Introduction
Diabetic nephropathy (DN), which occurs in 20-40% of
all diabetes cases, is one of the systemic microvascular
complications of diabetes as well as the leading cause of end-stage
renal disease (1). DN is a chronic
process whose early clinical symptoms are often not evident
(2). In addition, there is a large
risk of DN patients developing uremia, which is a key contributor
to diabetes-related disability or death (2). Hyperglycemia, in the long term, causes
hypertrophy, infiltration of inflammatory cells, thickening of the
basement membrane, glomerular sclerosis and tubular atrophy,
eventually leading to kidney failure (3). Once kidney failure is developed, DN is
even more difficult to treat than other kidney diseases (3). Thus, the early prevention of DN is
important for diabetes management. However, the current methodology
is still insufficient for timely identification of disease onset
and prognosis in DN.
Long non-coding RNAs (lncRNAs) are transcripts that
measure more than 200 nucleotides with limited protein-coding
ability (4). The effects of lncRNAs
have been revealed in the pathogenesis of various human diseases
including diabetes (5). Several
lncRNAs, including lnc-Rpph1, lnc-MEG3 and
lnc-CYP4B1-PS1-001, are known to regulate fibrosis and
inflammation in DN (3,6,7). Our
previous study selected five candidate lncRNAs from lncRNA
microarray data and found that lncRNA Dlx6os1 was highly
expressed in high glucose-treated mouse mesangial cells (MMCs)
(8). The next stage of the
investigation observed that inhibition of lncRNA Dlx6os1
suppressed proliferation and fibrosis but promoted apoptosis in
MMCs of the DN cellular model (8).
lncRNA Dlx6os1 is a novel gene that has not been studied in
DN, despite the aforementioned evidence from our previous study
(8). The present study further
analyzed the effect of lncRNA Dlx6os1 suppression on the
mRNA expression profile in MMCs of a DN cellular model to explore
the mechanism of lncRNA Dlx6os1 inhibition in the
pathogenesis of DN.
Materials and methods
Construction of DN cellular model
lncRNA Dlx6os1 exhibits good homology between
mice and humans. A DN cellular model was constructed according to
the method described in our previous study (8). In brief, SV40 MES13 cells (MMCs) were
purchased from Shanghai Institutes for Biological Sciences and were
cultured in Dulbecco modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) in a humidified incubator with 5% CO2
at 37˚C. Then 30 mmol/l glucose was added to treat SV40 MES13 cells
for 96 h to construct the DN cellular model.
Construction and transfection of short
hairpin (sh)RNA plasmids
Negative control (NC) shRNA and lncRNA
Dlx6os1 shRNA plasmids were constructed by Genewiz, Inc. and
transfected into a DN cellular model (SV40 MES13 cells under 30
mmol/l glucose culture), then termed the sh-NC group and sh-lncRNA
group. Subsequently, i) lncRNA Dlx6os1 expression in the two
groups was detected by reverse transcription-quantitative (RT-q)
PCR at 48 h; ii) proliferation ability was detected by Cell
Counting Kit-8 (Sigma-Aldrich; Merck KGaA) at 0, 24, 48 and 72 h
according to the manufacturers' instructions; iii) the mRNA and
protein expression levels of proliferative markers (cyclin D1 and
proliferating cell nuclear antigen) as well as fibrosis markers
(fibronectin and collagen I) were detected by RT-qPCR and western
blotting at 48 h; iv) the cell apoptosis rate was detected by FITC
Annexin V Apoptosis Detection kit II with propidium iodide (AV/PI;
BD Biosciences) at 48 h according to the manufacturer's protocol
(Fig. S1). Data were analyzed using
a flow cytometer (BD FACSCalibur) with FlowJo 7.6 software (FlowJo,
LLC). A wavelength of 488/535 nm was used.
mRNA sequencing
mRNA sequencing was performed by Genergy Bio Company
(http://www.genenergy.cn/). In brief, cells were
harvested at 48 h after transfection, and total RNA was extracted
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), then RNA concentration, purity and integrity
were assessed. Subsequently, mRNA was captured by Dynabeads Oligo
(dT)25 (Thermo Fisher Scientific, Inc.), then first and
second strands of the cDNA were synthesized and library fragments
were purified using AMPure XP system (Beckman Coulter, Inc.). PCR
assay and assessment of library quality were conducted using
Bioanalyzer 2100 system (Agilent Technologies, Inc.), then
clustering of index-coded samples was performed using HiSeq PE
Cluster kit v4 cBot (Illumina, Inc.), and the libraries were
sequenced on Illumina Hiseq X10 platform (Illumina, Inc.). A total
of 150 bp paired-end reads was produced after cluster generation
for mRNA. Automated quality control and adapter trimming were
conducted using Trim Galore, Cutadapt and FastQC (9). Then the trimmed reads were mapped to
the mouse genome mm10 by Hisat2 (Version 2.1.0; http://ccb.jhu.edu/software/hisat2/manual.shtml), and
mapping quality control was conducted using RSeQC (Version 3.0.0;
http://rseqc.sourceforge.net/). Finally,
the read counts of mRNA were then calculated using FeatureCounts
(Rsubread; http://www.bioconductor.org). The top 20 dysregulated
mRNAs are presented in Table I as a
reference for future studies that investigate specific mRNA
targeted by lncRNA D1x6os1 in DN.
 | Table ITop 20 dysregulated mRNAs (10
upregulated and 10 downregulated) in sh-lncRNA cells vs. sh-NC
cells. |
Table I
Top 20 dysregulated mRNAs (10
upregulated and 10 downregulated) in sh-lncRNA cells vs. sh-NC
cells.
| Gene symbol | ID | Chromosome | Log2
FC | P-value | Trend |
|---|
| Ramp2 |
ENSMUSG00000001240 | 11 | 2.078303 |
4.52x10-6 | Up |
| Tagln |
ENSMUSG00000032085 | 9 | 2.016348 |
6.7x10-6 | Up |
| Krt7 |
ENSMUSG00000023039 | 15 | 1.749297 |
2.09x10-5 | Up |
| Aifm3 |
ENSMUSG00000022763 | 16 | 1.757504 |
3.1x10-5 | Up |
| Sult1c2 |
ENSMUSG00000023122 | 17 | 1.781638 |
3.4x10-5 | Up |
| 1700001P01Rik |
ENSMUSG00000018543 | 11 | 5.796657 |
3.42x10-5 | Up |
| Foxp2 |
ENSMUSG00000029563 | 6 | 2.688633 |
4.09x10-5 | Up |
| Sirt2 |
ENSMUSG00000015149 | 7 | 2.458172 |
4.93x10-5 | Up |
| Sep15 |
ENSMUSG00000037072 | 3 | 2.153584 |
7.67x10-5 | Up |
| Slc39a4 |
ENSMUSG00000063354 | 15 | 3.04629 |
8.55x10-5 | Up |
| Ccdc160 |
ENSMUSG00000073207 | X | -6.79158 |
4.98x10-7 | Down |
| Myom1 |
ENSMUSG00000024049 | 17 | -2.09334 |
6.97x10-6 | Down |
| Lmbr1l |
ENSMUSG00000022999 | 15 | -2.19777 |
8.64x10-6 | Down |
| Ddx43 |
ENSMUSG00000070291 | 9 | -1.72587 |
1.94x10-5 | Down |
| Vwa5b2 |
ENSMUSG00000046613 | 16 | -2.63226 |
2.39x10-5 | Down |
| Slc11a1 |
ENSMUSG00000026177 | 1 | -5.77347 |
2.84x10-5 | Down |
| Ccdc7b |
ENSMUSG00000056018 | 8 | -4.83542 |
4.45x10-5 | Down |
| Fmn1 |
ENSMUSG00000044042 | 2 | -1.85445 |
4.85x10-5 | Down |
| Fer1l4 |
ENSMUSG00000013338 | 2 | -5.69906 |
5.37x10-5 | Down |
| As3mt |
ENSMUSG00000003559 | 19 | -1.94909 |
5.43x10-5 | Down |
Bioinformatics
The mRNAs which were identified in ≥50% samples were
included in the analysis and raw read counts were normalized and
logarithmically transformed for bioinformatics analysis. The
bioinformatics analysis was conducted using R software (Version
3.3.3; http://www.R-project.org/) (10). In brief, i) differentially expressed
mRNAs were detected using DeSeq2 package (Version 1.12.3;
http://www.bioconductor.org/), and
statistical significance was defined as adjusted P-value <0.05;
the biological significance was defined as a difference of at least
abs [log2 (fold change)]>2, which was presented as a
Volcano plot; ii) Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) enrichment analysis were performed using
Database for Annotation, Visualization and Integrated Discovery web
servers; iii) gene-set enrichment analysis (GSEA) on apoptosis and
inflammation pathways was performed using the phenoTest package
(11). The datasets of the present
study were uploaded on Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) for public
availability. The GSE number is GSE145301.
Statistical analysis
Statistical analysis and figures were performed
using GraphPad Prism 7.00 (GraphPad Software, Inc.). Comparison
between two groups was determined by paired t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
lncRNA D1x6os1 expression following
transfection
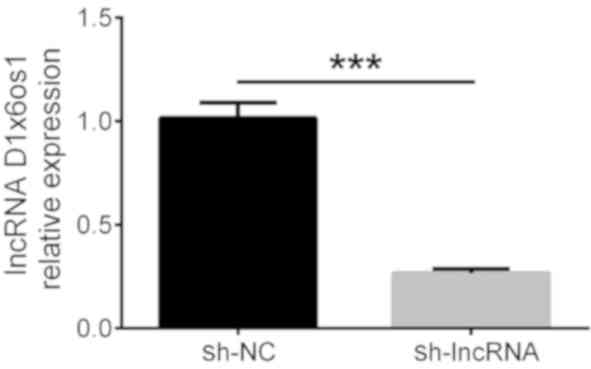
In order to detect the relative expression of lncRNA
D1x6os1 after transfection, qPCR was conducted at 48 h.
lncRNA D1x6os1 expression was decreased in the sh-lncRNA
group compared with the sh-NC group following transfection
(P<0.001), which indicated successful transfection under high
glucose exposure (Fig. 1).
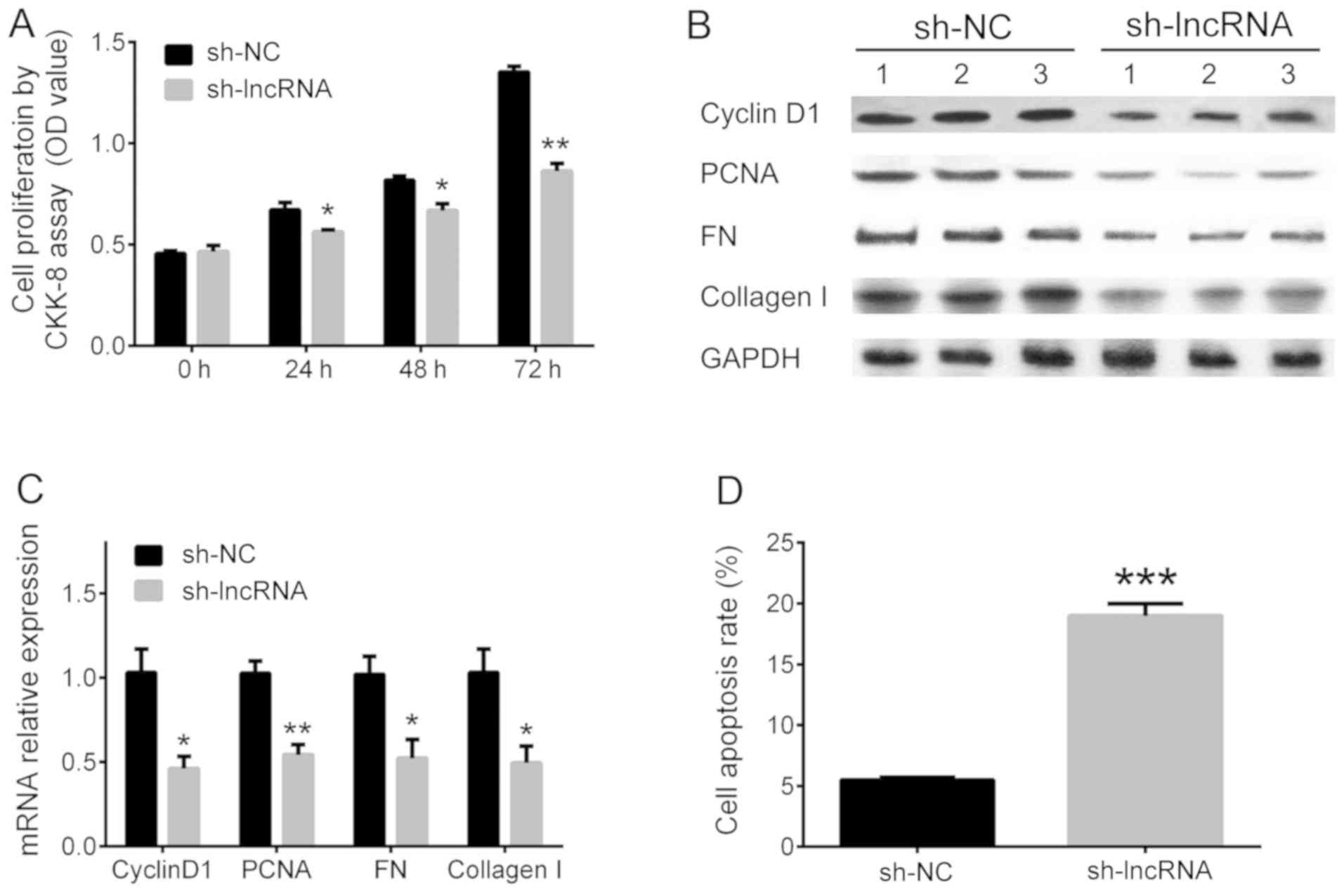
Effect of lncRNA D1x6os1 silencing on
cell proliferation, fibrosis and apoptosis in MMCs of a DN cellular
model
Cell proliferation, fibrosis and apoptosis were
determined to assess the influence of lncRNA D1x6os1
silencing. Cell proliferation was reduced in the sh-lncRNA group
compared with the sh-NC group at 24 (P<0.05), 48 (P<0.05),
and 72 h (P<0.01; Fig. 2A). The
relative expression levels of cyclin D1 (P<0.05), PCNA
(P<0.01), FN (P<0.05) and collagen I (P<0.05) were
suppressed in the sh-lncRNA group compared with the sh-NC group
(Fig. 2B and C). In addition, the cell apoptosis rate was
increased in the sh-lncRNA group compared with the sh-NC group
(P<0.001; Fig. 2D).
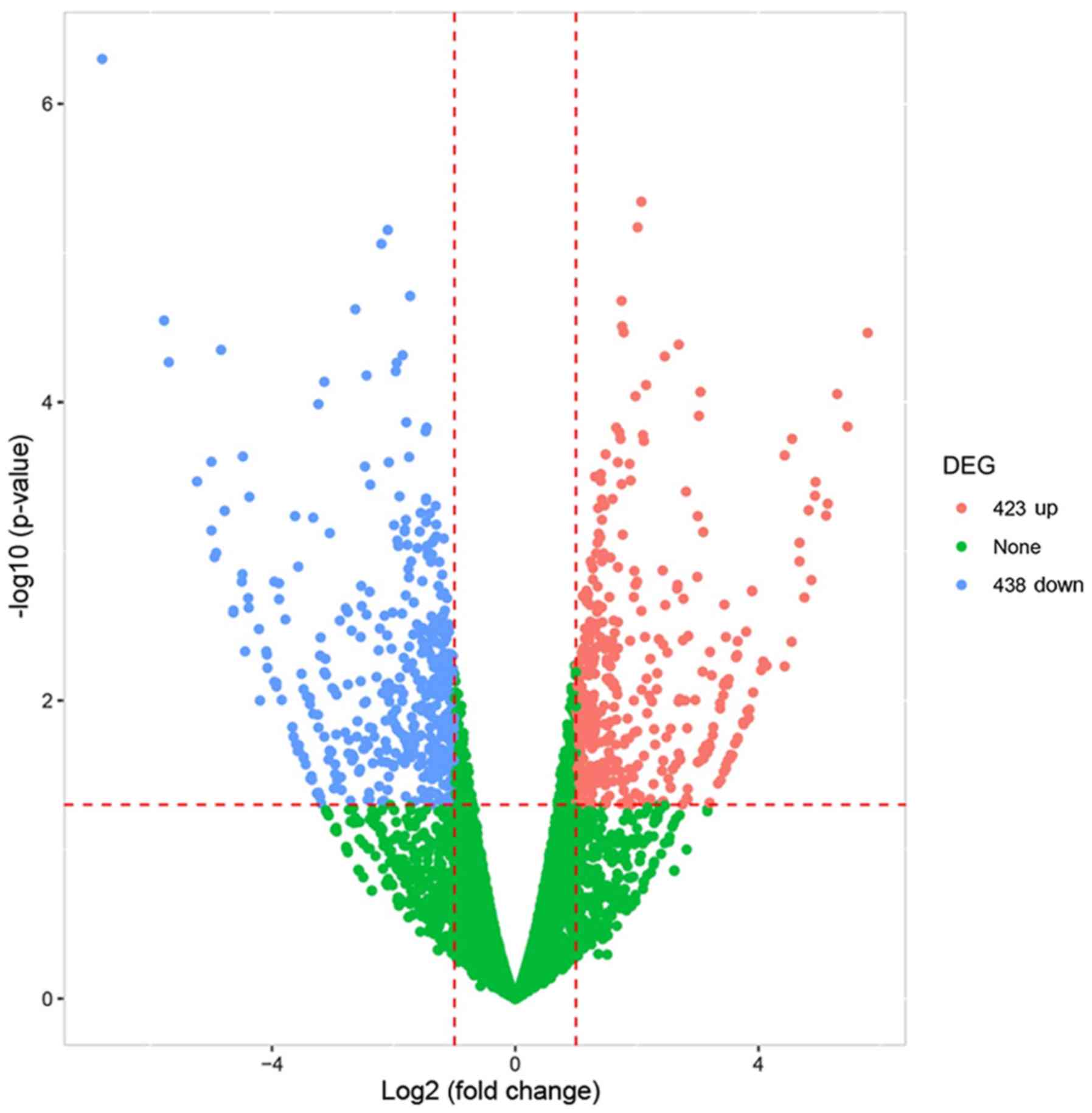
Volcano plot of differentially
expressed mRNAs
A Volcano plot is an intuitive plot that screens out
the differentially expressed genes between the two group of
samples. In the present study, it revealed 423 upregulated mRNAs
and 438 downregulated mRNAs in the sh-lncRNA group compared with
the sh-NC group in MMCs of a DN cellular model (absolute value of
FC>2, Padj<0.05; Fig.
3). The top 20 dysregulated mRNAs are presented in Table I.
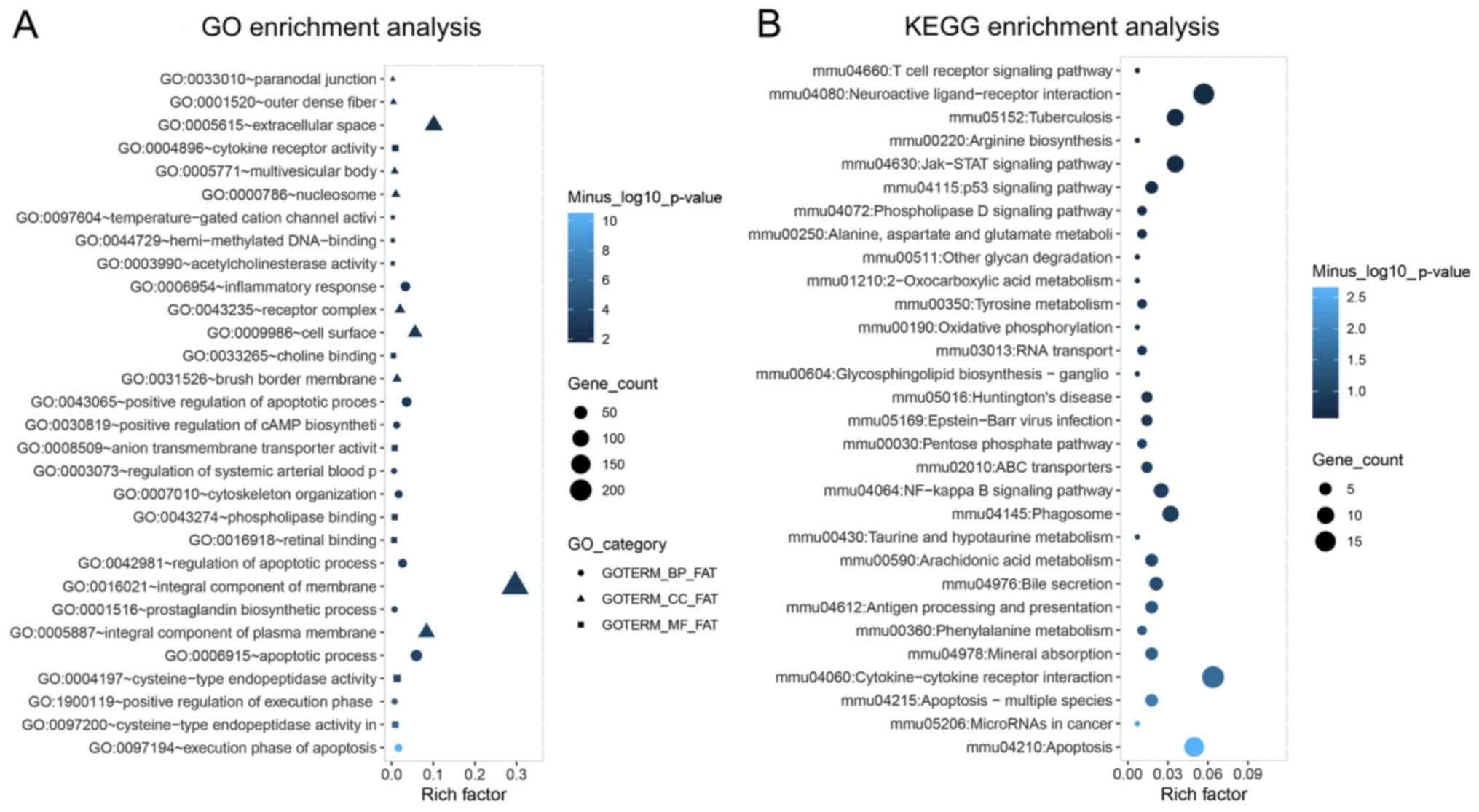
GO and KEGG enrichment analysis of
differentially expressed mRNAs
To evaluate the possible mechanisms of dysregulated
mRNAs in response to lncRNA D1x6os1 silencing, GO and KEGG
enrichment analyses were conducted. GO enrichment analysis revealed
that the differentially expressed mRNAs were mainly enriched in
biological processes including ‘apoptotic process’, ‘inflammatory
response’ and ‘cytoskeleton organization’; and were mainly present
in cellular components such as ‘integral component of membrane’,
‘cell surface’ and ‘extracellular space’; in addition, they were
involved in molecular functions including ‘cytokine receptor
activity’, ‘phospholipase binding’ and ‘cysteine-type endopeptidase
activity’ (Fig. 4A). According to
KEGG enrichment analysis, the dysregulated mRNAs were mainly
enriched in inflammation and apoptosis-related pathways such as
‘NF-κB signaling pathway’, ‘cytokine-cytokine receptor interaction’
and apoptosis pathways (Fig.
4B).
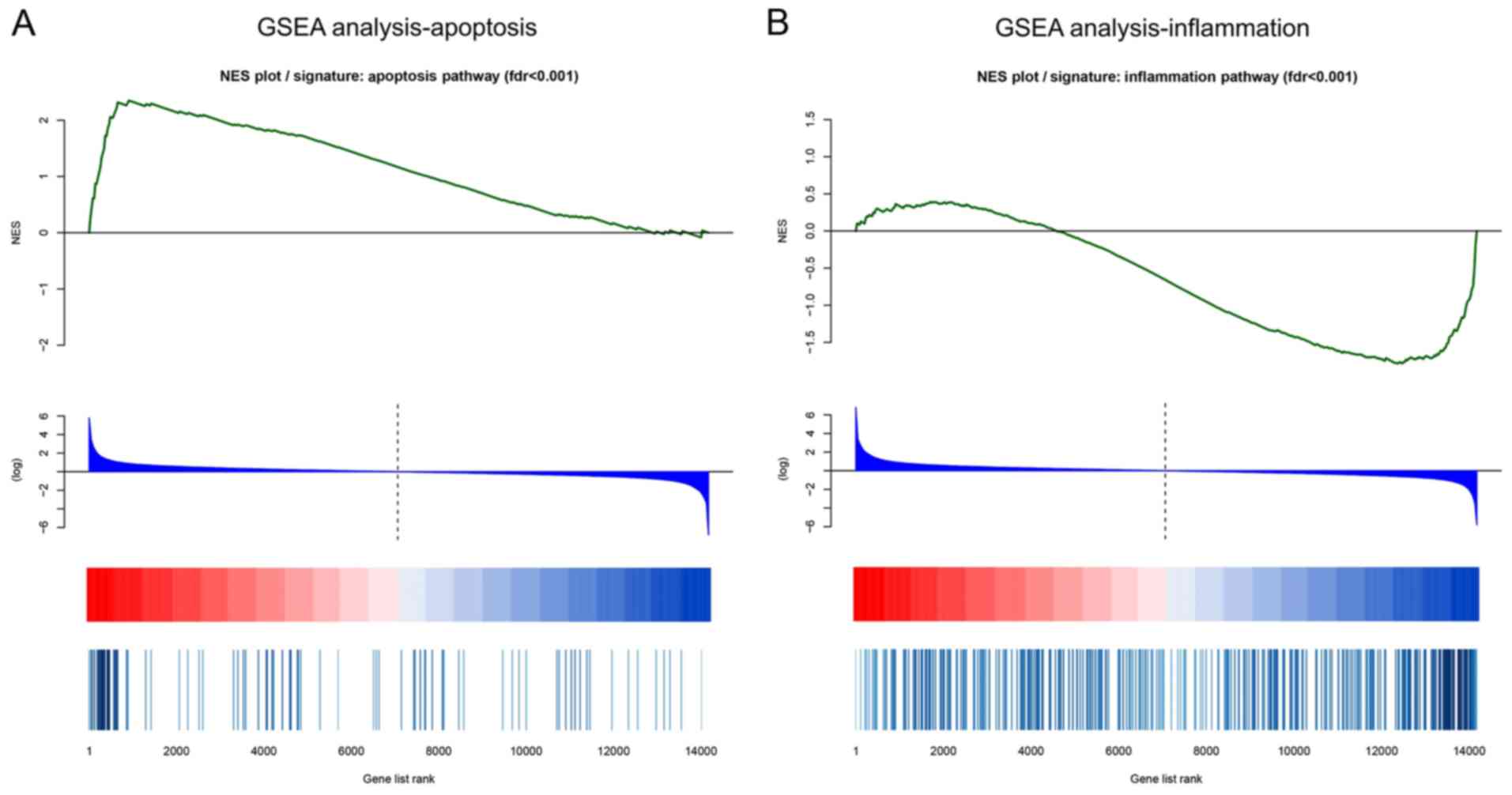
GSEA
Since apoptosis and inflammation are the two main
pathological mechanisms of DN, GSEA of apoptosis and inflammation
was performed. In GSEA analysis of apoptosis, the leading-edge
subset of mRNAs were overexpressed in sh-lncRNA cells compared with
sh-NC cells according to the location of the maximum enrichment
score (Fig. 5A). While in GSEA
analysis of inflammation, the leading-edge subset of mRNAs were
downregulated in sh-lncRNA cells compared with sh-NC cells
(Fig. 5B). These results indicated
that lncRNA D1x6os1 inhibition promoted cell apoptosis and
suppressed inflammation in MMCs of a DN cellular model.
Discussion
As a progressive microvascular complication, DN is
heralded by a subsequent reduction in glomerular filtration rate
and marked by microalbuminuria (12). The renal inflammation underlies the
pathogenic mechanism of DN (13).
Principle inflammatory cells such as macrophages are responsible
for inflammation response via the release of cytokines, chemokines
and reactive oxygen species, which eventually increase the
production of extracellular matrix in glomeruli as well as
progressive tubulointerstitial fibrosis (13). In addition, T-lymphocyte activation
induced by hyperglycemia induces kidney damage in the early stages
by disturbing albumin glomerular excretion as well as renal
infiltration (14).
Apoptosis-induced dysfunction of podocytes and damage to mesangial
cells and renal tubular epithelial cells are indicators of
glomerular filtration dysfunction (15-18).
Although the pathology and pathophysiology of DN have been
investigated for some time and there has been progress, the complex
molecular network is still not fully understood and identification
of genes or molecules by conventional tools remains deficient.
Therefore, screening of genes and molecules by novel strategies,
such as high-throughput platforms are necessary for determining
valuable information on the numerous molecules involved in DN
pathology.
From the published microarray data, lncRNA
expression profile is critical in the pathogenesis and progression
of DN (3,19). Previous studies report that several
lncRNAs regulate inflammation and cell proliferation in DN
(3,19). For instance, lncRNA Rpph1 was
demonstrated to facilitate inflammation and promote proliferation
of MMCs by interacting with DN-related factor galectin-3 in a DN
cellular model (3). Knockdown of
lncRNA Gm4419 was revealed to inhibit pro-inflammatory
cytokine expression and renal fibrosis biomarkers and reduce cell
proliferation of MMCs under high glucose conditions (19). lncRNA Dlx6os1 was upregulated
in the kidneys of diabetic mice and its expression was induced
under high glucose conditions (20).
Therefore, in order to mimic the DN environment, SV40 MES13 cells
(MMCs) cultured under high glucose conditions were used for the
transfection of lncRNA Dlx6os1 inhibitor plasmids in our
previous study (8). In the present
study, the same DN cellular model derived from MMCs was
constructed, and it was confirmed that lncRNA Dlx6os1
silencing suppressed cell proliferation and fibrosis and promoted
cell apoptosis in MMCs of the DN cellular model. The following
theories may account for these effects: i) lncRNA Dlx6os1
silencing may influence the expression of target mRNAs and
attenuate the cell cycle to suppress cell proliferation and
facilitate apoptosis. It was demonstrated in the following GSEA
analysis of apoptosis that lncRNA Dlx6os1 silencing promoted
cell apoptosis in MMCs of the DN cellular model. The reduced cell
apoptosis may attenuate the mesangial expansion as well as
formation of glomerulosclerosis in DN (21). ii) Inhibition of lncRNA
Dlx6os1 may suppress inflammation by blocking signaling
pathways such as the NF-κB pathway, interfering with
cytokine-cytokine receptor interaction (as demonstrated in the KEGG
enrichment analysis) and reducing the inflammatory cytokines, as
well as reactive oxygen species, leading to reduced secretion of
the extracellular matrix and subsequent fibrosis.
Since the pathogenic mechanism of DN is complex and
involves multiple molecular pathways, a more refined screening of
the genetic background of lncRNA Dlx6os1 will certainly
provide a greater insight into the disease. Therefore, additional
RNA sequencing for the mRNA expression profile in lncRNA
Dlx6os1-silenced MMCs of the DN cellular model was
performed. The mRNA expression profile revealed that lncRNA
Dlx6os1 silencing led to 423 upregulated mRNAs and 438
downregulated mRNAs in MMCs of the DN cellular model. The KEGG
enrichment analyses revealed that the differentially expressed
mRNAs were enriched in apoptotic and inflammatory
responses/pathways. GSEA analysis of apoptosis and inflammation
pathways was also performed and suggested that lncRNA
Dlx6os1 silencing promoted cell apoptosis and inhibited
inflammation in MMCs of the DN cellular model. These observations
supplemented the potential molecular mechanism of lncRNA
Dlx6os1 in the pathogenesis of DN to some extent. However,
deeper exploration of downstream mRNAs of lncRNA Dlx6os1 in
regulating cell proliferation, fibrosis and cell apoptosis in DN is
still required.
There were several limitations to the present study:
i) The effect of lncRNA Dlx6os1 silencing on the mRNA
expression profile was evaluated by RNA sequencing, which is
high-throughput and lacks accuracy (22). Therefore, detecting individual mRNA
expression using qPCR and corresponding protein expression using
western blotting may be helpful to validate the influence of lncRNA
Dlx6os1 on mRNA expression in MMCs of the DN cellular model.
ii) Although commonly used in DN research, knocking down an lncRNA
in a short-term as well as an acute kidney damage cellular model
does not substantially represent the real DN condition as a chronic
disease. Therefore, further animal study is required to validate
the results of the present study. iii) lncRNA Dlx6os1 was
revealed to regulate apoptosis and inflammation in MMCs of the DN
cellular model, whereas the clinical implications, including its
correlation with disease risk, disease progression or prognosis of
DN patients, remains to be elucidated, which may be a valuable
research direction for diabetic diseases in the future. iv) The
present study only reported general information concerning lncRNA
Dlx6os1 silencing on biological processes and molecular functions
such as apoptosis and inflammation in DN, whereas the detailed
pathways or the protein markers were not investigated.
In conclusion, lncRNA Dlx6os1 silencing
attenuated disease progression by regulating cell apoptosis and
inflammation-related pathways in MMCs of a DN cellular model,
suggesting that it could assist with developing a novel treatment
target for DN.
Supplementary Material
Annexin V/propidium iodide analysis
for cell apoptosis. A total of three repetitions were performed for
sh-NC cells and sh-lncRNA cells. sh, short hairpin; NC, negative
control; lnc, long non-coding.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Foundation of
Guizhou Science and Technology Plan Project [Guizhou Science and
Technology platform talents; grant no. (2017) 5735-03] and the
Dominant Discipline of the Second Affiliated Hospital of Guizhou
University of Traditional Chinese Medicine, Changning District
Committee of Science and Technology (grant no. CNKW2017Y06).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SH conceived and designed the study. LC and JC
collected the data. WP and XJ performed the experiments and drafted
the manuscript. All authors reviewed and edited the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sun HJ, Wu ZY, Cao L, Zhu MY, Liu TT, Guo
L, Lin Y, Nie XW and Bian JS: Hydrogen Sulfide: Recent progression
and perspectives for the treatment of diabetic nephropathy.
Molecules. 24(2857)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fried LF, Emanuele N, Zhang JH, Brophy M,
Conner TA, Duckworth W, Leehey DJ, McCullough PA, O'Connor T,
Palevsky PM, et al: Combined angiotensin inhibition for the
treatment of diabetic nephropathy. N Engl J Med. 369:1892–1903.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhang P, Sun Y, Peng R, Chen W, Fu X,
Zhang L, Peng H and Zhang Z: Long non-coding RNA Rpph1 promotes
inflammation and proliferation of mesangial cells in diabetic
nephropathy via an interaction with Gal-3. Cell Death Dis.
10(526)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Reichelt-Wurm S, Wirtz T, Chittka D,
Lindenmeyer M, Reichelt RM, Beck S, Politis P, Charonis A, Kretz M,
Huber TB, et al: Glomerular expression pattern of long non-coding
RNAs in the type 2 diabetes mellitus BTBR mouse model. Sci Rep.
9(9765)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Moran I, Akerman I, van de Bunt M, Xie R,
Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J,
Rodríguez-Seguí S, et al: Human β cell transcriptome analysis
uncovers lncRNAs that are tissue-specific, dynamically regulated,
and abnormally expressed in type 2 diabetes. Cell Metab.
16:435–448. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zha F, Qu X, Tang B, Li J, Wang Y, Zheng
P, Ji T, Zhu C and Bai S: Long non-coding RNA MEG3 promotes
fibrosis and inflammatory response in diabetic nephropathy via
miR-181a/Egr-1/TLR4 axis. Aging (Albany NY). 11:3716–3730.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang S, Chen X, Wang M, Yao D, Chen T, Yan
Q and Lu W: Long non-coding RNA CYP4B1-PS1-001 inhibits
proliferation and fibrosis in diabetic nephropathy by interacting
with nucleolin. Cell Physiol Biochem. 49:2174–2187. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cheng J, Cheng L, Tang Y, Li H, Peng W and
Huang S: Inhibition of lncRNA Dlx6os1 decreases cell proliferation
and fibrosis and increases cell apoptosis in diabetic nephropathy.
Int J Clin Exp Pathol. 11:3302–3309. 2018.PubMed/NCBI
|
|
9
|
Kim D, Paggi JM, Park C, Bennett C and
Salzberg SL: Graph-based genome alignment and genotyping with
HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
The R Foundation for Statistical
Computing, Vienna, Austria. URL http://www.R-project.org.
|
|
11
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zheng M, Lv LL, Cao YH, Liu H, Ni J, Dai
HY, Liu D, Lei XD and Liu BC: A pilot trial assessing urinary gene
expression profiling with an mRNA array for diabetic nephropathy.
PLoS One. 7(e34824)2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Barutta F, Bruno G, Grimaldi S and Gruden
G: Inflammation in diabetic nephropathy: Moving toward clinical
biomarkers and targets for treatment. Endocrine. 48:730–742.
2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Duran-Salgado MB and Rubio-Guerra AF:
Diabetic nephropathy and inflammation. World J Diabetes. 5:393–398.
2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gao L, Sun N, Xu Q, Jiang Z and Li C:
Comparative analysis of mRNA expression profiles in type 1 and type
2 diabetes mellitus. Epigenomics. 11:685–699. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yi W and OuYang Q: Adiponectin improves
diabetic nephropathy by inhibiting necrotic apoptosis. Arch Med
Sci. 15:1321–1328. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang T, Gao Y, Wang X, Shi Y, Xu J, Wu B,
He J and Li Y: Calpain-10 drives podocyte apoptosis and renal
injury in diabetic nephropathy. Diabetes Metab Syndr Obes.
12:1811–1820. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tsai YC, Kuo PL, Hung WW, Wu LY, Wu PH,
Chang WA, Kuo MC and Hsu YL: Angpt2 induces mesangial cell
apoptosis through the MicroRNA-33-5p-SOCS5 loop in diabetic
nephropathy. Mol Ther Nucleic Acids. 13:543–555. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yi H, Peng R, Zhang LY, Sun Y, Peng HM,
Liu HD, Yu LJ, Li AL, Zhang YJ, Jiang WH and Zhang Z:
LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3
inflammasome-mediated inflammation in diabetic nephropathy. Cell
Death Dis. 8(e2583)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang M, Wang S, Yao D, Yan Q and Lu W: A
novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation
and fibrosis in diabetic nephropathy. Mol Cell Endocrinol.
426:136–145. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Han F, Xue M, Chang Y, Li X, Yang Y, Sun B
and Chen L: Triptolide suppresses glomerular mesangial cell
proliferation in diabetic nephropathy is associated with inhibition
of PDK1/Akt/mTOR pathway. Int J Biol Sci. 13:1266–1275.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li W, Wang P, Li Y, Zhang K, Ding F, Nie
T, Yang X, Lv Q and Zhao L: Identification of MicroRNAs in response
to different day lengths in soybean using high-throughput
sequencing and qRT-PCR. PLoS One. 10(e0132621)2015.PubMed/NCBI View Article : Google Scholar
|