Introduction
Among adults in China, the estimated overall
prevalence of diabetes and pre-diabetes was 10.9 and 35.7% in 2013,
respectively (1). Although diabetes
causes severe complications and has been one of the most important
public health problems worldwide, its undetermined etiology means
that the prevention and treatment of the disease remains
challenging (2).
Diabetes is an inflammatory disease (3), in which pancreatic islet cells are
usually in a state of inflammation triggered by active macrophages
or lymphocytes (4,5). Leukocyte invasion and activation in
islet cells may inhibit insulin production or cause islet apoptosis
(6). However, to the best of our
knowledge, the molecular targets and main contributors, as well as
the reason inflammation affects islet function, remains
unknown.
MicroRNAs (miRNAs/miRs) target the 3'-untranslated
region of mRNAs and regulate protein translation (7). miRNAs are emerging as important tools
for understanding the molecular mechanisms and etiology of various
diseases (8). miRNAs serve
important roles in the dialogue between immune or inflammatory
systems and pancreatic endocrine cells (9). However, few systematic investigations
of this process have been conducted.
The present study performed proteomics and
miRNA-omics to investigate pancreatic islet cell inflammation.
Based on systematic changes of proteins and miRNAs, a network of
molecules involved in islet inflammation was constructed to
determine the potential molecular mechanisms underlying islet
inflammation and diabetes.
Materials and methods
Cell culture
Cell culture, inflammation induction and sample
collection were performed using methods as previously described
(10). RAW264.7 and Beta-TC-6 cells
were provided by the Cell Resource Centre of the Shanghai
Institutes for Biological Science, Chinese Academy of Sciences,
China. Each cell line was cultured in high glucose DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (PAN
Biotech UK, Ltd.) and 1% penicillin-streptomycin antibiotics
(Gibco; Thermo Fisher Scientific, Inc.). The cells were incubated
in a humidified atmosphere with 5% CO2 at 37˚C. The
cells were seeded into 6-well plates at a density of
2.5x105 cells/well.
Inflammation was induced in RAW264.7 cells by adding
2 µg/ml lipopolysaccharide (LPS; cat. no. L4391; Sigma-Aldrich;
Merck KGaA) after 12 h of incubation at 37˚C. Subsequently, the
supernatant of LPS-induced RAW264.7 cell medium (LRM) was
collected. LRM usually markedly increases levels of IL-1A (~20
pg/ml), IL-6 (~800 pg/ml) and TNF-α (~1,200 pg/ml) in cell medium
(10). In Beta-TC-6 cells,
inflammation was induced for 24 h by adding a mixture of LRM and
common medium (DMEM with 10% FBS) at a ratio of 1:3 (v:v).
To obtain protein samples, Beta-TC-6 cells in the
6-well plates were washed with ice-cold PBS twice. A 200 µl aliquot
of cell lysis buffer (50 mM Tris-HCl, 4 M urea and 1% Triton X-100;
pH 8.0) was subsequently added to the wells. Cell samples were
transferred into 1 ml Eppendorf tubes for further protein
extraction and immediately stored at -80˚C for cell protein
assays.
To obtain RNA samples, Beta-TC-6 cells in the 6-well
plates were collected at 24 h after LRM induction. Cells were
washed twice with ice-cold PBS and 1 ml TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) was added to the
wells. Subsequently, RNA extraction was performed in 1 ml Eppendorf
tubes according to the instruction manual of the TRIzol®
kit
Protein assay
Protein extraction, digestion, isobaric tags for
relative and absolute quantitation (iTRAQ) labeling and liquid
chromatography with tandem mass spectrometry (MS/MS) were performed
in LRM-treated Beta-TC-6 cells and untreated control cells after 24
h of LRM treatment at 37˚C by PTM Biolab LLC as previously
described (11). Raw data from
MS/MS were processed for peptide identification by searching the
Maxquant database (12), and the
resultant peptides were assembled as proteins as previously
described (13). The ion intensity
of the iTRAQ reporter in each sample was used for quantitation
analysis and comparison (14).
Proteins with a fold expression change >1.5 for upregulation or
<0.67 for downregulation between LRM-treated Beta-TC-6 cells and
LRM-untreated control cells were selected for further analysis.
Protein-protein interaction analysis was performed
using the Search Tool for the Retrieval of Interacting
Genes/Proteins database (v10; http://string-db.org/) as previously described
(15). Protein signaling pathways
were annotated using the Kyoto Encyclopedia of Genes and Genomes
(KEGG) database (https://www.kegg.jp/kegg/pathway.html). A two-tailed
Fisher's exact test was used to test the enrichment of the altered
proteins against all identified proteins. P<0.05 was considered
to indicate a statistically significant difference.
RNA isolation, library preparation and
sequencing
RNA isolation, library preparation and sequencing
were performed in LRM-treated Beta-TC-6 cells and untreated control
cells after 24 h of LRM treatment at 37˚C by Novogene Co., Ltd. as
previously described (16). After
total RNA was extracted, RNA degradation and contamination were
monitored on 1% agarose gels. RNA purity was checked using the
NanoPhotometer® spectrophotometer (Implen GmbH) and RNA
concentrations were measured using a Qubit® RNA assay
kit on a Qubit® 2.0 Flurometer (each, Thermo Fisher
Scientific, Inc.). RNA integrity was assessed using the RNA Nano
6000 assay kit and the Agilent Bioanalyzer 2100 system (each,
Agilent Technologies, Inc.).
Total RNA in quantities of 3 µg per sample was used
as input material for the small RNA library. Sequencing libraries
were generated using the NEBNext® Multiplex Small RNA
Library Prep Set for Illumina® (New England BioLabs,
Inc.) according to the manufacturer's protocol. Index codes were
added to attribute sequences to each sample.
The clustering of index-coded samples was performed
on a cBot Cluster Generation system (Illumina, Inc.) using the
TruSeq SR Cluster kit v3-cBot-HS (Illumina, Inc.) according to the
manufacturer's protocol. Following cluster generation, library
preparations were sequenced on an Illumina HiSeq 2500/2000 platform
(Illumina, Inc.) and 50 bp single-end reads were generated.
Data analysis
Raw data (raw reads) in the fastq format were first
processed using custom perl and python scripts. During this step,
clean data (clean reads) were obtained by removing those containing
poly-N with 5' adapter contaminants, without the 3' adapter or the
insert tag. Those containing poly A, T, G or C, and low-quality
reads from raw data were also removed. At the same time, Q20, Q30
and GC-content of the raw data were calculated. Subsequently, a
range of length was selected from the clean reads to perform all
downstream analyses. The small RNA tags were mapped to a reference
sequence using Bowtie (bowtie-0.12.9; http://bowtie-bio.sourceforge.net/index.shtml) without
mismatch to analyze their expression and distribution on the
reference (17). Mapped small RNA
tags were used to identify known miRNAs. miRbase 20.0 was used as
reference (http://www.mirbase.org). Modified
mirdeep2 software (mirdeep2_0_0_5; https://github.com/rajewsky-lab/mirdeep2) and
srna-tools-cli (http://srna-tools.cmp.uea.ac.uk/) were used to obtain
the potential miRNAs and draw the resultant secondary structures
(18). Custom scripts were used to
obtain the miRNA counts as well as base bias (on the first position
of the identified miRNA with a certain length and on each position
of all identified miRNAs), respectively. miRNA expression levels
were estimated in terms of transcript per million using the
following criteria (19):
Normalization formula: Normalized expression=mapped read
count/total reads x1,000,000. For samples without biological
replicates, differential expression analysis of two samples was
performed using the DEGseq (2010) R package (version 1.2.2)
(20). P-values were adjusted using
the q-value (21). A q-value
<0.01 and log2 (fold change) >1 was set as the threshold for
significant differential expression by default. miRNAs with
log2.fold change values ≥0.5 or ≤-0.5 between LRM-treated Beta-TC-6
cells and control cells were selected for further analysis. Taking
into account abundance and fold changes in miRNA expression, miRNAs
with an abundance value >1,000 and a fold change >2 were
selected for further reverse transcription-quantitative PCR
(RT-qPCR) validation.
NF-κB inhibition
As the NF-κB signaling pathway serves an important
role in the activation of inflammation (22), the present study used an NF-κB
inhibitor to investigate whether NF-κB mediated miRNA changes.
After Beta-TC-6 cells were incubated for 12 h, inflammation was
induced by LRM as aforementioned. Simultaneously, 10 µM NF-κB
inhibitor (MLN120B; MedChemExpress) was added. Following treatment
with MLN120B for 12 h at 37˚C, total RNA was extracted from
Beta-TC-6 cells for miRNA or mRNA detection according to the
aforementioned protocol.
miRNA transfection
The present study used miR-21a-5p and miR-146a-5p
inhibitors to determine whether these miRNAs contributed to islet
dysfunction as previously described (10). miRNA inhibitors (miR-21a-5p
inhibitor, 5'-UCAACAUCAGUCUGAUAAGCUA-3'; miR-146a-5p inhibitor,
5'-AACCCAUGGAAUUCAGUUCUCA-3') and negative inhibitor control
(5'-CAGUACUUUUGUGUAGUACAAA-3') were synthesized by Guangzhou
RiboBio Co., Ltd. Beta-TC-6 cells were seeded at a density of
2.5-5x105 per well into 6-well-plates and incubated for
12 h at 37˚C. Cells were transferred to fresh medium (DMEM with 10%
FBS) and transfected with miRNA inhibitors at a concentration of 60
pmol/well using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
After 6-12 h of transfection at 37˚C, inflammation was induced for
24 h at 37˚C by LRM as aforementioned. Total RNA was extracted from
cells for miRNA or mRNA detection according to the aforementioned
protocol after 24 h of inflammation induction.
RT-qPCR
miRNA and mRNA qPCR was performed as previously
described (10,23). To determine the expression of
miR-21a-5p and miR-146a-5p in Beta-TC-6 cells, an miRNA assay kit
(Shanghai GenePharma Co., Ltd.) was used according to the
manufacturer's protocol. The U6 gene was utilized as an internal
control for normalization. The primers used for miRNA analysis are
provided in Table I. RT was
conducted using a DNA Engine H Peltier Thermal Cycler (Bio Rad
Laboratories, Inc.) under the following temperature protocol: 25˚C
for 30 min, 42˚C for 30 min and 85˚C for 5 min, followed by a
holding step at 4˚C. The primers for the mRNA assay were according
to PrimerBank (https://pga.mgh.harvard.edu/primerbank/) and
synthesized by Genewiz, Inc. (Table
II). Actin was used as an internal control for normalization.
RT of mRNA samples extracted from Beta-TC-6 cells was performed
using a PrimeScript™ 1st Strand cDNA Synthesis kit (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
RT was conducted using a DNA Engine H Peltier Thermal Cycler with
the following conditions: 16˚C for 30 min, 42˚C for 30 min and 85˚C
followed by a hold step at 4˚C. All qPCR assays including miRNAs
and mRNAs were performed using SYBR® Green I dye (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol
(95˚C for 3 min, followed by 40 cycles of 95˚C for 12 sec and 60˚C
for 40 sec) using an ABI PRISM 7300 Real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). To determine the
unicity of the transcript analysis, dissociation curve analysis of
amplification products was performed and it was confirmed that only
one peak (PCR amplification product) was observed in each curve.
The fold change was calculated using the 2-ΔΔCq method
of relative quantification (24).
All experiments were performed in triplicate.
 | Table ImiRNA primers for reverse
transcription-quantitative PCR. |
Table I
miRNA primers for reverse
transcription-quantitative PCR.
| miRNAs | Primer sequence (5'
to 3') |
|---|
| mmu-miR-21a-5p | F:
TCGCCCGTAGCTTATCAGACT |
| | R:
CAGAGCAGGGTCCGAGGTA |
|
mmu-miR-146a-5p | F:
CTGCCGCTGAGAACTGAATT |
| | R:
CAGAGCAGGGTCCGAGGTA |
| U6 snRNA | F:
CGCTTCGGCAGCACATATAC |
| | R:
TTCACGAATTTGCGTGTCATC |
 | Table IImRNAs primers for reverse
transcription-quantitative PCR. |
Table II
mRNAs primers for reverse
transcription-quantitative PCR.
| Gene name | NCBI accession
No. | Primer sequence (5'
to 3') | Size (bp) |
|---|
| Mouse IL-1A | NP_034684 | Forward:
TCTGCCATTGACCATCTC | 182 |
| | | Reverse:
ATCTTCCCGTTGCTTGAC | |
| Mouse TNF-α | NP_038721 | Forward:
GGGCTTCCAGAACTCCA | 213 |
| | | Reverse:
GCTACAGGCTTGTCACTCG | |
| Mouse Nkx-2.2 | NP_035049 | Forward:
CCGGGCGGAGAAAGGTATG | 156 |
| | | Reverse:
CTGTAGGCGGAAAAGGGGA | |
| Mouse Pdx-1 | NP_032840 | Forward:
CCCCAGTTTACAAGCTCGCT | 177 |
| | | Reverse:
CTCGGTTCCATTCGGGAAAGG | |
| Mouse Gcg | NP_032126 | Forward:
TTACTTTGTGGCTGGATTGCTT | 149 |
| | | Reverse:
AGTGGCGTTTGTCTTCATTCA | |
| Mouse Pax-6 | NP_001231129 | Forward:
GCAGATGCAAAAGTCCAGGTG | 285 |
| | | Reverse:
CAGGTTGCGAAGAACTCTGTTT | |
| Mouse Snap25 | NP_035558 | Forward:
CAACTGGAACGCATTGAGGAA | 177 |
| | | Reverse:
GGCCACTACTCCATCCTGATTAT | |
| Mouse Mafa | NP_919331 | Forward:
AGGAGGGTCATCCGACTG | 113 |
| | | Reverse:
CTTCTCGCTCCAGAATGTG | |
| Mouse Ins2 | NP_001172013 | Forward:
GCTTCTTCTACACACCCATGTC | 147 |
| | | Reverse:
AGCACTGATCTACAATGCCAC | |
| Mouse Actin | NP_031419 | Forward:
GTGACGTTGACATCCGTAAAGA | 245 |
| | | Reverse:
GCCGGACTCATCGTACTCC | |
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5 software (version 5.01; GraphPad Software, Inc.) as
previously described (10). Data
are presented as the mean ± SD. A two tailed unpaired t test was
used to evaluate the statistical significance of data. P<0.05
was considered to indicate a statistically significant
difference.
Results
Protein assay
Based on the results of proteomics, proteins with a
fold expression change >1.5 for upregulation or <0.67 for
downregulation between LRM-treated Beta-TC-6 cells and control
cells were selected for further analysis. It was revealed that 87
proteins were upregulated and 42 proteins were downregulated in
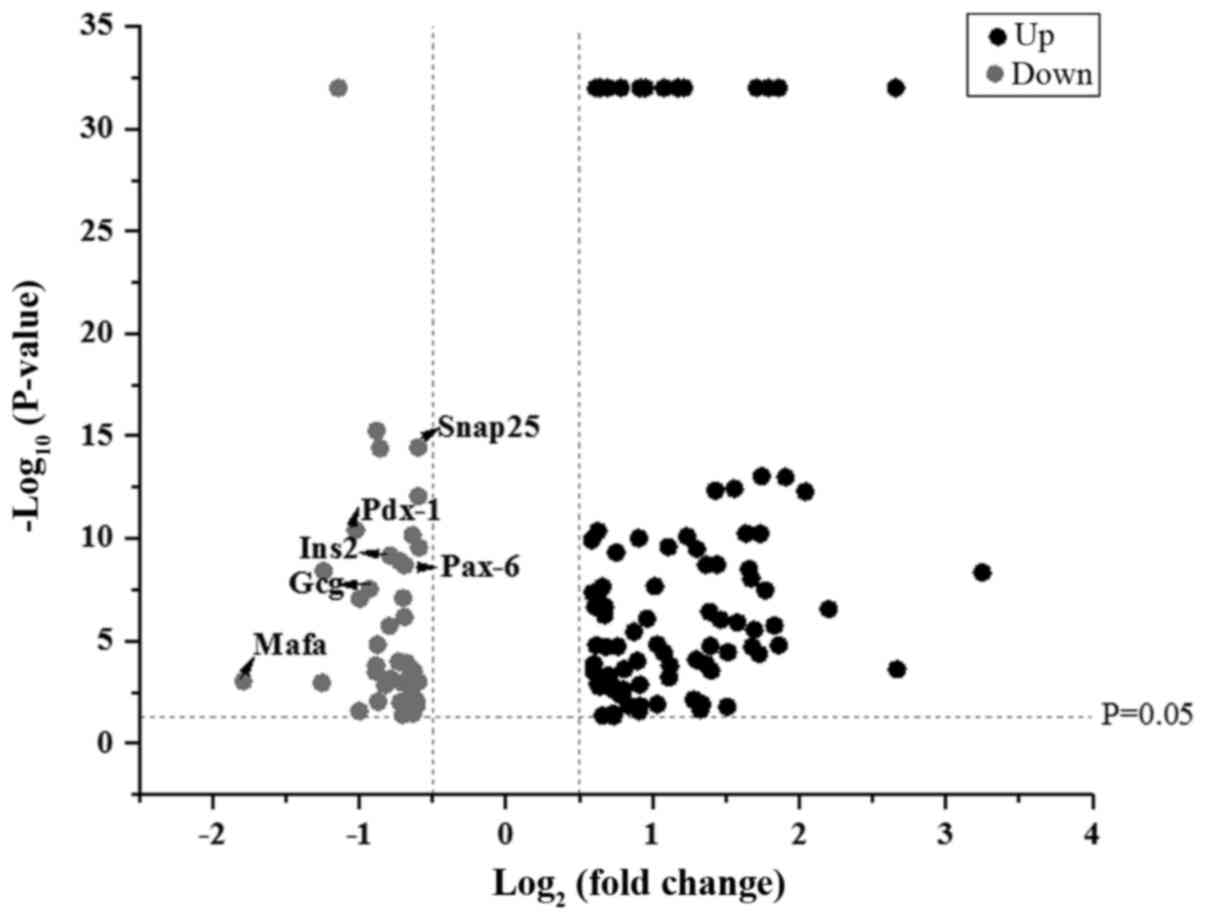
LRM-treated Beta-TC-6 cells compared with control cells (Fig. 1).
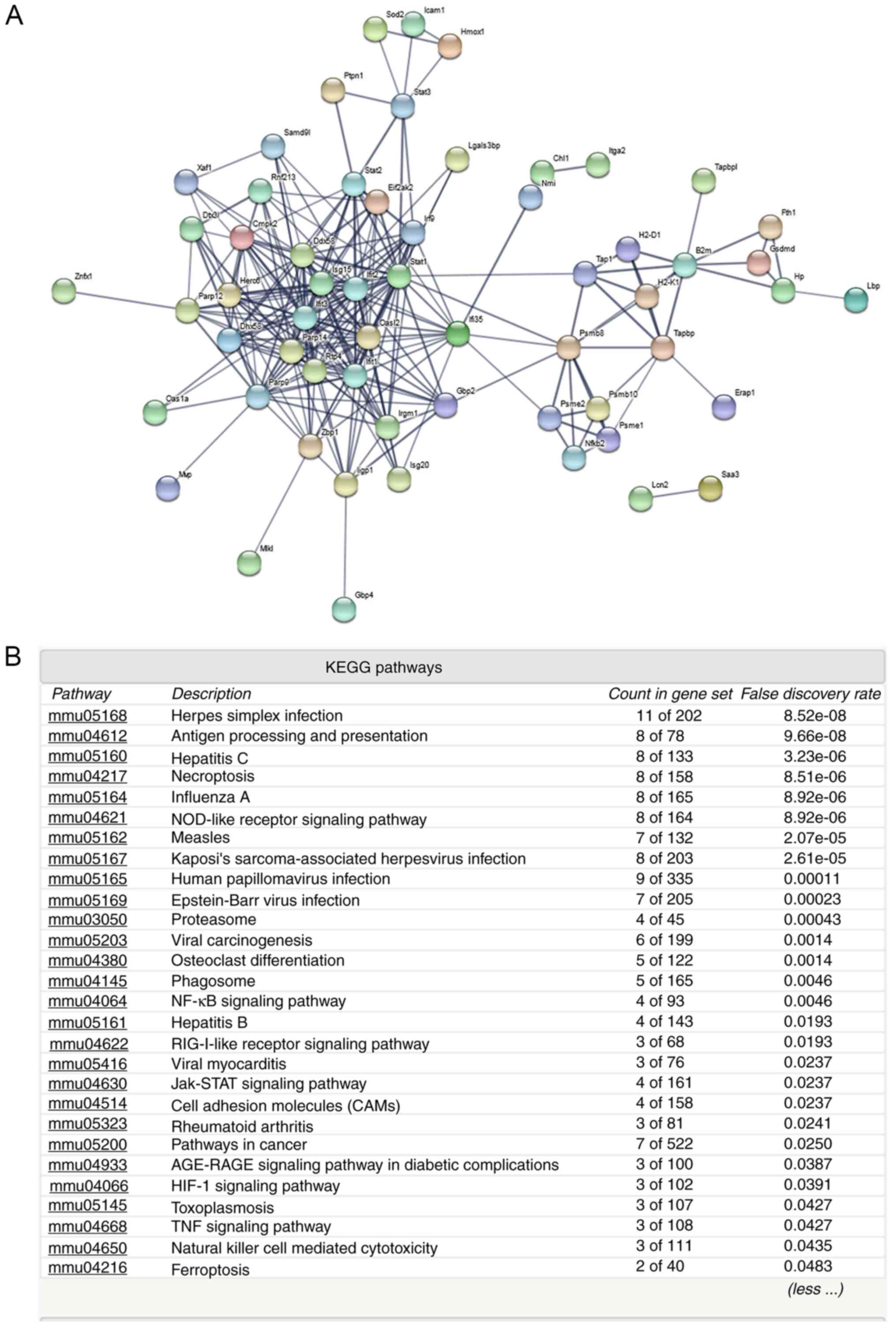
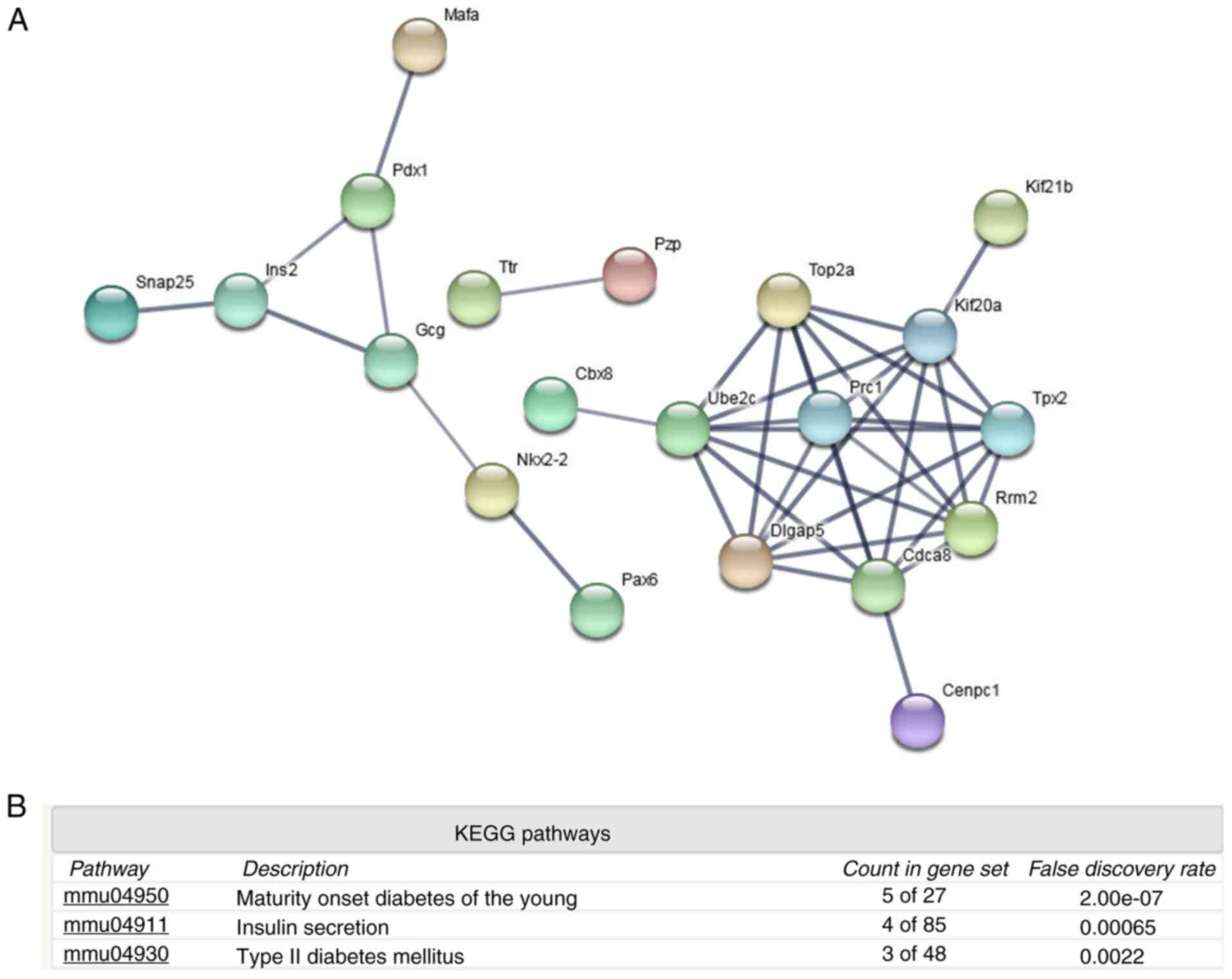
Protein-protein interaction and KEGG pathway
analyses (Figs. 2 and 3) of the upregulated proteins indicated
that various immune and inflammatory signaling pathways may be
activated, including ‘antigen processing and presentation’,
‘NF-kappa B signaling pathway’, ‘cell adhesion molecules’,
‘Jak-STAT signaling pathway’ and ‘hepatitis C and B’ (Fig. 2B). However, analysis of the
downregulated proteins indicated that islet function-related
certain signaling pathways may be attenuated, including ‘maturity
onset diabetes of the young’, ‘insulin secretion’ and ‘type II
diabetes mellitus’ (Fig. 3B). The
downregulated proteins included transcription factor MafA (Mafa),
pancreatic and duodenal homeobox 1 (Pdx-1), paired box 6 (Pax-6),
homeobox protein Nkx-2.2 (Nkx-2.2), synaptosomal-associated protein
25 (Snap25), glucagon (Gcg) and insulin-2 (Ins2). These proteins
may therefore mediate islet dysfunction.
miRNA expression profile
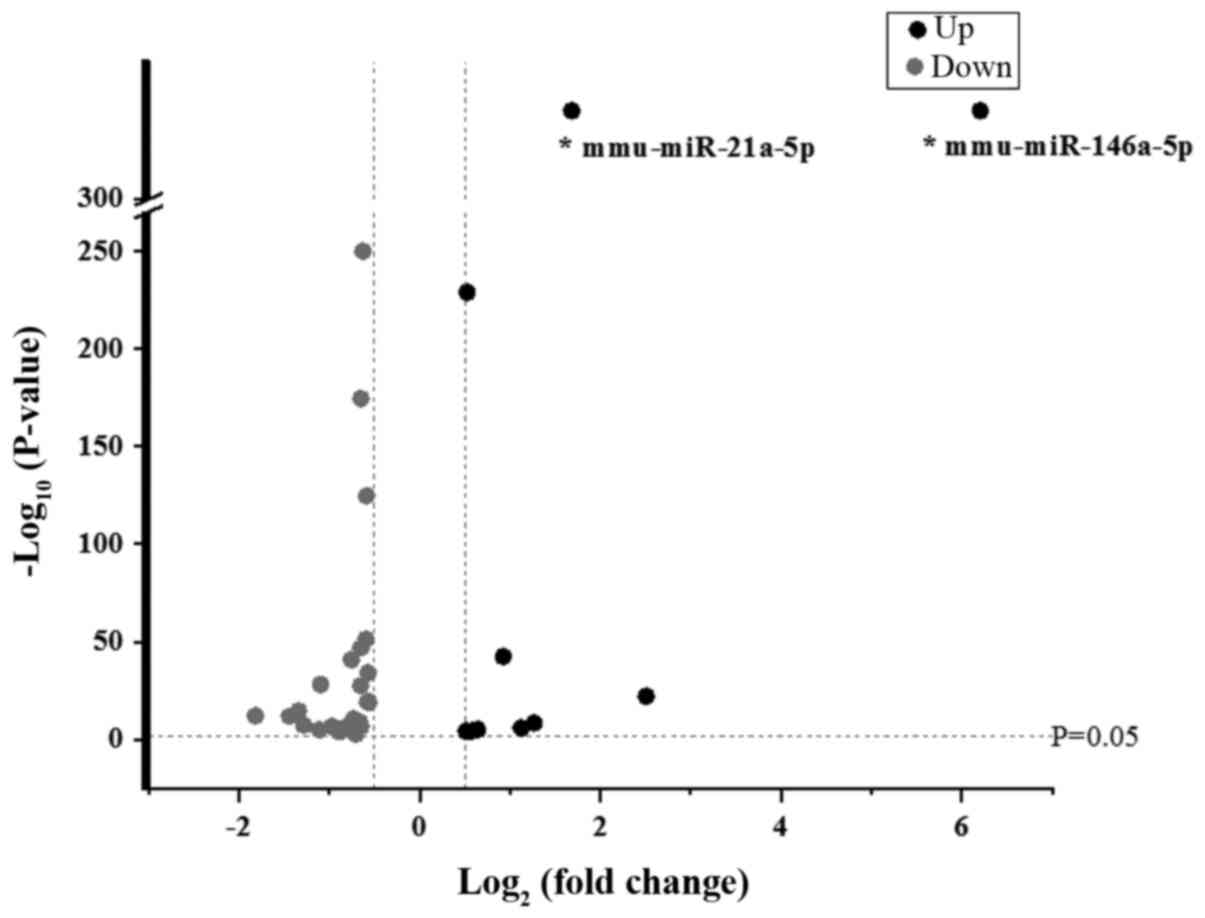
Based on the results of the miRNA assay, miRNAs with
log2.Fold change values ≥0.5 or ≤-0.5 between LRM-treated Beta-TC-6
cells and control cells were selected for further analysis. It was
revealed that 11 miRNAs were upregulated and 28 miRNAs were
downregulated (Fig. 4).
Taking into account abundance and fold changes in
miRNA expression, miRNAs with an abundance value >1,000 and a
fold change >2 were selected for further validation. Therefore,
only miR-146a-5p and miR-21a-5p were appropriate for further
analysis.
Validation of miRNAs and
protein-matched gene functions
The present study selected miR-21a-5p and
miR-146a-5p for further investigation as aforementioned. The
results of RT-qPCR indicated that the expression of miR-21a-5p and
miR-146a-5p, and inflammatory factors IL-1A and TNF-α were
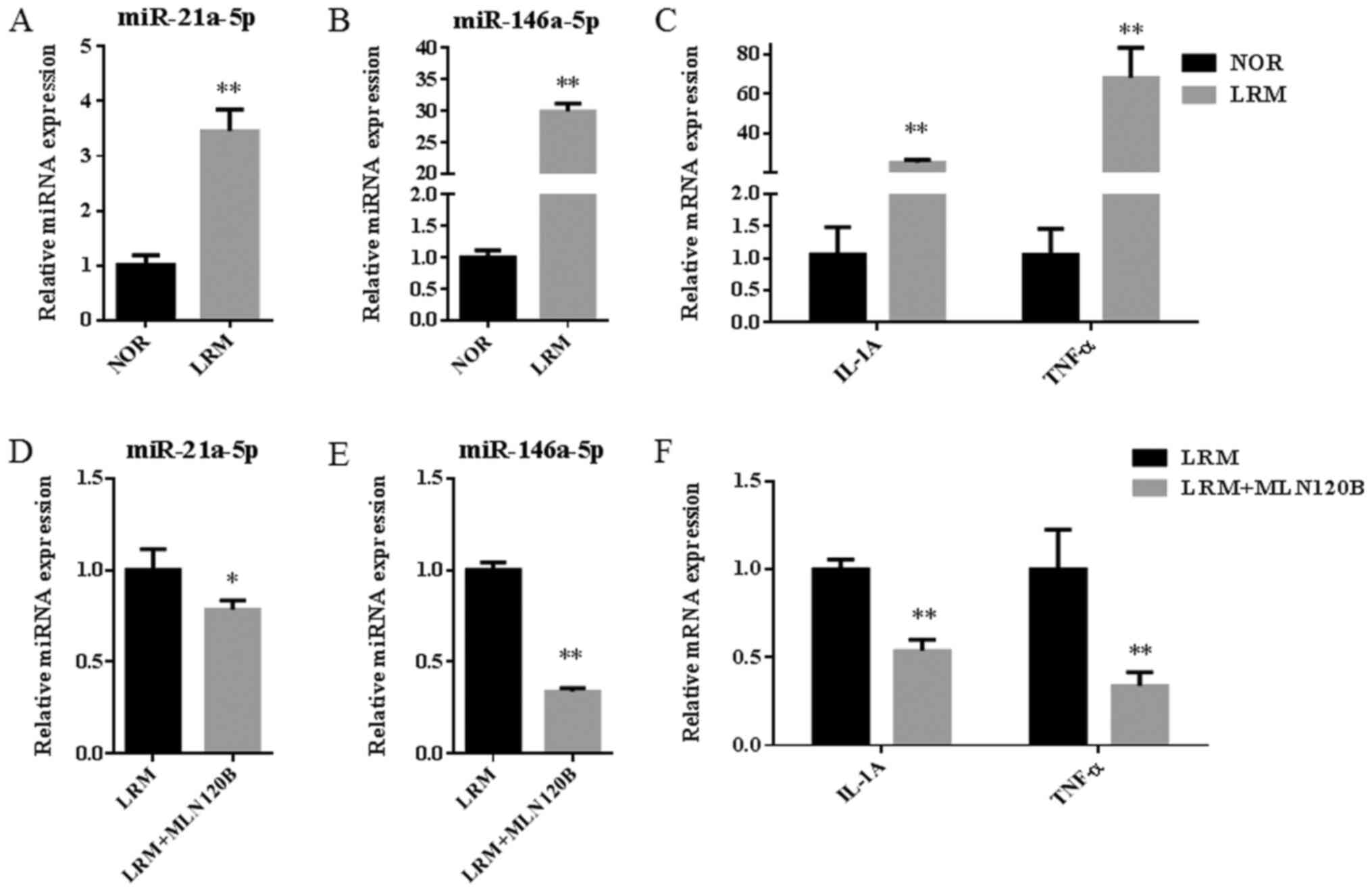
significantly increased following inflammation induction (Fig. 5A-C; all, P<0.01). Additionally,
miR-146a-5p was further upregulated when compared with miR-21a-5p
(~6-fold; P<0.01). Following treatment with the NF-κB inhibitor
MLN120B, the expression of miR-21a-5p and miR-146a-5p were
significantly decreased (P<0.05 and P<0.01, respectively;
Fig. 5D and E). Furthermore, levels of IL-1A and TNF-α
were significantly decreased following the same treatment (each,
P<0.01; Fig. 5F). The results
indicated that inflammation activation by NF-κB may contribute to
the upregulation of miR-21a-5p and miR-146a-5p expression.
It was not clear whether upregulated miR-21a-5p and
miR-146a-5p expression contributed to islet dysfunction. In the
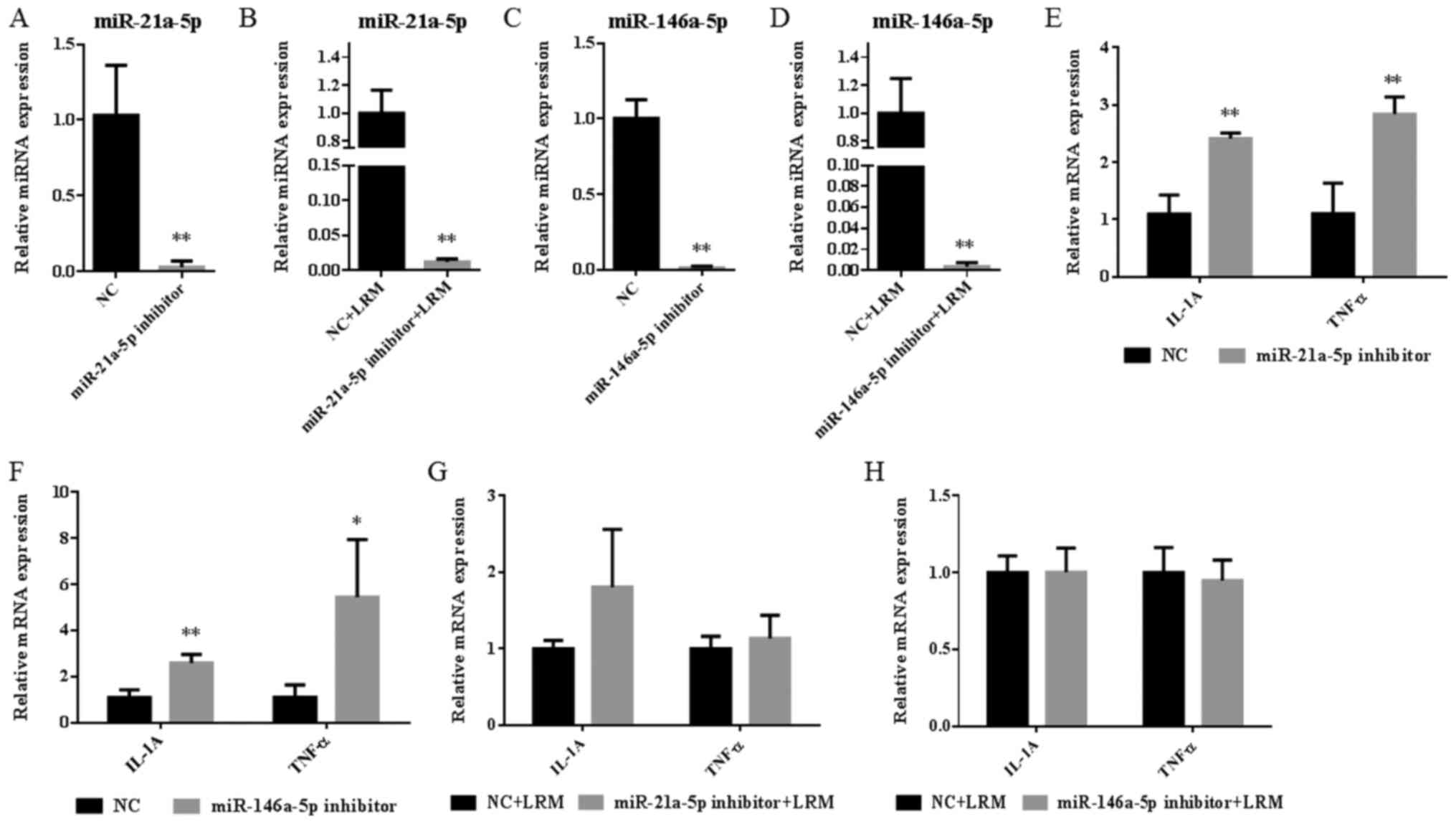
present study, the miR-21a-5p inhibitor was used to specifically
inhibit the expression of miR-21a-5p in the miR-21a-5p inhibitor
(decreased by 97%; Fig. 6A) and the
miR-21a-5p inhibitor+LRM groups (decreased by 99%; Fig. 6B). Additionally, the miR-146a-5p
inhibitor was used to specifically inhibit the expression of
miR-146a-5p in the miR-146a-5p inhibitor (decreased by 99%;
Fig. 6C) and miR-146a-5p
inhibitor+LRM groups (decreased by 100%; Fig. 6D).
After analyzing the expression of inflammatory
factors, the results revealed that treatment with the miR-21a-5p or
miR-146a inhibitor alone significantly increased the expression of
IL-A and TNF-α when compared with the NC group (all, P<0.01;
Fig. 6E and F). However, following the induction of
inflammation, the miR-21a-5p and miR-146a-5p inhibitor did not
significantly affect the levels of each inflammatory factor when
compared with the NC+LRM group (Fig.
6G and H). The results
indicated that miR-21a-5p and miR-146a-5p may serve a minor role in
the regulation of physiological inflammation homeostasis, but not
in severe pathological inflammatory dysfunction.
As expected, LRM significantly decreased the mRNA
expression of various islet functional factors when compared with
the NC group, including Nkx-2.2 (P<0.05), Gcg (P<0.05), Pax-6
(P<0.05), Snap25 (P<0.05), Pdx-1 (P<0.01), Mafa
(P<0.01) and Ins2 (P<0.01) (Fig.
7A). Additionally, it was revealed that miR-21a-5p inhibition
significantly reversed the decrease in Nkx-2.2 (P<0.01), Mafa
(P<0.01) and Ins2 (P<0.05) expression observed in the NC+LRM
group (Fig. 7C). However, the
miR-146 inhibitor did not exert the same effect (Fig. 7E). The results indicated that
inflammation-induced miR-21a-5p expression may serve an important
role in islet dysfunction. However, miR-146a-5p did not have a
major effect in terms of reversing islet dysfunction factors, but
may serve as a sensitive biomarker in islet cell inflammation.
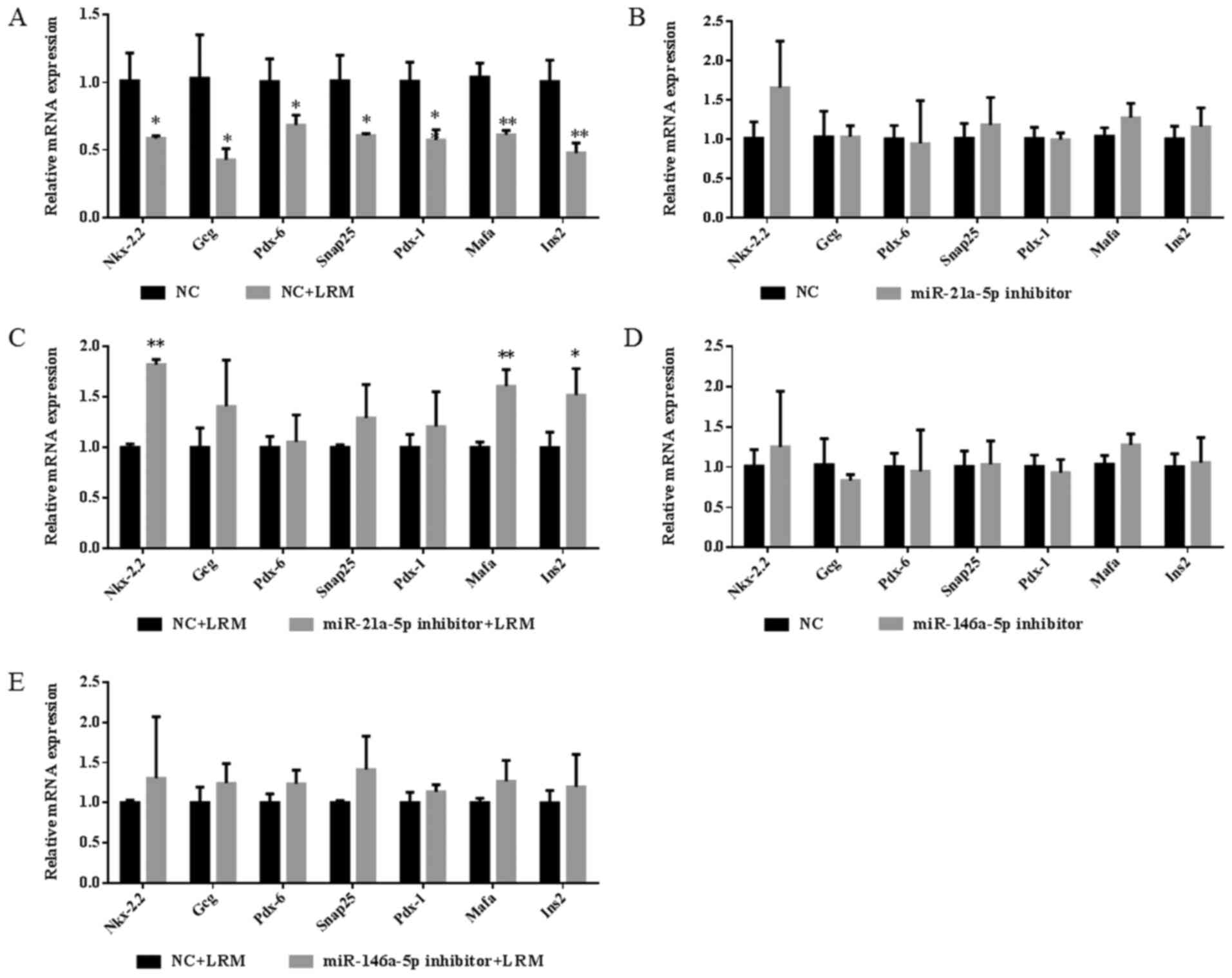 | Figure 7Expression of islet functional
factors in Beta-TC-6 islet cells. (A) Expression of islet
functional factors in LRM-treated and untreated Beta-TC-6 islet
cells. (B-E) Expression levels of islet functional factors after
the addition of inhibitors in LRM-treated and untreated Beta-TC-6
islet cells. *P<0.05 and **P<0.01 vs.
NC or LRM+NC. Data are presented as the mean ± SD (n=3). LRM,
lipopolysaccharide-induced macrophage cell medium; NC, negative
control; miR, microRNA; Nkx-2.2, homeobox protein Nkx-2.2; Gcg,
glucagon; Snap25, synaptosome-associated protein 25; Pdx-1,
pancreatic and duodenal homeobox 1; Pax-6, paired box 6; Mafa,
transcription factor MafA; Ins2, insulin-2. |
Discussion
Although it is known that the pancreatic islet cells
of patients with type 1 or 2 diabetes may be in a state of
inflammation, no ideal in vitro islet cell model is
available for humans (25). In
addition, in preliminary experiments using LPS alone, Beta-TC-6
cells could not be induced for evident inflammatory activation
compared with LRM (data not shown). Considering that >90% of
patients with diabetes demonstrate the type 2 subtype and that
macrophage cells accumulate in type 2 diabetic islets (26), the present study focused on the
cross-talk between macrophages and islets. In the present study,
LRM contained a large quantity of secreted inflammatory factors,
and following collection, LRM was used to simulate the complicated
microenvironment of inflammation around mouse pancreatic islet
cells as previously described (10).
The proteomics assay of the present study revealed
that inflammation induction by LRM downregulated the levels of key
proteins associated with islet function, including Mafa, Pdx-1,
Pax-6, Nkx-2.2, Gcg, Snap25 and Ins2, which mediate islet
development and insulin secretion. Pdx-1 and Mafa are key
transcription regulators of beta cell development and regeneration
(27). Pax-6 is a transcription
factor that has emerged as a key modulator of multiple steps in
pancreatic development and differentiation, serving a pivotal role
in the regulation of pancreatic islet hormone synthesis and
secretion (28). Nkx-2.2 is a
homeodomain transcription factor that is essential for the
differentiation of three of the pancreatic endocrine populations:
Alpha, beta and pancreatic polypeptide cells (29). The core proteins forming the SNARE
complex are Snap25, vesicle-associated membrane protein and
syntaxins (30), which primarily
serve exocytotic functions (31).
Snap25 is also associated with insulin secretion (32). Inflammation appears to affect
insulin production by causing the loss of islet identity and
inhibiting insulin secretion. The results of the present study
determined the molecules involved in inflammatory dysfunction
mechanisms and their pathological basis in islet cells. However,
the exact mechanisms underlying the downregulation of these
proteins remains unknown.
In humans, certain islet-specific miRNAs have been
identified, including miR-375, miR-184, miR-183-5p, miR-182-5p and
miR-127-3p (33). However, the
function of the majority of miRNAs remain undetermined.
Additionally, it has not yet been elucidated whether the function
of the aforementioned miRNAs exhibit significant changes when
subjected to inflammatory stimulation. In the present study, the
miRNA assays revealed that inflammation promoted a large change in
the miRNA profile of LRM-treated Beta-TC-6 cells. These miRNAs
(upregulated 11 and downregulated 28) may serve an important role
in the pathological process of inflammatory dysfunction in islet
cells. miR-21a-5p and miR-146a-5p may serve as effective targets
due to their significant fold changes and high abundances observed
following inflammatory stimulation in islet cells of the present
study. Furthermore, miR-21a-5p and miR-146a-5p may be regulated by
the NF-κB signaling pathway. miR-21 serves an important role in
pro-inflammatory and anti-inflammatory responses (34). Whilst miR-21 targets Bcl-2 mRNA and
promotes islet cell apoptosis (35), miR-21 silencing prolongs islet
allograft survival by inhibiting Th17 cells (36). Furthermore, miR-21 promotes cardiac
fibrosis after myocardial infarction by targeting smad7(37). miR-21 has also emerged as a key
mediator of the anti-inflammatory response, with inflammatory
stimuli additionally triggering miR-21 induction (34). The present results indicated that
miR-21a-5p could exert slight anti-inflammatory roles in a state of
low-grade inflammation. miR-146a-5p serves as an important negative
regulator of inflammation that can be upregulated by LPS (38). miR-21a-5p and miR-146a-5p appear to
serve an important role in immune response tolerance or the
homeostasis of inflammation stimulation (10,39,40).
In the present study, it was hypothesized that the upregulation of
these miRNAs may affect islet function in addition to inflammatory
regulation. However, this hypothesis requires further
validation.
Using miRNA target prediction software (http://c1.accurascience.com/miRecords/;
updated April 27, 2013), it was revealed that miR-21a-5p and
miR-146a-5p can target numerous genes. Although miR-21a-5p and
miR-146a-5p may not directly regulate the aforementioned
downregulated proteins, the proteins derived from their target
genes may interact with them instead. It was suggested by authors
that the upregulation of miR-21a-5p and miR-146a-5p may be involved
in the downregulation of proteins associated with islet dysfunction
induced by inflammation. Following mRNA validation, the
downregulation of IL-1A and TNF-α was partially reversed in islets
following treatment with an inhibitor of miR-21a-5p. However, the
same affect was not induced following miR-146a-5p inhibitor
treatment. Therefore, the regulatory mechanism underlying these
miRNAs may be complex. Using an inhibitor of one miRNA may not be
sufficient to validate its true function, since the cells exhibited
a variety of changes in numerous miRNAs. It is possible that a
single miRNA may only serve a limited role but likely exerts
stronger effects when working in unison with other miRNAs. However,
the coordinated function of all miRNAs with altered levels require
further investigation. It may be necessary to converge all these
altered miRNAs to validate their coordinated functions. However, it
is very difficult to simultaneously reverse all downregulated or
upregulated miRNAs in a single cellular system. Despite this, the
results of the present study indicated that the upregulation of
miR-21a-5p expression in inflammation may serve an important role
in inflammatory islet dysfunction.
The present study provided valuable information
regarding the altered expression of certain miRNAs and proteins.
However, only the expression of two miRNAs and several mRNAs were
validated. The remaining miRNAs and proteins involved should be
further validated in future studies since other factors may be
involved in the complex pathological system. Furthermore, primary
or human islet cells should be investigated, as only one mouse
islet cell line was utilized in the current study. In vivo
experiments should be also be conducted in animals and humans to
reflect true islet functions in the state of inflammation.
In conclusion, through proteomics and miRNA-omics,
the present study drafted a complete profile of protein and miRNA
changes that occur simultaneously in islet cells induced by
inflammation, which may further the understanding of the underlying
molecular mechanisms of diabetes and islet inflammation. miR-21a-5p
and miR-146a-5p may serve as targets or biomarkers for inflammatory
dysfunction in islet cells. Additionally, it was determined that
these miRNAs were mediated via the NF-κB signaling pathway. Changes
in proteins and miRNAs may form a large network to coordinate
changes in islet cell functions in a pathological state. However,
how miRNAs regulate target genes and proteins requires further
investigation.
Acknowledgements
The authors would like to thank Mr. Pengbo Sun and
Ms. Jingyi Luo (Shenzhen International Graduate School, Tsinghua
University) for technical help when constructing volcano plots.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81373460 and
91957110), the Natural Science Foundation of Guangdong Province
(grant no. 2014A030313744) and the Shenzhen Science and Technology
Innovation Committee (grant no. JCYJ20170307152357168).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YD, JZ and YW performed the experiments. YD and WX
analyzed the data. WX conceived the study and wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang L, Gao P, Zhang M, Huang Z, Zhang D,
Deng Q, Li Y, Zhao Z, Qin X, Jin D, et al: Prevalence and ethnic
pattern of diabetes and prediabetes in China in 2013. Jama.
317:2515–2523. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lung CW, Wu FL, Liao F, Pu F, Fan Y and
Jan YK: Emerging technologies for the prevention and management of
diabetic foot ulcers. J Tissue Viability. 29:61–68. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Xie W and Du L: Diabetes is an
inflammatory disease: Evidence from traditional Chinese medicines.
Diabetes Obes Metab. 13:289–301. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Collier JJ, Sparer TE, Karlstad MD and
Burke SJ: Pancreatic islet inflammation: An emerging role for
chemokines. J Mol Endocrinol. 59:R33–R46. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Morgan NG, Leete P, Foulis AK and
Richardson SJ: Islet inflammation in human type 1 diabetes
mellitus. IUBMB Life. 66:723–734. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Mathis D, Vence L and Benoist C: Beta-Cell
death during progression to diabetes. Nature. 414:792–798.
2001.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Lee RC, Feinbaum RL and Ambros V: The
C. Elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854.
1993.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhao C, Zhang Y and Popel AS: Mechanistic
computational models of MicroRNA-mediated signaling networks in
human diseases. Int J Mol Sci. 20(421)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ventriglia G, Nigi L, Sebastiani G and
Dotta F: MicroRNAs: Novel players in the dialogue between
pancreatic islets and immune system in autoimmune diabetes. Biomed
Res Int. 2015(749734)2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang X, Xu C, Lei F, Liao M, Wang W, Xu
N, Zhang Y and Xie W: MiR-30a targets IL-1α and regulates islet
functions as an inflammation buffer and response factor. Sci Rep.
7(5270)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang ZK, Wang J, Liu J, Ying SH, Peng XJ
and Feng MG: Proteomic and phosphoproteomic insights into a
signaling hub role for Cdc14 in asexual development and multiple
stress responses in beauveria bassiana. PLoS One.
11(e0153007)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tyanova S, Temu T and Cox J: The maxQuant
computational platform for mass spectrometry-based shotgun
proteomics. Nat Protoc. 11:2301–2319. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang H, Liu T, Zhang Z, Payne SH, Zhang
B, McDermott JE, Zhou JY, Petyuk VA, Chen L, Ray D, et al:
Integrated proteogenomic characterization of human high-grade
serous ovarian cancer. Cell. 166:755–765. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ross PL, Huang YN, Marchese JN, Williamson
B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et
al: Multiplexed protein quantitation in saccharomyces
cerevisiae using amine-reactive isobaric tagging reagents. Mol
Cell Proteomics. 3:1154–1169. 2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ouyang X, Jiang X, Gu D, Zhang Y, Kong SK,
Jiang C and Xie W: Dysregulated serum MiRNA profile and promising
biomarkers in dengue-infected patients. Int J Med Sci. 13:195–205.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Guan J, Long K, Ma J, Zhang J, He D, Jin
L, Tang Q, Jiang A, Wang X, Hu Y, et al: Comparative analysis of
the microRNA transcriptome between yak and cattle provides insight
into high-altitude adaptation. PeerJ. 5(e3959)2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10(R25)2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Friedlander MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: MiRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y,
Zhao X, Liang C, Wang Y, Sun L, et al: Integrated profiling of
microRNAs and mRNAs: MicroRNAs located on Xq27.3 associate with
clear cell renal cell carcinoma. PLoS One. 5(e15224)2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang L, Feng Z, Wang X, Wang X and Zhang X
and Zhang X: DEGseq: An R package for identifying differentially
expressed genes from RNA-seq data. Bioinformatics. 26:136–138.
2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Storey JD: The positive false discovery
rate: A Bayesian interpretation and the q-value. Ann Stat.
31:2013–2035. 2003.
|
|
22
|
Hu X, Liu S, Liu X, Zhang J, Liang Y and
Li Y: DPP-4 (CD26) inhibitor sitagliptin exerts anti-inflammatory
effects on rat insulinoma (RINm) cells via suppressing NF-κB
activation. Endocrine. 55:754–763. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Xie W, Li M, Xu N, Lv Q, Huang N, He J and
Zhang Y: MiR-181a regulates inflammation responses in monocytes and
macrophages. PLoS One. 8(e58639)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Green AD, Vasu S and Flatt PR: Cellular
models for beta-cell function and diabetes gene therapy. Acta
Physiol (Oxf). 2018(222)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Eguchi K and Manabe I: 2013. Macrophages
and islet inflammation in type 2 diabetes. Diabetes Obes Metab.
15:152–158. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhu Y, Liu Q, Zhou Z and Ikeda Y: PDX1,
neurogenin-3, and MAFA: Critical transcription regulators for beta
cell development and regeneration. Stem Cell Res Ther.
8(240)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Panneerselvam A, Kannan A,
Mariajoseph-Antony LF and Prahalathan C: PAX proteins and their
role in pancreas. Diabetes Res Clin Pract.
155(107792)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Sussel L, Kalamaras J, Hartigan-O'Connor
DJ, Meneses JJ, Pedersen RA, Rubenstein JL and German MS: Mice
lacking the homeodomain transcription factor Nkx2.2 have diabetes
due to arrested differentiation of pancreatic beta cells.
Development. 125:2213–2221. 1998.PubMed/NCBI
|
|
30
|
Cupertino RB, Kappel DB, Bandeira CE,
Schuch JB, da Silva BS, Muller D, Bau CH and Mota NR: SNARE complex
in developmental psychiatry: Neurotransmitter exocytosis and
beyond. J Neural Transm (Vienna). 123:867–883. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shin OH: Exocytosis and synaptic vesicle
function. Compr Physiol. 4:149–175. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhuang GQ, Wu W, Liu F, Ma JL, Luo YX,
Xiao ZX, Liu Y, Wang W and He Y: SNAP-25(1-180) enhances insulin
secretion by blocking Kv2.1 channels in rat pancreatic islet
beta-cells. Biochem Biophys Res Commun. 379:812–816.
2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
van de Bunt M, Gaulton KJ, Parts L, Moran
I, Johnson PR, Lindgren CM, Ferrer J, Gloyn AL and McCarthy MI: The
miRNA profile of human pancreatic islets and beta-cells and
relationship to type 2 diabetes pathogenesis. PLoS One.
8(e55272)2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sheedy FJ: Turning 21: Induction of miR-21
as a key switch in the inflammatory response. Front Immunol.
6(19)2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sims EK, Lakhter AJ, Anderson-Baucum E,
Kono T, Tong X and Evans-Molina C: MicroRNA 21 targets BCL2 mRNA to
increase apoptosis in rat and human beta cells. Diabetologia.
60:1057–1065. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang H, Fan H, Tao J, Shao Q and Ding Q:
MicroRNA-21 silencing prolongs islet allograft survival by
inhibiting Th17 cells. Int Immunopharmacol. 66:274–281.
2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yuan J, Chen H, Ge D, Xu Y, Xu H, Yang Y,
Gu M, Zhou Y, Zhu J, Ge T, et al: Mir-21 promotes cardiac fibrosis
after myocardial infarction via targeting smad7. Cell Physiol
Biochem. 42:2207–2219. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Nahid MA, Satoh M and Chan EK: Mechanistic
role of microRNA-146a in endotoxin-induced differential
cross-regulation of TLR signaling. J Immunol. 186:1723–1734.
2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xie W, Li Z, Li M, Xu N and Zhang Y:
MiR-181a and inflammation: MiRNA homeostasis response to
inflammatory stimuli in vivo. Biochem Biophys Res Commun.
430:647–652. 2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang S, Gu D, Ouyang X and Xie W:
Proinflammatory effects of the hemagglutinin protein of the avian
influenza A (H7N9) virus and microRNAmediated homeostasis response
in THP1 cells. Mol Med Rep. 12:6241–6246. 2015.PubMed/NCBI View Article : Google Scholar
|