Introduction
Glioma is the most common malignant tumor in the
brain and has a very poor prognosis as most patients with glioma
are diagnosed at advanced stages (1,2). While
increased effort has been made to investigate the etiology of
glioma in recent decades, the underlying molecular mechanisms
remain poorly understood (3,4).
Therefore, identifying these molecular mechanisms is of great
importance for developing novel strategies for the diagnosis and
treatment of glioma (3-5).
Long non-coding RNAs (lncRNAs), a type of single
strand non-coding RNA >200 nucleotides in length, have been
reported to serve key roles in various biological processes, such
as cell proliferation, migration, apoptosis and tumorigenesis
(6,7). For instance, the lncRNA steroid
receptor RNA activator promotes vascular smooth muscle cell
proliferation and neointimal hyperplasia via the MEK/ERK/cAMP
response element-binding protein pathway (8). It has also been shown that lncRNA H19
regulates trophoblastic spheroid adhesion (9). Moreover, a large number of lncRNAs
have been observed to be deregulated during the development and
progression of human cancer types, including glioma, and some have
been suggested as promising therapeutic targets (3,7,10). For
example, upregulation of lncRNA AFAP1-AS1 predicts poor prognosis
in patients with cervical cancer, as well as promotes the migration
and invasion of cervical cancer cells (11). Jiang et al (12) also reported that silencing the
lncRNA HOXA distal transcript antisense RNA suppressed prostate
cancer cell proliferation and enhanced cell sensitivity to
cisplatin by inhibiting the Wnt/β-catenin pathway.
MicroRNAs (miRNAs/miRs), a type of small non-coding
RNA with ~20 nucleotides, are regulators of gene expression and are
involved in a variety of physiological and pathological processes,
such as differentiation, development, cell proliferation and
motility and tumorigenesis (13).
lncRNAs can competitively bind miRNAs and negatively affect miRNA
expression levels (13,14). For instance, lncRNA ANRIL inhibits
the senescence of vascular smooth muscle cells via regulating
miR-181a (15), while lncRNA
LINC01234 promotes the proliferation of colon cancer cells by
regulating the expression of miR-642a-5p (16).
LINC00210, a newly discovered lncRNA, can drive
Wnt/β-catenin signaling activation and thus liver tumor progression
(17). However, to the best of our
knowledge, no previous study has focused on the expression and
function of LINC00210 in glioma. Therefore, the present study aimed
to evaluate the clinical significance of LINC00210 expression in
glioma. In addition, the role of LINC00210 in the regulation of
glioma cell proliferation and migration, as well as the potential
molecular mechanism were examined.
Materials and methods
Clinical tissue samples
The current study collected 54 glioma tissues and 14
healthy brain tissues (with 1 cm distance from the glioma tissues)
from 54 patients with primary glioma at The Third Xiangya Hospital
of Central South University (Changsha, China) between March 2011
and June 2013. These patients with glioma included 33 men and 21
women who were 32-66 years old, with a mean age of 48.6 years. No
patient with glioma received adjuvant treatment before surgery. The
tissues were stored at -80˚C until use. These patients were
followed-up for 5 years for survival analysis. This study was
approved by the Ethics Committee of The Third Xiangya Hospital of
Central South University, and informed written consent was obtained
from these patients with glioma.
Cell culture and transfection
Human glioma cell lines, including U251, T98G,
U-373MG Uppsala and U-87MG, were obtained from the Cell Bank of the
Chinese Academy of Sciences, and normal human astrocytes (NHAs)
were purchased from Lonza Group, Ltd. The U-87MG cell line is the
original glioblastoma cell line established in the University of
Uppsala. The authentication of these cell lines has been confirmed
via STR profiling. Cells were cultured in DMEM (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Thermo Fisher
Scientific, Inc.) at 37˚C with 5% CO2.
For cell transfection, Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.) was used to transfect U251
and T98G cells with 100 nM negative control (NC) small interfering
(si)RNA (cat. no. AM4611), LINC00210 siRNA1 (cat. no. n545578),
LINC00210 siRNA2 (cat. no. n545579), miR-NC (cat. no. 4464058),
miR-328 mimic (cat. no. 4464066) or miR-328 inhibitor (cat. no.
4464084; anti-miR-328 group), according to the manufacturer's
instructions. All of these transfects were obtained from Thermo
Fisher Scientific, Inc. At 48 h after cell transfection, reverse
transcription-quantitative PCR (RT-qPCR) was performed.
RT-qPCR
Total RNA was extracted from clinical tissue samples
and cell lines using TRIzol® reagent (Thermo Fisher
Scientific, Inc.). RNA was then reverse transcribed into cDNA using
High Capacity cDNA Reverse Transcription kit (cat. no. 4368814;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The reverse transcription was performed as follows:
16˚C for 30 min, followed by 42˚C for 30 min and 85˚C for 5 min.
Then, cDNA was used to conduct qPCR using a miScript
SYBR® Green PCR kit (Qiagen GmbH) on an ABI 7500 PCR
machine (Thermo Fisher Scientific, Inc.). The PCR conditions were
as follows: Initial denaturation at 95˚C for 3 min, followed by 40
cycles of 95˚C for 15 sec, 60˚C for 15 sec and 72˚C for 15 sec, and
final extension at 72˚C for 10 min. U6 was used as the internal
reference for miR-328, while GAPDH was used as the internal
reference for LINC00210. The 2-ΔΔCq method was used for
analyzing the expression (18). The
primer sequences were as follows: LINC00210 forward,
5'-AACACGTTAGCGGGTTCTCA-3' and reverse, 5'-TCAAAAACCACCGAGGGAGG-3';
GAPDH forward, 5'-CTGGGCTACACTGAGCACC-3' and reverse,
5'-AAGTGGTCGTTGAGGGCAATG-3'; miR-328 forward,
5'-AACGAGACGACGACAGAC-3' and reverse,
5'-GGGGGGGCAGGAGGGGCTCAGGG-3'; and U6 forward,
5'-CTCGCTTCGGCAGCACA-3' and reverse,
5'-AACGCTTCACGAATTTGCGT-3'.
Cell Counting Kit (CCK)-8 assays
For cell proliferation analysis, the transfected
U251 and T98G cells (5,000 cells/well) were seeded in a 96-well
plate and cultured at 37˚C for 0, 24, 48 or 72 h, as indicated.
Then, according to the manufacturer's instructions, 10 µl CCK-8
solution (Beyotime Institute of Biotechnology) was added and the
cells were incubated at 37˚C for 1 h. Next, the absorbance was
determined at a wavelength of 450 nm with a microplate reader
(Bio-Rad Laboratories, Inc.).
Wound healing assays
Transfected U251 and T98G cells (2x105
cells/well) were seeded in a 24-well plate and cultured at 37˚C to
90% confluence. A sterilized 200-µl pipette tip was then used to
scratch the cells to generate a line. Cells were washed twice with
DPBS (Thermo Fisher Scientific, Inc.), serum-free DMEM was added
and images were captured under a light microscope (magnification,
x40; DMI 4000B; Leica Microsystems GmbH). Next, the cells were
incubated at 37˚C for another 24 h and imaged under a light
microscope. The wounds were analyzed using ImageJ software version
1.48 (National Institutes of Health).
Luciferase reporter gene assays
The potential miR targets of LINC00210 were
predicted using the bioinformatics software miRcode version 11
(http://www.mircode.org/). Dual-luciferase Target
Vectors (Promega Corporation) containing wild-type (WT) and mutant
type (MT) LINC00210 binding sites with miR-328 were generated.
Lipofectamine 2000 was used to co-transfect U251 and T98G cells
with 100 nM miR-NC or miR-328 mimics and 100 ng WT-LINC00210 or
MT-LINC00210 luciferase reporter plasmid. After transfection for 24
h, a Dual-Luciferase Reporter assay system kit (Promega
Corporation) was used to examine the luciferase activity according
to the manufacturer's instructions. The ratio of firefly to
Renilla luciferase activity was determined.
Statistical analysis
The experiments were repeated three times. Data are
presented as the mean ± SD and were analyzed using SPSS 21.0
software (IBM Corp.). A two-tailed unpaired Student's t-test was
applied for analyzing differences between two groups, while a
one-way ANOVA followed by Tukey's post hoc test was used for
analyzing differences among ≥3 groups. The association between
LINC00210 expression and clinical factors in patients with glioma
was analyzed using a χ2 test. Survival analysis was
performed using a Kaplan-Meier survival curve and a log-rank test.
The Spearman rank correlation test was used to analyze the
correlation between the expression levels of LINC00210 and miR-328
in glioma tissues. P<0.05 was considered to indicate a
statistically significant difference.
Results
Upregulation of LINC00210 is
associated with glioma progression and poor prognosis
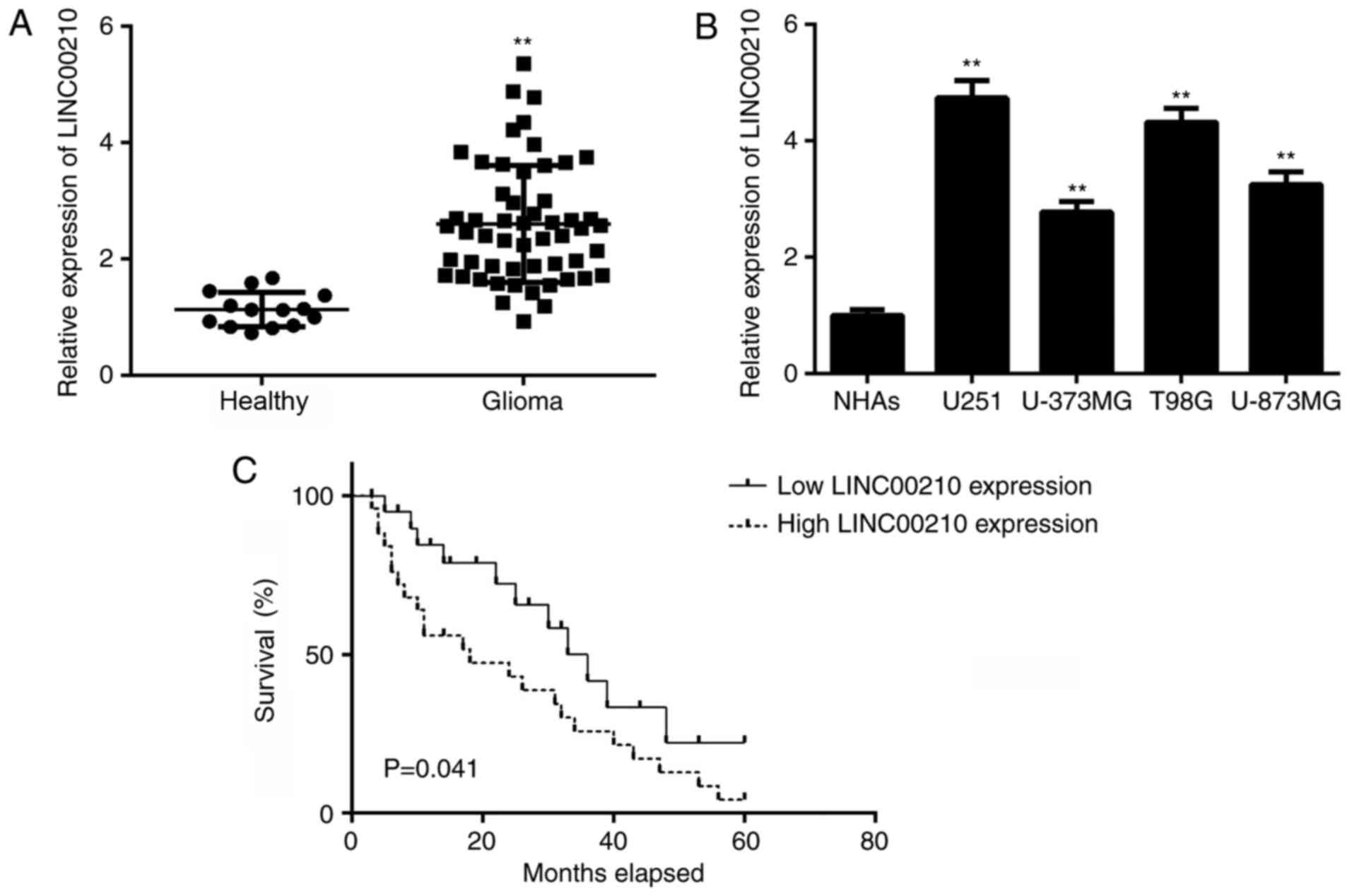
RT-qPCR was conducted to examine the expression of
LINC00210 in glioma tissues and healthy brain tissues, and it was
found that LINC00210 expression was significantly higher in glioma
tissues compared with healthy brain tissues (Fig. 1A). Consistent with the in
vivo data, the expression of LINC00210 was also significantly
upregulated in human glioma cell lines, including U251, T98G,
U-373MG Uppsala and U-87MG, compared with NHAs (Fig. 1B). Moreover, survival analysis
results demonstrated that patients with glioma with high LINC00210
expression had significantly shorter survival times compared with
patients with low LINC00210 expression (Fig. 1C). A χ2 test was
performed to analyze the association between LINC00210 expression
and clinical factors in patients with glioma. High expression of
LINC00210 was positively associated with an advanced clinical stage
in patients with glioma (Table I).
Thus, upregulation of LINC00210 was associated with disease
progression and poor prognosis in glioma.
 | Table IAssociation between LINC00210
expression and clinicopathological characteristics of patients with
glioma. |
Table I
Association between LINC00210
expression and clinicopathological characteristics of patients with
glioma.
| Variables | Total cases
(n=54) | Low LINC00210
(n=30) | High LINC00210
(n=24) |
χ2-value | P-value |
|---|
| Age, years | | | | | |
|
<50 | 28 | 16 | 12 | 0.059 | 0.808 |
|
≥50 | 26 | 14 | 12 | | |
| Sex | | | | | |
|
Male | 33 | 18 | 15 | 0.035 | 0.852 |
|
Female | 21 | 12 | 9 | | |
| WHO stage | | | | | |
|
I-II | 20 | 15 | 5 | 4.864 | 0.027a |
|
III-IV | 34 | 15 | 19 | | |
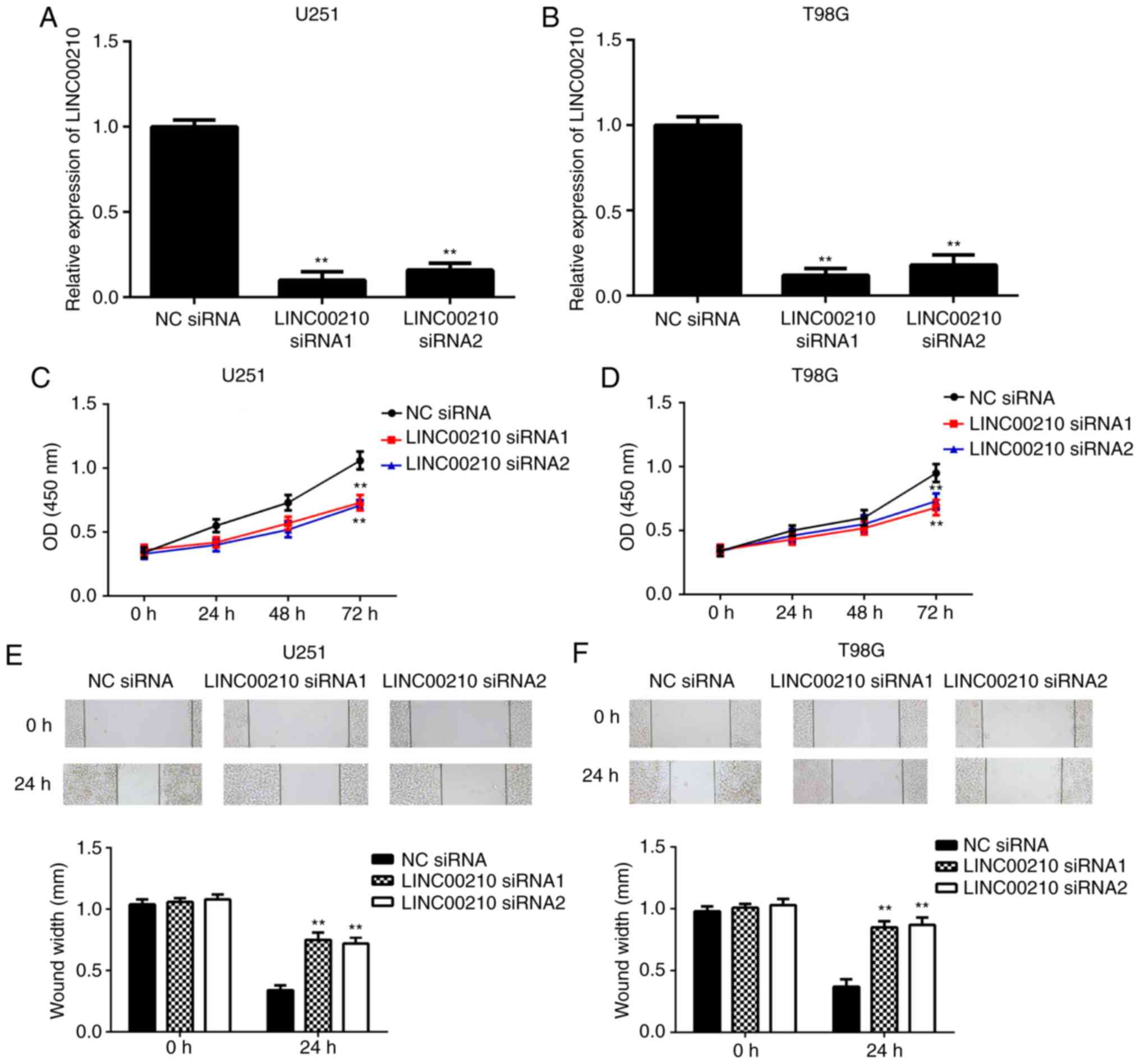
Knockdown of LINC00210 suppresses the
proliferation and migration of U251 and T98G cells
As U251 and T98G cell lines exhibited the highest
expression of LINC00210 among the four glioma cell lines examined,
these two cell lines were used in subsequent experiments. To
further examine the role of LINC00210 in glioma cell proliferation
and migration, U251 and T98G cells were transfected with NC siRNA
or two LINC00210 siRNAs. After transfection, the expression levels
of LINC00210 were significantly lower in the LINC00210 siRNA1 and
siRNA2 groups compared with the NC siRNA group (Fig. 2A and B). The CCK-8 assay results demonstrated
that the proliferation of U251 and T98G cells was significantly
lower in the LINC00210 siRNA1 and siRNA2 groups compared with the
NC siRNA group, indicating that knockdown of LINC00210
significantly inhibited glioma cell proliferation (Fig. 2C and D). Similarly, the migration of glioma
cells was significantly decreased after knocking down LINC00210
expression (Fig. 2E and F). These data suggested that LINC00210 may
have a promoting role in glioma cell proliferation and
migration.
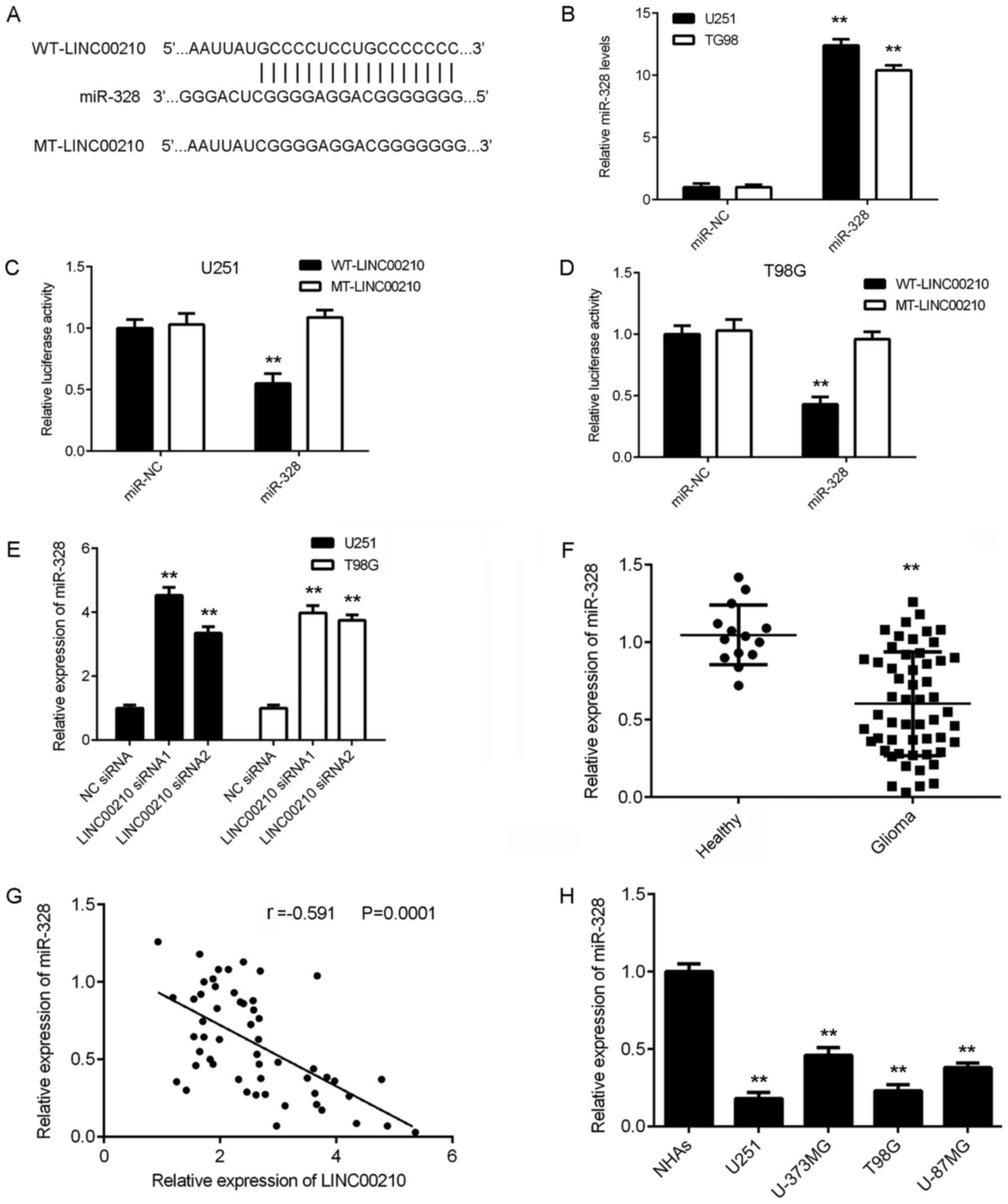
LINC00210 directly targets miR-328 in
glioma cells and tissues
It has been reported that lncRNAs can function by
regulating downstream miRNAs (13,14).
Bioinformatics analysis predicted that miR-328 was a direct target
of LINC00210, and to assess this prediction, luciferase reporter
plasmids containing WT and MT LINC00210 binding sites with miR-328
were generated (Fig. 3A). U251 and
T98G cells were also transfected with miR-NC or miR-328 mimic.
After transfection, RT-qPCR data indicated that miR-328 expression
was significantly increased in the miR-328 group compared with the
miR-NC group (Fig. 3B). Luciferase
reporter gene assays were then conducted in U251 and T98G cells.
Transfection with miR-328 mimic significantly decreased the
luciferase activity in the WT-LINC00210 group but did not affect
the luciferase activity in the MT-LINC00210 group (Fig. 3C and D). These data indicated that LINC00210
directly targeted miR-328 in U251 and T98G cells.
It was found that the expression of miR-328 was
significantly higher in U251 and T98G cells transfected with
LINC00210 siRNAs compared with cells transfected with NC siRNA
(Fig. 3E), suggesting that
silencing LINC00210 increased the expression of miR-328 in glioma
cells. To further investigate the relationship between miR-328 and
LINC00210 in glioma, RT-qPCR was performed to examine the
expression of miR-328 in glioma tissues. It was identified that
miR-328 expression was significantly lower in glioma tissues
compared with healthy brain tissues (Fig. 3F), and was also significantly
downregulated in glioma cell lines (Fig. 3H). In addition, the expression of
miR-328 was moderately, negatively correlated with the expression
of LINC00210 in glioma tissues (Fig.
3G). Thus, it was demonstrated that upregulation of LINC00210
in glioma contributed to downregulation of miR-328.
Knockdown of LINC00210 suppresses
glioma cell proliferation and migration by increasing the
expression of miR-328
Based on the aforementioned findings, it was
considered that miR-328 may be involved in LINC00210-mediated
glioma cell proliferation and migration. To further test this
hypothesis, U251 and T98G cells were transfected with NC inhibitor
or miR-328 inhibitor. After transfection, RT-qPCR data indicated
that miR-328 expression was significantly decreased in the
anti-miR-328 group compared with the anti-NC group (Fig. 4A).
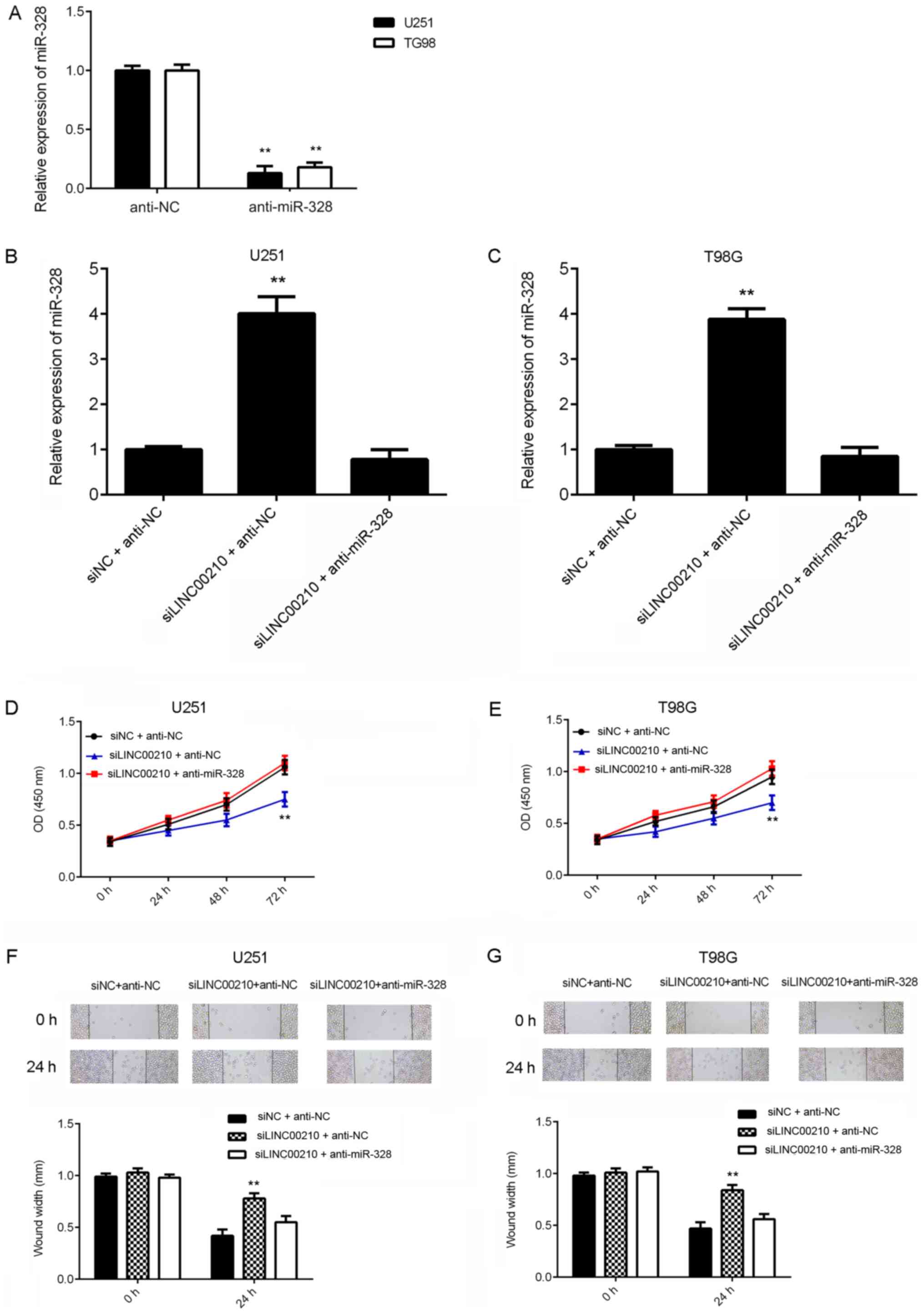 | Figure 4Knockdown of LINC00210 suppresses
glioma cell proliferation and migration by increasing the
expression of miR-328. (A) U251 and T98G cells were transfected
with NC inhibitor or miR-328 inhibitor. After transfection, RT-qPCR
was used to examine the expression of miR-328. Then, U251 and T98G
cells were transfected with NC siRNA and NC inhibitor (siNC +
anti-NC), LINC00210 siRNA1 and NC inhibitor (siLINC00210 + anti-NC)
or LINC00210 siRNA1 and miR-328 inhibitor (siLINC00210 +
anti-miR-328). After transfection, RT-qPCR was used to examine the
expression of LINC00210 in (B) U251 and (C) T98G cells. After
transfection, Cell Counting Kit-8 assays were used to determine (D)
U251 and (E) T98G cell proliferation. After transfection, wound
healing assays were used to determine (F) U251 and (G) T98G cell
migration. **P<0.01 vs. siNC + anti-NC. RT-qPCR,
reverse transcription-quantitative PCR; miR, microRNA; NC, negative
control; siRNA, small interfering RNA; OD, optical density. |
As LINC00210 siRNA1 was more efficient in
suppressing the expression of LINC00210 compared with siRNA2,
LINC00210 siRNA1 was used in subsequent experiments. U251 and T98G
cells were co-transfected with NC siRNA and NC inhibitor (siNC +
anti-NC), LINC00210 siRNA1 and NC inhibitor (siLINC00210 + anti-NC)
or LINC00210 siRNA1 and miR-328 inhibitor (siLINC00210 +
anti-miR-328). After transfection, RT-qPCR results demonstrated
that transfection with LINC00210 siRNA significantly increased
miR-328 expression in glioma cells, which was eliminated by
co-transfection with the miR-328 inhibitor (Fig. 4B and C).
CCK-8 and wound healing assays were conducted to
examine cell proliferation and migration. It was found that cell
proliferation was significantly inhibited in the siLINC00210 +
anti-NC group when compared with that in the siNC + anti-NC group,
but no significant difference in cell proliferation was observed
between the siNC + anti-NC group and the siLINC00210 + miR-328
inhibitor group (Fig. 4D and
E). These data indicated that
silencing miR-328 eliminated the inhibitory effects on glioma cell
proliferation induced by LINC00210 knockdown. Similarly, LINC00210
knockdown inhibited glioma cell migration, which was eliminated by
the miR-328 inhibitor (Fig. 4F and
G). Collectively, these data
suggested that knockdown of LINC00210 inhibited glioma cell
proliferation and migration by increasing the expression of
miR-328.
Discussion
To the best of our knowledge, the expression and
function of lncRNA LINC00210 in glioma had not previously been
studied. The present results suggested that LINC00210 was
significantly upregulated in glioma tissues, and high expression of
LINC00210 was significantly associated with advanced clinical stage
and poor prognosis in patients with glioma. It was identified that
knockdown of LINC00210 significantly inhibited the proliferation
and migration of glioma cells. Moreover, LINC00210 could directly
target miR-328 in glioma cells, and miR-328 expression was
negatively correlated with LINC00210 expression in glioma tissues.
Knocking down LINC00210 significantly promoted the expression of
miR-328 in glioma cells. Furthermore, silencing miR-328 impaired
the inhibitory effects of LINC00210 knockdown on the proliferation
and migration of U251 and T98G cells.
It has been well-established that lncRNAs
participate in regulating physiological and pathological processes,
such as tumor development and progression (6,7).
Numerous lncRNAs have been reported to exert important roles in
glioma. For instance, the lncRNA nuclear paraspeckle assembly
transcript 1 (NEAT1) is upregulated in glioma, and NEAT1 inhibition
reduces glioma cell viability, migration and invasion, indicating
that NEAT exerts an oncogenic role in glioma (5).
LINC00210 is a newly discovered lncRNA and has
rarely been studied. Fu et al (17) revealed that LINC00210 was frequently
upregulated in liver cancer tissues and liver tumor-initiating
cells (TICs). These authors also observed that LINC00210 promoted
the self-renewal and tumor initiating capacity of liver TICs by
interacting with β-catenin interacting protein 1 and activating
Wnt/β-catenin signaling (17).
However, to the best of our knowledge, the expression pattern and
function of LINC00210 in glioma have not been previously reported.
The present study revealed for the first time that the expression
of LINC00210 was significantly higher in glioma tissues and cell
lines compared with healthy brain tissues and NHAs. Moreover, it
was identified that high LINC00210 expression was associated with
advanced clinical stage and poor prognosis in patients, suggesting
that upregulation of LINC00210 may promote glioma progression. It
was also demonstrated that knocking down LINC00210 expression via
siRNA significantly inhibited glioma cell proliferation and
migration, indicating that targeting LINC00210 may be a promising
therapeutic strategy for glioma treatment in the future.
miRNAs, a class of small non-coding RNAs, have been
reported to function as key regulators of gene expression via
binding to the 3'-untranslated region of their target mRNAs and
causing translation suppression or RNA degradation (13,19).
Various miRNAs also possess promoting or suppressing roles in
different human cancer types, including glioma (10,20,21).
For instance, miR-365 inhibits proliferation, migration and
invasion of glioma by targeting phosphoinositide-3-kinase
regulatory subunit 3(22), while
miR-25 promotes glioma cell proliferation by targeting cyclin
dependent kinase inhibitor 1C (23).
Previous studies have reported that miR-328 acts as
a tumor suppressor in glioma (24,25).
For example, Wu et al (24)
revealed that miR-328 expression was decreased in high-grade glioma
and was associated with worse survival in primary glioblastoma. The
present results also indicated that the expression of miR-328 was
lower in glioma tissues compared with healthy brain tissues. In
addition, Yuan et al (25)
observed that miR-328 was a favorable prognostic marker in glioma
as it suppressed the invasive and proliferative phenotypes of
glioma cells. Wang et al (26) also reported that enhancer of zeste 2
polycomb repressive complex 2 subunit could promote β-catenin
signaling by inhibiting miR-328 expression, which further promoted
glioma cell proliferation and glucose metabolism. The present study
demonstrated that miR-328 was a target of LINC00210 in glioma
cells, and found a negative correlation between the expression
levels of LINC00210 and miR-328. These findings suggested that the
lower expression of miR-328 may be due to the upregulation of
LINC00210 in glioma. Moreover, it was identified that silencing
miR-328 eliminated the inhibitory effects on glioma cell
proliferation induced by LINC00210 knockdown, suggesting that
LINC00210 serves a promoting role in glioma cell proliferation and
migration by inhibiting miR-328. Similarly, Zhang et al
(27) reported that LINC00210
promoted nasopharyngeal carcinoma tumorigenesis by regulating
miR-328-5p expression and activating the NOTCH3 pathway. The
LINC00210/miR-328 axis may also serve important roles in cancer
types other than glioma and nasopharyngeal cancer, which should be
further studied in the future. However, the limitations of the
present study are that the findings were not confirmed in
vivo in animal models.
In conclusion, the present study demonstrated that
upregulation of LINC00210 promoted glioma progression and predicted
poor prognosis. Furthermore, it was suggested that LINC00210
exerted an oncogenic role in glioma cell proliferation and
migration via regulating miR-328 expression. The results of the
present study may facilitate the development of glioma
treatment.
Acknowledgements
Not applicable.
Funding
This study was supported by the Surface Project of
Hunan Natural Science Foundation (grant no. 2018JJ2607).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW designed this study. HW collected clinical
samples and performed analysis of clinical data. HY performed
statistical analysis. TC, JD and QL performed experiments. ZW and
QL wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Third Xiangya Hospital of Central South University (Hunan, China).
Written informed consents were obtained from patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Peng Z, Liu C and Wu M: New insights into
long noncoding RNAs and their roles in glioma. Mol Cancer.
17(61)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Anjum K, Shagufta BI, Abbas SQ, Patel S,
Khan I, Shah SAA, Akhter N and Hassan SSU: Current status and
future therapeutic perspectives of glioblastoma multiforme (GBM)
therapy: A review. Biomed Pharmacother. 92:681–689. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhou K, Zhang C, Yao H, Zhang X, Zhou Y,
Che Y and Huang Y: Knockdown of long non-coding RNA NEAT1 inhibits
glioma cell migration and invasion via modulation of SOX2 targeted
by miR-132. Mol Cancer. 17(105)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Smolle MA and Pichler M: The role of long
non-coding RNAs in osteosarcoma. Noncoding RNA. 4(7)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xu S, Kong D, Chen Q, Ping Y and Pang D:
Oncogenic long noncoding RNA landscape in breast cancer. Mol
Cancer. 16(129)2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang CJ, Liu C, Wang YX, Zhu N, Hu ZY,
Liao DF and Qin L: Long non-coding RNA-SRA promotes neointimal
hyperplasia and vascular smooth muscle cells proliferation via
MEK-ERK-CREB pathway. Vascul Pharmacol. 116:16–23. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
He D, Zeng H, Chen J, Xiao L, Zhao Y and
Liu N: H19 regulates trophoblastic spheroid adhesion by
competitively binding to let-7. Reproduction. 157:423–430.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ma Z: Downregulation of SETD8 by miR-382
is involved in glioma progression. Pathol Res Pract. 214:356–360.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bo H, Fan L, Gong Z, Liu Z, Shi L, Guo C,
Li X, Liao Q, Zhang W, Zhou M, et al: Upregulation and
hypomethylation of lncRNA AFAP1AS1 predicts a poor prognosis and
promotes the migration and invasion of cervical cancer. Oncol Rep.
41:2431–2439. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jiang H, Xiong W, Chen L, Lv Z, Yang C and
Li Y: Knockdown of the long noncoding RNA HOTTIP inhibits cell
proliferation and enhances cell sensitivity to cisplatin by
suppressing the Wnt/β-catenin pathway in prostate cancer. J Cell
Biochem. 120:8965–8974. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhou S, Yu L, Xiong M and Dai G: lncRNA
SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by
upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res
Commun. 495:1822–1832. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tan P, Guo YH, Zhan JK, Long LM, Xu ML, Ye
L, Ma XY, Cui XJ and Wang HQ: lncRNA-ANRIL inhibits cell senescence
of vascular smooth muscle cells by regulating miR-181a/Sirt1.
Biochem Cell Biol. 97:571–580. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lin C, Zhang Y, Chen Y and Bai Y: Long
noncoding RNA LINC01234 promotes serine hydroxymethyltransferase 2
expression and proliferation by competitively binding miR-642a-5p
in colon cancer. Cell Death Dis. 10(137)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fu X, Zhu X, Qin F, Zhang Y, Lin J, Ding
Y, Yang Z, Shang Y, Wang L, Zhang Q and Gao Q: Linc00210 drives
Wnt/β-catenin signaling activation and liver tumor progression
through CTNNBIP1-dependent manner. Mol Cancer.
17(73)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tan Z, Zhao J and Jiang Y: miR-634
sensitizes glioma cells to temozolomide by targeting CYR61 through
Raf-ERK signaling pathway. Cancer Med. 7:913–921. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xiao F, Li Y, Wan Y and Xue M:
MircroRNA-139 sensitizes ovarian cancer cell to cisplatin-based
chemotherapy through regulation of ATP7A/B. Cancer Chemother
Pharmacol. 81:935–947. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhu Y, Zhao H, Rao M and Xu S:
MicroRNA-365 inhibits proliferation, migration and invasion of
glioma by targeting PIK3R3. Oncol Rep. 37:2185–2192.
2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang J, Gong X, Tian K, Chen D, Sun J,
Wang G and Guo M: miR-25 promotes glioma cell proliferation by
targeting CDKN1C. Biomed Pharmacother. 71:7–14. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu Z, Sun L, Wang H, Yao J, Jiang C, Xu W
and Yang Z: miR-328 expression is decreased in high-grade gliomas
and is associated with worse survival in primary glioblastoma. PLoS
One. 7(e47270)2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yuan J, Zheng Z, Zheng Y, Lu X, Xu L and
Lin L: microRNA-328 is a favorable prognostic marker in human
glioma via suppressing invasive and proliferative phenotypes of
malignant cells. Int J Neurosci. 126:145–153. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang Y, Wang M, Wei W, Han D, Chen X, Hu
Q, Yu T, Liu N, You Y and Zhang J: Disruption of the
EZH2/miRNA/β-catenin signaling suppresses aerobic glycolysis in
glioma. Oncotarget. 7:49450–49458. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang S, Li P, Zhao L and Xu L: LINC00210
as a miR-328-5p sponge promotes nasopharyngeal carcinoma
tumorigenesis by activating NOTCH3 pathway. Biosci Rep.
38(BSR20181168)2018.PubMed/NCBI View Article : Google Scholar
|