Introduction
Ventilator-associated pneumonia (VAP) is a common
nosocomial infection in patients who require mechanical ventilation
in intensive care units (ICUs). VAP is associated with increased
mortality, prolonged hospital stay and an increased economic burden
(1-3).
Inappropriate therapy and delayed initiation of appropriate therapy
may lead to increased patient mortality (4).
Guidelines recommend instant empirical antibiotic
therapy after the diagnosis of VAP (5). Therapy choices for initial antibiotics
are mainly based on the local antibiogram, risk factors for
multiple drug-resistant infections and the onset time of VAP.
Intensive and combined initial therapy and de-escalating therapy
are recommended for VAP. Of note, administration of inappropriate
initial antibiotics may lead to failure to protect against multiple
drug-resistant microbes and may also select for multiple
drug-resistant strains. Determining the aetiology of VAP is
therefore important for initial antibiotic treatment and therapy
adjustment.
Novel DNA-based molecular techniques for
microorganism detection have been reported as alternative tools for
the aetiological diagnosis of bacteraemia and respiratory
infections. Conventional laboratory culture generally takes 2-3
days for testing and has a positive rate influenced by numerous
factors. Early recognition and subsequent appropriate antibiotic
therapy are beneficial. Existing diagnostic approaches delay the
start of targeted antibiotic therapy and result in unnecessary
antibiotic exposure. PCR coupled to electrospray ionization mass
spectrometry (PCR/ESI-MS) is a novel tool that provides automatic,
rapid and high-throughput pathogen detection and has been proven
successful in identifying bacterial and fungal pathogens in
clinical infections (6,7). However, whether PCR/ESI-MS is useful
for providing an accurate and rapid pathogenic diagnosis for
clinical antibiotic therapy in respiratory infections remains
controversial.
The aim of the present study was to evaluate the
capacity of PCR/ESI-MS to provide a pathogenic diagnosis for
patients with VAP in a short time (<6 h) and to determine
whether this technique is able to identify respiratory pathogen
alterations in mechanically ventilated (MV) patients to help
clinicians choose the appropriate initial empirical therapy when
VAP occurs. First, the concordance between PCR/ESI-MS and
conventional bacterial culture was investigated for identifying VAP
pathogens and subsequently, the correlation between bacterial
species identified by the sequential PCR/ESI-MS test and the final
pathogenic diagnosis of VAP was examined.
Materials and methods
Study design
Patients admitted to the surgical ICU of a
comprehensive teaching hospital between September 2011 and July
2012 who required mechanical ventilation for >48 h were
recruited. Patients between 18 and 85 years of age without any
infectious disease prior to mechanical ventilation were eligible
and were excluded if they had diffuse bronchiectasis, massive
haemoptysis, cystic fibrosis or end-stage malignant disease. They
were also excluded if they received any antibiotics or
immunosuppressive drugs within 1 week of ventilation or if they
were pregnant or breast-feeding. A total of 30 subjects were
eligible and enrolled in the present study. Of these subjects, 12
were diagnosed with VAP within 2 weeks after MV initiation. The
diagnosis of VAP was based on the following criteria: New-onset
fever of >38.5˚C, elevated peripheral blood white cell count
(>10x109/l), elevated procalcitonin level, purulent
secretions in mini-bronchoalveolar lavage fluid (BALF), new diffuse
or localized infiltration in a chest X-ray or CT scan and a
decreased ratio of partial pressure of arterial oxygen to the
fraction of inspired oxygen of <240. Data, including demographic
characteristics, initial clinical presentation, laboratory tests,
MV parameters and duration, antibiotic therapy management,
complications and outcomes, were collected prospectively.
Mini-BALF sampling
Mini-BALF samples were collected from each eligible
patient 0, 5 and 10 days after MV initiation by qualified doctors
using sterile tubes (with a diameter of 5 mm) as previously
described. The total volume of physiological saline for
bronchoalveolar lavage was 10-20 and 7-10 ml of fluid was
collected. Each sample was divided equally into two parts: One part
was sent for bacterial culture immediately and the other part was
stored at -80˚C. If the patient developed VAP within 10 days after
MV, the stored mini-BALF samples were tested using PCR/ESI-MS. In
addition to mini-BALF sampling, sputum samples were sent for
routine clinical culture when required.
Routine clinical microbiological
culture
Clinical cultures for fungi, aerobic bacteria and
anaerobic bacteria identification were performed by the hospital
laboratory using mini-BALF samples as previously described
(8). A culture was defined as
positive when bacteria were present at a concentration of
≥1x104 colony-forming units (CFU)/ml. Culture results
assisted in the therapeutic decision-making of the clinicians.
DNA extraction and PCR/ESI-MS
Total DNA was isolated from centrifuged cells of
each mini-BALF sample using the QIAamp DNA Mini kit (cat. no.
51304; Qiagen, Inc.) according to the manufacturer's protocol.
PCR/ESI-MS was performed using a PLEX-ID Broad Bacteria Assay plate
(Ibis Biosciences, Inc.), which was coated with primers, placed on
a plate vibrator at room temperature for 15 min and then
centrifuged at 450 x g for 1 min. Extracted nucleic acids from
mini-BALF samples were loaded into wells in the assay plate (5
µl/well). The broad bacterial assay uses primer pairs designed for
amplifying a variable region of common clinical gram-positive and
gram-negative bacteria, as well as Candida spp. The assay
plates were sealed and centrifuged (450 x g, 1 min, 4˚C) before PCR
was performed according to the manufacturer's protocol. PCR
products were immediately desalted and then submitted to a PLEX-ID
system for mass spectral analysis (9). In brief, electrospray ionization moves
charged amplicons into a mass spectrometer. Based on the resulting
spectrum, base compositions are algorithmically predicted and then
compared to a reference database for quantitative identification of
bacterial species (10).
PCR/ESI-MS analysis
For each microorganism identified, a Q-score ranging
from 0 to 1 was reported by the PLEX-ID system (Abbott
Laboratories). The Q-score is calculated using principal component
analysis and represents a relative measure of the strength of the
data supporting the identification of a certain pathogen.
Microorganisms reported by the system were considered to be
‘positive’ when the Q-score was ≥0.8. ‘Positive’ results from
PCR/ESI-MS were compared to routine cultures (within 30 days) of
paired samples as the gold standard (11). The clinical significance of positive
results for certain bacteria that were negative in paired culture
was evaluated by a microbiologist and an ICU physician who reviewed
the medical history and the results of cultures or PCR/ESI-MS of
specimens (e.g., nasal swab, throat swab, sputum or mini-BALF
within 7 days) from the patient (11).
Management of oral bacterial flora
detected in the respiratory tract
Bacterial species identified by culture or
PCR/ESI-MS that were clinically regarded as normal respiratory
flora (including alpha-haemolytic streptococci, coagulase-negative
staphylococci, Neisseria spp., and Candida spp.) were
considered to indicate a negative result in the data evaluation
process.
Statistical analysis
Data were presented as the % for categorical
variables and as the mean ± SD or median with interquartile range
(25-75%) for continuous variables. Data were analysed with SPSS
version 22.0 (IBM Corp.).
Results
Demographics and clinical information
of subjects diagnosed with VAP
A total of 34 mini-BALF samples from 12 patients
diagnosed with VAP within 10 days after MV were analysed in the
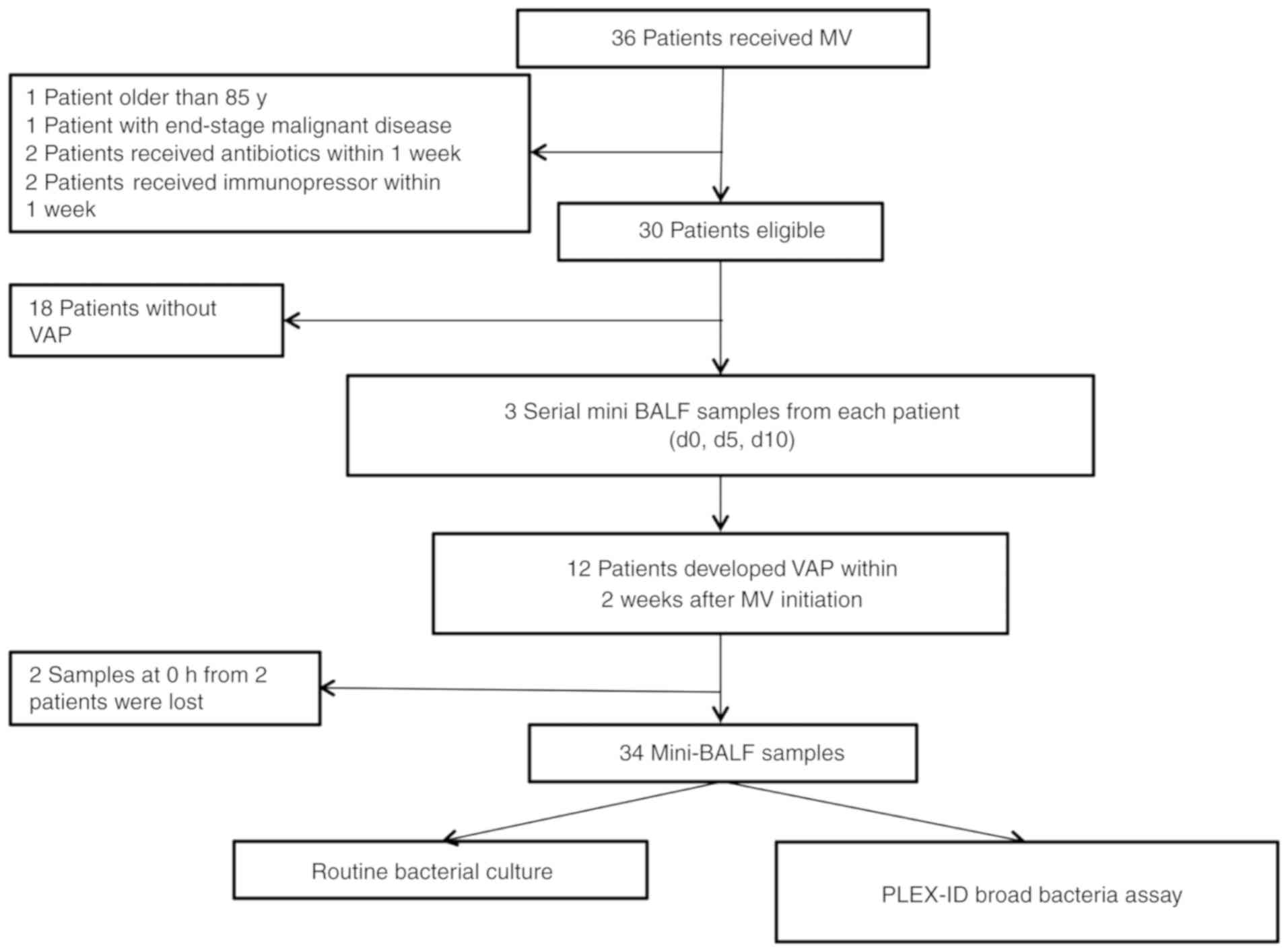
present study (see workflow chart in Fig. 1). The clinical characteristics of the
subjects are summarized in Table I.
The mean age of subjects was 62.8±12.2 years, and 16.7% awere
female. The most common comorbidities were hypertension and
malignant solid tumors.
 | Table IClinical characteristics of the
patients with VAP (n=12). |
Table I
Clinical characteristics of the
patients with VAP (n=12).
| Characteristic | Value |
|---|
| Age (years) | 62.8 (33-81) |
| Female gender | 2 (16.7) |
| Diagnosis | |
|
MT | 4 (30.8) |
|
Esophageal | 3 (23.1) |
|
Colon | 1 (7.7) |
|
Trauma | 2 (15.4) |
|
Gastrointestinal
perforation | 2 (15.4) |
|
Othersa | 4 (30.8) |
| Comorbidity | |
|
Diabetes
mellitus | 1 (7.7) |
|
Hypertension | 3 (23.1) |
|
Arrhythmia | 1 (7.7) |
| Surgery | 10 (76.9) |
| WBC
(106/l) | 12.98±5.45 |
| PCT (µg/l) | 10.13±8.85 |
| Chest X-ray | |
|
Infiltration | 11 (91.7) |
|
Diffuse
infiltration | 1 (8.3) |
| Time-point of VAP
diagnosis from hospitalization (days) | 7 (3-22) |
| Days of MV | 19.5 (10-80) |
| Antibiotics prior to
MV | 5 (38.5) |
| Outcome | |
|
Discharge | 8(62) |
|
Deceased | 4(31) |
Agreement of routine clinical culture
and PCR/ESI-MS in mini-BALF samples from VAP patients
At the specimen level, the concordance (positive as
well as negative) between the two methods was 50% (17/34). Routine
clinical culture identified at least 1 positive isolate in 13 of
the 34 mini-BALF specimens (positive rate, 38.2%), while the
PCR/ESI-MS method detected pathogenic bacteria in 30 of the 34
samples (positive rate, 88.2%) according to the criteria for
positivity (see Methods section). Of note, all of the positive
culture specimens were also identified by PCR/ESI-MS (matched
positive: 13/34, 38.2%) with a positive agreement rate of 100%.
However, except for the 4 specimens that were negative by the two
methods (matched negative: 4/34, 11.8%), PCR/ESI-MS identified
bacteria in 17 culture-negative specimens (17/34, 50%; negative
agreement rate: 19%; Table II).
These results imply that PCR/ESI-MS was sensitive in identifying
bacteria in mini-BALF samples from ventilated patients at the
specimen level.
 | Table IIConcordance of routine clinical
culture and PCR/ESI-MS in mini-bronchoalveolar lavage fluid samples
at the specimen level (n=34). |
Table II
Concordance of routine clinical
culture and PCR/ESI-MS in mini-bronchoalveolar lavage fluid samples
at the specimen level (n=34).
| Concordance | n (%) |
|---|
| Matched (+) | 13 (38.2) |
| Matched (-) | 4 (11.8) |
| Only culture
(+) | 0 (0) |
| Only PCR/ESI-MS
(+) | 17(50) |
| Overall agreement
(%) | 50 |
| Positive
agreementa (%) | 100 |
| Negative
agreementb (%) | 19.0 |
PCR/ESI-MS has high sensitivity for
identifying VAP bacteria at the species level
A total of 51 bacterial species were detected in 30
specimens using clinical culture, PCR/ESI-MS or the two methods
(Table III). Routine clinical
culture identified 16 bacterial isolates (16/51, 31.4%) from 13/34
specimens (Table III). Of these,
15 were also positive on PCR/ESI-MS (positive agreement: 15/16,
93.8%) and 1 was negative (1/16, 6.2%). The only isolate positive
in culture but negative in PCR/ESI-MS was from one of the 8
mixed-infection specimens, which included 3 distinct bacterial
isolates, while 2 isolates from another mixed-infection specimen
were correctly identified by PCR/ESI-MS. The positive agreements in
simple infection and mixed infection were 100% (11/11) and 80.0%
(4/5), respectively (Table IV),
which suggested that PCR/ESI-MS had notable sensitivity and it was
indicated that it is able to detect specimens with mixed
infections.
 | Table IIIBacterial species identified by
routine bacterial culture and/or PLEX-ID in 34 mini-bronchoalveolar
lavage fluid specimens from 12 patients diagnosed with
ventilator-associated pneumonia. |
Table III
Bacterial species identified by
routine bacterial culture and/or PLEX-ID in 34 mini-bronchoalveolar
lavage fluid specimens from 12 patients diagnosed with
ventilator-associated pneumonia.
| Bacterial
Species |
Culture/PCR/ESI-MS |
|---|
| | +/+ | +/- | -/+ (With clinical
significance) | -/+ (Without
clinical significance) |
|---|
| Acinetobacter
baumannii/calcoaceticus | 4 | 0 | 4 | 2 |
| Burkholderia
ambifaria/vietnamiensis | 0 | 0 | 5 | 1 |
| Enterobacter
aerogenes | 1 | 0 | 0 | 0 |
| Enterobacter
cloacae | 1 | 0 | 0 | 0 |
| Escherichia
coli | 1 | 0 | 0 | 0 |
| Klebsiella
pneumoniae | 1 | 0 | 3 | 2 |
| Pseudomonas
aeruginosa | 6 | 0 | 6 | 1 |
| Staphylococcus
aureus | 1 | 1 | 1 | 2 |
| Haemophilus
influenzae | 0 | 0 | 0 | 3 |
| Streptococcus
pneumoniae | 0 | 0 | 0 | 5 |
| Total | 15 | 1 | 19 | 16 |
 | Table IVConcordance of routine clinical
culture and PCR coupled to electrospray ionization mass
spectrometry for identified bacteria. |
Table IV
Concordance of routine clinical
culture and PCR coupled to electrospray ionization mass
spectrometry for identified bacteria.
| Concordance | By bacteria (all)
(n=51) | By bacteria (simple
infection)a n=43 | By bacteria (mixed
infection)b n=8 |
|---|
| Matched positive
(n) | 15 | 11 | 4 |
| Only
culture-positive (n) | 1 | 0 | 1 |
| Only PCR-positive
(n) | 35 | 32 | 3 |
| Overall agreement
(%) | 29.4 | 25.6 | 50.0 |
| Positive
agreementc (%) | 93.8 | 100.0 | 80.0 |
PCR/ESI-MS identifies bacteria
possibly missed by routine culture
PCR/ESI-MS identified 50 bacterial species from 30
mini-BALF specimens (Table III).
As previously mentioned, 15 of them (15/50, 30%) were positive on
the two methods. However, the other 35 (35/50, 70%) detected by
PCR/ESI-MS did not produce any positive results in routine culture.
It was then assessed whether the bacteria identified only by
PCR/ESI-MS had clinical significance, i.e. whether they were
false-positive on PCR/ESI-MS or false-negative in routine clinical
culture. The results of the routine culture were compared to those
of the other two of the three sequential mini-BALF specimens or
sputum specimens from the same patient sent by clinicians; 19 of
the PCR/ESI-MS-identified bacterial species (19/35, 54.3%) had
positive results according to the clinical criteria (specified in
the Methods section) and they represented 38.0% of all 50
PCR/ESI-MS reports. Thus, these bacteria were possibly missed by
the routine cultures.
A total of 16 bacterial species that had positive
results on PCR/ESI-MS (16/35, 45.7% of the bacterial species
identified only by PCR/ESI-MS; 16/50, 32% of the total bacterial
species identified by PCR/ESI-MS) were not clinically significant
(Table III). Streptococcus
pneumoniae (n=5) and Haemophilus influenzae (n=3), which
are two common pathogens of community-acquired pneumonia and are
less likely to cause VAP, represented 50% (8/16) of the bacterial
species. In addition, the bacterial species identified only by
PCR/ESI-MS and with clinical significance included Pseudomonas
aeruginosa (n=6), Burkholderia spp. (n=5),
Acinetobacter baumannii (n=4), Klebsiella pneumoniae
(n=3), and Staphylococcus aureus (n=1). The results suggest
that 81.0% (34/42) of the PCR/ESI-MS-identified species were common
VAP pathogens with clinical significance in mini-BALF samples of
mechanically ventilated patients.
Variations in the pathogenic spectrum
in patients during MV
It was attempted to track the alterations in
bacterial composition in the airways of subjects under MV using two
aetiological techniques and their efficiency in terms of providing
a quick pathogenic diagnosis was compared. Bacterial species
determined by either routine culture or PCR/ESI-MS were used in
this longitudinal analysis, while the results from 2 subjects whose
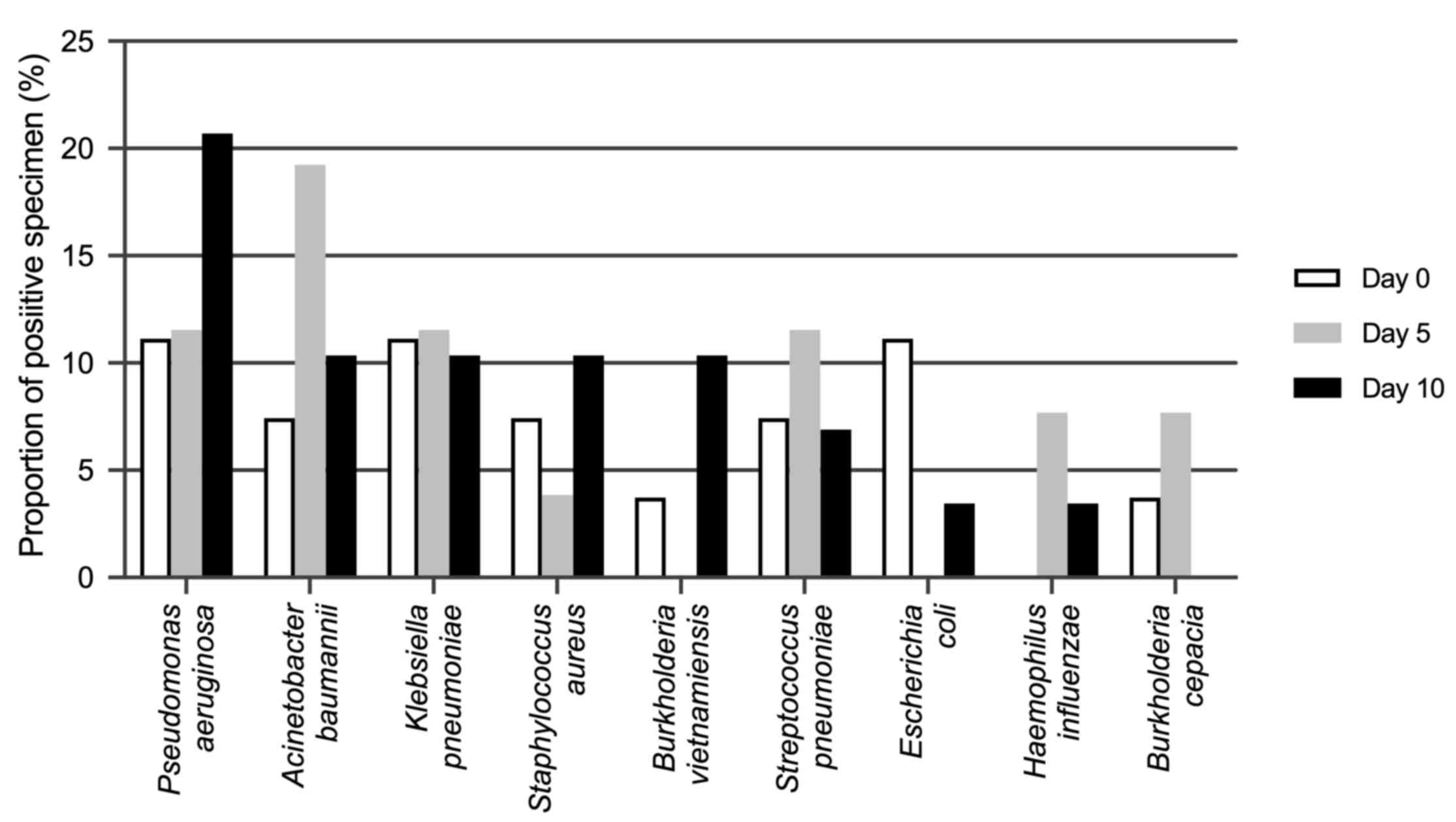
specimens were not complete were excluded (Fig. 2). When MV started, there were
relatively few positive results for bacteria and the most common
species detected were Streptococcus pneumoniae and
Pseudomonas aeruginosa. As MV continued, the positive rates
of Pseudomonas aeruginosa (7/10) increased significantly and
were highest on day 10, followed by Acinetobacter baumannii
(5/10), Staphylococcus aureus (3/10), Burkholderia
spp. (3/10) and Klebsiella pneumoniae (2/10). No
fungus other than Candida spp., which was classified
as normal flora, was detected by either of the two methods.
Non-cultivable pathogens, including Legionella pneumophila
or Mycoplasma pneumoniae, did not reach the positivity
criteria of PCR/ESI-MS, which was possibly due to their low
concentration.
A total of 16 bacteria from 12 subjects were
confirmed as pathogens that cause VAP according to mini-BALF or
sputum cultures (Table V). When the
results of sequential PCR/ESI-MS were examined on days 0, 5 and 10,
7 of the final VAP pathogens were detected by PCR/ESI-MS 5-20 days
prior to the report of routine culture; the other 7 VAP pathogens
were detected simultaneously from specimens taken on the same day
by using the two methods. Only 2 VAP pathogens were detected first
by culture of sputum specimens sent by clinicians on days 7 and 8
after the second PCR/ESI-MS test, but the specimens were also
positive in the third PCR/ESI-MS test on day 10. Thus, sequential
PCR/ESI-MS tests on days 0, 5 and 10 were able to identify the
pathogens of VAP in certain subjects earlier than routine culture
(Table V). Of note, in 8 subjects,
PCR/ESI-MS even confirmed VAP pathogens on day 0 and day 5 before
any clinical manifestations occurred. These bacteria already
existed in the respiratory tract of ventilated subjects in the
early days of ventilation and caused pulmonary infection later. By
contrast, routine culture predicted such pathogens in only 1
patient. In conclusion, sequential PCR/ESI-MS may not only lead to
a pathogenic diagnosis of VAP that is faster than routine culture
but also provide references for clinicians to select an appropriate
initial antibiotic therapy at the time VAP is diagnosed.
 | Table VTime-points of VAP diagnosis and
identification VAP pathogens by routine clinical culture and
PCR/ESI-MS. |
Table V
Time-points of VAP diagnosis and
identification VAP pathogens by routine clinical culture and
PCR/ESI-MS.
| Case ID | Day of
diagnosisa | Confirmed VAP
pathogenb | Days of positive
culture | Day of PCR/ESI-MS
positivity |
|---|
| A | 4 | Pseudomonas
aeruginosa | 10 | 10 |
| C | 9 | Staphylococcus
aureus | 10 | 10 |
| D | 9 | Pseudomonas
aeruginosa | 10 | 0 |
| E | 5 | Acinetobacter
baumannii | 5 | 0 |
| F | 8 | Acinetobacter
baumannii | 5 | 5 |
| | | Pseudomonas
aeruginosa | 5 | 5 |
| G | 8 | Pseudomonas
aeruginosa | 20 | 0 |
| I | 13 | Burkholderia
spp. | 14 | 0 |
| | | Pseudomonas
aeruginosa | 17 | 5 |
| J | 6 | Pseudomonas
aeruginosa | 7 | 0 |
| K | 3 | Acinetobacter
baumannii | 10 | 10 |
| | | Pseudomonas
aeruginosa | 10 | 10 |
| | | Escherichia
coli | 10 | 10 |
| L | 5 | Pseudomonas
aeruginosa | 7 | 10 |
| M | 3 | Klebsiella
pneumoniae | 10 | 5 |
| O | 6 | Staphylococcus
aureus | 6 | 10 |
Discussion
Early and appropriate antibiotic therapy is key to
improving outcomes, reducing side effects and lowering treatment
costs in patients with VAP. The identification of bacterial
isolates from clinical specimens by routine culture usually takes
2-3 days. To overcome the technical delay, novel strategies with
high efficiency and sensitivity are required to provide an
aetiological diagnosis of VAP. Culture-independent microbiological
techniques developed based on PCR have impressive potential in
pathogen identification for infectious diseases, particularly
life-threatening infections including sepsis, bloodstream infection
and pneumonia (12-15).
As one of these techniques, PCR/ESI-MS has been reported to improve
microbiological detection in blood and respiratory specimens
(11,15,16), as
well as samples from patients already receiving antibiotics
(17). However, in contrast to those
on blood samples, prospective studies on the clinical value of
PCR/ESI-MS for evaluating BALF specimens in patients with suspected
VAP are limited.
In the present study, the effectiveness of
PCR/ESI-MS in examining mini-BALF samples of patients with
suspected VAP was evaluated. The results confirmed the high
sensitivity of PCR/ESI-MS in that all routine culture-positive
specimens were also positive on PCR/ESI-MS with high accuracy for
the bacterial species identified (93%). Only one exception was
observed with a positive culture result paired with a negative
PCR/ESI-MS result, which was observed with a sample containing two
other species mixed in the same specimen, suggesting that there may
be interference from two coexisting bacterial species. Other
studies of mini-BALF samples reported a sensitivity of 91.7% in
patients suspected to have VAP (17), and the types of microorganisms
identified in 79-100% of patients with suspected pneumonia or other
diseases requiring mini-BALF sampling exhibited a variation
(16,18). Certain organisms had a higher
‘missed’ rate by PCR/ESI-MS, including Pneumocystis jirovecii,
Actinomyces odontolyticus and Enterobacter cloacae
(17,18), while it was rare that bacterial
species that are common pathogens of VAP were missed by PCR/ESI-MS.
Thus, a better bacterial detection performance may be achieved by
screening particular patient groups, e.g. patients with suspected
VAP in whom the causative pathogens are relatively focused. In
summary, the good sensitivity and negative predictive value
confirmed PCR/ESI-MS to be valuable for ruling out
ventilation-associated infection.
In addition to the long time required by routine
cultures, the negative results from routine microbiological
cultures in patients with suspected VAP remain a challenge for
clinicians. Routine cultures tend to be negative when the
concentration or activity of microorganisms is low. Treatment with
antibiotics prior to sampling affects the sensitivity of the
culture. By contrast, PCR-based methods specifically detect nucleic
acids in specimens, regardless of the microbial activity. The
diagnostic value of PCR-based techniques in culture-negative cases
has been reported (14,19,20).
According to previous studies, the limit of detection using
PCR/ESI-MS was ~16 CFU/ml and 1x103 to 1x104
genome copies per ml, indicating that the sensitivity is much
higher than that of culture-based quantitative microbiology
(21) and matrix-assisted laser
desorption ionization-time of flight/MS techniques (9). In the present study, PCR/ESI-MS
detected ≥1 bacterial species from 50% of the total specimens.
Further investigation revealed that 68% (34/50) of bacteria only
positive on PCR/ESI-MS were confirmed by paired mini-BALF cultures
or routine culture of other mini-BALF or sputum specimens from the
same patient within 7 days and clinical manifestations. Of note, in
contrast to the low positive prediction (0/8) of common community
acquired pneumonia pathogens (Streptococcus pneumoniae and
Haemophilus influenzae), the percentage of common VAP
bacteria identified by PCR/ESI-MS was 81% (34/42). It may be
speculated that common VAP bacteria detected by PCR/ESI-MS with
negative culture results were missed in routine clinical culture
despite their proliferation activity and their pathogenicity cannot
be confirmed. These results highlight the complementary role of
PCR/ESI-MS as an informative microbiological test in patients with
suspected VAP, particularly in culture-negative cases.
The present study differs from previous studies, as
serial mini-BALF specimens were collected from ventilated patients
and compared the microorganisms reported by PCR/ESI-MS prior to the
onset of VAP, and the final aetiological diagnosis of VAP was
confirmed by routine culture. It was hypothesized that due to its
high-throughput feature, PCR/ESI-MS may rapidly detect variations
in the respiratory microbiota of ventilated patients; thus, regular
monitoring of mini-BALF is able to predict the pathogens that may
cause VAP. In the patient cohort, the presence of several bacterial
species, including Pseudomonas aeruginosa, Acinetobacter
baumannii, and Staphylococcus aureus, was increased and
these species were reported to mutually exclude other community
members (21). In 10/12 subjects,
PCR/ESI-MS reported VAP pathogens no later than routine culture. Of
note, in 5 subjects, the final VAP pathogen was identified by
PCR/ESI-MS before VAP occurred. The results are consistent with a
previous study indicating the presence of a microbiologically
positive pathogen burden prior to the clinical diagnosis of VAP
(20). Monitoring mini-BALF using
highly sensitive methods in ventilated patients may improve patient
outcomes.
The present study has certain limitations. First,
ventilated patients who did not develop VAP were not included.
Thus, it cannot be confirmed whether VAP pathogens detected using
PCR/ESI-MS indicate an elevated risk for VAP. Initiation of
antibiotic therapy based on the PCR/ESI-MS results in subjects
without any manifestations of VAP may lead to overmedication.
Further evidence is needed to determine if a prophylactic
antibiotic is beneficial in asymptomatic ventilated patients with
highly pathogenic species detectted in BALF by PCR/ESI-MS. Second,
pathogen isolation by routine microbiological cultures is regarded
as the gold standard, however, it is not very sensitive. Some
pathogens detected by RT-PCR/ESI-MS specimens detected by could not
be confirmed or thoroughly ruled out. Furthermore, the PCR/ESI-MS
tests were performed together at the end of the study, and
therefore, it was not possible to observe the possible benefits of
rapid PCR/ESI-MS reports. Additional prospective studies are
required to evaluate its effects on clinical outcomes.
In conclusion, PCR/ESI-MS has the potential to
accelerate the aetiological diagnosis of VAP within 6 h and predict
the bacterial species that tend to cause VAP prior to clinical
diagnosis. Regular respiratory specimen monitoring using PCR/ESI-MS
has possible clinical benefits for ventilated subjects by guiding
appropriate and adequate initial antibiotic therapy to achieve
better outcomes and decrease the use of broad-spectrum antibiotics
to reduce costs and side effects.
Acknowledgements
The authors would like to thank Ms. Cuicui Chen, Dr
Linlin Wang and Dr Nana Feng (Department of Pulmonary and Critical
Care Medicine, Zhongshan Hospital, Fudan University, Shanghai,
China) for data collection.
Funding
This research was supported by the National Natural
Science Foundation of China (81490533, 81770075, 81800008,
91543114), National Program on Key Research Projects of China
(2017YFC1310602), Shanghai Sailing Program (18YF1404300), Shanghai
Clinical Key Disciplines Construction Project, Shanghai
Top-Priority Clinical Key Disciplines Construction Project
(2017ZZ02013), and The State Key Basic Research Program (973)
project of China (2015CB553404).
Availability of data and materials
The datasets used or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DH and MJ analysed the data and wrote the draft of
the manuscript. YW, D Zhang, CZ did laboratory work. D Zhu, MZ, YS
and XC designed the study, supervised the study and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Zhongshan Hospital, Fudan University (Shanghai, China;
approval no. 2011-212). Informed consent was obtained from all
individual participants included in the study.
Patient consent for publication
Written informed consent for publication was
obtained from all participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Melsen WG, Rovers MM, Groenwold RH,
Bergmans DC, Camus C, Bauer TT, Hanisch EW, Klarin B, Koeman M,
Krueger WA, et al: Attributable mortality of ventilator-associated
pneumonia: A meta-analysis of individual patient data from
randomised prevention studies. Lancet Infect Dis. 13:665–671.
2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Rello J, Ollendorf DA, Oster G,
Vera-Llonch M, Bellm L, Redman R and Kollef MH: VAP Outcomes
Scientific Advisory Group. Epidemiology and outcomes of
ventilator-associated pneumonia in a large US database. Chest.
122:2115–2121. 2002.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chastre J and Fagon JY:
Ventilator-associated pneumonia. Am J Respir Crit Care Med.
165:867–903. 2002.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kuti EL, Patel AA and Coleman CI: Impact
of inappropriate antibiotic therapy on mortality in patients with
ventilator-associated pneumonia and blood stream infection: A
meta-analysis. J Crit Care. 23:91–100. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kalil AC, Metersky ML, Klompas M,
Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O'Grady NP,
Bartlett JG, Carratala J, et al: Management of adults with
hospital-acquired and ventilator-associated pneumonia: 2016
clinical practice guidelines by the infectious diseases society of
America and the American Thoracic Society. Clin Infect Dis.
63:e61–e111. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wu CJ, Chen YP, Wang HC, Su IJ, Ko WC,
Chen JS, Cheng CN, Lee NY, Sun HS, Chi CY and Chen TY:
Identification of fungal pathogens from clinical specimens using
multi-locus PCR coupled with electrospray ionization mass
spectrometry. Diagn Microbiol Infect Dis. 78:141–143.
2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Peeters B, Herijgers P, Beuselinck K,
Peetermans WE, Herregods MC, Desmet S and Lagrou K: Comparison of
PCR-electrospray ionization mass spectrometry with 16S rRNA PCR and
amplicon sequencing for detection of bacteria in excised heart
valves. J Clin Microbiol. 54:2825–2831. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ruiz M, Torres A, Ewig S, Marcos MA, Alcon
A, Lledo R, Asenjo MA and Maldonaldo A: Noninvasive versus invasive
microbial investigation in ventilator-associated pneumonia:
Evaluation of outcome. Am J Respir Crit Care Med. 162:119–125.
2000.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kaleta EJ, Clark AE, Johnson DR, Gamage
DC, Wysocki VH, Cherkaoui A, Schrenzel J and Wolk DM: Use of PCR
coupled with electrospray ionization mass spectrometry for rapid
identification of bacterial and yeast bloodstream pathogens from
blood culture bottles. J Clin Microbiol. 49:345–353.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jacob D, Sauer U, Housley R, Washington C,
Sannes-Lowery K, Ecker DJ, Sampath R and Grunow R: Rapid and
high-throughput detection of highly pathogenic bacteria by Ibis
PLEX-ID technology. PLoS One. 7(e39928)2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jordana-Lluch E, Gimenez M, Quesada MD,
Rivaya B, Marco C, Dominguez MJ, Armestar F, Martro E and Ausina V:
Evaluation of the broad-range PCR/ESI-MS technology in blood
specimens for the molecular diagnosis of bloodstream infections.
PLoS One. 10(e0140865)2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sircar M, Ranjan P, Gupta R, Jha OK, Gupta
A, Kaur R, Chavhan N, Singh M and Singh SK: Impact of
bronchoalveolar lavage multiplex polymerase chain reaction on
microbiological yield and therapeutic decisions in severe pneumonia
in intensive care unit. Crit Care. 31:227–232. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Toma I, Siegel MO, Keiser J, Yakovleva A,
Kim A, Davenport L, Devaney J, Hoffman EP, Alsubail R, Crandall KA,
et al: Single-molecule long-read 16S sequencing to characterize the
lung microbiome from mechanically ventilated patients with
suspected pneumonia. J Clin Microbiol. 52:3913–3921.
2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Guembe M, Marin M, Martin-Rabadan P,
Echenagusia A, Camunez F, Rodriguez-Rosales G, Simo G, Echenagusia
M and Bouza E: GEIDI Study Group. Use of universal 16S rRNA gene
PCR as a diagnostic tool for venous access port-related bloodstream
infections. J Clin Microbiol. 51:799–804. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Morozumi M, Nakayama E, Iwata S, Aoki Y,
Hasegawa K, Kobayashi R, Chiba N, Tajima T and Ubukata K:
Simultaneous detection of pathogens in clinical samples from
patients with community-acquired pneumonia by real-time PCR with
pathogen-specific molecular beacon probes. J Clin Microbiol.
44:1440–1446. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ullberg M, Luthje P, Molling P, Stralin K
and Ozenci V: Broad-range detection of microorganisms directly from
bronchoalveolar lavage specimens by PCR/electrospray
ionization-mass spectrometry. PLoS One. 12(e0170033)2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Stralin K, Ehn F, Giske CG, Ullberg M,
Hedlund J, Petersson J, Spindler C and Ozenci V: The IRIDICA
PCR/electrospray ionization-mass spectrometry assay on
bronchoalveolar lavage for bacterial etiology in mechanically
ventilated patients with suspected pneumonia. PLoS One.
11(e0159694)2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Huttner A, Emonet S, Harbarth S, Renzi G,
Kaiser L and Schrenzel J: Polymerase-chain reaction/electrospray
ionization-mass spectrometry for the detection of bacteria and
fungi in bronchoalveolar lavage fluids: A prospective observational
study. Clin Microbiol Infect. 20:O1059–O1066. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zakharkina T, Martin-Loeches I, Matamoros
S, Povoa P, Torres A, Kastelijn JB, Hofstra JJ, de Wever B, de Jong
M, Schultz MJ, et al: The dynamics of the pulmonary microbiome
during mechanical ventilation in the intensive care unit and the
association with occurrence of pneumonia. Thorax. 72:803–810.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Douglas IS, Price CS, Overdier KH, Wolken
RF, Metzger SW, Hance KR and Howson DC: Rapid automated microscopy
for microbiological surveillance of ventilator-associated
pneumonia. Am J Respir Crit Care Med. 191:566–573. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bacconi A, Richmond GS, Baroldi MA,
Laffler TG, Blyn LB, Carolan HE, Frinder MR, Toleno DM, et al:
Improved sensitivity for molecular detection of bacterial and
Candida infections in blood. Journal of clinical microbiology
52(9):3164-3174. doi:10.1128/JCM.00801-14, 2014.
|