Introduction
Colorectal cancer (CRC) is the third most common
malignancy worldwide (1). Despite
advances in therapeutic strategies, the overall survival rate of
patients (15-50%) with CRC remains poor (2,3). CRC
is a complex process characterized by multiple genomic variations
and an aberrant biological microenvironment (4). Although previous studies have reported
that alterations in numerous oncogenes and cancer-suppressor genes
are correlated with CRC, the biological molecular mechanism
underlying CRC development and progression is not completely
understood (5-8).
Therefore, developing an effective strategy for the early diagnosis
and treatment of CRC is important.
Only 2% of the human genome is protein encoding,
whereas the remaining genome consists of non-coding RNA (9,10).
Previous studies have identified the role of non-protein-coding
genes in normal physiological processes and the etiopathogenesis of
diseases, such as cancer (11-13).
Long non-coding RNAs (lncRNAs), which are >200 nucleotides in
length, lack protein coding abilities (14). Numerous lncRNAs are involved in
regulating gene expression, cell differentiation, ontogenetical
processes and epigenetic modification, as well as other cellular
biological processes (15-18).
lncRNAs have also been identified as diagnostic and prognostic
biomarkers, and serve critical roles in cancer progression-related
signaling pathways. For example, lncRNA-linc00152 inhibited by
microRNA (miR)-376c-3p suppresses colorectal cancer cell
proliferation and induces apoptosis (19).
lncRNA-activated by transforming growth factor β
(lncRNA-ATB) has been identified as an oncogene (20). Abnormal expression of lncRNA-ATB is
observed in various malignant tumors, such as breast, colon and
lung cancer, as well as hepatocellular carcinoma (20). For instance, Wei et al
(21) demonstrated that lncRNA-ATB
increased lung cancer cell proliferation and metastasis by
stimulating the p38 signaling pathway. In colon cancer, lncRNA-ATB
promotes cancer metastasis via inhibition of E-cadherin, and serves
as a predictor of poor prognosis (22). Previous studies have reported the
biological functions of lncRNA-ATB in CRC. For example, Gao et
al (23) investigated the role
of lncRNA-ATB in CRC cell proliferation and apoptosis, and
indicated that lncRNA-ATB promotes CRC cell proliferation and
suppresses apoptosis via downregulation of miR-200c. Moreover, Yang
et al (24) reported that
lncRNA-ATB maintained CRC cell stemness by suppressing the
β-catenin signaling pathway. A further previous study identified
that higher expression of lncRNA-ATB was correlated with CRC
metastasis (25). However, to the
best of our knowledge, the mechanism underlying lncRNA-ATB-mediated
promotion of CRC metastasis is not completely understood.
The present study aimed to investigate the
association between the expression of lncRNA-ATB and the clinical
characteristics and prognosis of patients with CRC. The biological
function of lncRNA-ATB in CRC in vitro and in vivo
was also investigated. Therefore, the results of the present study
might provide novel insight into the biological function of
lncRNA-ATB in CRC.
Materials and methods
Tissue samples
A total of 50 tumor and adjacent non-cancerous
tissues (~2 cm distance from the tumor margin) were obtained from
patients (mean age, 58) who underwent tumor resection surgery at
Shenzhen People's Hospital, The Second Clinical Medical College of
Jinan University between February 2017 and September 2019. Patients
did not receive any radiotherapy or chemotherapy prior to surgery,
which was the inclusion criteria. The exclusion criteria were
patients who received radiotherapy or chemotherapy prior to
surgery. All tissues were pathologically confirmed via
histopathology. Written informed consent was obtained from all
patients. The present study was approved by Shenzhen People's
Hospital Medical Ethics Committee (approval no. HH-TD-2017388).
Cell lines
A total of five CRC cell lines (CaCO-2, HT-29,
SW480, LoVo and HCT116 cells) and a normal intestinal mucous cell
line (CCC-HIE-2) were purchased from The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences. The HT-29 cell line
was authenticated as a colorectal cancer cell line by STR
profiling, which was performed by the supplier. Cells were cultured
in DMEM (Sigma-Aldrich; Merck KGaA) supplemented with 10% FBS
(Thermo Fisher Scientific, Inc.), and 1% penicillin and
streptomycin (Sigma-Aldrich; Merck KGaA) at 37˚C with 5%
CO2.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from samples using
TRIzol® (Sigma-Aldrich; Merck KGaA). Total RNA was
reversed transcribed into cDNA using PrimeScript™ RT Master Mix
(Takara Biotechnology Co., Ltd.) according to the manufacturer's
instructions. The temperature protocol using for RT was as follows:
37˚C for 15 min and 85˚C for 5 sec. Subsequently, qPCR was
performed using SYBR-Green Supermix (Bio-Rad Laboratories, Inc.)
and the Bio-Rad CFX 96 RT PCR system (Bio-Rad Laboratories, Inc.).
The thermocycling conditions used for qPCR were as follows: Step 1:
95˚C for 30 sec; step 2: 95˚C for 5 sec, 60˚C for 30 sec (repeated
for 39 cycles); step 3: 95˚C for 10 sec followed by 65˚C to 95˚C in
0.5˚C/5 sec increments. The following primers were used for qPCR:
miR-141-3p (26) forward,
5'-CGTCGCTAACACTGTCTGGTAA-3' and reverse,
5'-GTGCAGGGTCCGAGGTATTC-3'; U6 forward,
5'-GCTTCGGCAGCACATATACTAAAAT-3' and reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3'; lncRNA-ATB (27) forward, 5'-ACAAGCTGTGCAGTCTCAGG-3'
and reverse, 5'-CTAGGCCCAAAGACAATGGA-3'; and GAPDH forward,
5'-AGCAAGAGCACAAGAGGAAG-3' and reverse, 5'-GGTTGAGCACAGGGTACTTT-3'.
mRNA and miRNA expression levels were normalized to the internal
reference genes GAPDH and U6, respectively. Quantification of mRNA
and miRNA expression was performed by using the
2-ΔΔCq method (28).
Transfection
Cells were seeded (2x105 cells/well) into
6-well plates and cultured for 24 h at 37˚C with 5% CO2.
HT-29 cells were transfected with 100 nM of lncRNA-ATB small
interfering (si)RNA (si-ATB; 5'-ATAAGAGCCCTTGGTCCTTAA-3') or
negative control (NC; si-NC; 5'-GATTTACCAGAGAATAATCTA-3'; both
provided by Sigma-Aldrich; Merck KGaA) using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.).
HCT116 cells were transfected with 0.5 µg/µl of lncRNA-ATB
overexpression plasmid or pcDNA3.1 vector (Both overexpression
plasmid and empty vector were purchased from Shanghai GenePharma
Co., Ltd.). HCT116 cells (2x105 cells/well in 6-well
plate) were transfected with 50 nM miR-141-3p mimics
(5'-UAACACUGUCUGGUAAAGAUGG-3') or mimics NC
(5'-AGCCGCACUGUACGAUGCUAUGA-3'; both obtained from Guangzhou
RiboBio Co., Ltd.) using Lipofectamine 3000. At 48 h
post-transfection, cells were used for subsequent experiments.
Cell proliferation assay
Transfected cells were seeded (4x103
cells/well) into 96-well plates. To assess cell proliferation, a
Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of
Biotechnology) was performed according to the manufacturer's
protocol.
For the colony formation assay, transfected cells
were seeded (4x102 cells/well) into 6-well plates and
cultured at 37˚C with 5% CO2 for 2 weeks. Following
staining with 0.2% crystal violet for 30 min at room temperature,
visible colonies (>50 cells) were counted under a light
microscope (x40 magnification).
Migration and invasion assay
Transwell assays were performed to assess CRC cell
migration and invasion. In the Transwell chamber (8-µm pore size;
EMD Millipore), cells (1x104) in 100 µl serum-free
medium were plated in the upper chambers, which were pre-coated
with Matrigel at 37˚C with 5% CO2 for 30 min for the
invasion assay. In the lower chamber, 600 µl culture medium
supplemented with 10% FBS was plated. Following incubation for 24 h
at 37˚C with 5% CO2, cells on the upper surface of the
membrane were removed with a cotton swab. Invading and migratory
cells were fixed with 4% formaldehyde for 15 min at room
temperature and stained with 0.2% crystal violet for 30 min at room
temperature. Stained cells were visualized using a light microscope
(x200 magnification).
Western blotting
Total protein was extracted from cells using RIPA
buffer (Thermo Fisher Scientific, Inc.) and quantified using a
Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific,
Inc.). A total of 50 µg of protein was loaded per lane. Proteins
were separated via 10% SDS-PAGE and transferred to PVDF membranes
(EMD Millipore), which were blocked with 5% skim milk at room
temperature for 1 h. Subsequently, the membranes were incubated
overnight at 4˚C with primary antibodies targeted against:
E-Cadherin (cat. no. 14472; 1:1,000; Cell Signaling Technology,
Inc.), Vimentin (cat. no. 5741; 1:1,000; Cell Signaling Technology,
Inc.) and β-Actin (cat. no. 4970; 1:1,000; Cell Signaling
Technology, Inc.). Following primary incubation, the membranes were
incubated for 1 h at room temperature with HRP-conjugated
anti-mouse IgG (cat. no. 7076; 1:2,000; Cell Signaling Technology,
Inc.) and anti-rabbit IgG (cat. no. 7074; 1:2,000; Cell Signaling
Technology, Inc.) secondary antibodies. Protein bands were
visualized using an enhanced chemiluminescence reagent (Bio-Rad
Laboratories, Inc.) with ChemiDoc MP imaging system (Bio-Rad
Laboratories, Inc.). Densitometry was measured using Image Lab
Touch Software, version 2.4 (Bio-Rad Laboratories, Inc.). β-actin
was used as the loading control.
Tumor formation in nude mice
A total of 12 BALB/c-nu/nu nude mice (male; age, 4-6
weeks; weight, ~20 g) were supplied by SLAC National Accelerator
Laboratory. Mice were housed in climate-controlled (18-23˚C)
research laboratory with 40-60% humidity and 12-h light/dark cycle.
Food and water were accessible at all times. The animal experiments
were approved by Shenzhen People's Hospital Medical Ethics
Committee (approval no. SZRMYY20190211). All animal experiments
were performed under the Guidelines for the Care and Use of
Laboratory Animals of Shenzhen People's Hospital, The Second
Clinical Medical College of Jinan University. The mice were
randomly divided into two groups (n=5 per group), an si-ATB group
injected with si-lncRNA-ATB-transfected HT-29 cells and the other
group was negative control (NC) group, injected with
si-lncRNA-NC-transfected HT-29 cells. si-lncRNA-ATB- or
si-NC-transfected HT-29 cells (5x106) were mixed with
0.2 ml Matrigel Matrix (BD Biosciences) and injected into the
subcutaneous tissue of the back near the right forelimb of the
mouse. After 12 days, the tumors were palpable (~100
mm3). Body weight and tumor size were measured every 3
days using an electronic balance and caliper, respectively. Tumor
volume was calculated according to the following formula:
lengthxwidth2/2. On day 33, mice were anesthetized with
an intraperitoneal injection of ketamine (100 mg/kg) and xylazine
(10 mg/kg), and sacrificed by cervical dislocation. Death was
verified by the absence of vital signs. The tumors were excised for
further analysis.
lncRNA target prediction and dual
luciferase reporter gene assay
lncRNA-ATB target prediction was performed using
StarBase v2.0 (starbase.sysu.edu.cn) (29,30).
pmirGLO-ATB-wild-type (WT) and pmirGLO-ATB-mutant (Mut) were
constructed by Guangzhou RiboBio Co., Ltd. 293 cells were purchased
from the Chinese Academy of Sciences Cell Bank. Cells were seeded
into 96-well plate (1x104 cells/well) and co-transfected
with 200 ng of pmirGLO-ATB-WT or pmirGLO-ATB-Mut and 50 nM of
miR-141-3p mimics (5'-UAACACUGUCUGGUAAAGAUGG-3') or miR-NC
(5'-AGCCGCACUGUACGAUGCUAUGA-3'; both obtained from Guangzhou
RiboBio Co., Ltd.) using Lipofectamine® 3000. At 48 h
post-transfection, relative luciferase activity was measured using
the Dual Luciferase Reporter Assay system (Promega Corporation).
Renilla luciferase activity was used to normalize firefly
luciferase activity.
RNA immunoprecipitation assays
(RIP)
The Ago antibody (cat. no. ab186733, 1:30) and the
normal mouse IgG antibody (cat. no. ab188776; 1:30) were purchased
from Abcam. The RIP experiment was performed using a Magna RIP
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore)
according to the manufacturer's protocol. The normal mouse IgG was
used as the NC.
Immunohistochemistry
Tumors were fixed by using 4% paraformaldehyde at
room temperature for 12 h. An ascending alcohol series was used for
dehydration. Subsequently, tumors were embedded with paraffin.
Xenograft tumors were sliced into ~5 µm thick sections. Sections
were blocked with Immunol staining blocking buffer (cat. no. P0102;
Beyotime Institute of Biotechnology) at room temperature for 1 h
and then incubated with an anti-Ki67 primary antibody (cat. no.
12202; 1:400; Cell Signaling Technology, Inc.) at 4˚C overnight.
Subsequently, sections were incubated with a secondary antibody
(cat. no. 31822; 1:1,000; Invitrogen; Thermo Fisher Scientific,
Inc.) at room temperature for 1 h. A DAB kit (cat. no. P0203;
Beyotime Institute of Biotechnology) was used to identify the
positive rate of Ki67. Images were obtained under a light
microscope (x100 magnification) and analyzed using Image Pro-Plus
software (version 6.0; Media Cybernetics, Inc.).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software (version 7.0; GraphPad Software, Inc.). Data are
presented as the mean ± SD (n=3). Comparisons between tumor and
adjacent non-cancerous tissues were analyzed using a paired
Student's t-test. Comparisons between two groups were analyzed
using an unpaired Student's t-test. Comparisons among multiple
groups were analyzed using one-way ANOVA followed by Tukey's post
hoc test. Patient characteristics were analyzed using the
χ2 test. The relationship between lncRNA-ATB expression
and overall survival was assessed via Kaplan Meier analysis, and
survival curves were compared using the log-rank test. The
correlation between lncRNA-ATB and miR-141-3p expression in CRC
tissues was analyzed using Spearman's correlation analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Upregulation of lncRNA-ATB in CRC
tissues and its implication in cancer progression
RT-qPCR was performed to determine lncRNA-ATB
expression levels in 50 paired CRC tissues and corresponding
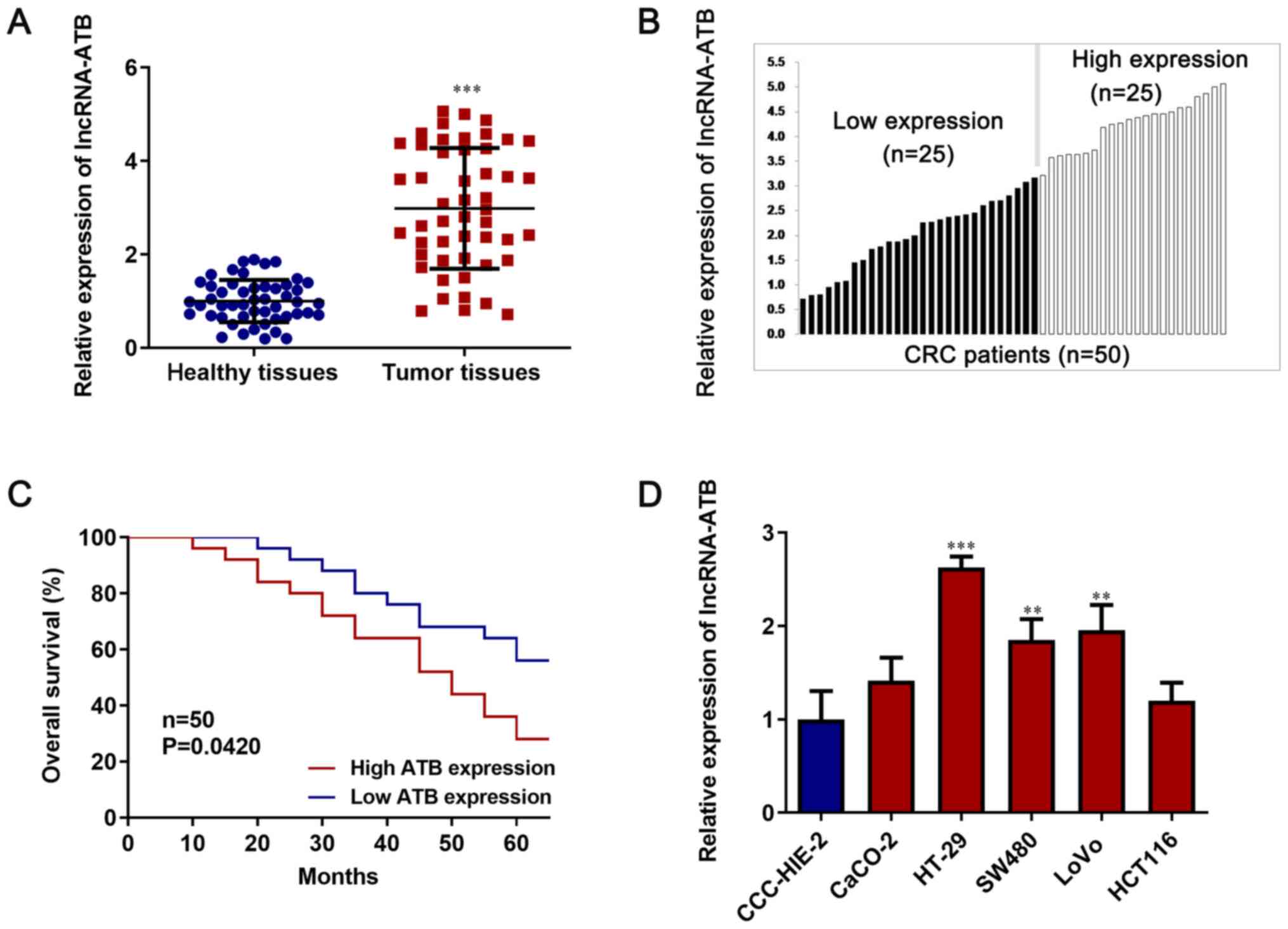
healthy tissues. lncRNA-ATB expression was significantly increased
in CRC tissues compared with healthy tissues (P<0.05; Fig. 1A). Subsequently, patients were
divided into high ATB expression (n=25) and low ATB expression
(n=25) groups, according to the median value (equal to 2.88) of
lncRNA-ATB expression (Fig. 1B).
The association between lncRNA-ATB and several clinical factors of
CRC was analyzed (Table I). Higher
expression of lncRNA-ATB was observed in patients with advanced TNM
stages (P=0.0235) and metastasis (P=0.0235). However, there was no
significant association between lncRNA-ATB and other clinical
parameters, including sex, age and histological
type-differentiation (P>0.05). In addition, patients with higher
lncRNA-ATB expression displayed shorter overall survival, whereas
patients with lower lncRNA-ATB expression displayed longer overall
survival (P<0.05; Fig. 1C).
 | Table IAssociation between lncRNA-ATB
expression and clinicopathological features. |
Table I
Association between lncRNA-ATB
expression and clinicopathological features.
| | Relative lncRNA-ATB
expression level | |
|---|
| Variable | High (n=25) | Low (n=25) | P-value |
|---|
| Gender | | | 0.0801 |
|
Male | 15 | 11 | |
|
Female | 10 | 14 | |
| Age | | | 0.5920 |
|
≤60 | 13 | 14 | |
|
>60 | 12 | 11 | |
| TNM stage | | | 0.0235 |
|
I, II | 8 | 16 | |
|
III, IV | 17 | 9 | |
| Metastasis | | | 0.0227 |
|
No | 7 | 15 | |
|
Yes | 18 | 10 | |
| Histological
type-differentiation | | | 0.2575 |
|
Well | 10 | 14 | |
|
Moderate or
poor | 15 | 11 | |
The relative expression of lncRNA-ATB was examined
among the five CRC cell lines (CaCO-2, HT-29, SW480, LoVo and
HCT116 cells) and the normal intestinal mucous cell line
(CCC-HIE-2). CRC cell lines displayed markedly higher lncRNA-ATB
expression levels compared with the CCC-HIE-2 normal cell line
(P<0.05; Fig. 1D). Moreover,
among the five CRC cell lines, HT-29 cells displayed the highest
expression levels of lncRNA-ATB and HCT116 cells displayed the
lowest lncRNA-ATB expression levels. Therefore, lncRNA-ATB
knockdown was performed in HT-29 cells and lncRNA-ATB
overexpression was established in HCT116 cells.
lncRNA-ATB facilitates CRC cell
proliferation
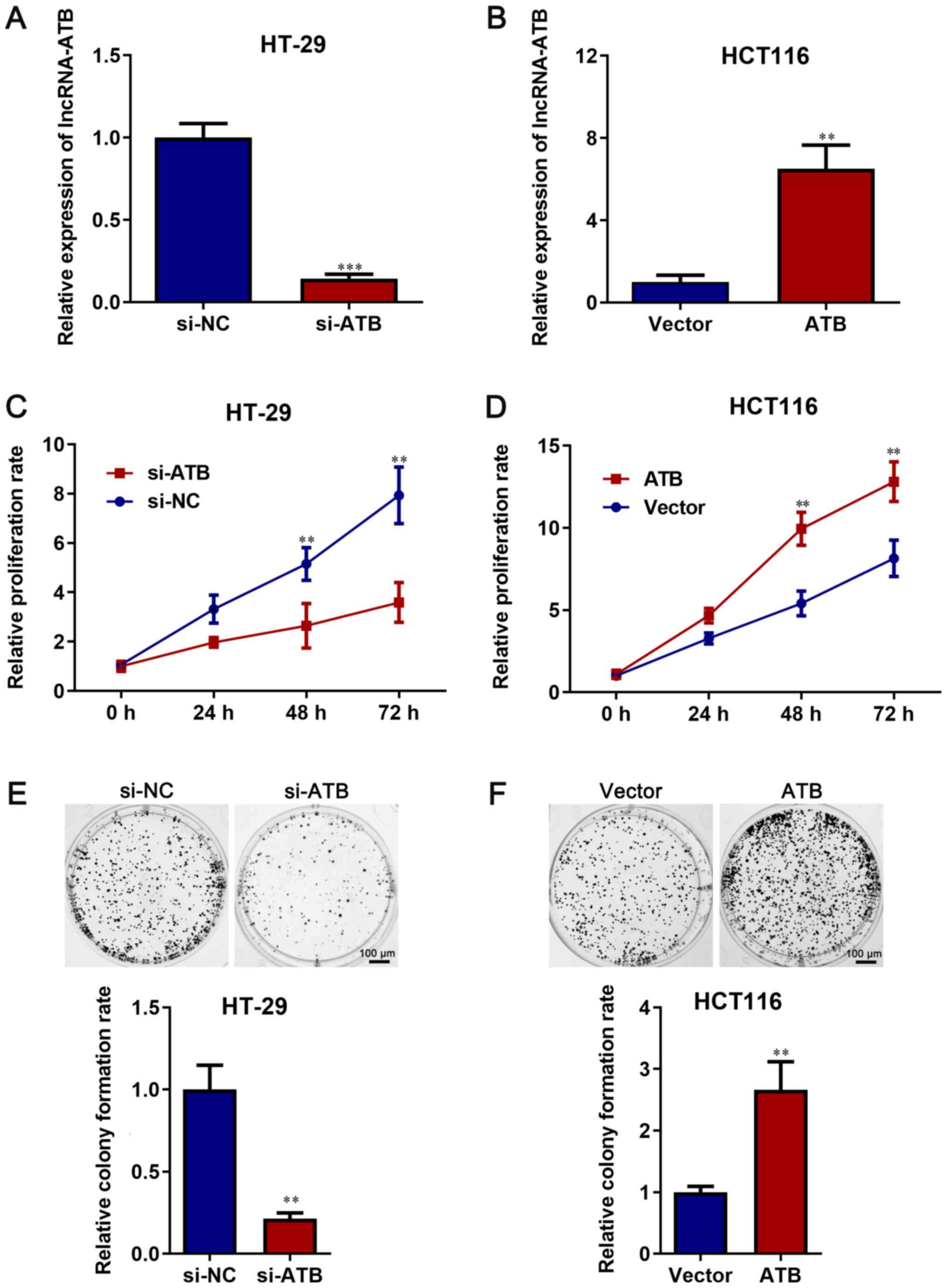
Compared with si-NC, si-lncRNA-ATB significantly
decreased lncRNA-ATB expression in HT-29 cells (P<0.05; Fig. 2A). Compared with vector, lncRNA-ATB
overexpression plasmid significantly increased lncRNA-ATB
expression in HCT116 cells (P<0.05; Fig. 2B).
A CCK-8 assay was performed to assess the role of
lncRNA-ATB in cell proliferation. The results demonstrated that
lncRNA-ATB knockdown significantly decreased HT-29 cell
proliferation compared with si-NC (P<0.05; Fig. 2C). However, lncRNA-ATB
overexpression significantly increased cell proliferation compared
with vector (P<0.05; Fig. 2D).
Moreover, the colony formation rate was significantly reduced by
lncRNA-ATB knockdown in HT-29 cells compared with si-NC, but the
colony formation rate was significantly increased by lncRNA-ATB
overexpression in HCT116 cells compared with vector (P<0.05;
Fig. 2E and F).
lncRNA-ATB increases CRC cell
migration and invasion
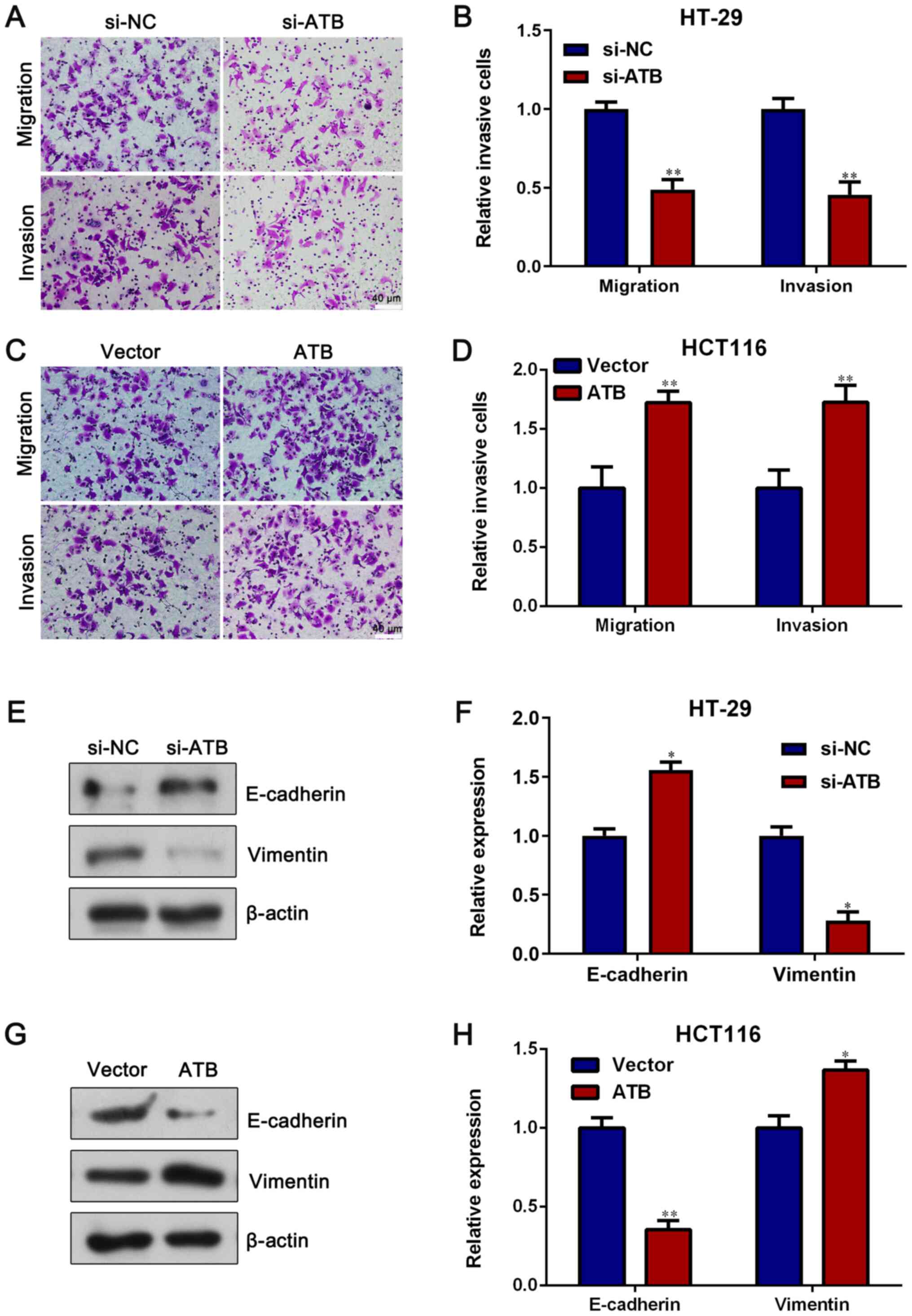
Transwell assays were conducted to assess the role
of lncRNA-ATB in CRC cell metastasis. lncRNA-ATB knockdown
significantly reduced HT-29 cell migration and invasion compared
with si-NC (P<0.05; Fig. 3A and
B). However, lncRNA-ATB
overexpression significantly increased HCT116 cell migration and
invasion compared with vector (P<0.05; Fig. 3C and D). Furthermore, in HT-29 cells, si-ATB
significantly increased the expression of the epithelial protein
E-cadherin, but significantly decreased the expression of the
mesenchymal protein Vimentin compared with si-NC (P<0.05;
Fig. 3E and F). An opposite result was observed in
HCT116 cells transfected with pcDNA3.1-lncRNA-ATB, as lncRNA-ATB
overexpression significantly decreased E-cadherin expression but
significantly increased Vimentin expression compared with vector
(P<0.05; Fig. 3G and H). The results suggested that lncRNA-ATB
had a positive effect on CRC cell migration, invasion and
epithelial-mesenchymal transition.
lncRNA-ATB facilitates xenograft tumor
growth
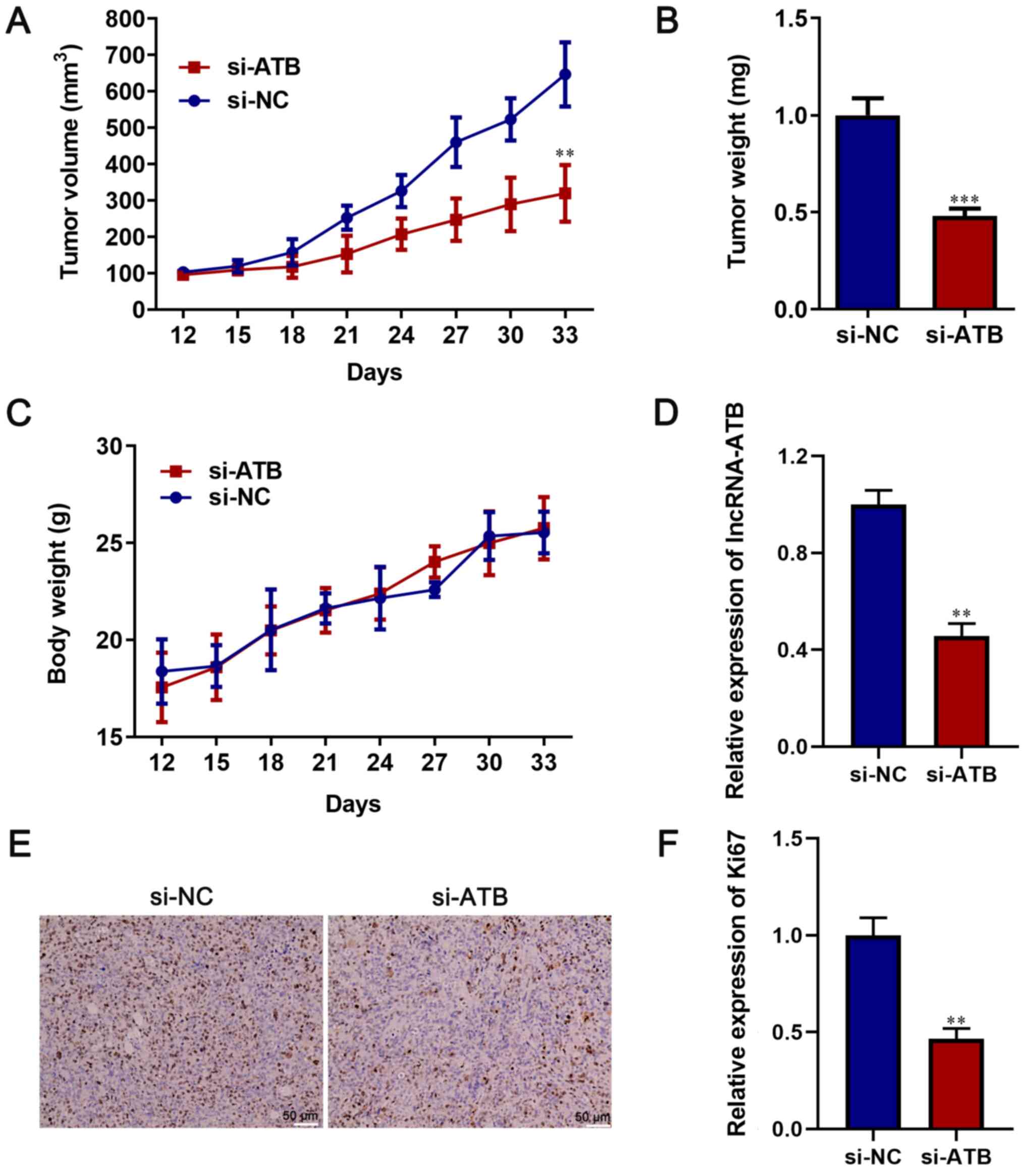
An in vivo study was conducted to assess the
tumorigenic effect of lncRNA-ATB. lncRNA-ATB-knockdown HT-29 cells
significantly decreased tumor diameter, volume and weight compared
with the si-NC group (P<0.05; Figs.
4A, B and S1). However, there was no significant
difference in body weight between the si-lncRNA-ATB and si-NC
groups (P>0.05; Fig. 4C). In
addition, the expression of lncRNA-ATB was measured in the
xenograft tumors via RT-qPCR. lncRNA-ATB expression in the si-ATB
group was significantly decreased compared with the si-NC group
(P<0.05; Fig. 4D). Moreover, the
immunohistochemistry results indicated that the positive rate of
Ki67 was significantly lower in the si-ATB group compared with the
si-NC group, indicating that lncRNA-ATB knockdown inhibited tumor
growth in vivo (P<0.05; Fig.
4E and F). Therefore, the
results suggested that lncRNA-ATB may serve as an oncogene in
CRC.
lncRNA-ATB directly binds to
miR-141-3p
To identify the molecular mechanism underlying
lncRNA-ATB in promoting CRC cell proliferation and metastasis, a
miRNA-lncRNA interaction module was selected. The comprehensive
score of miR-141-3p was higher compared with other miRNAs, and
miR-141-3p has been reported to regulate cancer cell proliferation
and invasion (31-35).
Therefore, miR-141-3p was selected for further study. lncRNA-ATB
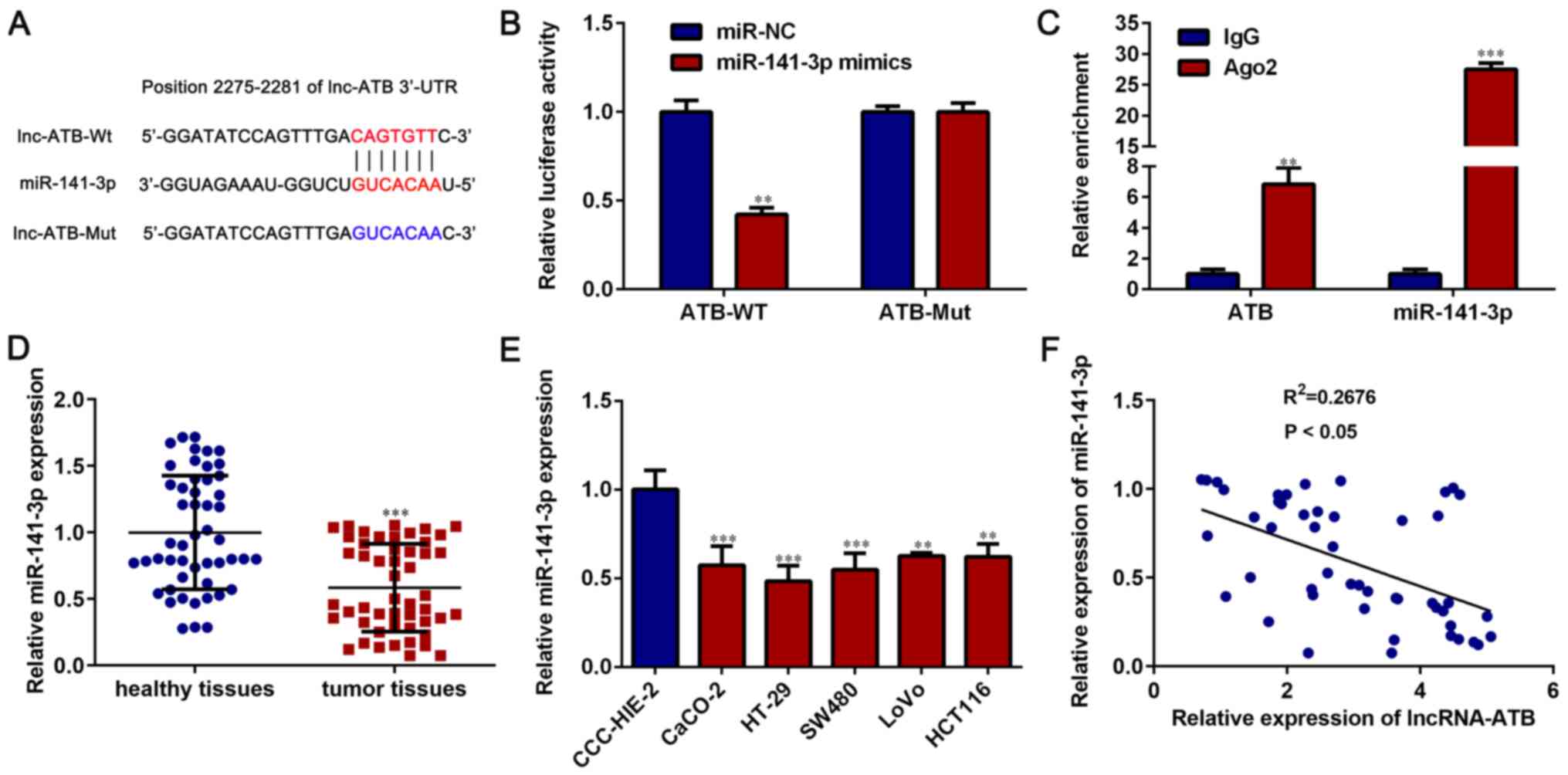
was predicted to bind with miR-141-3p (Fig. 5A). The dual luciferase reporter gene
and RIP assays were performed to examine the interaction between
lncRNA-ATB and miR-141-3p. Compared with miR-NC, miR-141-3p
overexpression significantly downregulated the luciferase activity
of lncRNA-ATB-WT (P<0.05), but had no significant effect on the
luciferase activity of lncRNA-ATB-Mut vector (P>0.05; Fig. 5B). The RIP assay results indicated
that the enrichment of lncRNA-ATB and miR-141-3p by the Ago2
antibody was significantly higher compared with the IgG antibody
(P<0.05; Fig. 5C).
miR-141-3p was significantly downregulated in CRC
tissues compared with paired healthy tissues (P<0.05; Fig. 5D). Moreover, miR-141-3p expression
was significantly lower in CRC cell lines (CaCO-2, HT-29, SW480,
LoVo and HCT116 cells) compared with the normal cell line CCC-HIE-2
(P<0.05; Fig. 5E). The
correlation between lncRNA-ATB and miR-141-3p expression was
assessed in CRC tissues, and the results indicated that lncRNA-ATB
expression was negatively correlated with miR-141-3p expression
(P<0.05; Fig. 5F). The results
indicated that lncRNA-ATB directly bound to miR-141-3p, and
miR-141-3p may serve a vital role in lncRNA-ATB-mediated CRC cell
proliferation and metastasis.
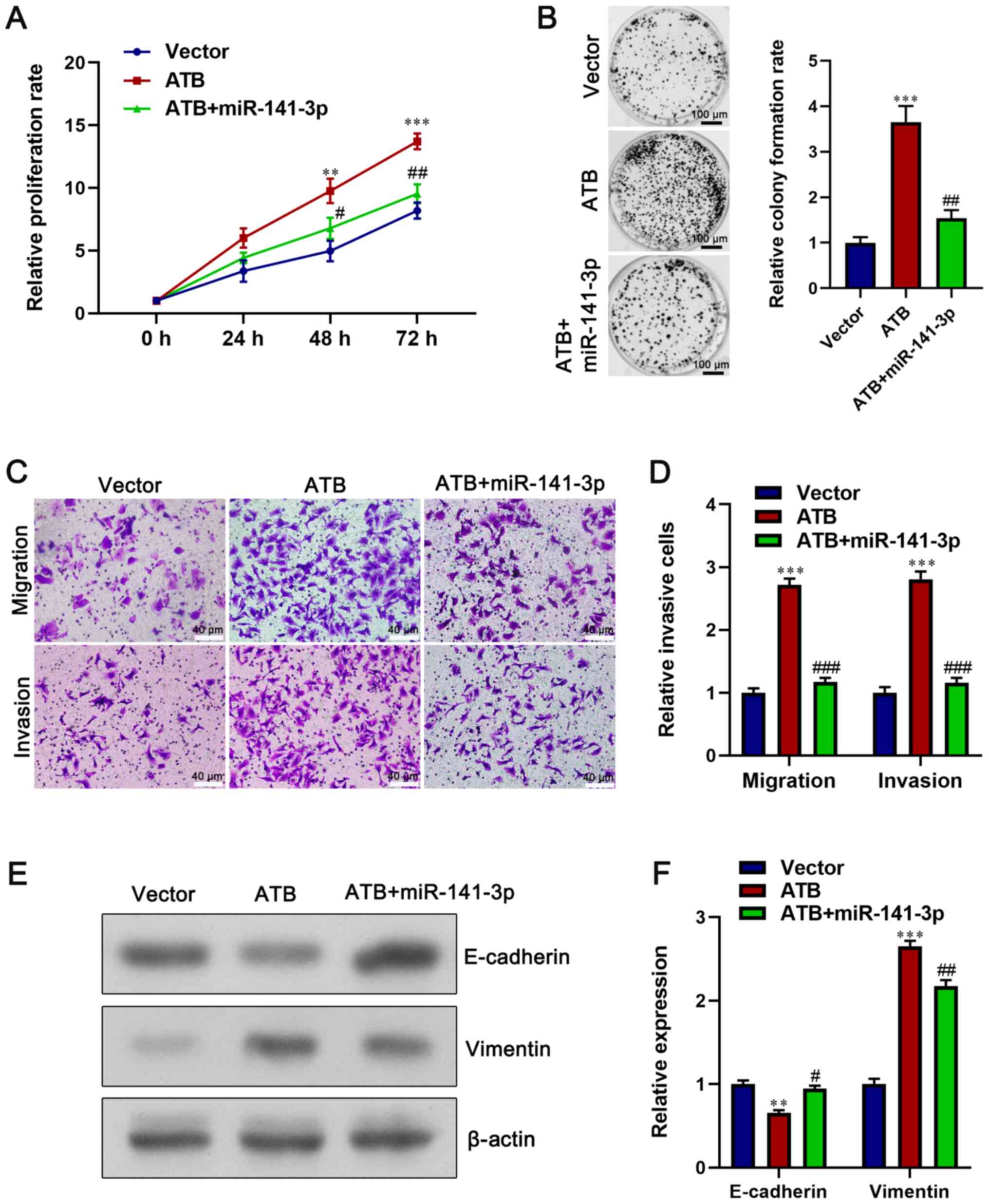
miR-141-3p overexpression reverses the
oncogenic effect of lncRNA-ATB
To further examine the association between
lncRNA-ATB and miR-141-3p, rescue experiments were performed by
transfecting lncRNA-ATB-overexpression HCT116 cells with miR-141-3p
mimics. We confirmed miR-141-3p mimics can significantly increase
the expression of miR-141-3p in HCT116 cells compared with mimics
NC (Fig.S2). The CCK-8 and colony
formation assay results demonstrated that miR-141-3p overexpression
in lncRNA-ATB-overexpression HCT116 cells significantly decreased
cell proliferation compared with lncRNA-ATB-overexpression HCT116
cells (P<0.05; Fig. 6A and
B). In addition, miR-141-3p
overexpression significantly inhibited the migration and invasion
of lncRNA-ATB-overexpression HCT116 cells (P<0.05; Fig. 6C and D). Moreover, miR-141-3p mimics
significantly increased E-cadherin expression and decreased
Vimentin expression in lncRNA-ATB-overexpression HCT116 cells
(P<0.05; Fig. 6E and F). Collectively, the results suggested
that miR-141-3p may serve as an anticancer gene against the
oncogenic effect of lncRNA-ATB in CRC cells.
Discussion
As CRC remains prevalent in China (36), identifying the molecular mechanism
underlying CRC development and progression is an important clinical
challenge. lncRNAs have gained increased research attention
worldwide (37,38), and there is increasing evidence that
lncRNAs are related to cancer development and progression (39,40).
However, the role of lncRNAs in tumors is different, as some serve
as tumor suppressor genes, whilst others serve as oncogenes
(41). A variety of lncRNAs have
been identified to serve a crucial role in CRC (25,42,43),
thus, identifying novel lncRNAs in CRC is necessary.
A previous study reported that the expression of
lncRNA-ATB was upregulated in ovarian cancer tissues and cell lines
compared with adjacent tissue and normal cell lines, and a higher
expression of lncRNA-ATB predicted poorer prognosis of ovarian
cancer (44). Cai et al
(45) demonstrated that lncRNA-ATB
expression in breast cancer tissues was higher compared with
corresponding adjacent healthy tissues. Furthermore, the underlying
molecular mechanism study indicated that lncRNA-ATB competitively
binds miR-98 with E2F transcription factor 5 and facilitates breast
cancer cell migration. However, the value of lncRNA-ATB in the
clinical diagnosis of CRC is not completely understood.
The present study aimed to evaluate the biological
role of lncRNA-ATB in CRC. Firstly, the expression of lncRNA-ATB
was examined in clinical specimens of CRC. The clinical data
demonstrated that lncRNA-ATB was significantly upregulated in CRC
tissues compared with healthy tissues, and higher lncRNA-ATB
expression indicated shorter overall survival in patients with CRC.
In addition, high lncRNA-ATB expression was significantly
associated with advanced TNM stages and tumor metastasis in
patients with CRC.
To further investigate the carcinogenesis role of
lncRNA-ATB in CRC, lncRNA-ATB knockdown and overexpression
experiments were conducted. lncRNA-ATB knockdown inhibited HT-29
cell proliferation, colony formation, migration and invasion
compared with si-NC. However, lncRNA-ATB overexpression facilitated
HCT116 cell proliferation, colony formation, migration and invasion
compared with vector. Moreover, the results indicated that aberrant
E-cadherin and Vimentin expression levels facilitated tumor
metastasis. Therefore, the results of the present study suggested
that lncRNA-ATB suppressed E-cadherin expression and elevated
Vimentin expression, indicating that lncRNA-ATB may serve as an
oncogene in CRC.
Based on the increasing evidence of the competitive
endogenous RNA regulatory network and the role of lncRNAs in
regulatory loops (25-27),
it was hypothesized that lncRNA-ATB may function as a competing
endogenous RNA to modulate cell-associated biological process.
Therefore, bioinformatics analysis and dual luciferase reporter
gene assays were performed to predict the possible target of
lncRNA-ATB. The results indicated that miR-141-3p bound to the
3'-untranslated region of lncRNA-ATB. In addition, the RIP assay
results identified the endogenous interaction between lncRNA-ATB
and miR-141-3p.
miR-141-3p downregulation has been reported in a
variety of tumors, such as prostate cancer and non-small cell lung
cancer, and facilitates cancer development and progression via
various mechanisms (31-35).
For example, Huang et al (31) revealed that miR-141-3p expression
was lower in prostate cancer with bone metastasis compared with
prostate cancer with non-bone metastasis, and restoration of
miR-141-3p reduced prostate cancer metastasis by inactivating the
NF-κB signaling pathway. Moreover, Sun and Zhang (46) reported that lncRNA-X inactive
specific transcript could facilitate pancreatic cancer cell
proliferation, migration and invasion via targeting miR-141-3p. It
has also been reported that miR-141-3p suppresses CRC cell
proliferation, mobility and invasion by inhibiting TNF receptor
associated factor-5(47). In the
present study, miR-141-3p was significantly downregulated in CRC
tissues and cell lines compared with healthy tissues and cells,
respectively. In addition, a negative correlation between
miR-141-3p and lncRNA-ATB expression was identified in CRC tissues,
suggesting that miR-141-3p may serve as an anticancer gene in
CRC.
In conclusion, the results of the present study
indicated that lncRNA-ATB was upregulated in CRC tissues and cell
lines compared with healthy tissues and cells, respectively. The
results also suggested that lncRNA-ATB promoted cancer cell
proliferation, migration and invasion. Therefore, the present study
suggested a critical role of lncRNA-ATB in CRC development and
progression, and provided novel insights into the potential
development of lncRNA-based targeted therapeutic strategies for CRC
therapy.
Supplementary Material
Figure S1. Alterations in the long and
short tumor diameter from day 12 to day 33.
***P<0.001 vs. NC-long diameter;
###P<0.001 vs. NC-short diameter. NC, negative
control; si, small interfering RNA.
Figure S2. Transfection efficiency of
miR-141-3p mimics in HCT116 cells. ***P<0.001 vs.
mimics NC. miR, microRNA; NC, negative control.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and CW wrote the manuscript, performed the
experiments, collected clinical samples and analyzed the data. CW
designed the study and revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent. The
present study was approved by the Institutional Review Boards of
the Shenzhen People's Hospital, the Second Clinical Medical College
of Jinan University (approval nos. HH-TD-2017388 and
SZRMYY20190211).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Roncucci L and Mariani F: Prevention of
colorectal cancer: How many tools do we have in our basket? Eur J
Intern Med. 26:752–756. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Brody H: Colorectal cancer. Nature.
521(S1)2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Angelova M, Charoentong P, Hackl H,
Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik
B, Galon J and Trajanoski Z: Characterization of the
immunophenotypes and antigenomes of colorectal cancers reveals
distinct tumor escape mechanisms and novel targets for
immunotherapy. Genome Biol. 16(64)2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Obuch JC and Ahnen DJ: Colorectal cancer:
Genetics is changing everything. Gastroenterol Clin North Am.
45:459–476. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang JY, Hsieh JS, Chang MY, Huang TJ,
Chen FM, Cheng TL, Alexandersen K, Huang YS, Tzou WS and Lin SR:
Molecular detection of APC, K- ras, and p53 mutations in the serum
of colorectal cancer patients as circulating biomarkers. World J
Surg. 28:721–726. 2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wu C, Zhu X, Tao K, Liu W, Ruan T, Wan W,
Zhang C and Zhang W: MALAT1 promotes the colorectal cancer
malignancy by increasing DCP1A expression and miR203
downregulation. Mol Carcinog. 57:1421–1431. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Yu Y, Liu D, Liu Z, Li S, Ge Y, Sun W and
Liu B: The inhibitory effects of COL1A2 on colorectal cancer cell
proliferation, migration, and invasion. J Cancer. 9:2953–2962.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen LL and Carmichael GG: Long noncoding
RNAs in mammalian cells: What, where, and why? Wiley Interdiscip
Rev RNA. 1:2–21. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151.
2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Arase M, Horiguchi K, Ehata S, Morikawa M,
Tsutsumi S, Aburatani H, Miyazono K and Koinuma D: Transforming
growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse
breast cancer JygMC(A) cells. Cancer Sci. 105:974–982.
2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
An X, Sarmiento C, Tan T and Zhu H:
Regulation of multidrug resistance by microRNAs in anti-cancer
therapy. Acta Pharm Sin B. 7:38–51. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wu Z, Liu X, Liu L, Deng H, Zhang J, Xu Q,
Cen B and Ji A: Regulation of lncRNA expression. Cell Mol Biol
Lett. 19:561–575. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ghazal S, McKinnon B, Zhou J, Mueller M,
Men Y, Yang L, Mueller M, Flannery C, Huang Y and Taylor HS: H19
lncRNA alters stromal cell growth via IGF signaling in the
endometrium of women with endometriosis. EMBO Mol Med. 7:996–1003.
2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li
YJ, Yan B and Jiang Q: Pathogenic role of lncRNA-MALAT1 in
endothelial cell dysfunction in diabetes mellitus. Cell Death Dis.
5(e1506)2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rodríguez-Malavé NI, Fernando TR, Patel
PC, Contreras JR, Palanichamy JK, Tran TM, Anguiano J, Davoren MJ,
Alberti MO, Pioli KT, et al: BALR-6 regulates cell growth and cell
survival in B-lymphoblastic leukemia. Mol Cancer.
14(214)2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang YH, Fu J, Zhang ZJ, Ge CC and Yi Y:
LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability
and promotes apoptosis of colorectal cancer cells. Am J Transl Res.
8:5286–5297. 2016.PubMed/NCBI
|
|
20
|
Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y
and Jiang X: LncRNA-ATB: An indispensable cancer-related long
noncoding RNA. Cell Prolif. 50(e12381)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wei L, Wu T, He P, Zhang JL and Wu W:
LncRNA ATB promotes the proliferation and metastasis of lung cancer
via activation of the p38 signaling pathway. Oncol Lett.
16:3907–3912. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu
F, Peng Z and Yan D: LncRNA-ATB mediated E-cadherin repression
promotes the progression of colon cancer and predicts poor
prognosis. J Gastroenterol Hepatol. 31:595–603. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gao Z, Zhou H, Wang Y, Chen J and Ou Y:
Regulatory effects of lncRNA ATB targeting miR-200c on
proliferation and apoptosis of colorectal cancer cells. J Cell
Biochem. 121:332–343. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yang X, Tao H, Wang C, Chen W, Hua F and
Qian H: lncRNA-ATB promotes stemness maintenance in colorectal
cancer by regulating transcriptional activity of the β-catenin
pathway. Exp Ther Med. 19:3097–3103. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Iguchi T, Uchi R, Nambara S, Saito T,
Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, et
al: A long noncoding RNA, lncRNA-ATB, is involved in the
progression and prognosis of colorectal cancer. Anticancer Res.
35:1385–1388. 2015.PubMed/NCBI
|
|
26
|
Zhang Y, Li J, Jia S, Wang Y, Kang Y and
Zhang W: Down-regulation of lncRNA-ATB inhibits
epithelial-mesenchymal transition of breast cancer cells by
increasing miR-141-3p expression. Biochem Cell Biol. 97:193–200.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ma CC, Xiong Z, Zhu GN, Wang C, Zong G,
Wang HL, Bian EB and Zhao B: Long non-coding RNA ATB promotes
glioma malignancy by negatively regulating miR-200a. J Exp Clin
Cancer Res. 35(90)2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li S, Hao J, Hong Y, Mai J and Huang W:
Long non-coding RNA NEAT1 promotes the proliferation, migration,
and metastasis of human breast-cancer cells by inhibiting
miR-146b-5p expression. Cancer Manag Res. 12:6091–6101.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42 (Database Issue):D92–D97. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yang JH, Li JH, Shao P, Zhou H, Chen YQ
and Qu LH: starBase: A database for exploring microRNA-mRNA
interaction maps from Argonaute CLIP-Seq and Degradome-Seq data.
Nucleic Acids Res. 39 (Database Issue):D202–D209. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Huang S, Wa Q, Pan J, Peng X, Ren D, Huang
Y, Chen X and Tang Y: Downregulation of miR-141-3p promotes bone
metastasis via activating NF-κB signaling in prostate cancer. J Exp
Clin Cancer Res. 36(173)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhou Y, Zhong JH, Gong FS and Xiao J:
MiR-141-3p suppresses gastric cancer induced transition of normal
fibroblast and BMSC to cancer-associated fibroblasts via targeting
STAT4. Exp Mol Pathol. 107:85–94. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li W, Cui Y, Wang D, Wang Y and Wang L:
MiR-141-3p functions as a tumor suppressor through directly
targeting ZFR in non-small cell lung cancer. Biochem Biophys Res
Commun. 509:647–656. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu CZ, Ye ZH, Ma J, He RQ, Liang HW, Peng
ZG and Chen G: A qRT-PCR and gene functional enrichment study
focused on downregulation of miR-141-3p in hepatocellular carcinoma
and its clinicopathological significance. Technol Cancer Res Treat.
16:835–849. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Fang M, Huang W, Wu X, Gao Y, Ou J, Zhang
X and Li Y: MiR-141-3p suppresses tumor growth and metastasis in
papillary thyroid cancer via targeting Yin Yang 1. Anat Rec
(Hoboken). 302:258–268. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yin J, Bai Z, Zhang J, Zheng Z, Yao H, Ye
P, Li J, Gao X and Zhang Z: Burden of colorectal cancer in China,
1990-2017: Findings from the Global Burden of Disease Study 2017.
Chin J Cancer Res. 31:489–498. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Han D, Wang M, Ma N, Xu Y, Jiang Y and Gao
X: Long noncoding RNAs: Novel players in colorectal cancer. Cancer
Lett. 361:13–21. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y and
Bai XZ: LncRNA-ATB promotes trastuzumab resistance and
invasion-metastasis cascade in breast cancer. Oncotarget.
6:11652–11663. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao
Z, Zhi H, Wang T, Guo Z and Li X: Identification of
lncRNA-associated competing triplets reveals global patterns and
prognostic markers for cancer. Nucleic Acids Res. 43:3478–3489.
2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Guzel E, Okyay TM, Yalcinkaya B,
Karacaoglu S, Gocmen M and Akcakuyu MH: Tumor suppressor and
oncogenic role of long non-coding RNAs in cancer. North Clin
Istanb. 7:81–86. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liao Q, He W, Liu J, Cen Y, Luo L, Yu C,
Li Y, Chen S and Duan S: Identification and functional annotation
of lncRNA genes with hypermethylation in colorectal cancer. Gene.
572:259–265. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Shi J, Li X, Zhang F, Zhang C, Guan Q, Cao
X, Zhu W, Zhang X, Cheng Y, Ou K, et al: Circulating lncRNAs
associated with occurrence of colorectal cancer progression. Am J
Cancer Res. 5:2258–2265. 2015.PubMed/NCBI
|
|
44
|
Yang XS, Wang GX and Luo L: Long
non-coding RNA SNHG16 promotes cell growth and metastasis in
ovarian cancer. Eur Rev Med Pharmacol Sci. 22:616–622.
2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Cai C, Huo Q, Wang X, Chen B and Yang Q:
SNHG16 contributes to breast cancer cell migration by competitively
binding miR-98 with E2F5. Biochem Biophys Res Commun. 485:272–278.
2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Sun J and Zhang Y: LncRNA XIST enhanced
TGF-β2 expression by targeting miR-141-3p to promote pancreatic
cancer cells invasion. Biosci Rep. 39(BSR20190332)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liang Z, Li X, Liu S, Li C, Wang X and
Xing J: MiR-141-3p inhibits cell proliferation, migration and
invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys
Res Commun. 514:699–705. 2019.PubMed/NCBI View Article : Google Scholar
|