessed in the GWAS using the frequentist
association test of the SNPTEST analysis software (version 2.5.2;
University of Oxford). In the GWAS analysis, a linear regression
model was fitted to test for additive effects of SNPs (genotype
dosage) adjusting for sex and population stratification using four
principal components. Sex and the first four principal components
were used as covariates.
The post-GWAS QC included the following steps: Minor
allele frequency <0.05, deviation from Hardy-Weinberg
equilibrium P-value <1x10-7. The generally accepted
limit of P<5x10-8 for genome-wide significance and
P<5x10-5 for suggestive evidence of association were
used. Under the null hypothesis, the threshold where one false
positive is expected per genome scan is called the suggestive level
(32). Manhattan and
Quantile-Quantile (Q-Q) plots of the results were generated using R
(version 3.5.0) and the qqman package (version 0.1.7) (33). The closest coding and non-coding
genes in the identified loci were determined using the University
of California Santa Cruz Genome Browser (December 2013), which is a
graphical viewing tool of the human genome assembly. The genes were
visualized using the LocusZoom visualization software (version 1.4;
University of Michigan).
After GWAS, the lead SNPs were analyzed using the
public databases GWAS database (GWASdb) (34), GWAS catalog (35), The Genotype-Tissue Expression (GTEx)
Portal (36) and RegulomeDB
(37). GWASdb and GWAS catalog are
databases of genetic variants identified in published GWASs. GTEx
Portal is database of tissue-specific gene expression. RegulomeDB
is used to annotate variants to known and predicted regulatory
elements. Database searches were performed for the locus
surrounding the lead SNP (chr10:11491165-12291165). Associated loci
were compared with summary statistics from two studies available on
the Genetic Factors for Osteoporosis Consortium (GEFOS) website
(38,39). Trajanoska et al (38) included 25 cohorts and had discovery
dataset of 37,857 fracture cases and 227,116 controls composed. All
the subjects were adults and predominantly of European descent
(38). Medina-Gomez et al
(39) was meta-analysis of total
body bone mineral density and included 66,628 individuals, both
children and adults of mostly European descent (39).
Results
Fracture demographics
In total, 48 patients had a fracture prior to the
age of 7 and one of these patients had two fractures. The
characteristics of the study population are presented in Table II. Of the 48 subjects with
fractures, 16 (33.3%) were female and 32 (66.7%) were male. The
average age at the time of fracture was 4.05 years (range 0-6
years). Furthermore, the fractures included one cervical spine
fracture (ICD-9 806), two skull fractures (ICD-9 800), eight lower
limb fractures (ICD-9 821, 823) and 37 upper limb fractures (ICD-9
810, 812, 813 and 816).
 | Table IICharacteristics of the study
population. |
Table II
Characteristics of the study
population.
| Characteristic | Value |
|---|
| Sex
(female/male) |
|
Fracture | 16/32 |
|
No
fracture | 1,648/1,534 |
| Average age at the
time of the fracture (years) | 4.05 (0-6) |
| Average BMI at 7
years of agea |
|
Fracture | 16.6
(13.1-20.8) |
|
No
fracture | 16.2
(10.4-34.8) |
| Location of
fracture |
|
Skull | 1 |
|
Facial
bones | 1 |
|
Vertebral
column | 1 |
|
Clavicle | 2 |
|
Humerus | 10 |
|
Radius or
ulna | 14 |
|
One or more
phalanges of the hand | 4 |
|
Other or
unspecified parts of femur | 4 |
|
Tibia or
fibula | 12 |
Genetics of fractures
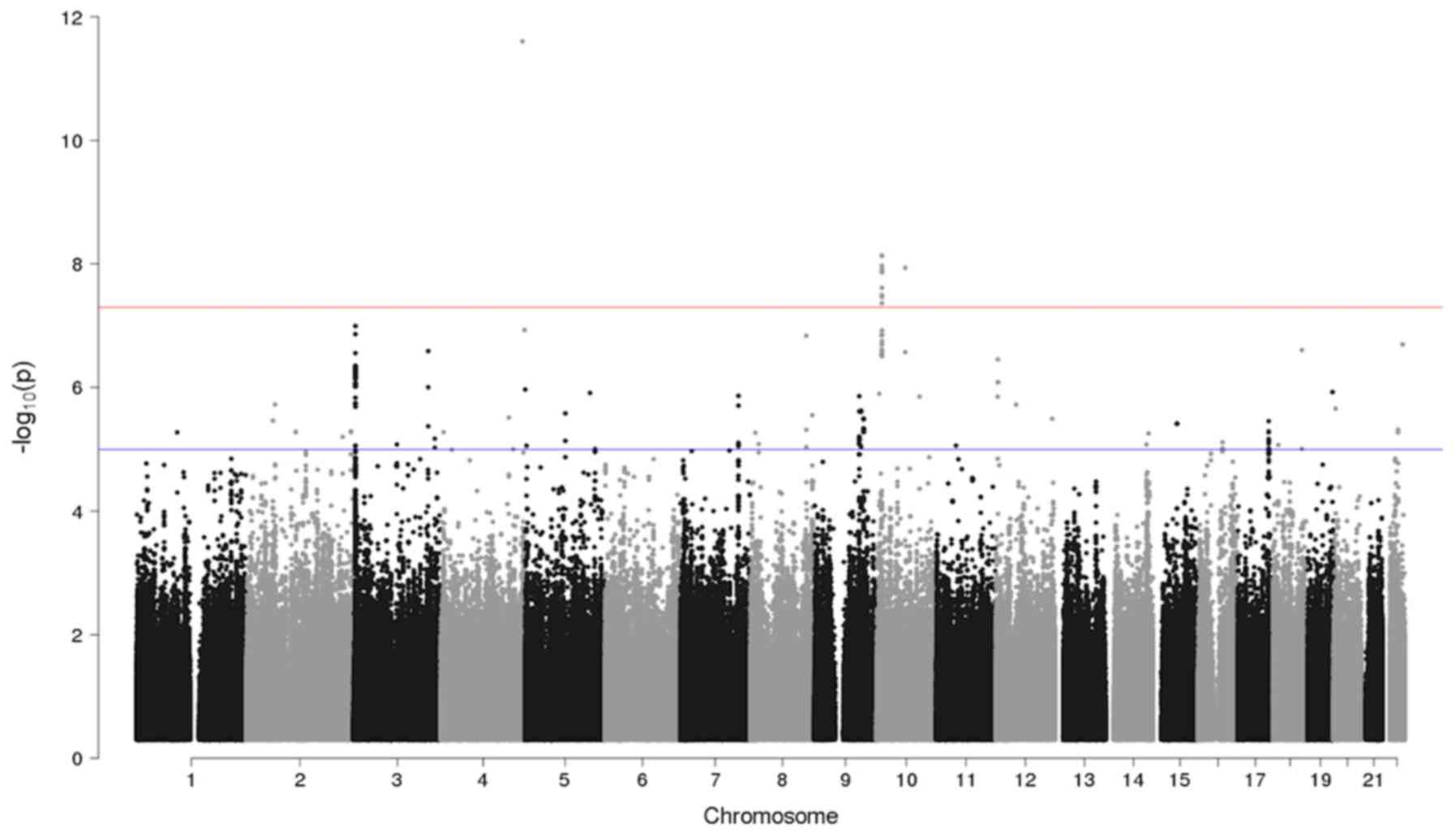
The GWAS analysis identified one locus with a
significant association and six loci with a suggestive result
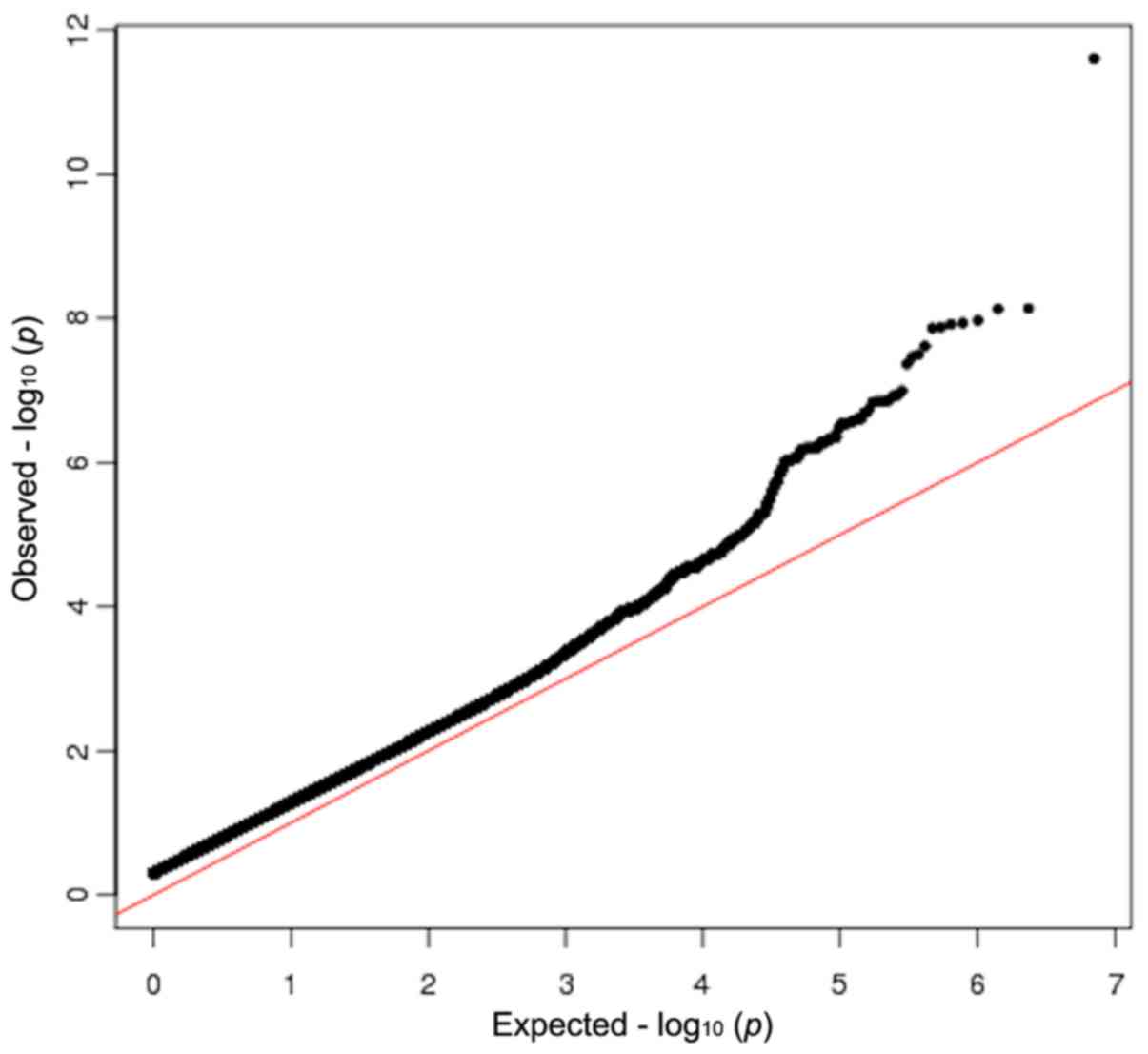
(Table III; Fig. 2). The Q-Q plot of the GWAS is
presented in Fig. 3.
 | Table IIIGenetic variants identified in the
genome-wide association study for childhood fractures performed in
the Northern Finland Birth Cohort 1986. |
Table III
Genetic variants identified in the
genome-wide association study for childhood fractures performed in
the Northern Finland Birth Cohort 1986.
| SNP ID | Chr | Position | EA | NEA | EAF | BETA | SE | P-value | Adjacent
gene(s) |
|---|
| rs112635931 | 10 | 11891165 | A | G | 0.0512 | 3.187 | 0.551 |
7.28x10-9 | PROSER2,
PROSER2-AS1 |
| rs9827298 | 3 | 4023053 | G | C | 0.0635 | 2.325 | 0.437 |
1.01x10-7 | LRRN1, SETMAR,
SUMF1 |
| rs374077976 | 8 | 126506632 | C | CTT | 0.1462 | 1.752 | 0.333 |
1.46x10-7 | TRIB1, NSMCE2 |
| rs41316954 | 9 | 101767382 | T | C | 0.0890 | 1.809 | 0.375 |
1.38x10-6 | COL15A1,
TGFBR1 |
| rs17762577 | 14 | 97517062 | T | C | 0.4670 | -0.955 | 0.210 |
5.53x10-6 | VRK1 |
| rs35417231 | 7 | 131772615 | T | C | 0.0703 | 1.988 | 0.412 |
1.37x10-6 | PLXNA4 |
| rs111299584 | 17 | 69935057 | A | AT | 0.1007 | 1.715 | 0.370 |
3.50x10-6 | SOX9, SOX9-AS1 |
The lead SNP rs112635931 with the most significant
GWA was located within the proline- and serine-rich 2 (PROSER2) and
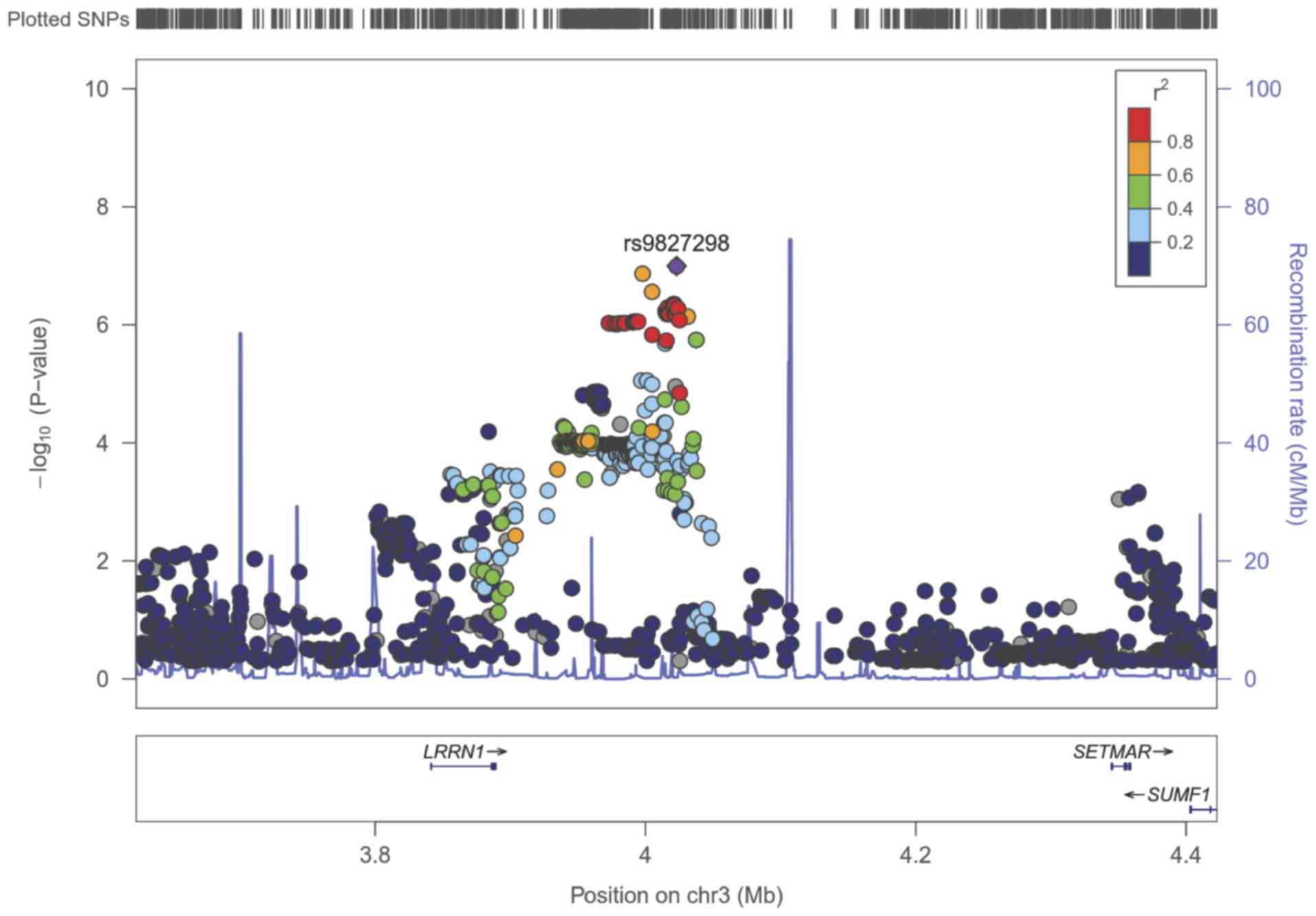
PROSER2-antisense RNA 1 (AS1) genes (Fig. 4). The lead SNP (rs9827298) with
results suggestive of an association with fracture risk is located
in the intergenic region near the leucine-rich repeat neuronal 1
(LRRN1), SET domain and mariner transposase fusion gene (SETMAR)
and sulfatase-modifying factor 1 (SUMF1) genes (Fig. 5). According to GWASdb and GWAS
catalog analyses, no genome-wide significant associations with any
other trait were reported for either of the SNPs. They were also
not present in the RegulomeDB. In addition, the present study did
not identify any SNPs associated with other phenotypes that may be
in linkage disequilibrium with the lead SNP.
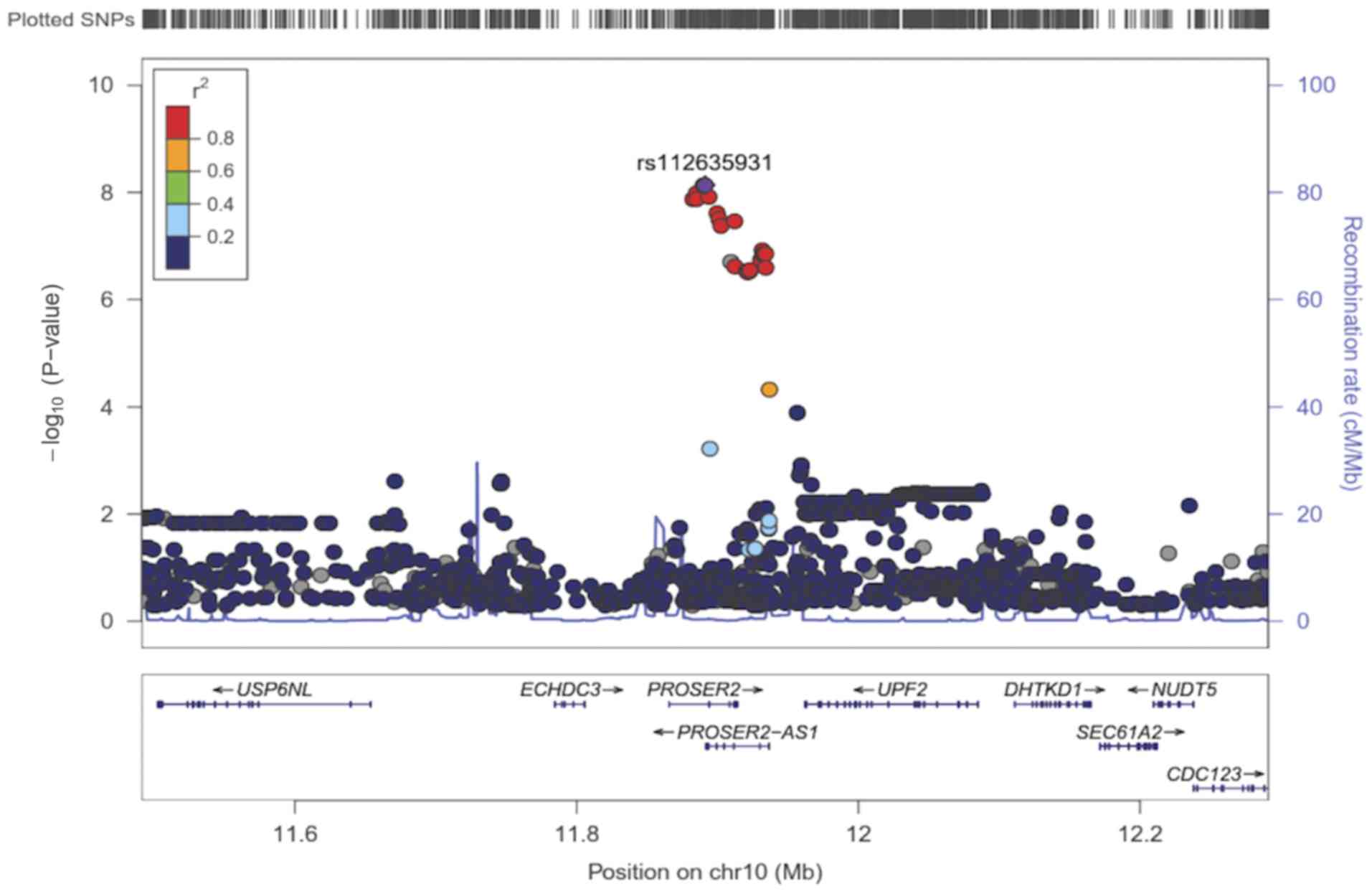 | Figure 4LocusZoom of the rs112635931, Chr
10:11891165, P=7.28x10-9. Each dot represents the
P-value of the SNP obtained from the genome-wide association study.
The color of the dot represents the linkage disequilibrium
(r2), which means that the frequency of association of
the different alleles is higher than what would be expected if the
loci were independent and associated randomly. Chr, chromosome;
SNP, single-nucleotide polymorphism; PROSER2-AS1, proline- and
serine-rich 2-antisense RNA 1; USP6NL, Ubiquitin Specific Peptidase
6 N-terminal Like; ECHDC3, Enoyl-CoA Hydratase Domain Containing 3;
UPF2, up-frameshift 2; DHTKD1, Dehydrogenase E1 And Transketolase
Domain Containing 1; SEC61A2, Selenocystein 61 Translocon Subunit
Alpha 2; NUDT5, Nudix Hydrolase 5, Nudix Hydrolase 5; CDC123, Cell
Division Cycle 123. |
The SNP rs374077976 was present in GWASdb and this
SNP has been associated with total cholesterol in a European
population (40). Furthermore, the
SNP rs111299584 in chr 3 is considered to likely affect the binding
of transcription factors to DNA according to the RegulomeDB.
Furthermore, SNPs rs374077976, rs17762577, rs41316954 and
rs35417231 were categorized to have minimal evidence of DNA binding
of transcription factors in the RegulomeDB. In addition, none of
the studied loci were replicated in the summary statistics of the
two GEFOS studies (35,38).
Discussion
The results of the present study are important, as
the understanding of the genetic associations with childhood
fractures is limited, despite the fact that these are common
injuries. Recent studies have identified certain risk factors for
pediatric fractures both at the micro-level (for example, maternal
smoking associates with childhood fractures) (41) and the macro-level (for example,
weather conditions associates with upper extremity fractures)
(42). However, further studies
examining childhood fractures are required (43). Furthermore, previous studies focusing
on the possible genetic origins of fractures in pediatric patients
without diseases affecting the bone are limited; to the best of our
knowledge, the present study provides novel GWAS data on fractures
in early childhood.
Previous studies have reported that BMC and BMD are
highly heritable (7-11).
Furthermore, GWAS have revealed ≥11 different chromosomes that
affect BMD in adults (15-21).
Chesi et al (8) identified
five loci that influence pediatric BMD at multiple skeletal sites.
Duren et al (7) identified
three genomic regions (2p25.2, 3p25.3 and 17q21.2) with linkage to
cortical bone thickness in 10-year-old children (7). These regions contain candidate genes,
certain of which have been associated with adult bone mass in other
studies (17,44-46).
Chesi et al (47) performed a
GWAS of areal BMD and BMC and indicated that two loci had a
statistically significant association with the BMD and BMC of the
distal radius. Furthermore, the bone mass phenotype has been
revealed to influence the development of osteoporosis and it is
suggested that genetics may impact osteoporotic fracture risk
(22).
The primary result of the present study was the
statistically significant association between the lead SNP
rs112635931 and bone fractures in preschool pediatric patients.
Furthermore, SNP rs112635931 was located within the PROSER2 gene,
and according to expression quantitative trait loci (eQTL) of the
GTEx database, it may regulate the expression of the PROSER2-AS1
gene. Furthermore, the PROSER2 protein is abundant throughout
different organs in the human body, including the bone marrow and
muscle tissues, based on the GTEx and Human Protein atlas databases
(48,49). The expression of PROSER2 protein was
also reported to be present in two mouse osteoblast cell lines
according to data in the Gene Expression Omnibus database (50,51);
however, the exact function of PROSER2 remains to be fully
elucidated.
In addition to the associative gene loci, the
present study identified six loci that may have an association with
fracture risk. The most adjacent genes to the observed locus in chr
3 (lead SNP rs9827298) were LRRN1, SUMF1 and SETMAR. Furthermore,
based on the GTEx database, it was speculated that rs9827298 may
regulate the expression of LRRN1 in the thyroid, lung and blood;
however, the database does not include bone tissue or cells.
Furthermore, this SNP may affect the SUMF1 gene, which encodes the
formylglycine-generating enzyme (FGE). FGE was detected in the
endoplasmic reticulum of bone cells, but also in the brain and skin
(33). In addition, FGE converts
cysteine into C-α-formylglycine and this activates type I
sulfatases (52). Previous studies
have indicated that there are ≥8 pathologies caused by disruption
of sulfatases, including chondrodysplasia punctata type 1 (53,54). The
identified SNP is also in the region of the SETMAR gene, which
encodes a fusion protein that binds DNA and functions in DNA repair
activities (55). Furthermore, the
lead SNP (rs374077976) on chr 8 has previously been associated with
total cholesterol (40) and high
cholesterol levels have also been suggested to correlate with low
BMD in adults (56,57). In addition, there are indications
that this may have a relevance in pediatric patients and
adolescents (58). The SNP
rs41316954 on chr 9 is located within the COL15A1 gene, which
encodes the α chain of type XV collagen, a member of the
fibril-associated collagens with interrupted helices (FACIT)
collagen family. Furthermore, type XV collagen has been revealed to
be secreted by human osteoblasts (59). SNP rs111299584 on chr 17 is located
upstream of the SOX9 gene, a key transcription factor involved in
chondrocyte differentiation. In addition, it has been reported that
SOX9 deficiency leads to campomelic dysplasia (60).
A total of two out of the six suggestive loci that
were indicated in the GWAS are located in the vicinity of genes
that do not have a definitive connection to bone. The first, SNP
rs17762577 (chr 14), is located near the vaccinia-related kinase
(VRK) serine/threonine kinase 1 gene, which encodes the protein
VRK1(61). The expression of this
gene is increased in actively dividing cells and it is widely
expressed in cells throughout the body (61). The second, SNP rs35417231 on chr 7,
encodes a protein (Plexin-A4, a receptor protein) that is necessary
for signaling of semaphorins (62).
The major strengths of the present study are the
large size of the birth cohort and the valid fracture data from the
NHDR. Furthermore, the laboratory and statistical methods used to
analyze the potential association were validated and of a high
standard. Prior to initiation of the present study, it was
determined that less severe childhood fractures may not provide
reliable information on underlying disorders of bone development.
Thus, only fractures treated in hospital were included to provide
data with increased reliability regarding the genetic background
associated with the risk of fractures. However, the authors
acknowledge that in the future, it may be important to perform
another study with a population including conventional
out-of-hospital treated low-energy fractures and high-energy
fractures. Prior to initiation of the study, it was also decided
that only fractures among pre-school aged pediatric patients would
be included; in later childhood, individual behavioral factors and
recreational activities have a greater impact on the individual
risk of conventional bone fractures (63,64).
However, there were certain limitations to the
present study. For instance, the genetic data were not available
for all patients in the cohort, and thus, the study group did not
include all the pediatric patients with fractures treated in
hospital prior to the age of 7 years. Furthermore, a larger study
population with larger amounts of available genetic material would
have increased the strength of the GWAS. However, the present
sample of 3,230 is still satisfactory for genetic research to
assess the preliminary hypothesis of the study. As another
limitation, no subgroup analyses were performed, which may have
provided further information on differences between sexes or
differences between fractures suffered at different ages (63,64). In
addition, the number of cases was modest (n=48) and no further
sub-group analyses were performed at this stage. Therefore, to
validate the present results, they require to be verified in other
populations in future studies.
Bone density measurements were not available in the
present study. Therefore, it was not possible to perform any
analysis regarding BMC or BMD. The fractures that had been caused
by a high-energy injury, e.g. a motor vehicle collision, were not
excluded, as information on the mechanism of injury was not
available for all fractures. The potential effect of the injury
energy on each individual fracture, as a confounding factor,
remains elusive and the results of the present study should be
interpreted with this limitation in mind. Furthermore, the healing
outcomes of the fractures were not available and this was another
limitation of the study.
An additional limitation of the present study is the
lack of information regarding the other potential confounding
factors in early childhood, including mental or physical health
conditions or abnormal weight gain. Furthermore, cases with
diseases clearly affecting the bone, including malignancies of the
bone and osteogenesis imperfecta, were removed from the study, but
other types, e.g. growth retardations, were not excluded. In future
studies, it may be useful to consider mental deficits, including
Attention Deficit Hyperactivity Disorder or developmental
retardations, which may increase the risk of fracture (65). However, two essential factors
affecting the pediatric fracture risk in general, sex and age, were
assessed in the present analyses.
The nutritional status of pediatric patients over
the duration of the in-hospital stay has not been previously
documented. In Finland, in general, if a pediatric patient had
their first or second fracture and there is an obvious explanation,
including a fall, vitamin D levels or other laboratory parameters
are not assessed. In cases where chronic disease is suspected,
further investigations are performed as a common policy in Finnish
hospitals. Among the patients with fracture in the present study,
no such cases were present. It should also be noted that vitamin D
supplementation has been provided to Finnish children since the
1940s (66,67). In addition, normal growth and
development of all children in Finland are followed up by mandatory
and regular nurse or doctor out-of-hospital visits at a maternity
clinic from birth until the age of graduation from junior high
school (68). Due to these measures,
rickets and other illnesses caused by nutritional deficiencies are
rare (66,67). In future studies, assessment of the
possible implication of the vitamin D levels may be beneficial, as
variations in these levels may be a confounding factor.
In future studies it would be beneficial to also
take into account recurrent fractures. In the present study only
one patient had suffered from >one fracture in the first six
years of life and therefore the recurrence of the fractures was not
taken into account in the analysis.
To the best of our knowledge, the present study was
the first to evaluate the potential association between pediatric
fractures and genetics in this particular setting. The present
results suggested that genes may have a role in the fracture risk
of children, even in cases without a disease affecting the bones.
However, further research in this field is required and future
studies should be performed with an additional, larger dataset. In
addition, further investigations are required to identify the
underlying biological mechanisms of the reported association.
Acknowledgements
The authors would like to acknowledge the
biostatistician Mrs. Eeva Vaaramo, M.Sc. (University of Oulu,
Faculty of Medicine, Northern Finland Birth Cohort Studies), who
was involved in planning the study, and gathering and analyzing the
data. The authors would also like to thank Dr Fiona Hanlon-Dearman
(University of Alberta, Edmonton AB, Canada) who performed the
proofreading of the article.
Funding
No funding was received.
Availability of data and materials
The data that support the findings of this study are
available from the NFBC (www.oulu.fi/nfbc/), but restrictions apply to the
availability of these data, which were used under license for the
present study, and so are not publicly available. The remaining
datasets used and/or analzyed are available from the corresponding
author on reasonable request and with permission of the NFBC.
Authors' contributions
RP conceived the and designed the present study, and
drafted and revised the manuscript. SS acquired, analyses and
interpretated the data. SS was also involved in drafting and
revising the manuscript. LK was involved in the conception and
design of the study, and also took part in the drafting and
revising the manuscript. WS was involved in the conception and
design of the study as well as revising the manuscript critically.
MM was involved in the conception and design of the study. MM also
revised the manuscript critically. JJS responsible for the
conception and design of the study, and JJS was also involved in
drafting and revising the manuscript. All authors read and approved
the final manuscript
Ethics approval and consent to
participate
The Ethics Committee of the Northern Ostrobothnia
Hospital District approved the study (approval no. 12/2003; 27th of
Feb 2003). Participation was voluntary and all participants signed
a written informed consent form. Personal information was replaced
with an identification code and the data were handled at a group
level only. The study followed the principles of the Declaration of
Helsinki (Oulu, Finland).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Melton LJ III, Atkinson EJ, O'Fallon WM,
Wahner HW and Riggs BL: Long-term fracture prediction by bone
mineral assessed at different skeletal sites. J Bone Miner Res.
8:1227–1233. 1993.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jones IE, Williams SM, Dow N and Goulding
A: How many children remain fracture-free during growth? A
longitudinal study of children and adolescents participating in the
Dunedin Multidisciplinary Health and Development Study. Osteoporos
Int. 13:990–995. 2002.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Manias K, McCabe D and Bishop N: Fractures
and recurrent fractures in children; varying effects of
environmental factors as well as bone size and mass. Bone.
39:652–657. 2006.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Goulding A, Jones IE, Williams SM, Grant
AM, Taylor RW, Manning PJ and Langley J: First fracture is
associated with increased risk of new fractures during growth. J
Pediatr. 146:286–288. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Grabala P: Epidemiology of forearm
fractures in the population of children and adolescents: Current
data from the typical polish city. Orthop Muscular Syst.
4(203)2015. View Article : Google Scholar
|
|
6
|
Ma DQ and Jones G: Clinical risk factors
but not bone density are associated with prevalent fractures in
prepubertal children. J Paediatr Child Health. 38:497–500.
2002.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Duren DL, Blangeroc J, Sherwood RJ, Šešelj
M, Dyer T, Cole SA, Lee M, Choh AC, Chumlea WC, Siervogel RM, et
al: Cortical bone health shows significant linkage to chromosomes
2p, 3p, and 17q in 10-year-old children. Bone. 49:1213–1218.
2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chesi A, Mitchell JA, Kalkwarf HJ,
Bradfield JP, Lappe JM, Cousminer DL, Roy SM, McCormack SE, Gilsanz
V, Oberfield SE, et al: A genomewide association study identifies
two sex-specific loci, at SPTB and IZUMO3, influencing pediatric
bone mineral density at multiple skeletal sites. J Bone Miner Res.
32:1274–1281. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pocock NA, Eisman JA, Hopper JL, Yeates
MG, Sambrook PN and Eberl S: Genetic determinants of bone mass in
adults. A twin study. J Clin Invest. 80:706–710. 1987.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Arden NK, Baker J, Hogg C, Baan K and
Spector TD: The heritability of bone mineral density, ultrasound of
the calcaneus and hip axis length: a study of postmenopausal twins.
J Bone Miner Res. 11:530–534. 1996. View Article : Google Scholar
|
|
11
|
Harris M, Nguyen TV, Howard GM, Kelly PJ
and Eisman JA: Genetic and environmental correlations between bone
formation and bone mineral density: a twin study. Bone. 22:141–145.
1998.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mora S and Gilsanz V: Establishment of
peak bone mass. Endocrinol Metab Clin North Am. 32:39–63.
2003.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Goulding A, Jones IE, Taylor RW, Manning
PJ and Williams SM: More broken bones: A 4-year double cohort study
of young girls with and without distal forearm fractures. J Bone
Miner Res. 15:2011–2018. 2000.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cook SD, Harding AF, Morgan EL, Doucet HJ,
Bennett JT, O'Brien M and Thomas KA: Association of bone mineral
density and pediatric fractures. J Pediatr Orthop. 7:424–427.
1987.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kemp JP, Morris JA, Medina-Gomez C,
Forgetta V, Warrington NM, Youlten SE, Zheng J, Gregson CL,
Grundberg E, Trajanoska K, et al: Identification of 153 new loci
associated with heel bone mineral density and functional
involvement of GPC6 in osteoporosis. Nat Gen. 49:1468–1475.
2017.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Estrada K, Styrkarsdottir U, Evangelou E,
Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP,
et al: Genome-wide meta-analysis identifies 56 bone mineral density
loci and reveals 14 loci associated with risk of fracture. Nat
Genet. 44:491–501. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Wynne F, Drummond FJ, Daly M, Brown M,
Shanahan F, Molloy MG and Quane KA: Suggestive linkage of 2p22-25
and 11q12-13 with low bone mineral density at the lumbar spine in
the Irish population. Calcif Tissue Int. 72:651–658.
2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Karasik D, Cupples LA, Hannan MT and Kiel
DP: Age, gender, and body mass effects on quantitative trait loci
for bone mineral density: the framingham study. Bone. 33:308–316.
2003.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wilson SG, Reed PW, Bansal A, Chiano M,
Lindersson M, Langdown M, Prince RL, Thompson D, Thompson E, Bailey
M, et al: Comparison of genome screens for two independent cohorts
provides replication of suggestive linkage of bone mineral density
to 3p21 and 1p36. Am J Hum Genet. 72:144–155. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Shen H, Zhang YY, Long JR, Xu FH, Liu YZ,
Xiao P, Zhao LJ, Xiong DH, Liu YJ, Dvornyk V, et al: A genome-wide
linkage scan for bone mineral density in an extended sample:
Evidence for linkage on 11q23 and Xq27. J Med Genet. 41:743–751.
2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li GH, Cheung CL, Xiao SM, Lau KS, Gao Y,
Bow CH, Huang QY, Sham PC and Kung A: Identification of QTL genes
for BMD variation using both linkage and gene-based association
approaches. Hum Genet. 130:539–546. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Richards JB, Zheng HF and Spector TD:
Genetics of osteoporosis from genome-wide association studies:
advances and challenges. Nat Rev Genet. 13:576–588. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Goulding A, Cannan R, Williams SM, Gold
EJ, Taylor RW and Lewis-Barned NJ: Bone mineral density in girls
with forearm fractures. J Bone Miner Res. 13:143–148.
1998.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Goulding A, Jones IE, Taylor RW, Manning
PJ and Williams SM: More broken bones: A 4-year double cohort study
of young girls with and without distal forearm fractures. J Bone
Miner Res. 15:2011–2018. 2000.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Statistics Finland: Population development
in independent Finland-greying Baby Boomers. https://www.stat.fi/tup/suomi90/joulukuu_en.html.
Accessed May 12, 2007.
|
|
26
|
Järvelin MR, Elliott P, Kleinschmidt I,
Martuzzi M, Grundy C, Hartikainen AL and Rantakallio P: Ecological
and individual predictors of birthweight in a northern Finland
birth cohort 1986. Paediatr Perinat Epidemiol. 11:298–312.
1997.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Northern Finland Cohorts. University of
Oulu. https://www.oulu.fi/nfbc/.
|
|
28
|
Schierding W, Antony J, Karhunen V,
Vääräsmäki M, Franks S, Elliott P, Kajantie E, Sebert S, Blakemore
A, Horsfield JA, et al: GWAS on prolonged gestation (post-term
birth): Analysis of successive Finnish birth cohorts. J Med Genet.
55:55–63. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Finnish Institute for Health and Welfare:
Care Register for Health Care. https://thl.fi/en/web/thlfi-en/statistics/information-on-statistics/register-descriptions/care-register-for-health-care.
|
|
30
|
Parkkari J, Mattila V, Niemi S and Kannus
P: Injury-related deaths among Finnish children, 1971-2001. JAMA.
289:702–703. 2003.PubMed/NCBI View Article : Google Scholar
|
|
31
|
World Health Organization: Basic
tabulation list with alphabetic index. In: International
Classification of Diseases Ninth revision. World Health
Organization, Switzerland, 1978.
|
|
32
|
Lander E and Kruglyak L: Genetic
dissection of complex traits: guidelines for interpreting and
reporting linkage results. Nat Genet. 11:241–247. 1995.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Turner SD: qqman: An R package for
visualizing GWAS results using Q-Q and manhattan plots. J Open
Source Softw. 3(731)2018. View
Article : Google Scholar
|
|
34
|
Li MJ, Wang P, Liu X, Lim EL, Wang Z,
Yeager M, Wong MP, Sham PC, Chanock SJ and Wang J: GWASdb: A
database for human genetic variants identified by genome-wide
association studies. Nucleic Acids Res.
40(D1047-5104)2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
GWAS Catalog. The NHGRI-EBI Catalog of
published genome-wide association studies. https://www.ebi.ac.uk/gwas/.
|
|
36
|
GTEx Consortium. Human genomics. The
Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene
regulation in humans. Science. 348:648–660. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Boyle AP, Hong EL, Hariharan M, Cheng Y,
Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et
al: Annotation of functional variation in personal genomes using
RegulomeDB. Genome Res. 22:1790–1797. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Trajanoska K, Morris JA, Oei L, Zheng HF,
Evans DM, Kiel DP, Ohlsson C, Richards JB and Rivadeneira F:
Assessment of the genetic and clinical determinants of fracture
risk: Genome wide association and mendelian randomisation study.
BMJ. 362(k3225)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Medina-Gomez C, Kemp JP, Trajanoska K,
Luan J, Chesi A, Ahluwalia TS, Mook-Kanamori DO, Ham A, Hartwig FP,
Evans DS, et al: Life-course genome-wide association study
meta-analysis of total body BMD and assessment of age-specific
effects. Am J Hum Genet. 102:88–102. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Asselbergs FW, Guo Y, van Iperen EP,
Sivapalaratnam S, Tragante V, Lanktree MB, Lange LA, Almoguera B,
Appelman YE, Barnard J, et al: Large-scale gene-centric
meta-analysis across 32 studies identifies multiple lipid loci. Am
J Hum Genet. 91:823–838. 2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Parviainen R, Auvinen J, Pokka T, Serlo W
and Sinikumpu JJ: Maternal smoking during pregnancy is associated
with childhood bone fractures in offspring - A birth-cohort study
of 6718 children. Bone. 101:202–205. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sinikumpu JJ, Pokka T, Sirnio K, Ruuhela R
and Serlo W: Population-based research on the relationship between
summer weather and paediatric forearm shaft fractures. Injury.
44:1569–1573. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Sinikumpu JJ: Too many unanswered
questions in children's forearm shaft fractures: High-standard
epidemiological and clinical research in pediatric trauma is
warranted. Scand J Surg. 104:137–138. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Pan F, Xiao P, Guo Y, Liu YJ, Deng HY,
Recker RR and Deng HW: Chromosomal regions 22q13 and 3p25 may
harbor quantitative trait loci influencing both age at menarche and
bone mineral density. Hum Genet. 123:419–427. 2008.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ioannidis JP, Ng MY, Sham PC, Zintzaras E,
Lewis CM, Deng HW, Econs MJ, Karasik D, Devoto M, Kammerer CM, et
al: Meta-analysis of genome-wide scans provides evidence for sex-
and site-specific regulation of bone mass. J Bone Miner Res.
22:173–183. 2007.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Brunkow ME, Gardner JC, Van NJ, Paeper BW,
Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, et al:
Bone dysplasia sclerosteosis results from loss of the SOST gene
product, a novel cystine knot-containing protein. Am J Hum Genet.
68:577–589. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
47
|
Chesi A, Mitchell JA, Kalkwarf HJ,
Bradfield JP, Lappe JM, McCormack SE, Gilsanz V, Oberfield SE,
Hakonarson H, Shepherd JA, et al: A trans-ethnic genome-wide
association study identifies gender-specific loci influencing
pediatric aBMD and BMC at the distal radius. Hum Mol Genet.
24:5053–5059. 2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gene Page. https://www.gtexportal.org/home/gene/PROSER2.
Accessed March 5, 2019.
|
|
49
|
The Human Protein Atlas. https://www.proteinatlas.org/ENSG00000148426-PROSER2/tissue.
Accessed March 5, 2019.
|
|
50
|
GEO Gene Expression Omnibus Profile.
https://www.ncbi.nlm.nih.gov/geoprofiles/94126705.
Accessed March 5, 2019.
|
|
51
|
GEO Gene Expression Omnibus Profile.
https://www.ncbi.nlm.nih.gov/geoprofiles/35462505.
Accessed March 5, 2019.
|
|
52
|
Dierks T, Schmidt B, Borissenko LV, Peng
J, Preusser A, Mariappan M and von Figura K: Multiple sulfatase
deficiency is caused by mutations in the gene encoding the human
Cα-formylglycine generating enzyme Cell. 113:435–444.
2003.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Appel MJ and Bertozzi CR: Formylglycine, a
post-translationally generated residue with unique catalytic
capabilities and biotechnology applications. ACS Chem Biol.
10:72–84. 2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Diez-Roux G and Ballabio A: Sulfatases and
human disease. Annu Rev Genomics Hum Genet. 6:355–379.
2005.PubMed/NCBI View Article : Google Scholar
|
|
55
|
GeneCards Human Gene Database. uriwww.genecards.org/cgi-bin/carddisp.pl?gene=SETMARhttps://www.genecards.org/cgi-bin/carddisp.pl?gene=SETMAR
Accessed March 5, 2019.
|
|
56
|
Mandal CC: High Cholesterol deteriorates
bone health: New insights into molecular mechanisms. Front
Endocrinol (Lausanne). 6(165)2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Ghosh-Choudhury N, Mandal CC and Choudhury
GG: Statin-induced Ras activation integrates the
phosphatidylinositol 3-kinase signal to Akt and MAPK for bone
morphogenetic protein-2 expression in osteoblast differentiation. J
Biol Chem. 282:4983–4993. 2007.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Lim HH: Effect of serum cholesterol on
bone mineral density in normal-weight children and adolescents. J
Pediatr Endocrinol Metab. 28:1313–1319. 2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Lisignoli G, Codeluppi K, Todoerti K,
Manferdini C, Piacentini A, Zini N, Grassi F, Cattini L, Piva R,
Rizzoli V, et al: Gene array profile identifies collagen type XV as
a novel human osteoblast-secreted matrix protein. J Cell Physiol.
220:401–409. 2009.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Matsushita M, Kitoh H, Kaneko H, Mishima
K, Kadono I, Ishiguro N and Nishimura G: A novel SOX9 H169Q
mutation in a family with overlapping phenotype of mild campomelic
dysplasia and small patella syndrome. Am J Med Genet.
161A:2528–2534. 2013.PubMed/NCBI View Article : Google Scholar
|
|
61
|
GeneCards Human Gene Database. https://www.genecards.org/cgi-bin/carddisp.pl?gene=VRK1.
Accessed March 5, 2019.
|
|
62
|
Sijaona A, Luukko K, Kvinnsland IH and
Kettunen P: Expression patterns of Sema3F, PlexinA4, -A3,
Neuropilin1 and -2 in the postnatal mouse molar suggest roles in
tooth innervation and organogenesis. Acta Odontol Scand.
70:140–148. 2012.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Javaid MK and Cooper C: Prenatal and
childhood influences on osteoporosis. Best Pract Res Clin
Endocrinol Metab. 16:349–367. 2002.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Godfrey K, Walker-Bone K, Robinson S,
Taylor P, Shore S, Wheeler T and Cooper C: Neonatal bone mass:
Influence of parental birthweight, maternal smoking, body
composition, and activity during pregnancy. J Bone Miner Res.
16:1694–1703. 2001.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Guo NW, Lin CL, Lin CW, Huang MT, Chang
WL, Lu TH and Lin CJ: Fracture risk and correlating factors of a
pediatric population with attention deficit hyperactivity disorder:
A nationwide matched study. J Pediatr Orthop B. 25:369–74.
2016.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lehtonen-Veromaa M, Möttönen T, Irjala K,
Kärkkäinen M, Lamberg-Allardt C, Hakola P and Viikari J: Vitamin D
intake is low and hypovitaminosis D common in healthy 9- to
15-year-old Finnish girls. Eur J Clin Nutr. 53:746–751.
1999.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Hallman N, Hultin H and Visakorpi JK:
Prophylactic use of vitamin d in Finland. Duodecim.
80(185-9)1964.(In Swedish). PubMed/NCBI
|
|
68
|
Ministry of Social Affairs and Health.
Maternity and child health clinics. https://stm.fi/en/maternity-and-child-health-clinics.
|