Introduction
Soft tissue sarcomas (STS) are a rare type of
malignant tumor that account for 1% of all cancers in adults. They
are a heterogeneous group, comprising >50 different types
(1,2). The diagnosis of STS is based on the
morphologic pattern and immunohistochemistry (IHC), in addition to
traditional H&E staining. An increasing number of specific
genetic aberrations, including chromosomal translocations, gene
amplification and mutations, have been identified in various types
of sarcoma. These results led to a recommendation to perform
molecular tests, including reverse transcription PCR, fluorescence
in situ hybridization and next-generation sequencing
(3,4). Histologic diagnosis is important in
predicting the outcome of STS. Sarcoma grading is also widely used
as a predictor of outcome in the majority of cases of STS and
several different grading systems have been developed (2,5,6).
Irrespective of the system used, high-grade STS is associated with
a higher rate of metastasis and a lower survival rate, and
identification of the grade may influence clinical decisions made
during primary tumor treatment (7).
The 5-year survival rate (5YSR) of STS is ~65%, but the 5YSR of
high-grade, advanced-stage STS is only 10-20% (1). Although surgical resection is the
primary treatment method, conventional chemotherapy is generally
used in patients with high-grade and/or advanced-stage STS. In
high-risk patients, the tumor is frequently not completely resected
and the prognosis is poor, despite additional treatment. Patients
also frequently suffer from treatment-associated toxicity (8). These data emphasize the urgent
requirement for novel therapies (9).
Numerous types of immune-associated cell detected in
the tumor microenvironment, as well as tumor cells themselves,
exhibit genetic mutations that are essential for the
prognostication or treatment of the different subtypes of STS
(10). Studies have indicated that
immunotherapy is effective for the treatment of solid tumors.
Immune checkpoint inhibitors targeting the programmed cell death-1
(PD-1)/PD-1 ligand 1 (PD-L1) signaling axis are being explored as a
novel treatment modality in STS, and evaluation of PD1- and
PD-L1-positivity in STS may provide the criteria necessary for
determining the suitability of patients for PD-1-based
immunotherapy (11,12).
Analysis of immune-associated genes in the tumor
microenvironment in STS may thus be helpful in identifying markers
for the prognostication and management of STS. In the present
study, the expression levels of immune-associated genes in
high-grade STS were evaluated using the NanoString
nCounter® PanCancer Immune Profiling Panel, which is a
novel multiplex gene expression panel designed to quantitate 770
genes from different immune cell types and populations covering
both adaptive and innate immune responses, common checkpoint
inhibitors, tumor-specific antigens and housekeeping genes
(13), and tumor-specific immune
targets were identified. The selected genes were subsequently
confirmed/further evaluated through additional immunohistochemistry
using high-grade STS samples embedded in paraffin blocks and their
clinicopathological significance was assessed.
Materials and methods
Patients and tissue samples
High-grade STS tissue samples were obtained by
surgical resection between January 1998 and December 2013 at Pusan
National University Hospital (Busan, Korea). The included cases
were of high-grade STS, comprising pleomorphic tumor cells and
advanced tumor stages. Diagnoses were confirmed by pathological
analysis using the diagnostic criteria defined by the World Health
Organization. Each case was evaluated according to the French
Federation of Cancer Centers (FNCLCC) sarcoma group grading system
and the staging system of the American Joint Committee on Cancer
(2). A total of 35 formalin-fixed,
paraffin-embedded blocks were selected for immunohistochemistry and
12 were subjected to gene expression analysis using the NanoString
nCounter® System (Nanostring Technologies, Inc.). All
cases were FNCLCC grade 2 or 3 high-grade STS; the subtypes were as
follows: Undifferentiated pleomorphic sarcoma (n=27), high-grade
myxofibrosarcoma (n=5), leiomyosarcoma (n=2) and rhabdomyosarcoma
(n=1). Clinical information was obtained from the patients' medical
records. Chemotherapy was given following surgical resection,
consisting of mesna, doxorubicin and ifosfamide (mesna 1.2
g/m2/day, doxorubicin 25 mg/m2/day and
intravenous ifosfamide 2.0 g/m2/day on days 1-3;
repeated every 3 weeks). Overall survival (OS) was calculated from
the date of surgery to the date of death or the last follow-up
visit. For the comparative analysis, patients were divided into two
categories, a good prognosis group and a poor prognosis group,
depending on survival of the follow-up period.
Written informed consent for the use of the resected
tissues and clinical data was obtained from the patients and
approval was obtained from the Institutional Ethics Board of Pusan
National University Hospital (Busan, Korea), prior to use of any
materials. The biospecimens and data used for this study were
provided by the Biobank of Pusan National University Hospital, a
member of the Korea Biobank Network.
NanoString nCounter® PanCancer
Immune Profiling Panel for gene expression analysis.
Formalin-fixed paraffin blocks were selected from 12 high-grade STS
cases for gene expression analysis using the nCounter®
PanCancer Immune Profiling Panel (Nanostring Technologies, Inc.), a
unique 770-multiplex gene expression panel that determines the
human immune response in all cancer types. The 12 high-grade STS
cases included 7 cases belonging to the good prognosis group and 5
cases in which death occurred during the follow-up period.
Total
RNA was extracted using the RNeasy mini prep kit (Qiagen, Inc.).
RNA yield and purity were assessed using a DS 11 Spectrophotometer
(Denovix Inc.) and an RNA quality check was performed using
Fragment Analyzer™ (Advanced Analytical Technologies). Total RNA
(100 ng) was assayed using the nCounter Digital Analyzer
(Nanostring Technologies, Inc.) according to the manufacturer's
protocol. Total RNA was processed using the digital multiplexed
nCounter® human mRNA expression assay and the Human
Pancancer Immune Profiling Panel Kit (Nanostring Technologies,
Inc.). Hybridizations were performed by combining 5 µl of each RNA
sample with 8 µl of nCounter® Reporter probes in the
hybridization buffer and 2 µl of nCounter® Capture
probes (for a total reaction volume of 15 µl) overnight at 65˚C for
18 h. Excess probes were removed by two-step magnetic bead-based
purification with the nCounter® Prep Station. Specific
target molecules were then quantified on the nCounter®
Digital Analyzer by counting the individual fluorescent barcodes
and assessing the target molecules. For each assay, a high-density
scan encompassing 280 fields of view was performed. The data were
collected using the nCounter® Digital Analyzer after
images of the immobilized fluorescent reporters were acquired in
the sample cartridge with a charge-coupled device camera.
mRNA data anaylsis was performed using the freely
available nSolver™ analysis software (version 4.0.70; NanoString
Technologies, Inc.) with the mRNA profiling data normalized to the
expression of housekeeping genes. R software was used for the
analysis. Changes of >2-fold (upregulation or downregulation)
were considered significant. Genes that exhibited a fold change of
>2 and P<0.05 between the two groups were selected. A heat
map of gene expression for genes that were differentially expressed
between the good and poor prognostic groups was then plotted.
Immunohistochemical staining and
analysis
Sections were transferred to poly-L-lysine-coated
glass slides, dewaxed in xylene and rehydrated in ethanol. Staining
was performed using BondMax autostainer and reagents (Vision
Biosystems). Deparaffinization was performed automatically in the
autostainer with BondWash solution at 72˚C for 30 min. Slides were
then sequentially incubated with Epitope Retrieval Solution 1
(Leica Microsystems) for 20 min at 100˚C, peroxide-blocked for 5
min and incubated with primary antibody for 15 min, post-primary
reagent for 8 min and polymer for 8 min. Antibodies to human
leukocyte antigen (HLA)-DQA1 (rabbit monoclonal; ab128959; Abcam),
HLA-DQB1 (rabbit polyclonal; ab224600; Abcam) and HLA-DRB4 (rabbit
monoclonal; ab140612; Abcam) were used as primary antibodies. Human
tonsil tissue was used as a positive control, as it is known to
express these markers.
For all primary antibodies, the samples were
considered positive if cells exhibited cytoplasmic and/or
membranous staining. As the immunohistochemical scoring method, the
combined positive score (CPS) was determined. The CPS was estimated
by adding up the number of positively stained cells (tumor cells,
lymphocytes, macrophages), dividing the result by the total number
of viable tumor cells and multiplying by 100. Immunohistochemical
results were considered positive for expression of HLA-DQA1,
HLA-DQB1 and HLA-DRB4 if the CPS was ≥1 and considered negative
when the CPS was <1(14).
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM, Corp.). The associations between OS and the
expression of HLA-DQA1, HLA-DQB1 and HLA-DRB4 were assessed using
Pearson's χ2 test, as well as Kaplan-Meier curves with
the log-rank test. P<0.05 was considered to indicate a
statistically significant difference.
Results
NanoString nCounter®
PanCancer Immune Profiling
The 12 patients selected for gene expression
analysis ranged in age from 50 to 86 years (mean, 56 years). The
male:female ratio was 7:5. Patient follow-up was conducted from the
date of surgery until final follow-up date identified in medical
records; the date of death or the last visit to the hospital. The
follow-up period ranged from 4 to 122 months (mean, 68.3 months).
In total, 7 patients survived during the follow-up period and were
classified as the good prognosis group, while 5 died and were
classified as the poor prognosis group (Table I).
 | Table ITwelve cases applied to NanoString
nCounter® PanCancer Immune Profiling. |
Table I
Twelve cases applied to NanoString
nCounter® PanCancer Immune Profiling.
| Case no. | Prognosis group | Histological
diagnosis | Site | AJCC stage | FU data |
|---|
| 1 | Good | UPS | Upper
extremity | II | Survival |
| 2 | Good | UPS | Lower
extremity | II | Survival |
| 3 | Good | UPS | Lower
extremity | III | Survival |
| 4 | Good | UPS | Lower
extremity | II | Survival |
| 5 | Good | UPS | Lower
extremity | III | Survival |
| 6 | Good | UPS | Lower
extremity | II | Survival |
| 7 | Good | UPS | Lower
extremity | III | Survival |
| 8 | Poor | HG
myxofibrosarcoma | Upper
extremity | II | Death |
| 9 | Poor | UPS | Lower
extremity | III | Death |
| 10 | Poor | UPS | Lower
extremity | III | Death |
| 11 | Poor | HG
myxofibrosarcoma | Lower
extremity | II | Death |
| 12 | Poor | UPS | Upper
extremity | III | Death |
Nanostring nCounter® Analysis was
performed on all samples and gene expression levels were compared
between the two groups. In the panel of 770 immune-associated genes
in the nCounter® Analysis, a number of genes was
determined to be differentially expressed between the two groups
and 13 representative genes were selected based on gene expression
levels and fold changes (Fig. 1).
Of the 13 candidate genes, 7 [HLA-DRB4, neural cell adhesion
molecule (NCAM)1, CD36, CD3, FOS, Fc fragment of IgE receptor II
(FCER2) and dedicator of cytokines (DOCK)9] exhibited a significant
increase in the poor prognosis group (Table II). By contrast, the expression of
6 immune-associated genes [HLA-DQA1, HLA-DQB1, lymphocyte
activation gene 3 (LAG3), forkhead box P3 (FOXP3), baculoviral IAP
repeat containing 5 (BIRC5) and dual specificity phosphatase
(DUSP)4] was increased in the good prognosis group (Table III). The associated molecules are
classified according to the primary function of each gene: Antigen
processing and presentation (HLA-DRB4, HLA-DQA1 and HLA-DQB1),
adhesion molecule (NCAM1), transporter function (CD36), regulation
of immune response (C3, FCER2 and LAG3), cytokine (FOXP3), cell
cycle (BIRC5) and others (FOS, DOCK9 and DUSP4).
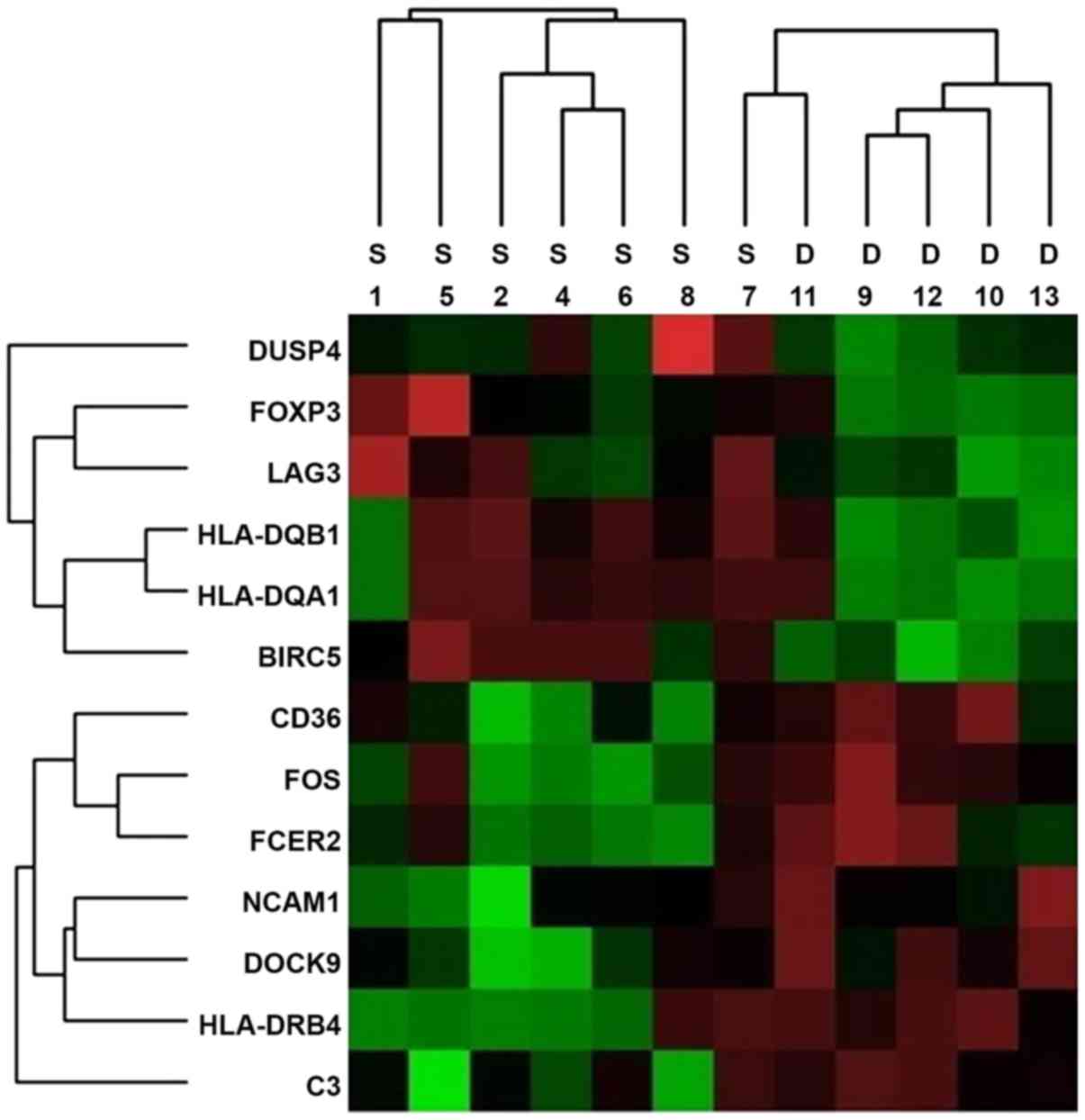 | Figure 1Heat map generated from mRNA
expression data reflecting 13 representative genes that were
differentially expressed between the S and D prognostic groups. Red
color indicates highly expressed genes. Groups: S, good prognosis;
D, poor prognosis. HLA-DRB4, human leukocyte antigen; NCAM1, neural
cell adhesion molecule; CD36; CD3, FOS, FCER2, Fc fragment of IgE
receptor II; DOCK9, dedicator of cytokines 9; HLA-DQA1, HLA-DQB1,
LAG3, lymphocyte antivation gene 3; FOXP3, forkhead box P3; BIRC5,
baculoviral IAP repeat containing 5; DUSP4, dual specificity
phosphatase 4. |
 | Table IISelected genes with increased
expression in the poor prognostic group according to the NanoString
nCounter® analysis. |
Table II
Selected genes with increased
expression in the poor prognostic group according to the NanoString
nCounter® analysis.
| Gene name | mRNA/probe ID | Fold change | Gene
annotation | P-value |
|---|
| HLA-DRB4 |
NM_021983.4:194 | 56.93 | Antigen processing
and presentation | 0.011 |
| NCAM1 |
NM_000615.5:1620 | 6.12 | Adhesion
molecules | 0.046 |
| CD36 |
NM_001001548.2:705 | 5.24 | Transporter
function | 0.016 |
| C3 |
NM_000064.2:4396 | 3.61 | Immune response
regulation | 0.046 |
| FOS |
NM_005252.2:1475 | 3.51 | Transcription
factor | 0.010 |
| FCER2 |
NM_002002.4:420 | 3.06 | Immune response
regulation | 0.043 |
| DOCK9 |
NM_001130048.1:1020 | 2.17 | Immune cell
function | 0.016 |
 | Table IIISix genes showing significant
expression in the good prognostic group by the NanoString
nCounter® analysis. |
Table III
Six genes showing significant
expression in the good prognostic group by the NanoString
nCounter® analysis.
| Gene name | mRNA/probe ID | Fold change | Gene
annotation | P-value |
|---|
| HLA-DQA1 |
NM_002122.3:261 | 77.78 | Antigen processing
and presentation | 0.026 |
| HLA-DQB1 |
NM_002123.3:384 | 28.94 | Antigen processing
and presentation | 0.022 |
| LAG3 |
NM_002286.5:1735 | 2.91 | Immune response
regulation T-cell function | 0.015 |
| FOXP3 |
NM_014009.3:1230 | 2.41 | Transcriptional
regulation T-cell function | 0.020 |
| BIRC5 |
NM_001168.2:1215 | 2.38 | G2 phase and G2/M
transition | 0.001 |
| DUSP4 |
NM_057158.2:3115 | 2.18 | Innate immune
response | 0.043 |
Immunohistochemical analysis
The subjects for immunohistochemical confirmation
included 35 patients with high-grade STS, including 19 males and 16
females with an average age of 66.2 years (range, 39-86 years). The
tumors were located in a lower limb in 25 patients, an upper limb
in 8 patients and the trunk in 2 patients. A total of 26 patients
had tumor sizes of ≤10 cm, while the other patients had tumor sizes
of >10 cm. The average follow-up period was 69.1 months (range,
4-150 months). As the clinical information was obtained from the
patients' medical records, the OS was calculated from the date of
surgery to the date of mortality or last follow-up visit. In total,
15 patients survived the follow-up period and were classified as
the good prognosis group, while 20 patients who died of the disease
were classified as the poor prognosis group. The
clinicopathological data of this cohort are summarized in Table IV.
 | Table IVClinicopathological features of 35
patients whose high-grade STS tumors were subjected to
immunohistochemical analysis. |
Table IV
Clinicopathological features of 35
patients whose high-grade STS tumors were subjected to
immunohistochemical analysis.
| Item | Total (n=35) | Good prognosis
group (n=15) | Poor prognosis
group (n=20) | P-value |
|---|
| Age (years, mean ±
SD) | 66.2±12.5 | 62.3±9.7 | 67.9±13.9 | 0.275 |
| Sex | | | | 0.182 |
|
Male | 16 | 6 | 13 | |
|
Female | 19 | 9 | 7 | |
| Tumor location | | | | 0.296 |
|
Upper
extremity | 8 | 0 | 8 | |
|
Lower
extremity | 25 | 14 | 11 | |
|
Trunk | 2 | 1 | 1 | |
| Tumor size
(cm) | | | | 0.244 |
|
≤10 | 26 | 13 | 13 | |
|
>10 | 9 | 2 | 7 | |
| AJCC stage | | | | 0.489 |
|
Ⅱ | 23 | 11 | 12 | |
|
Ⅲ | 11 | 4 | 7 | |
|
IV | 1 | 0 | 1 | |
| Histological
diagnosis | | | | 0.696 |
|
UPS | 27 | 11 | 16 | |
| High-grade
myxofibrosarcoma | 5 | 3 | 2 | |
|
Leiomyosarcoma | 2 | 1 | 1 | |
|
Rhabdomyosarcoma | 1 | 0 | 1 | |
Based on data obtained from the Nanostring
nCounter® analysis, HLA-DQA1, HLA-DQB1 and HLA-DRB4
exhibited the greatest differences in fold change and were selected
for further immunohistochemical study using paraffin-embedded
blocks of samples from the 35 patients. The immunohistochemical
data were considered positive if the cells were either cytoplasmic
or membranous stained; the results were analyzed by determining the
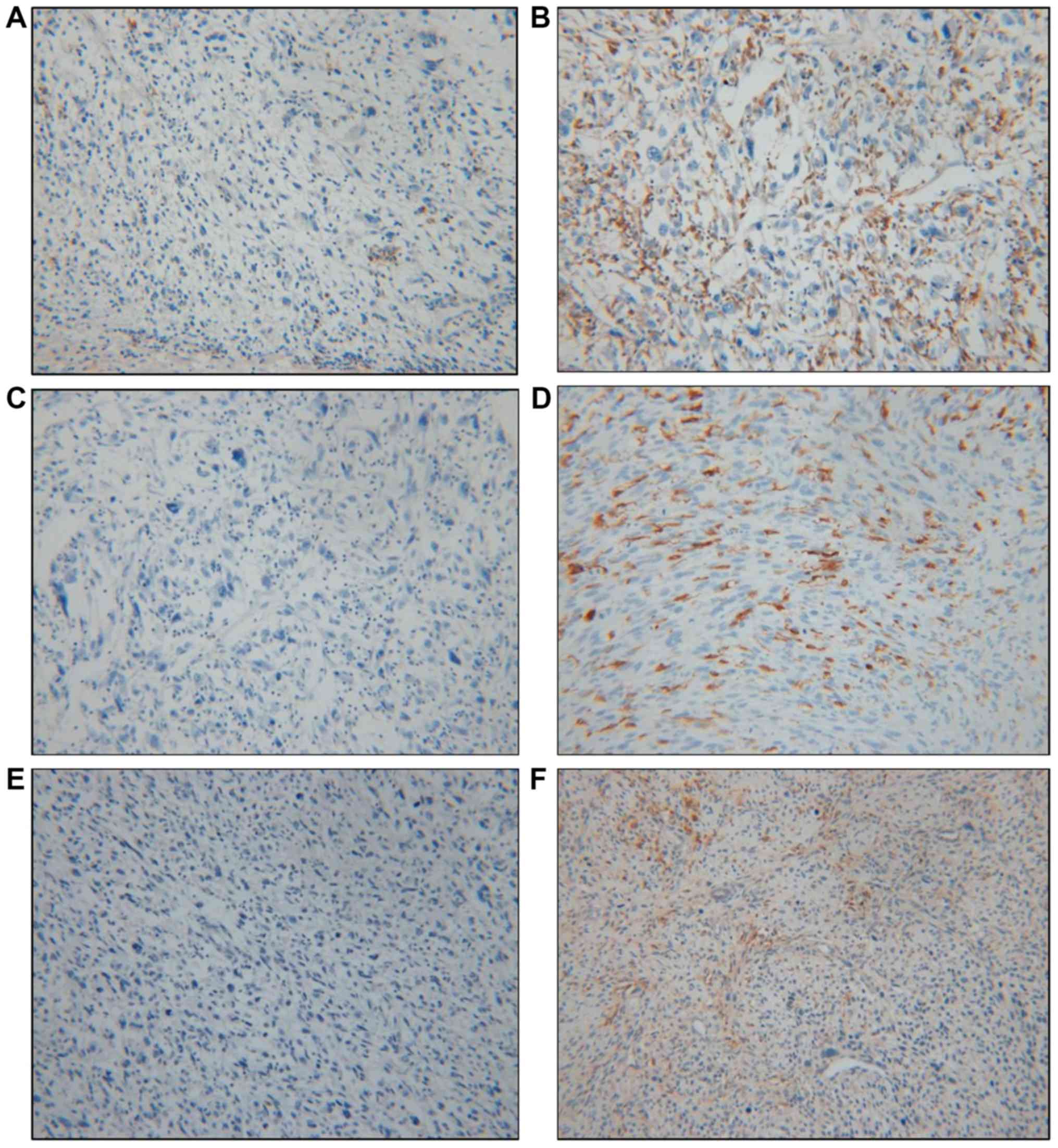
CPS (12) (Fig. 2). Positive expression of HLA-DQA1,
HLA-DQB1 and HLA-DRB4 was observed in 74.3% (26/35), 34.3% (12/35)
and 48.6% (17/35) of tumors, respectively (Table V). The immunohistochemical results
of the 12 cases subjected to the Nanostring nCounter®
Analysis were consistent with the results of the gene expression
analysis. For HLA-DQA1, 26 (74.3%) patients exhibited positive
expression in their tumor tissues, comprising 14 cases (14/26,
53.9%) in the good prognosis group and 12 cases (12/26, 46.1%) in
the poor prognosis group. In the group with negativity for
HLA-DQA1, only 1 (1/9, 11.1%) patient survived, while 8 (8/9,
88.9%) died of their disease. There was a statistically significant
difference between the groups stratified by HLA-DQA1 expression and
prognosis, with an increased survival rate in the positive
expression group (P=0.028). HLA-DQB1 exhibited positive expression
in 12 (34.3%) patients, 4 (33.3%) in the good prognosis group and 8
(66.7%) in the poor prognosis group, but there was no significant
difference between the groups stratified by HLA-DQB1 expression and
prognosis (P=0.489). HLA-DRB4 positivity was observed in 17 (48.6%)
patients, with no statistically significant difference between the
6 (35.3%) in the good prognosis group and 11 (64.7%) in the poor
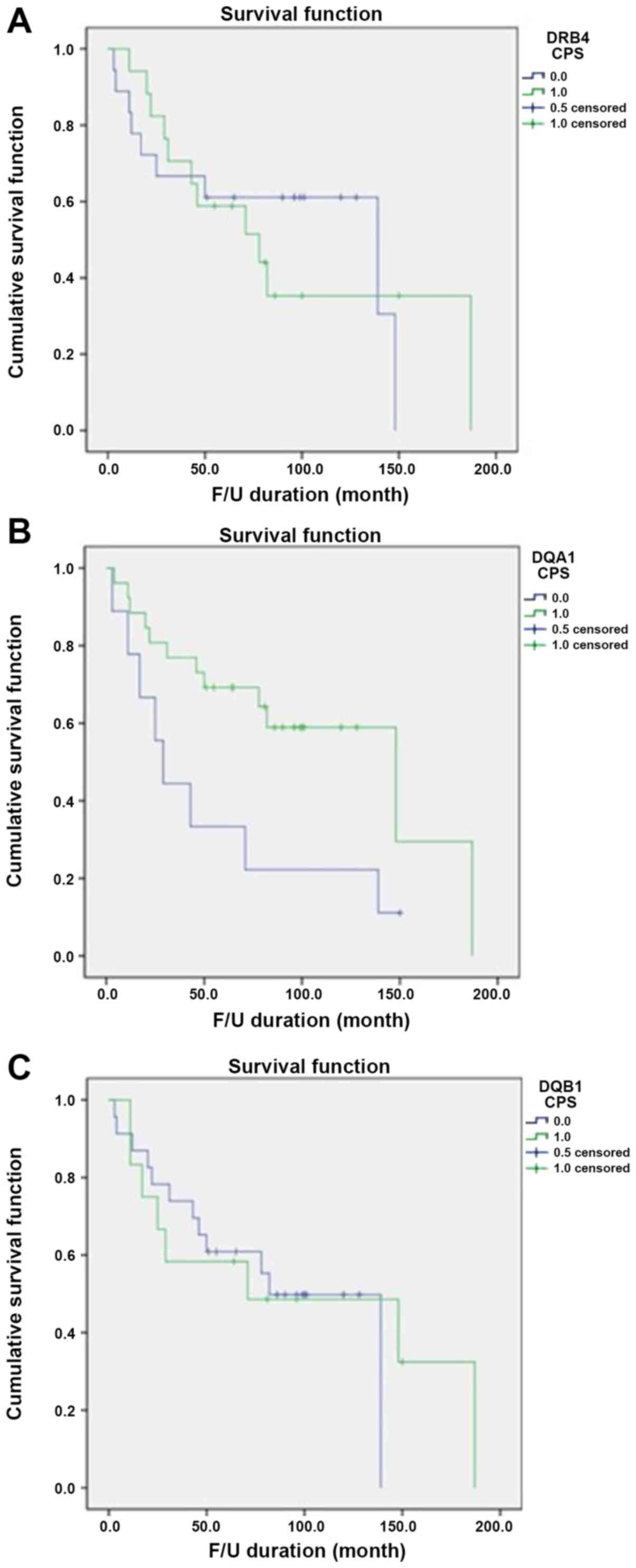
prognosis group (P=0.500). Only positive expression of HLA-DQA1 was
significantly associated with favorable OS (Fig. 3).
 | Table VSurvival analysis of
immunohistochemistry results for HLA-DQA1, HLA-DQB1 and
HLA-DRB4. |
Table V
Survival analysis of
immunohistochemistry results for HLA-DQA1, HLA-DQB1 and
HLA-DRB4.
| Gene/expression
status | Good prognosis
group (n=15) | Poor prognosis
group (n=20) | Total (n=35) | P-value |
|---|
| HLA-DQA1 | | | | 0.028 |
|
CPS (-) | 1 (11.1) | 8 (88.9) | 9 | |
|
CPS (+) | 14 (53.9) | 12 (46.1) | 26 | |
| HLA-DQB1 | | | | 0.489 |
|
CPS (-) | 11 (47.8) | 12 (52.2) | 23 | |
|
CPS (+) | 4 (33.3) | 8 (66.7) | 12 | |
| HLA-DRB4 | | | | 0.500 |
|
CPS (-) | 9(50) | 9(50) | 18 | |
|
CPS (+) | 6 (35.3) | 11 (64.7) | 17 | |
Discussion
High-grade STS has a mortality rate ranging from 40
to 60%. The predictors of survival time in patients with high-grade
STS include tumor size, histology, grading, margin status at
resection and the presence of pre-surgical metastasis (5). It has been previously indicated that
old age affects the prognosis of STS (15), but in the cohort of the present
study, with an average age of 66.2 years (range, 39-86 years), the
age difference between the two study groups was not statistically
significant. The standard treatment of high-grade STS includes
surgical resection, with radiation therapy as a supplementary
treatment and chemotherapy added if required. Based on the genetic
mutations detected in each case, targeted therapy may also be
performed (16,17). A previous study reported that
co-administration of the monoclonal antibody Olaratumab, which acts
against platelet-derived growth factor receptor α, and the
traditional chemotherapy agent doxorubicin, increase the overall
long-term survival rate (18). A
better understanding of prognostic factors is required to determine
the prognosis and select the most appropriate treatment regimen for
each patient.
A number of studies have reported changes in the
immune response and the activity of immune checkpoint inhibitors in
STS (9-12).
Recent years have seen the emergence of PD-1 and PD-L1 as novel
targets in cancer immunotherapy. PD-1 and PD-L1 expression has been
detected in tumor tissue and the microenvironment in certain types
of STS. The association between their expression levels and
clinical outcomes demonstrates the importance of immune checkpoint
inhibitors in patients with high-grade STS (11).
In the present study, the expression of diverse
immune-associated genes in high-grade STS according to prognosis
was investigated and the association between immune-associated
genes and survival was further examined using 35 samples of
formalin-fixed, paraffin-embedded tissue. The small number of
samples may be a limitation of the present study. However, it was
attempted to obtain high-grade STS samples with pleomorphic tumor
cells and advanced stages, as previous studies using STS samples
comprised numerous different histological types of tumor and it
affected their results (5,6). This was achieved on the fully
automated and highly precise Nanostring nCounter® system
designed for gene expression analysis, which accurately detects and
quantifies 770 transcripts from 24 different types of immune cell.
The system measures transcripts related to the adaptive and innate
immune response, common checkpoint inhibitors, tumor-specific
antigens and housekeeping genes; it does so using a small amount of
mRNA and is based on digital color-coded barcode technology
(13). The Nanostring
nCounter® Sarcoma Fusion CodeSet was recently introduced
in the field of STS research to assess fusion transcripts in
formalin-fixed, paraffin-embedded material (19). The custom-designed NanoString
nCounter®-based sarcoma assay is highly sensitive and
specific in a clinical setting for molecular diagnosis of
sarcoma.
The present results revealed significant genetic
variations between the survival group (good prognosis group) and
the poor prognosis group. The criteria applied to identify
significantly differentially expressed genes were P<0.05 and a
fold change of >2 between the two groups. A total of 13 genes
were selected. Of these, 7 immune-associated genes (C3, CD36,
DOCK9, FCER2, FOS, HLA-DRB4 and NCAM1) were significantly increased
in the poor prognosis group, while the expression of 6
immune-associated genes (BIRC5, DUSP4, FOXP3, HLA-DQA1, HLA-DQB1
and LAG3) was increased in the good prognosis group.
NCAM1 encodes a cell adhesion protein that is a
member of the immunoglobulin superfamily. The encoded protein was
reported to be involved in the expansion of T cells and dendritic
cells, which have an important role in immune surveillance. Certain
studies have indicated that NCAM1 is associated with therapeutic
resistance in cancer (20,21). CD36 has a role in immune signaling
and is also a scavenger receptor for fatty acid uptake that
modulates cell-to-extracellular matrix attachment and TGF-β
activation. CD36 has increasingly emerged as a prognostic marker
associated with the metastatic process (22). Complement C3 is useful in the
development of novel strategies to improve the effectiveness of
cancer immunotherapy (23). FCER2
is a B-cell specific antigen with essential roles in B-cell growth
and differentiation, as well as the regulation of IgE production,
although it remains unknown how this gene affects the development
or treatment of cancer (24). DOCK
is a family of guanine-nucleotide exchange factors for Rho GTPases
and FOS is a subunit of the transcription factor activator
protein-1. Their functions in the immune system and cancer remain
to be fully elucidated.
BIRC5, also called survivin, is a well-known cancer
therapeutic target. BIRC5 has multiple mechanisms of action in
immune responses, as well as utilities in molecular cancer
diagnostics and therapeutics (25).
Survivin peptide immunogen-reactive antibodies exert an additional
advantage for survivin immunotherapy. Survivin-specific T-cell
reactivity strongly correlates with tumor response and patient
survival (26). The present study
revealed reduced levels of survivin in the poor prognosis group;
hence, the possibility of survivin as a therapeutic target should
be considered in cases of high-grade STS. FOXP3 is a member of the
forkhead/winged-helix family of transcriptional regulators and is
associated with T-cell function. Regulatory T cells expressing the
transcription factor FOXP3 have a critical role in the maintenance
of immune homeostasis and prevention of autoimmunity and cancer
pathogenesis (27). Due to their
ability to suppress self-antigen responses, regulatory T cells with
FOXP3 may have anti-tumor immune function, whereas increased FOXP3
expression in tumor tissue was reported to be associated with
favorable prognosis in certain cancer types. This discrepancy was
identified particularly in colorectal cancer (28,29).
In the present study, FOXP3 expression was increased in the good
prognosis group and decreased in the poor prognosis group of
high-grade STS cases. These results emphasize the requirement for
accurate assessment of FOXP3 expression in tumor immunity. DUSP4
negatively regulates members of the MAPK superfamily, which are
associated with cellular proliferation and differentiation. DUSP4
promotes doxorubicin resistance in gastric cancer through its
effects on the epithelial-mesenchymal transition (30).
The proteins encoded by HLA-DRB4, HLA-DQA1 and
HLA-DQB1 are associated with antigen processing and presentation.
HLA, the human leukocyte antigen, is a group of proteins that are
encoded by the major histocompatibility complex (MHC) in humans and
is a cell membrane glycoprotein that is expressed on the surface of
human nucleated cells. It induces an adaptive immune response
against invading antigens by presenting an antigen to T lymphocytes
and protects normal cells from the apoptotic function of natural
killer cells. The HLA class II gene locus includes 9 types of HLA
(HLA-DRA, -DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1 and
-DPB1), and HLA class II DR, DQ and DP molecules are expressed by
antigen-presenting cells. HLA genes have been investigated over a
period of several decades, and accordingly, relatively much is
known about them. HLA-DQA1, HLA-DQB1 and DRB4 are associated with
numerous inflammatory and autoimmune diseases, such as celiac
disease, Addison disease, idiopathic inflammatory myopathy and
Hashimoto's thyroiditis. Previous studies on the associations
between HLA class II molecules and human cancers have indicated
diverse results. Mahmoodi et al (31) reported that the HLA-DQA1*0301 allele
is mainly associated with an increased risk of breast cancer
development, whereas HLA-DQB1*0602 appears to protect against
early-onset breast cancer. Shen et al (32) recently demonstrated that HLA-DQA1
has an important role in esophageal squamous cell carcinoma (ESCC)
progression and may be a biomarker for ESCC diagnosis and
prognosis, as well as a potential target for treatment of patients
with ESCC.
In the present study, to validate the association
between alterations in immune-related genes and patient prognosis,
cases of high-grade STS were selected and immunohistochemical
analysis was performed for HLA-DQA1, HLA-DQB1 and HLA-DRB4, which
indicated the maximum fold-change differences between the two
groups. The Nanostring nCounter® system analysis
revealed that the expression of HLA-DRB4 was markedly increased in
the poor prognosis group, whereas the expression of HLA-DQA1 and
HLA-DQB1 was increased in the good prognosis group.
Immunohistochemistry further demonstrated positive expression of
HLA-DQA1, HLA-DQB1 and HLA-DRB4 in 74.3% (26/35), 34.3% (12/35) and
48.6% (17/35) of cases, respectively. However, only the expression
of HLA-DQA1 was significantly associated with survival (P=0.028),
suggesting the possibility of its application as a good prognostic
factor for high-grade STS. This may be due to increased HLA-DQA1
expression being able to induce a good immune response in tumors or
increase the sensitivity to immune checkpoint blockade. The
mechanism remains elusive and requires further research.
The results provided by the present study may be
meaningful in the research area of immune-biomarker expression in
high-grade STS. Although formalin-fixed, paraffin-embedded
materials were used, good results were obtained from the Nanostring
nCounter® system. Certain immune-related genes were
identified and immunohistochemistry was performed to validate the
prognostic significance of HLA-DQA1, HLA-DQB1 and HLA-DRB4. The
present results indicated that the expression of HLA-DQA1 was
significantly associated with long-term survival, suggesting its
potential as an immune biomarker of good prognosis in high-grade
STS.
Acknowledgements
Not applicable.
Funding
This study was supported by a Biomedical Research
Institute Grant, Pusan National University Hospital (grant no.
2017-31).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JYB and KUC contributed to the conception and design
of the study. AK and SJL collected the patients' clinical data. KK,
JYK and ISL contributed significantly to the analysis and
interpretation of data. SHC helped perform the analysis and was
involved in constructive discussions. JIK supervised the study and
wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Approval for this study was obtained from the
Institutional Ethics of Pusan National University Hospital (Busan,
Korea). Informed consent was obtained from all the patients whose
tissues were used in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Weiss SW, Goldblum JR and Folpe AL:
Enzinger and Weiss's Soft Tissue Tumors. 6th edition. Elsevier,
Philadelphia, pp1176, 2014.
|
|
2
|
WHO classification of tumors: Soft Tissue
and Bone Tumors. 5th edition. IARC, 2020.
|
|
3
|
Meis-Kindblom JM, Bjerkehage B, Böhling T,
Domanski H, Halvorsen TB, Larsson O, Lilleng P, Myhre-Jensen O,
Stenwig E, Virolainen M, et al: Morphologic review of 1000 soft
tissue sarcomas from the Scandinavian Sarcoma Group (SSG) Register:
The peer-review committee experience. Acta Orthop Scand Suppl.
285:18–26. 1999.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Italiano A, Di Mauro I, Rapp J, Pierron G,
Auger N, Alberti L, Chibon F, Escande F, Voegeli AC, Ghnassia JP,
et al: Clinical effect of molecular methods in sarcoma diagnosis
(GENSARC): A prospective, multicentre, observational study. Lancet
Oncol. 17:532–538. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ottaiano A, De Chiara A, Fazioli F,
Talamanca AA, Mori S, Botti G, Milano A and Apice G: Biological
prognostic factors in adult soft tissue sarcoma. Anticancer Res.
25:4519–4526. 2005.PubMed/NCBI
|
|
6
|
Italiano A, Le Cesne A, Mendiboure J, Blay
JY, Piperno-Neumann S, Chevreau C, Delcambre C, Penel N, Terrier P,
Ranchere-Vince D, et al: Prognostic factors and impact of adjuvant
treatments on local and metastatic relapse of soft-tissue sarcoma
patients in the competing risks setting. Cancer. 120:3361–3369.
2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Coindre JM: Grading of soft tissue
sarcomas: Review and update. Arch Pathol Lab Med. 130:1448–1453.
2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gronchi A, Frustaci S, Mercuri M, Martin
J, Lopez-Pousa A, Verderio P, Mariani L, Valagussa P, Miceli R,
Stacchiotti S, et al: Short, full-dose adjuvant chemotherapy in
high-risk adult soft tissue sarcomas: A randomized clinical trial
from the Italian Sarcoma Group and the Spanish Sarcoma Group. J
Clin Oncol. 30:850–856. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
van der Graaf WT, Blay JY, Chawla SP, Kim
DW, Bui-Nguyen B, Casali PG, Schöffski P, Aglietta M, Staddon AP,
Beppu Y, et al: Pazopanib for metastatic soft-tissue sarcoma
(PALETTE): A randomised, double-blind, placebo-controlled phase 3
trial. Lancet. 379:1879–1886. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Raj S, Miller LD and Triozzi PL:
Addressing the adult soft tissue sarcoma microenvironment with
intratumoral immunotherapy. Sarcoma. 12(9305294)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Budczies J, Mechtersheimer G, Denkert C,
Klauschen F, Mughal SS, Chudasama P, Bockmayr M, Johrens K, Endris
V, Lier A, et al: PD-L1 (CD274) copy number gain, expression, and
immune cell infiltration as candidate predictors for response to
immune checkpoint inhibitors in soft-tissue sarcoma.
Oncoimmunology. 6(e1279777)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sorbye SW, Kilvaer T, Valkov A, Donnem T,
Smeland E, AlShibli K, Bremnes RM and Busund LT: Prognostic impact
of lymphocytes in soft tissue sarcomas. PLoS One.
6(e14611)2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cesano A: nCounter(®) pancancer
immune profiling panel (Nanostring technologies, Inc., Seattle WA).
J Immunother Cancer. 3(42)2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kulangara K, Zhang N, Corigliano E,
Guerrero L, Waldroup S, Jaiswal D, Ms MJ, Shah S, Hanks D, Wang J,
et al: Clinical utility of the combined positive score for
programmed death ligand-1 expression and the approval of
Pembrolizumab for treatment of gastric cancer. Arch Pathol Lab Med.
143:330–337. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hashimoto K, Nishimura S, Hara Y, Oka N,
Tanaka H, Iemura S and Akagi M: Clinical outcomes of patients with
primary malignant bone and soft tissue tumor aged 65 years or
older. Exp Ther Med. 17:888–894. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wardelmann E, Schildhaus HU,
Merkelbach-Bruse S, Hartmann W, Reichardt P, Hohenberger P and
Büttner R: Soft tissue sarcoma: From molecular diagnosis to
selection of treatment. Pathological diagnosis of soft tissue
sarcoma amid molecular biology and targeted therapies. Ann Oncol.
21 (Suppl 7):vii265–vii269. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Reichardt P: Soft tissue sarcomas, a look
into the future: Different treatments for different subtypes.
Future Oncol. 10 (Suppl 8):S19–S27. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tap WD, Jones RL, Van Tine BA, Chmielowski
B, Elias AD, Adkins D, Agulnik M, Cooney MM, Livingston MB, Pennock
G, et al: Olaratumab and doxorubicin versus doxorubicin alone for
treatment of soft-tissue sarcoma: An open-label phase 1b and
randomised phase 2 trial. Lancet. 388:488–497. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chang KTE, Goytain A, Tucker T, Karsan A,
Lee CH, Nielsen TO and Ng TL: Development and evaluation of a
pan-sarcoma fusion gene detection assay using the NanoString
nCounter platform. J Mol Diagn. 20:63–77. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sykes SM: NCAM1 supports therapy
resistance and LSC function in AML. Blood. 133:2247–2248.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jiang C, Zhao W, Qin M, Jin M, Chang L and
Ma X: CD56-chimeric antigen receptor T-cell therapy for
refractory/recurrent rhabdomyosarcoma: A 3.5-year follow-up case
report. Medicine (Baltimore). 98(e17572)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Enciu AM, Radu E, Popescu ID, Hinescu ME
and Ceafalan LC: Targeting CD36 as biomarker for metastasis
prognostic: How far from translation into clinical practice? Biomed
Res Int. 4(7801202)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Peng W, McKenzie JA and Hwu P:
Complementing T-cell function: An inhibitory role of the complement
system in T-cell-mediated antitumor immunity. Cancer Discov.
6:953–955. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Laitinen T, Ollikainen V, Lázaro C, Kauppi
P, de Cid R, Antó JM, Estivill X, Lokki H, Mannila H, Laitinen LA
and Kere J: Association study of the chromosomal region containing
the FCER2 gene suggests it has a regulatory role in atopic
disorders. Am J Respir Crit Care Med. 161:700–706. 2000.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li F, Aljahdali I and Ling X: Cancer
therapeutics using survivin BIRC5 as a target: What can we do after
over two decades of study? J Exp Clin Cancer Res.
38(368)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Becker JC, Andersen MH, Hofmeister-Müller
V, Wobser M, Frey L, Sandig C, Walter S, Singh-Jasuja H, Kämpgen E,
Opitz A, et al: Survivin-specific T-cell reactivity correlates with
tumor response and patient survival: A phase-II peptide vaccination
trial in metastatic melanoma. Cancer Immunol Immunother.
61:2091–2103. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wing JB, Tanaka A and Sakaguchi S: Human
FOXP3+ regulatory T cell heterogeneity and function in
autoimmunity and cancer. Immunity. 50:302–316. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
deLeeuw RJ, Kost SE, Kakal JA and Nelson
BH: The prognostic value of FoxP3+ tumor-infiltrating
lymphocytes in cancer: A critical review of the literature. Clin
Cancer Res. 18:3022–3029. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Salama P, Phillips M, Grieu F, Morris M,
Zeps N, Joseph D, Platell C and Iacopetta B: Tumor-infiltrating
FOXP3+ T regulatory cells show strong prognostic
significance in colorectal cancer. J Clin Oncol. 27:186–192.
2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kang X, Li M, Zhu H, Lu X, Miao J, Du S,
Xia X and Guan W: DUSP4 promotes doxorubicin resistance in gastric
cancer through epithelial-mesenchymal trantision. Oncotarget.
8:94028–94039. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mahmoodi M, Nahvi H, Mahmoudi M, Kasaian
A, Mohagheghi MA, Divsalar K, Nahavandian B, Jafari A, Ansarpour B,
Moradi B, et al: HLA-DRB1,-DQA1 and -DQB1 allele and haplotype
frequencies in female patients with early onset breast cancer.
Pathol Oncol Res. 18:49–55. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shen FF, Pan Y, Li JZ, Zhao F, Yang HJ, Li
JK, Gao ZW, Su JF, Duan LJ, Lun SM, et al: High expression of
HLA-DQA1 predicts poor outcome in patients with esophageal squamous
cell carcinoma in Northern China. Medicine (Baltimore).
98(e14454)2019.PubMed/NCBI View Article : Google Scholar
|