Introduction
The intervertebral disc (IVD) is composed of nucleus
pulposus (NP), anulus fibrosus and cartilage endplate (1). Back pain is mostly attributable to the
degeneration of IVD, which may cause nerve damage, poor labor
capacity and low quality of life in some severe cases (1). Despite intervertebral disc
degeneration (IDD) development being a natural process that comes
with aging, some evidence suggests that the development of IDD
stems from both genetic and environmental factors (2). For example, certain factors, including
the decrease of NP cells, imbalance between the synthesis and
degradation of the extracellular matrix (ECM) and overexpression of
inflammatory cytokines, are known to be responsible for
accelerating the development of IDD (3).
NP cell-produced collagen type II alpha 1 chain
(COL2A1), aggrecan (ACAN) and some ECM components are believed to
be crucial to maintain the integrity of IVD (4,5). It
has been reported that some molecules, such as insulin-like growth
factor-1, epidermal growth factor and transforming growth factor-β
(TGF-β), can promote anabolic metabolism of ECM (6,7). Tumor
necrosis factor (TNF)-α, interleukins (IL), matrix
metalloproteinases (MMPs) and a disintegrin and metalloproteinase
with thrombospondin type I motifs (ADAMTS) work synergistically and
facilitate the catabolism of the ECM (8-10).
Catabolic activities of MMPs and ADAMTSs were leveraged by
inhibitory actions and through tissue inhibitors of
metalloproteinases (TIMPs) (10).
In NP cells, TGF-β can delay the process of IDD via promoting the
synthesis of ACAN and COL2A1, which enhances cell proliferation and
inhibits MMPs expression (6,11). IDD
NP cells also produced diverse inflammatory mediators which caused
the aggravation of IDD, such as TNF-α, IL-1 α/β, IL-6 and
IL-17(12).
As a major component of activator protein-1 (AP-1),
c-Jun expression is increased when cells enter the logarithmic
phase of growth in the presence of growth-factor-induced
stimulation (13). Wisdom et
al (14) demonstrated that
c-Jun stimulates fibroblast growth and inhibits its apoptosis.
Furthermore, c-Jun was found to protect cells from TNF-α-induced
apoptosis, which is required for cell proliferation (14). Conversely, the presence and
phosphorylation-based activation of c-Jun are necessary for the
execution of apoptosis in both neuronal cells and thymocytes
(15). In the intestinal
ischemia-reperfusion injured autograft model, activation of both
c-Fos and c-Jun genes can trigger cell proliferation and apoptosis
(16).
Behrens et al (17) reported that a lack of c-Jun in mice
can lead to the impairment of hepatocyte proliferation and liver
regeneration. Notochordal cells in the absence of c-Jun have also
been demonstrated to experience an increase in apoptosis, leading
to impairment of IVD formation (18). Subsequently, it was speculated that
c-Jun expression may have an essential role in IDD process.
However, the function and underlying mechanisms of c-Jun in IDD
remain unknown. Because the present study aimed to explore the role
of c-Jun in NP cells, NP cells were transduced with a
c-Jun-overexpressing lentivirus, and changes in IVD-related genes
on a molecular level were detected. This study attempted to further
elucidate the pathogenesis of IDD and provide a novel addition to
valuable clinical information for the treatment of IDD.
Materials and methods
IVD tissue collection
IVD tissues were collected as surgical waste from 10
patients with IDD (age, 35-58 years). In addition, IVD tissues from
10 patients with lumbar fractures (age, 26-52 years), excluding
those with spinal tumors, infections and rheumatic immune diseases,
were collected as controls. This study was approved by the
Institutional Review Board of Tongji Medical College and followed
the Declaration of Helsinki. Written informed consent was obtained
from each patient. According to the MRI scanning techniques
reported by Pfirrmann et al (19), the obtained IVD tissues were graded
by T2-weighted images to determine degrees of degeneration.
Relative normal nondegenerated discs from patients with lumbar
fractures were graded I-II (Control), whereas degenerative discs
from patients with IDD were graded III-V. Subsequently, NP cells
were isolated from IVD tissues of patients with IDD and control
subjects.
Isolation and culture NP cells
Primary NP cells were isolated and cultured as
previously reported (5). The IDD
and control NP tissue samples were washed three times with D-Hanks
solution under aseptic conditions. These specimens were cut into
1-mm3 pieces, and digested with 0.25% trypsin (Beyotime
Institute of Biotechnology) and 0.2% collagenase II (Beyotime
Institute of Biotechnology) for 3 h at 37˚C. NP cells were filtered
through a 200-mesh sieve, washed three times with PBS and the
supernatant was discarded following centrifugation at 2,000 x g for
5 min (37˚C). Cells were washed with DMEM-F12 (Gibco; Thermo Fisher
Scientific, Inc.) medium containing 10% FBS to terminate digestion.
After centrifugation at 2,000 x g for 5 min (37˚C), NP cells were
counted and seeded into 25 cm2 culture dishes. DMEM-F12
medium was supplemented with 15% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 10 µg/ml insulin and 1% penicillin/streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to culture NP
cells under conventional incubation conditions (37˚C, 5%
CO2 and 95% humidity), and the medium was refreshed
twice a week. When NP confluency reached 80%, cells were passaged
at a ratio of 1:2. Cells in passage P2 were used for subsequent
experiments.
Lentiviral vector construction and
lentivirus infection of NP cells
The c-Jun gene was inserted into green fluorescence
protein (GFP)-labeled LV5 plasmids obtained from Boaimaidisen
Biotechnology Co., Ltd. and the lentivirus was packaged by four
plasmid systems namely LV5-c-Jun, PG-p1-VSVG, PG-P2-REV and
PG-P3-RRE (Boaimaidisen Biotechnology Co., Ltd.). Transfections
were performed into 293T packaging cell lines (American Type
Culture Collection). Lentiviral packaging enrichment was completed
by Chongqing Biomedicine Biotechnology Co., Ltd. The GFP-labeled
blank LV5-empty vector was used as the negative control. The viral
titer of LV5-c-Jun and LV5-empty vector lentivirus was
1x108 TU/ml.
Before lentiviral transfection, a preliminary test
was carried out to estimate the efficiency of lentiviral infection
in target cells. The multiplicity of infection (MOI) refers to the
proportion of infectious viruses per cells and is calculated as
follows: MOI=(virus titer x virus volume)/number of cells. NP cells
were seeded into 24-well culture plate at the density of
3x104/well. 293T cells served as the control cells in a
parallel experiment to determine the affinity of lentivirus to
target cells. Transfection was conducted when the confluency
reached 70%. The infectious lentiviruses were incubated with the NP
cells and control cells at a final MOI of 0, 10, 20, 40 and 100.
The mixed lentivirus and cells were incubated in a incubator (37˚C,
5% CO2) overnight. After 24 h, culture medium containing
lentivirus was replaced with normal culture medium. At 4 days
following infection, the transduction efficiency was tested via
flow cytometry according to GFP-positive cells (BD Diagnostics).
GFP expression was observed in cellSens 1.12 software (Olympus
Corporation) at a magnification of x100 using an inverted
fluorescence microscope (CKX53, Olympus Corporation).
Western blotting
NP cells were lysed in RIPA reagent (Beyotime
Institute of Biotechnology), and the concentration of extracted
proteins was measured by BCA standard method (Takara Biotechnology
Co., Ltd.). Proteins (30 µg) were separated by 10% SDS-PAGE and
transferred onto PVDF membranes (EMD Millipore). PVDF membranes
were washed with TBST (0.05% Tween-20) three times before being
blocked with 5% skimmed milk at room temperature for 2 h. Primary
antibodies (1:500) were incubated overnight at 4˚C. Subsequently,
the HRP-conjugated goat anti-rabbit antibody was incubated for 1.5
h at room temperature (1:1,500, ab205718; Abcam). The proteins were
visualized by chemiluminescence (Thermo Fisher Scientific, Inc.,).
Densitometry was semi-quantified with ImageJ 1.8.0 software
(National Institutes of Health). Results were normalized by GAPDH.
The following primary antibodies were used: Anti-c-Jun (cat. no.
ab40766), anti-TGF-β (cat. no. ab31013), anti-TIMP-1 (cat. no.
ab211926), anti-TIMP-3 (cat. no. ab39184), anti-ADAMTS-4 (cat. no.
ab185722), anti-ADAMTS-5 (cat. no. ab41037), anti-ACAN (cat. no.
ab3778), anti-COL1A1 (cat. no. ab34710), anti-COL2A1 (cat. no.
ab34712), anti-IL-1β (cat. no. ab2105), anti-IL-6 (cat. no.
ab6672), anti-IL-17 (cat. no. ab79056), anti-TNF-α (cat. no.
ab6671), anti-GAPDH (cat. no. ab181602) (all from Abcam).
Reverse transcription-quantitative PCR
(RT-qPCR) assay
Total RNA was isolated from NP cells using
TRIzol® reagent (Beyotime Institute of Biotechnology)
according to the manufacturer's instructions. Reverse transcription
was carried out using the PrimeScript™ RT reagent kit with gDNA
Eraser (Takara Bio, Inc.), according to the manufacturer's
instructions. The reverse transcription reactions occurred as
follows: 37˚C for 15 min, 85˚C for 5 sec, 4˚C for 5 min. RT-qPCR
analysis was performed on an ABI 7500 instrument (Thermo Fisher
Scientific, Inc.). All reaction systems and procedures of RT-qPCR
were conducted following the manufacturer's protocol of TB Green
Premix Ex Taq II (Takara Bio, Inc.). Reaction protocol was set with
three-step cycling conditions: 95˚C for 30 sec, followed by 40
cycles of 95˚C for 5 sec, 60˚C for 30 sec and 72˚C for 15 sec. The
melting curve stage was included, ramping from 65 to 95˚C
(increment 0.5˚C/5 sec) to verify the specificity of the primer
amplification based on the presence of a single and sharp peak. All
primer sequences are listed in Table
SI. The 2-ΔΔCq method was used to calculate the
relative expression levels (20).
ELISA assay
After the c-Jun- and empty vector-transfected NP
cells were cultured, the supernatants were collected into sterile
centrifuge tube. The expression levels of inflammatory factors,
including TNF-α (cat. no. P01P0087; Shanghai BlueGene Biotech Co.,
Ltd.), IL-1β (cat. no. E01I0010; Shanghai BlueGene Biotech Co.,
Ltd.), IL-6 (cat. no. E01I0006; Shanghai BlueGene Biotech Co.,
Ltd.) and IL-17 (cat. no. E01I0362; Shanghai BlueGene Biotech Co.,
Ltd.) were measured by ELISA according to the manufacturer's
instructions.
Cell Counting Kit-8 (CCK8) and TUNEL
assay
According to the CCK-8 (Sigma-Aldrich; Merck KGaA)
manufacturer's instructions, 100 µl/well cell suspension was seeded
into a 96-well plate (1x104 cells/well). After being
cultured for 24 h at 37˚C, c-Jun-mediated cells were treated with
TGF-β neutralizing antibody (2 µg/ml; cat. no. AB-100-NA; R&D
Systems, Inc.) and isotype-matched control IgG (2 µg/ml; cat. no.
AB-108-C; R&D Systems, Inc.). Upon 12 h of incubation, 10 µl
CCK8 solution was added to each well and for 2 h. The absorbance
was measured at 450 nm on a microplate reader.
For the TUNEL assay, cells were fixed with 4%
paraformaldehyde for 20 min at room temperature. Cells were washed
three times with PBS and cells were stained according to the
manufacturer's instructions of the TUNEL kit [cat. no. 11684817910;
Roche Diagnostics (Shanghai) Co., Ltd.]. Apoptotic cell was
observed at a magnification of x100 using an inverted fluorescence
microscope (cat. no. CKX53; Olympus Corporation), each sample was
randomly counted in four fields, with red representing apoptotic
cells and blue representing nucleus pulposus. The apoptosis rate
was the percentage of red fluorescent cells to blue fluorescent
cells, and the average value was taken as the final apoptosis rate
of nucleus pulposus cells.
Statistical analysis
Data were presented as the means ± standard
deviation. Each experiment was repeated three times independently.
Statistical analyses were performed using SPSS 23.0 (IBM Corp.).
Comparisons among multiple groups were made with one-way analysis
of variance (ANOVA) followed by a Tukey's post hoc test and
differences between two groups were identified with a Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Cell isolation and lentivirus
transfection
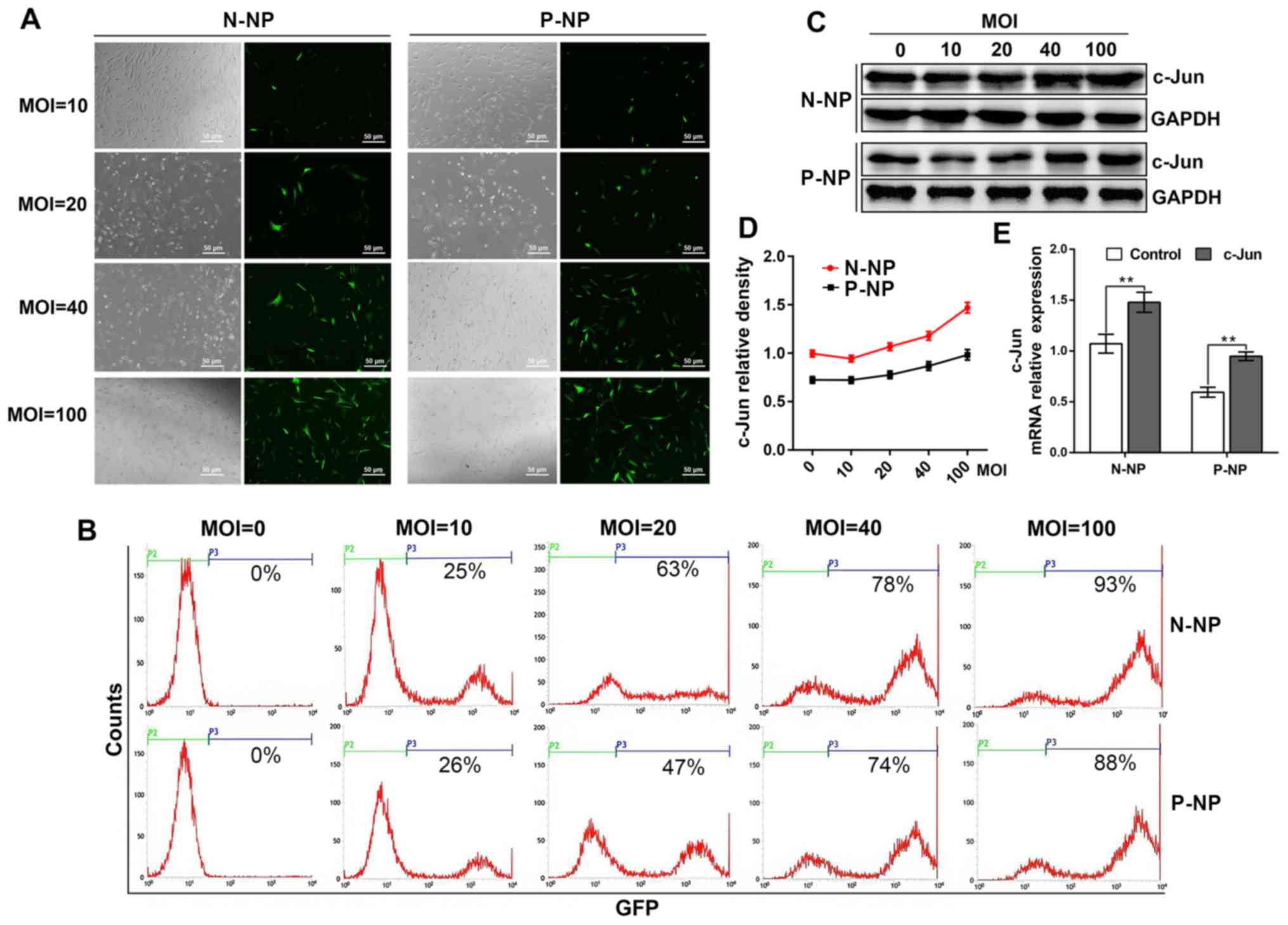
The present study demonstrated that NP cells were
successfully isolated from both normal and IDD tissues. Briefly,
following c-Jun overexpression, GFP expression in normal NP cells
displayed 25, 63, 78 and 93% efficiencies at MOI values of 10, 20,
40 and 100, respectively. When MOI values were set at 10, 20, 40
and 100, the GFP expression efficiency in the IDD NP cells
exhibited 26, 47, 74 and 88%, respectively. These results indicated
that cell expressed the transfected GFP most efficiently when the
MOI value was set at 100 (Fig. 1A
and B).
Detection of c-Jun expression by
western blotting and RT-qPCR
The results from western blotting indicated that
c-Jun expression was the highest when the MOI was set at 100 for
both normal and IDD NP cells (Fig.
1C and D). A similar increase
in c-Jun expression was observed by RT-qPCR between the normal and
IDD groups (Fig. 1E). These results
demonstrated a successful overexpression of c-Jun in NP cells.
Notably, the data demonstrated that c-Jun expression was lower in
IDD NP cells compared with normal NP cells (Fig. 1C-E).
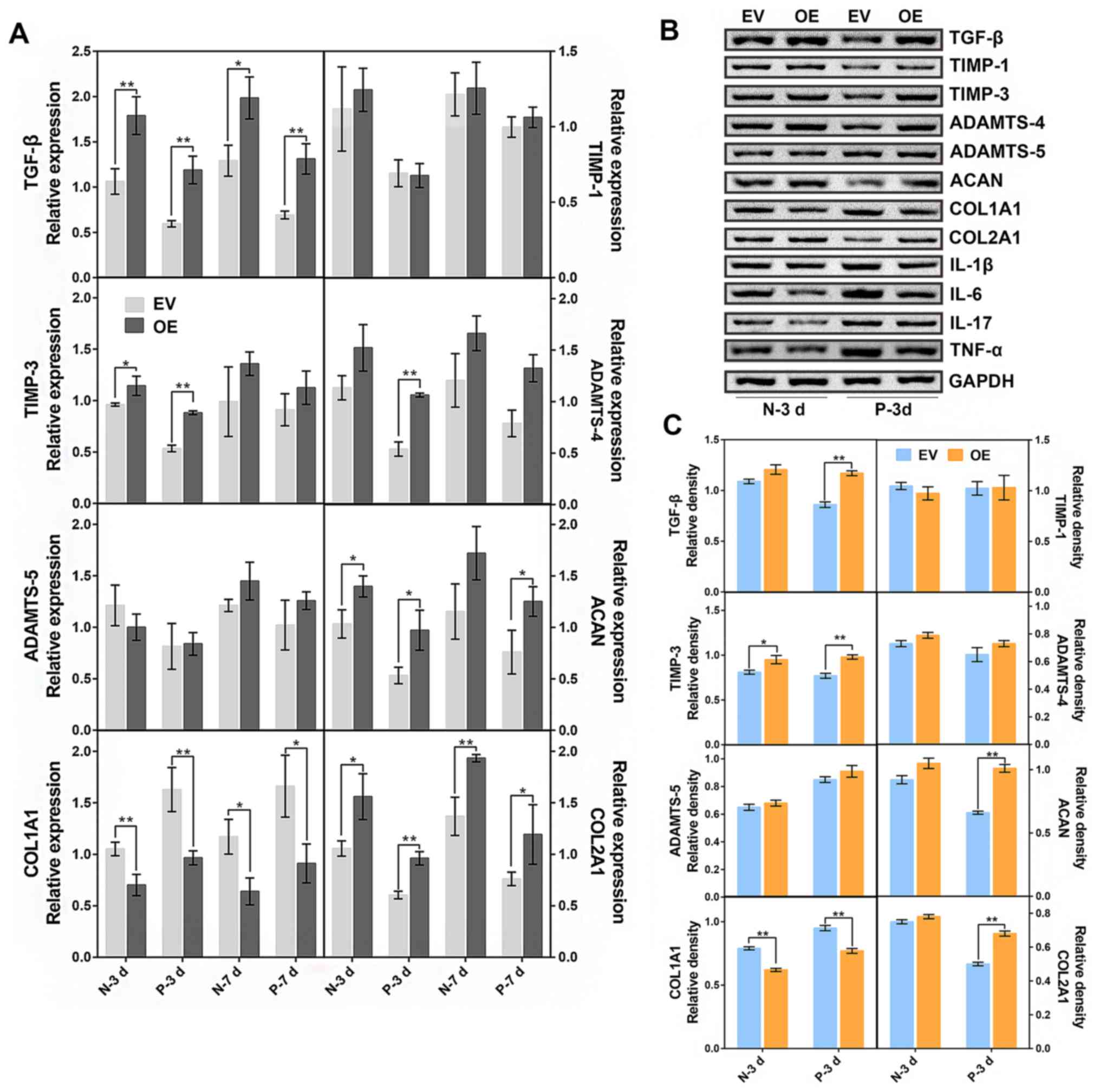
c-Jun overexpression upregulates the
genes associated with ameliorating IDD
The expression levels of IDD-related factors were
detected by western blotting and RT-qPCR following transduction
(MOI 100 PFU/cell) and subsequent cell culture for 3 and 7 days in
the normal and IDD NP cells. On day 3, a comparison was made
between the c-Jun-overexpressed IDD NP cells and the empty
vector-transfected cells. The results demonstrated that
c-Jun-overexpression groups exhibited significant increases in the
mRNA expression of TGF-β, TIMP-3, ADAMTS-4, ACAN and COL2A1, and a
significant decrease in COL1A1 expression, while no significant
changes were detected in TIMP-1 and ADAMTS-5 expression (Fig. 2A). On day 7, expression levels of
TGF-β, ACAN and COL2A1 were significantly upregulated after c-Jun
overexpression when compared with the control. The effect of c-Jun
on NP cells was most evident on the third day; consequently, the
cells transfected for 3 days were selected for subsequent protein
detection (Fig. 2B and C). The results suggested that expression
levels of TGF-β, TIMP-3, ACAN and COL2A1 were significantly
increased after c-Jun overexpression in the degenerative NP cells.
Taken together, these data demonstrated that c-Jun overexpression
may promote the expression of proteins associated with IDD
amelioration.
c-Jun overexpression downregulates the
expression of inflammatory cytokines
The expression levels of TNF-α, IL-1β, IL-6 and
IL-17 were detected by RT-qPCR, western blotting and ELISA. The
results indicated a significant decrease in mRNA and protein
expression of TNF-α, IL-1β, IL-6 and IL-17 following c-Jun
overexpression in normal and IDD NP cells (Figs. 2B, 3A-E). In addition, the expression levels
of inflammatory cytokines in IDD NP cells were higher than those in
normal healthy NP cells. Similar concentration-dependent trends
were observed in the ELISA assays. Furthermore, both in normal and
IDD NP cells, the concentration of inflammatory factors was
significantly lower than those in the empty-vector transfected
cells following c-Jun overexpression (Fig. 3F-I). These findings suggested that
the overexpression of c-Jun inhibited the expressions of
inflammatory cytokines.
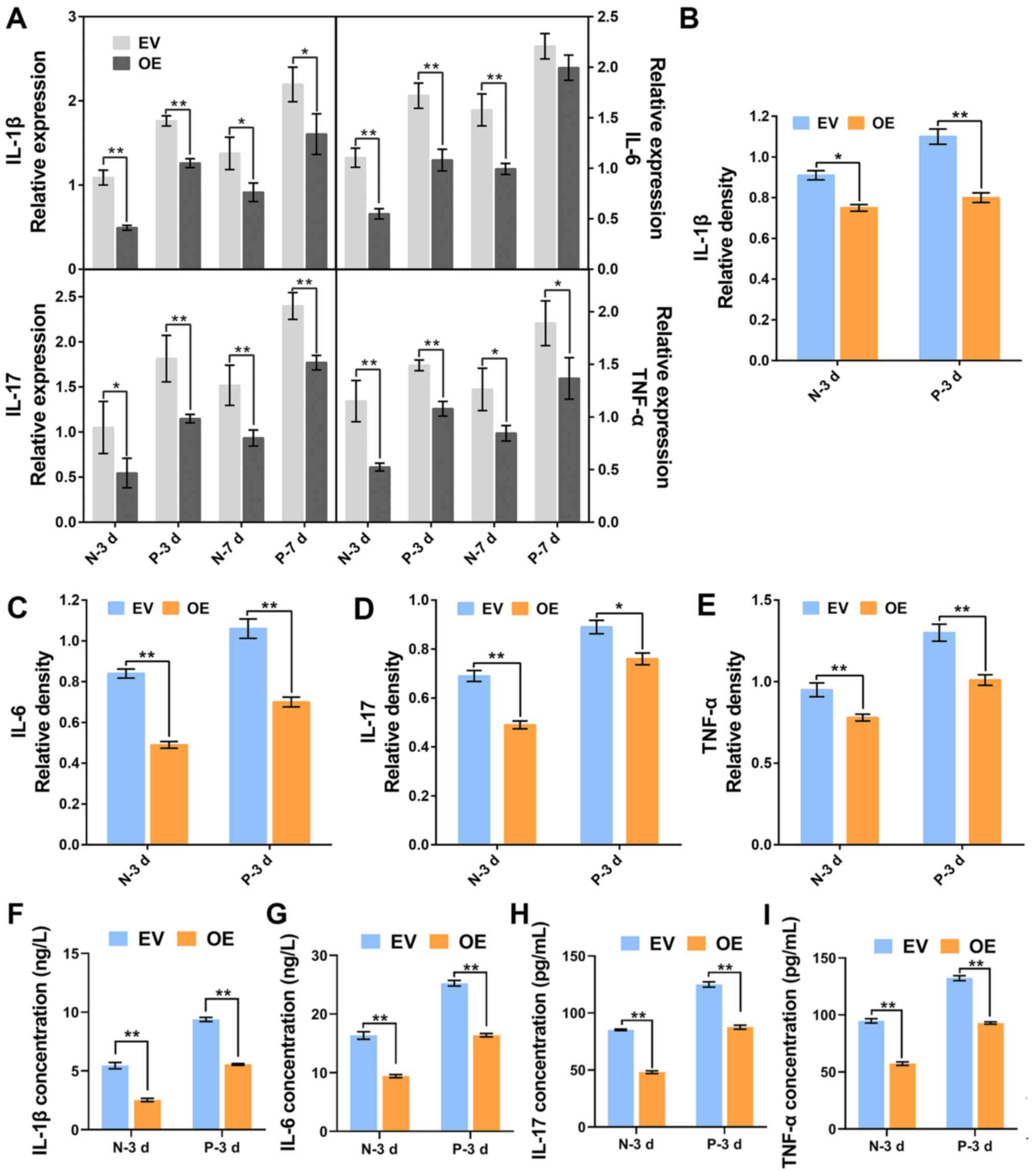 | Figure 3Expression of inflammatory cytokines
in normal and degenerative NP cells followingc-Jun overexpression.
(A) mRNA expression of inflammatory cytokines were detected by
reverse transcription-quantitative PCR in cells cultured for 3 and
7 days. (B-E) Relative density of (B) IL-1β, (C) IL-6, (D) IL-17
and (E) TNF-α was analyzed by ImageJ. (F-I) Concentrations of
IL-1β, IL-6, IL-17 and TNF-α in cell supernatant were analyzed by
ELISA kit. *P<0.05, **P<0.01. EV, empty
vector-transfected cells; OE, c-Jun-overexpressed cells; N-3 d, -7
d, normal cells were cultured for 3 days or 7 days; D-3 d, -7 d,
degenerative cells were cultured for 3 days or 7 days. |
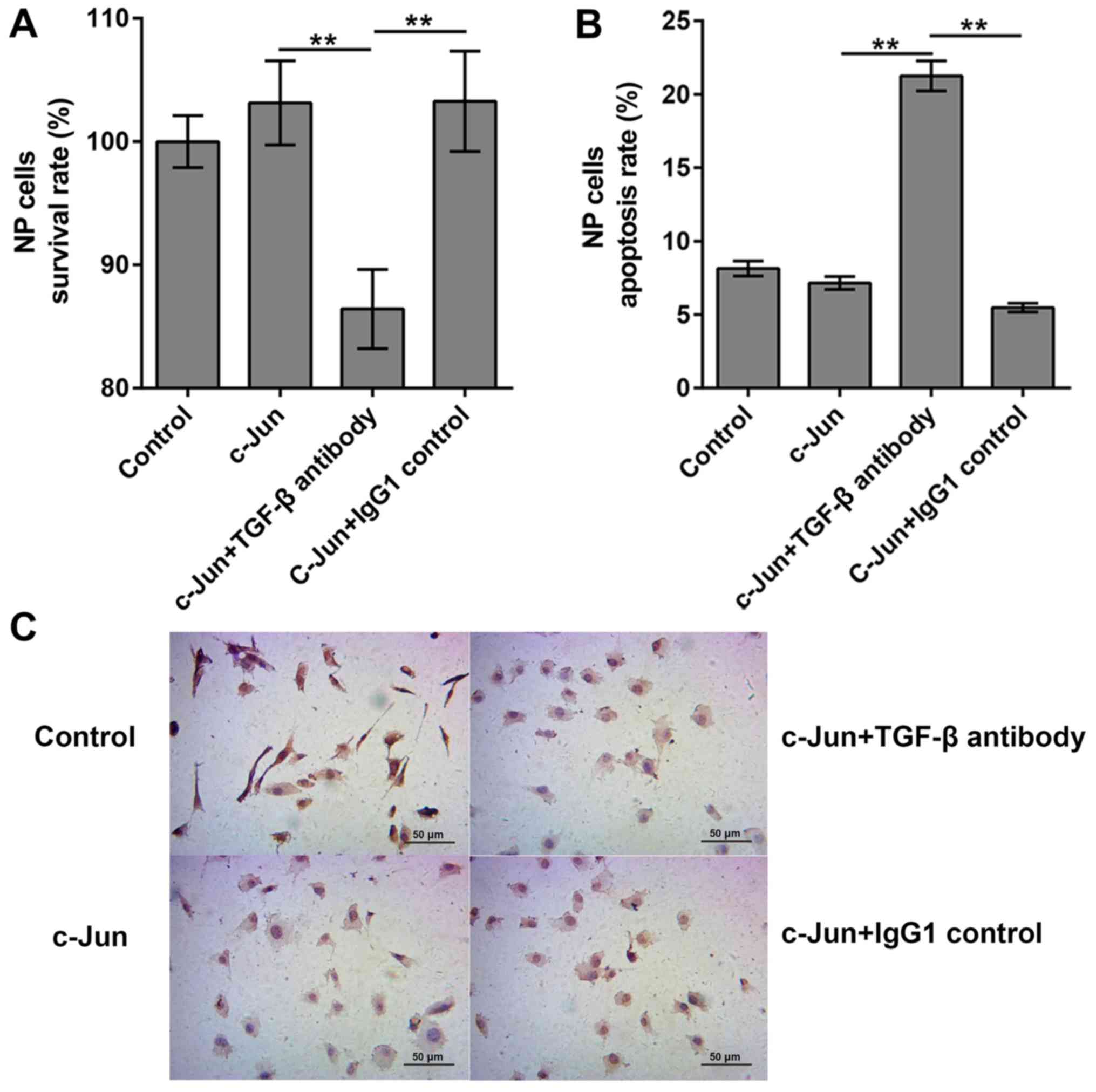
Effects of TGF-β antibody on
c-Jun-regulated cell
A previous study reported that c-Jun can activate
TGF-β-induced transcription (21).
To clarify the roles of TGF-β in c-Jun-regulated cell proliferation
and apoptosis, cells were treated with TGF-β antibody to inhibit
TGF-β activity, and the the survival and apoptosis rates of
degenerated cells were analyzed by CCK8 and TUNEL assays,
respectively. Survival rates in the empty vector-transfected and
the c-Jun transfected cells were 100 and 103.15%, respectively. The
survival rates manifested differently following the addition of
TGF-β antibody, which significantly decreased cell survival to
86.42%, compared with 103.28% in the c-Jun + IgG1 group (Fig. 4A). These results indicated that
TGF-β antibody was capable of decreasing cell proliferation and
inducing cell death in c-Jun-overexpressed NP cells. Consequently,
the apoptosis rates of empty vector-transfected group, c-Jun
transfected group, c-Jun + TGF-β antibody group and c-Jun + IgG1
control group were evaluated, and the results demonstrated that the
apoptosis rates were 8.15, 7.15, 21.25 and 5.48%, respectively
(Fig. 4B and C). The apoptosis rates increased
significantly following TGF-β antibody addition, indicating that
TGF-β could inhibit the apoptosis following c-Jun overexpression
within IDD NP cells.
Discussion
As a member of the AP-1 transcription factor family,
c-Jun has a functional role in various cellular responses to
extracellular stimuli (13,22). By functioning as a key transcription
factor, AP-1 was reported to regulate cell survival and cell death
pathways, initiate gene transcription as a molecular switch, and
was demonstrated to be involved in relevant cellular processes
including proliferation, apoptosis and inflammation (22). In c-Jun-deficient mice, the IVD
formation was impaired (18). In
addition, IVD cell clusters produce certain ECM components,
allowing the repair of damaged tissues with c-Jun as a major
protein expressed (23). It was
therefore hypothesized that c-Jun could have crucial effects on IVD
tissues. However, the role of c-Jun in the progression of IDD
remains unknown, in particular the its effects on NP cell
proliferation, apoptosis and target gene regulation in
vitro. The results from the present study demonstrated that the
mRNA and protein expression of c-Jun in normal NP cells was higher
compared with that in IDD NP cells. Furthermore, c-Jun had a
positive effect on NP cells in promoting cell proliferation,
reducing cell apoptosis, increasing ECM synthesis and
downregulating inflammatory factors.
A previous study demonstrated that inflammatory
mediators serve crucial roles in IVD and could contribute to the
IDD process (24). The expression
levels of inflammatory factors IL-1β, IL-6, IL-17 and TNF-α in the
degenerated disc tissue were distinctly increased compared with
normal disc tissue, and their expressions was positively correlated
with the degree of disc degeneration, including IL-1β, IL-6, IL-17
and TNF-α (25). In addition, AP-1
is considered as a pro-inflammatory factor that can directly
regulate the expression of ILs and MMPs (26). The results from the present study
suggested that NP cells overexpressin c-Jun exhibited a dramatic
decrease in the expression of TNF-α, IL-1β, IL-6 and IL-17,
suggesting that c-Jun may exert an anti-inflammatory function in NP
cells. However, AP-1 dimers composed of different protein subunits
processed different biological functions, and Jun family members
exhibit different functional properties as transcription factors
(27). Li et al (28) reported that IL-17a can enhance the
expression of cyclooxygenase 2 and prostaglandin E2 in NP cells,
leading to the regulation of inflammatory responses through
p38/c-Fos and c-Jun n-terminal kinase (JNK)/c-Jun pathways. Duval
et al (29) reported that
the JNK pathway could downregulate the production of TNF-α. JNK is
mainly known to regulate the phosphorylation of c-Jun, which
regulates the production of more c-Jun, along with other genes
(30). Inflammatory cytokines
induce the expression of c-Jun; however, the specific mechanism of
how the inflammatory cytokines control the overexpression of c-Jun
requires further research.
Previous studies reported that the cytokine TGF-β
can delay and repair IDD by upregulating ACAN and COL2A1
expression, therefore promoting NP cell proliferation and
increasing ECM synthesis (11,31,32).
Furthermore, the proto-oncogene c-myc serves an active role
in NP cell proliferation and cycle progression under TGF-β
stimulation (33). Interestingly,
c-Jun belongs to proto-oncogene family of nuclear transcription
factors, and it forms a complex with promoters of different genes
to regulate transcription (23).
Numerous growth-related cytokines contain
12-O-tetradecanoylphorbol-13-acetate response element (TRE)
binding sites, including basic fibroblast growth factor and TGF-β,
the dephosphorylated AP-1 is able to recognize the TRE binding
sites and initiate transcription of these cytokines (34). The Smad3-Smad4 heterodimeric complex
cooperate with c-Jun/c-Fos to induce transcriptional activation in
response to TGF-β (21,35). Significant increase in TGF-β mRNA
and protein expression was observed in NP cells following c-Jun
overexpression, and an upregulation was also observed for the
cytokines TIMP-3, ACAN and COL2A1, which are associated with
amelioration of IDD. It has been reported that TIMP3 can increase
synthesis of ECM (36). COL2A1 and
ACAN, which act as main components of matrix of NP (37), are essential to maintaining the
integrity of IVD (4). The results
from the present study suggested that c-Jun was positively related
to IDD in NP cells, and that TGF-β may act as a key regulator in
c-Jun signaling pathway. Hiyama et al (38) reported that when NP cells are
stimulated by TGF-β, there is a concomitant increase in c-Jun
expression and activity (38). In
the present study, when patients' NP cells were transduced to
overexpress c-Jun, cell survival rate was increased and apoptosis
rate was decreased. However, there was no significant difference
between the control group and the overexpressed c-Jun group.
Treatment with antibody against TGF-β significantly increased the
rate of apoptosis and decreased the survival rate, suggesting that
c-Jun might promote cell proliferation and inhibit cell apoptosis,
thereby minimizing the function of c-Jun after inhibition of TGF-β.
This result was consistent with previous studies showing that c-Jun
promotes cell proliferation and protects cells from apoptosis
(39-42).
In the present study, c-Jun increased the expression
of TGF-β, TIMP-3, ACAN and COL2A1 which may promote ECM synthesis,
and suppressed the inflammatory response by decreasing the
expression of TNF-α, IL-1β, IL-6 and IL-17. Furthermore, c-Jun
facilitated cell proliferation and decreased cell apoptosis by
upregulating TGF-β expression. These results suggested that c-Jun
may alleviate IDD by regulating TGF-β. The results from this study
allowed a better understanding of the molecular mechanisms of IDD,
which may help the development of novel treatment of IDD disease.
To the best of our knowledge, only a few studies have elucidated
the role of c-Jun in IDD or additional degenerative diseases. The
present study evaluated the effects of c-Jun on NP cells in
vitro; however, whether c-Jun could delay disc degeneration
in vivo, or directly interact with TGF-β remain unknown.
Further investigation is therefore required.
Supplementary Material
Sequences of the primers used for
reverse transcription quantitative PCR.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ML performed the experiments, collected the results
and wrote the manuscript. KW, SL, KZ, WH and XW contributed to data
analysis and manuscript revision. CY conceived the study and
contributed to reviewing/editing the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The study complied with the Declaration of Helsinki
and was approved by the Institutional Review Board Ethics Committee
of Union Hospital of Tongji Medical College. Written informed
consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Freemont AJ: The cellular pathobiology of
the degenerate intervertebral disc and discogenic back pain.
Rheumatology (Oxford). 48:5–10. 2009.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hanaei S, Abdollahzade S, Khoshnevisan A,
Kepler CK and Rezaei N: Genetic aspects of intervertebral disc
degeneration. Rev Neurosci. 26:581–606. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zheng L, Jianxiong S, Wu WK, Xin Y,
Jinqian L, Guixing Q and Jiaming L: The role of leptin on the
organization and expression of cytoskeleton elements in nucleus
pulposus cells. J Orthop Res. 31:847–857. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Li Z, Liang J, WK WK, Yu X, Yu J, Weng X
and Shen J: Leptin activates RhoA/ROCK pathway to induce
cytoskeleton remodeling in nucleus pulposus cells. Int J Mol Sci.
15:1176–1188. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Pattison ST, Melrose J, Ghosh P and Taylor
TKF: Regulation of gelatinase-a (mmp-2) production by ovine
intervertebral disc nucleus pulposus cells grown in alginate bead
culture by transforming growth factor-β and insulin like growth
factor-1. Cell Biol Int. 25:679–689. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gruber HE and Hanley EN Jr: Recent
advances in disc cell biology. Spine (Phila Pa 1976). 28:186–193.
2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jianru W, Dessislava M, D Greg A, Zhaomin
Z, Irving MS and Makarand VR: TNF-α and IL-1β promote a
disintegrin-like and metalloprotease with thrombospondin type I
motif-5-mediated aggrecan degradation through syndecan-4 in
intervertebral disc. J Biol Chem. 286:39738–39749. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang WJ, Yu XH, Wang C, Yang W, He WS,
Zhang SJ, Yan YG and Zhang J: MMPs and ADAMTSs in intervertebral
disc degeneration. Clin Chim Acta. 448:238–246. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Vo NV, Hartman RA, Yurube T, Jacobs LJ,
Sowa GA and Kang JD: Expression and regulation of
metalloproteinases and their inhibitors inintervertebral disc aging
and degeneration. Spine J. 13:331–341. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jin H, Shen J, Wang B, Wang M, Shu B and
Chen D: TGF-β signaling plays an essential role in the growth and
maintenance of intervertebral disc tissue. FEBS Lett.
585:1209–1215. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Feng C, Liu H, Yang M, Zhang Y, Huang B
and Zhou Y: Disc cell senescence in intervertebral disc
degeneration: Causes and molecular pathways. Cell Cycle.
15:1674–1684. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Angel P and Karin M: The role of Jun, Fos
and the AP-1 complex in cell-proliferation and transformation.
Biochim Biophys Acta. 1072:129–157. 1991.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wisdom R, Johnson RS and Moore C: c-Jun
regulates cell cycle progression and apoptosis by distinct
mechanisms. EMBO J. 18:188–197. 1999.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Behrens A, Sibilia M and Wagner EF:
Amino-terminal phosphorylation of c-Jun regulates stress-induced
apoptosis and cellular proliferation. Nat Genet. 21:326–329.
1999.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Santos MM, Tannuri AC, Coelho MC,
Gonçalves JD, Serafini S, Da Silva LF and Tannuri U: Immediate
expression of c-Fos and c-jun mRNA in a model of intestinal
autotransplantation and ischemia-reperfusion in situ. Clinics (Sao
Paulo). 70:373–379. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Behrens A, Sibilia M, David JP,
Möhle-Steinlein U, Tronche F, Schütz G and Wagner EF: Impaired
postnatal hepatocyte proliferation and liver regeneration in mice
lacking c-jun in the liver. EMBO J. 21:1782–1790. 2002.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Behrens A, Haigh J, Mechta-Grigoriou F,
Nagy A, Yaniv M and Wagner EF: Impaired intervertebral disc
formation in the absence of Jun. Development. 130:103–109.
2003.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila Pa 1976).
26:1873–1878. 2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang Y, Feng XH and Derynck R: Smad3 and
Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced
transcription. Nature. 394:909–913. 1998.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Ye N, Ding Y, Wild C, Shen Q and Jia Z:
Small molecule inhibitors targeting activator protein 1 (AP-1). J
Med Chem. 57:6930–6948. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tolonen J, Grönblad M, Virri J, Seitsalo
S, Rytömaa T and Karaharju E: Oncoprotein c-Fos and c-Jun
immunopositive cells and cell clusters in herniated intervertebral
disc tissue. Eur Spine J. 11:452–458. 2002.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Monchaux M, Forterre S, Spreng D, Karol A,
Forterre F and Wuertz-Kozak K: Inflammatory processes associated
with canine intervertebral disc herniation. Front Immunol.
8(1681)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liu XG, Hou HW and Liu YL: Expression
levels of IL-17 and TNF-α in degenerated lumbar intervertebral
discs and their correlation. Exp Ther Med. 11:2333–2340.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu Y, Du Y, Wang H, Li D and Feng WH:
Porcine reproductive and respiratory syndrome virus (PRRSV)
up-regulates IL-8 expression through TAK-1/JNK/AP-1 pathways.
Virology. 506:64–72. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Shaulian E and Karin M: AP-1 as a
regulator of cell life and death. Nat Cell Biol. 4:E131–E136.
2002.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li JK, Nie L, Zhao YP, Zhang YQ, Wang X,
Wang SS, Liu Y, Zhao H and Cheng L: IL-17 mediates inflammatory
reactions via p38/c-Fos and JNK/c-Jun activation in an
AP-1-dependent manner in human nucleus pulposus cells. J Transl
Med. 14(77)2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Duval H, Mbatchi SF, Grandadam S, Legendre
C, Loyer P, Ribault C, Piquet-Pellorce C, Guguen-Guillouzo C,
Boudjema K and Corlu A: Reperfusion stress induced during
intermittent selective clamping accelerates rat liver regeneration
through JNK pathway. J Hepatol. 52:560–569. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Koba S, Pakala R, Watanabe T, Katagiri T
and Benedict CR: Vascular smooth muscle proliferation-a protein
kinase stimulated by UV light and Ha-Ras that binds and
phosphorylates the c-Jun activation domain. J Am Coll Cardiol.
34:1644–1651. 1999.
|
|
31
|
Lee YJ, Kong MH, Song KY, Lee KH and Heo
SH: The relation between Sox9, TGF-beta1, and proteoglycan in human
intervertebral disc cells. J Korean Neurosurg Soc. 43:149–154.
2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang SL, Yu YL, Tang CL and Lv FZ: Effects
of TGF-β1 and IL-1β on expression of ADAMTS enzymes and TIMP-3 in
human intervertebral disc degeneration. Exp Ther Med. 6:1522–1526.
2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Nakai T, Mochida J and Sakai D:
Synergistic role of c-Myc and ERK1/2 in the mitogenic response to
TGF beta-1 in cultured rat nucleus pulposus cells. Arthritis Res
Ther. 10(R140)2008.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Iwata A, Masago A and Yamada K: Expression
of basic fibroblast growth factor mRNA after transient focal
ischemia: Comparison with expression of c-fos, c-jun, and hsp 70
mRNA. J Neurotrauma. 14:201–210. 1997.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Qing J, Zhang Y and Derynck R: Structural
and functional characterization of the transforming growth
factor-beta-induced Smad3/c-Jun transcriptional cooperativity. J
Biol Chem. 275:38802–38812. 2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yan L, Kang L, Han X, Mao C, Kai Z, Zhao T
and Jie Z: The imbalance between TIMP3 and matrix-degrading enzymes
plays an important role in intervertebral disc degeneration.
Biochem Biophys Res Commun. 469:507–514. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chung SA, Khan SN and Diwan AD: The
molecular basis of intervertebral disk degeneration. Orthop Clin
North Am. 34:209–219. 2003.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hiyama A, Gogate SS, Gajghate S, Mochida
J, Shapiro IM and Risbud MV: BMP-2 and TGF-beta stimulate
expression of beta 1,3-glucuronosyl transferase 1 (GlcAT-1) in
nucleus pulposus cells through AP1, TonEBP, and Sp1: Role of MAPKs.
J Bone Miner Res. 25:1179–1190. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gaudet P, Livstone MS, Lewis SE and Thomas
PD: Phylogenetic-based propagation of functional annotations within
the gene ontology consortium. Brief Bioinform. 12:449–462.
2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Qian Z, Li Y, Chen J, Li X and Gou D:
MiR-4632 mediates PDGFBB-induced proliferation and anti-apoptosis
of human pulmonary artery smooth muscle cells via targeting cJUN.
Am J Physiol Cell Physiol. 313:C380–C391. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Khachigian LM, Fahmy RG, Zhang G,
Bobryshev YV and Kaniaros A: c-Jun regulates vascular smooth muscle
cell growth and neointima formation after arterial injury.
Inhibition by a novel DNA enzyme targeting c-Jun. J Biol Chem.
277:22985–22991. 2002.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kappelmann M, Bosserhoff A and Kuphal S:
AP-1/c-Jun transcription factors: Regulation and function in
malignant melanoma. Eur J Cell Biol. 93:76–81. 2014.PubMed/NCBI View Article : Google Scholar
|