Introduction
Liver fibrosis has long been considered as a healing
response to various chronic liver injuries, including viral
hepatitis, immune compounds and toxic agents, among others
(1). Hepatitis B virus (HBV)
remains the most important cause of liver fibrosis in China, which
is an endemic area (2). During the
life cycle, HBV generates covalently closed circular DNA (cccDNA),
which acts as the model of HBV replication (3). Additionally, cccDNA intergrates into
the genome of hepatocyte, which makes it hard to eliminate HBV from
infected cells (3). A recent study
from China also showed that Persistent Low Level of Hepatitis B
Virus (LLV) widely existed in HBV infected patients despite
antiviral therapy (4). Moreover,
despite the widespread use of strong antiviral treatments, patients
with HBV infection still inevitably progress to liver fibrosis, or
even liver cirrhosis (5). However,
the mechanism underlying HBV-induced liver fibrosis remains poorly
understood.
Over the past decades, accumulative studies have
focused on the cascades of the TGF-β1 pathway, which takes part in
multiple biological progresses, including cell activation (6), tissue differentiation (7), fibrosis (8) and cancer progression (9). As expected, TGF-β1 has a strong
association with HBV replication and HBV related liver diseases
(10,11). TGF-β1 has been demonstrated to serve
important roles in hepatic stellate cell (HSC) activation, which is
a key event in liver fibrosis (11,12).
Accumulating evidence suggested that HBV infection may promote
TGF-β1 production by hepatocytes, which in turn activates HSCs and
accelerates liver fibrosis (13-15).
HSCs is the predominant cell type responsible for liver fibrosis.
In physiological conditions, quiescent-like HSCs exhibit necessary
effects on lipid metabolism, maintaining the balance of
extracellular matrix (ECM) production and degradation (16). When liver injury occurs, HSCs are
activated by cytokines including TGF-β1 secreted by a variety of
cells in the liver (17). These
activated HSCs produce excessive ECM deposited in liver tissue
disrupting liver structure leading to liver fibrogenesis (17).
MicroRNAs (miRNAs) are a group of small and
non-coding RNAs, which regulates gene expression by binding to
specific mRNA targets. miRNAs can promote degradation and/or
translational inhibition of target gene by post-transcription
modification (18). Interestingly,
a big cluster of miRNAs has been demonstrated to accelerate HSC
activation, fibrogenesis and cancer by augmenting
fibrosis-associated signaling pathways including transforming
growth factor-β/Smad (19). It was
reported that microRNA (miR/miRNA)-21-5p acts as a key mediator in
the TGF-β1 signaling pathway through assisting in the inhibition of
Smad7 expression (20,21). However, the role of the
TGF-β1/miR-21-5p pathway in HSC activation and liver fibrosis
remains to be elucidated. The present study was designed to
evaluate the role of the TGF-β1/miR-21-5p pathway in liver fibrosis
induced by HBV infection.
Materials and methods
Patients
Liver tissue samples were collected from 29 patients
with HBV infection. These patients were antiviral treatment-naive
and were admitted to the Department of Infectious Disease of the
Anhui Provincial Hospital (Hefei, China) between Jan 2018 and
December 2019. All patients were diagnosed with chronic hepatitis B
according to the guidelines of the Chinese Medical Association
(22). Inclusion criteria is the
positive of S antigen for at least 6 months. Exclusion criteria
includes co-infection with other viruses such as HCV/HIV/EBV/CMV;
suffering from alcohol, drug or antoimmune related hepatitis;
cancer and other disease receiving drugs which may affect host
immune system. A total of 5 patients without HBV infection were
enrolled as controls. The patients in the present study underwent
liver biopsy. All data from medical records of these patients were
collected at Anhui Provincial Hospital. All patients provided
written informed consent for the present study, and the study
protocol was approved by the Ethics Committee of Anhui Provincial
Hospital.
Biochemical examinations and HBV DNA
quantification
Serum alanine transaminase (ALT), aspartate
aminotransferase (AST), total bilirubin (TBil) and albumin (ALB)
levels were measured by routine laboratory tests at the Department
of Laboratory Science of Anhui Provincial Hospital (Hefei, China).
In addition, serology tests were performed on the ARCHITECT i2000SR
immunoassay analyzer (Abbott Pharmaceutical Co., Ltd.) and serum
HBV DNA levels were measured using the COBAS TaqMan HBV test (Roche
Diagnostics).
Histological assessment of liver
fibrosis
Liver tissues were collected from patients by liver
biopsy using a Bard biopsy needle (18G, 910 cm; BD Biosciences) as
previously described (23). Liver
tissue samples were fixed for 24 h at room temperature in 10%
formalin, embedded in paraffin and cut into 5-µm sections.
Hematoxylin and eosin (HE) and Masson's staining
were performed to assess the alterations in liver architecture and
the presence of liver fibrosis. The liver fibrosis score was
evaluated by two independent pathologists in a blinded manner
according to the METAVIR system (24) as follows: S0, no collagen
deposition; S1, minimal collagen deposition;
S2, extended fibrosis; S3, bridging fibrosis
and S4, cirrhosis. Smad7 expression in liver tissue was
observed by immunohistochemistry (IHC) and calculated using the
following formula: Positive index (PI)=mean optical density x
positive area percentage.
Cell culture and viral stocks
The human HSC line LX2, and Huh7.5.1 and HepAD38
hepatocellular carcinoma cells, were maintained in our laboratory.
Cells were grown in DMEM (HyClone; Cytiva) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) with 100 U/ml
penicillin and 100 µg/ml streptomycin under a humidified atmosphere
at 37˚C with 5% CO2 (v/v). HBV viral stock was collected
from the supernatant of HepAD38 cells and stored at -20˚C. HBV
copies were quantified by quantitative PCR (qPCR) analysis.
Entecavir (ETV) (Sigma-Aldrich; Merck KGaA) was diluted into DMEM
at 20 µM as the working concentration. Cells maintained in DMEM
without HBV were used as the mock group (mock infection). Human
TGF-β1 was purchased from R&D System (240-B-010/CF) and was
diluted into DMEM at 10 ng/ml as the working concentration.
Bioinformatics analysis
TargetScan (http://www.targetscan.org), PicTar (http://pictar.mdc-berlin.de/), miRanda (http://microrna.sanger.ac.uk) and KEGG Pathway
analysis (www.genome.jp/kegg) to explore the
target genes for miR-21-5p.
Plasmid preparation and
transfection
Sequences of miR-21-5p were collected from miRBase.
miR-21-5p mimic, inhibitor (antagomir-21) and their corresponding
negative control were all obtained from Guangzhou RiboBio Co.,
Ltd.
U6 small nucleolar RNA (Guangzhou RiboBio Co., Ltd.)
was used as the endogenous control. Plasmids were amplified and
purified using the HiSpeed Plasmid Midi Kit (cat. no. 12643;
Qiagen, Inc.) and stored in ddH2O at 1 µg/µl before use.
All plasmids were transfected into cells using
Lipofectamine® LTX with Plus™ reagent
(Invitrogen; Thermo Fisher Scientific, Inc.).
Co-culture system
A co-culture system containing sodium taurocholate
co-transporting polypeptide (NTCP)-Huh7.5.1 and LX2 cells were used
in the present study. First, pCMV-NTCP (RC210241; OriGene
Technologies, Ins.) was transfected into Huh 7.5.1 cells to
construct NTCP-Huh7.5.1 cells. Then, NTCP-Huh7.5.1 cells were
infected with HBV viral stock (MOI=1.0) (25) and LX2 cells were transfected with
miR-21-5p mimic inhibitor or the negative control, respectively.
After 72 h, HBV-infected NTCP-Huh7.5.1 cells were seeded in a
12-well plate at a density of 2x105 cells/well and LX2
cells were plated on 0.4-µM 12-well Transwell inserts (Costar;
Corning, Inc.). After overnight maintenance at 37˚C, LX2 Transwells
were loaded into NTCP-Huh7.5.1 Transwells. After another 72 h of
incubation, the cells were harvested for mRNA and protein
analysis.
Reverse transcription (RT)-qPCR
analysis
Total RNA was extracted from liver tissues and cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Subsequently, first-strand cDNA was reverse-transcribed from RNA
using the PrimeScript RT reagent (Takara Bio, Inc.) according to
the manufacturer's instructions. qPCR was performed with SYBR Green
Assay kit (Thermo Fisher Scientific, Inc.) on an ABI 7500 PRISM
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
following thermocycling conditions were used for the qPCR: Initial
denaturation at 95˚C for 3 min, followed by 40 cycles of 94˚C for
20 sec, 60˚C for 30 sec and 72˚C for 20 sec. The expression of each
mRNA was calculated against U6/GAPDH and quantified using the
2-ΔΔCq method (26). The
primers used in the present study are listed in Table I.
 | Table IList of primers used in reverse
transcription-quantitative PCR analysis. |
Table I
List of primers used in reverse
transcription-quantitative PCR analysis.
| Target | Species | Forward primer
(5'-3') | Reverse primer
(5'-3') |
|---|
| α-SMA | Human |
AAAAGACAGCTACGTGGGTGA |
GCCATGTTCTATCGGGTACTTC |
| TIMP-1 | Human |
ACTTCCACAGGTCCCACAAC |
GCTAAGCTCAGGCTGTTCCA |
| CoL1A1 | Human |
CAGCCGCTTCACCTACAGC |
TCAATCACTGTCTTGCCCCA |
| miR-21-5p | Human |
TAGCTTATCAGACTGATGTTGA |
CTGAAGTCGCCATGCAGATA |
| Smad7 | Human |
GCTCCCATCCTGTGTGTTAA |
TAGGTGTCAGCCTAGGATGGT |
| U6 | Human |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| GAPDH | Human |
ACCTTCCCCATGGTGTCTGA |
GCTCCTCCTGTTCGACAGTCA |
Western blot assay
Total protein was collected from cultured cells
using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific,
Inc.). Equal amounts (20 µg) of protein were separated by SDS-PAGE
4-12% Bis-Tris gradient gels and transferred to PVDF membranes.
Subsequently, membranes were blocked with 5% BSA (Sigma-Aldrich;
Merck KGaA) dissolved in TBS-Tween 20 (TBS-T; 0.05% Tween-20) for 2
h at room temperature. The membranes were incubated with primary
antibodies overnight at 4˚C including mouse anti-α-smooth muscle
actin (α-SMA; cat. no. ab7817; Abcam; 1:1,000), rabbit anti-tissue
inhibitor of metalloproteinase (TIMP)-1 (cat. no. ab211926; Abcam;
1:1,000), rabbit anti-collagen type 1 α 1 (CoL1A1; cat. no. 84336;
Cell Signaling Technology, Inc. 1:400), rabbit anti-Smad7 (cat. no.
ab216428; Abcam; 1:1,000) and rabbit anti-β-actin (cat. no. 4967;
Cell Signaling Technology, Inc.; 1:3,000). The blots were then
incubated with HRP-labeled goat-anti-rabbit IgG (cat. no. NA934V)
and sheep-anti-mouse IgG (NA931) secondary antibodies (Amersham;
Cytiva) diluted in 5% BSA at room temperature (25˚C) for
2 h. The blots were washed with TBS-T for 40 min visualization
using ECL (cat. no. NCI4106; Beijing Solarbio Science &
Technology Co., Ltd.) for 10 min. Protein expression was calculated
with β-actin as a reference using Image J software (v1.8.0;
National Institutes of Health).
Statistical analysis
All data in the present study are expressed as the
mean ± SD. All experiments were repeated three times. Data analyses
were performed by t-test or one-way ANOVA with Tukey's post-hoc
test with SPSS version 17.0 (SPSS, Inc.). The correlation between
miR-21-5p and liver fibrosis stage was assessed by Spearman's
correlation coefficient. P<0.05 was considered to indicate a
statistically significant difference.
Results
HBV replication enhances TGF-β1
expression in hepatocellular carcinoma cells
To observe the effects of HBV replication on TGF-β1
expression, NTCP-Huh7.5.1 cells were incubated with HBV stock from
the supernatant of HepAD38 cells. Entecavir (ETV) was used to
inhibit HBV replication in HBV-infected NTCP-Huh7.5.1 cells. The
supernatant from HBV-infected NTCP-Huh7.5.1 cells was collected at
different timepoints for HBV detection. At 72 h after incubation,
cells were harvested to examine TGF-β1 mRNA expression by qPCR
analysis.
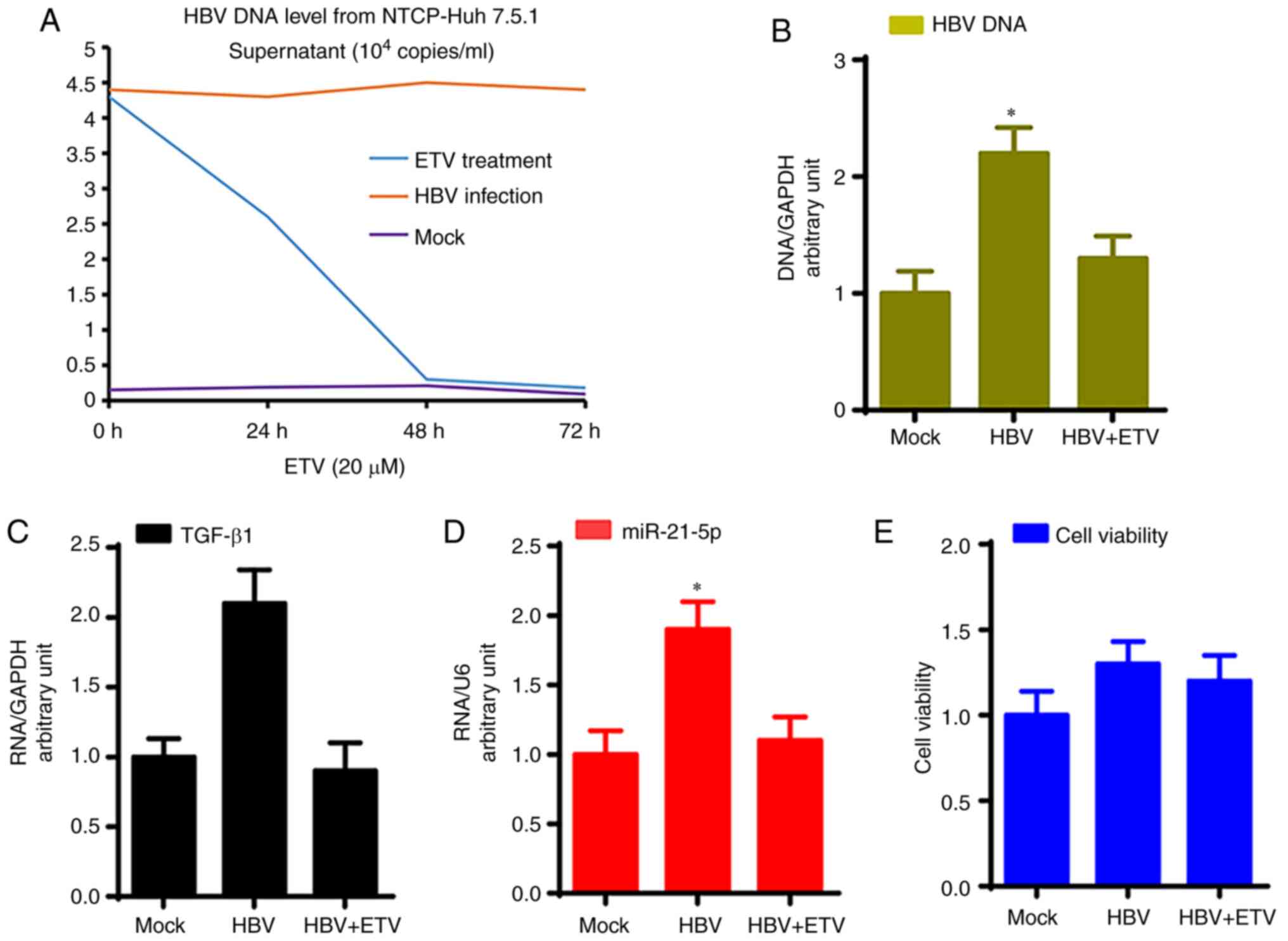
As shown in Fig. 1A,
NTCP-Huh7.5.1 cells can stably support HBV replication; HBV DNA
levels in the supernatant of HBV-infected NTCP-Huh7.5.1 cells were
4.6x104 copies/ml at 72 h after infection and remained
at a stable level over a further 72 h, while ETV treatment
significantly reduced HBV DNA levels in the supernatant of
HBV-infected NTCP-Huh7.5.1 cells (P<0.05). Furthermore, HBV DNA
levels of total cell DNA from HBV-infected NTCP-Huh7.5.1 cells were
significantly higher compared with mock infection, which was
significantly reduced by ETV treatment (Fig. 1B; P<0.05).
Moreover, HBV-infected NTCP-Huh7.5.1 cells showed
significantly higher TGF-β1 mRNA and miR-21-5p levels compared with
NTCP-Huh7.5.1 cells that underwent the mock infection, while ETV
treatment significantly reduced TGF-β1 mRNA and miR-21-5p levels
compared with HBV-infected NTCP-Huh7.5.1 cells (Fig. 1C and D; P<0.05). HBV infection and ETV
treatment did not affect NTCP-Huh7.5.1 cell viability (Fig. 1E).
TGF-β1 induces liver fibrosis gene
expression in LX2 cells through miR-21-5p
TGF-β1 was reported to be the key cytokine in the
process of HSC activation and liver fibrosis (17). The present study investigated the
role of miR-21-5p in TGF-β1-induced liver fibrosis. Following
incubation with 10 ng/ml TGF-β1 for 72 h, LX2 cells were collected
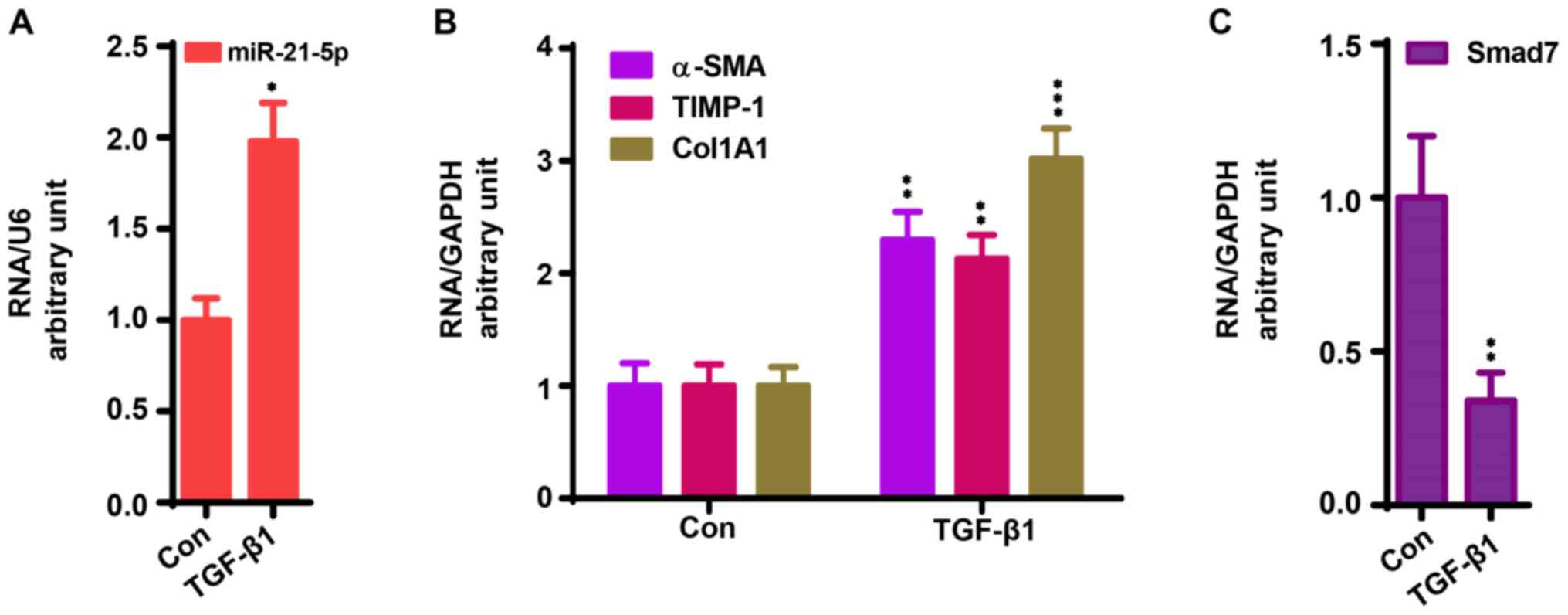
for qPCR assay as previously reported (27). As shown in Fig. 2A and B, TGF-β1 treatment significantly enhanced
the mRNA expression of miR-21-5p, α-SMA, TIMP-1 CoL1A1, while this
treatment significantly decreased the mRNA expression of Smad7
(Fig. 2C) compared with the control
group (P<0.05).
In addition, antagomiR-21 was used to knock down
miR-21-5p expression to further investigate the role of miR-21-5p
in TGF-β1-induced liver fibrosis. LX2 cells were transfected with
antagomiR-21 or miR-21 mimic together with TGF-β1. As shown in
Fig. 3A, miR-21-5p binds to the
Smad7 3'-untranslated region (UTR). miR-21-5p had no effect on
TGF-β1 expression (Fig. 3B). miR-21
mimic enhanced the upregulation of miR-21-5p induced by TGF-β1,
which was counteracted by antagomiR-21 (Fig. 3C; P<0.05). By contrast, miR-21
mimic aggravated the downregulation of Smad7 gene expression
induced by TGF-β1, which was alleviated by antagomiR-21 (Fig. 3D, F
and G; P<0.05). miR-21 mimic
significantly promoted TGF-β1-induced liver fibrosis gene
expression, which was significantly reduced by antagomiR-21
(Fig. 3E-G; P<0.05). Neither
miR-21 mimic nor antagomiR-21 affected the viability of LX2 cells
(Fig. 3H).
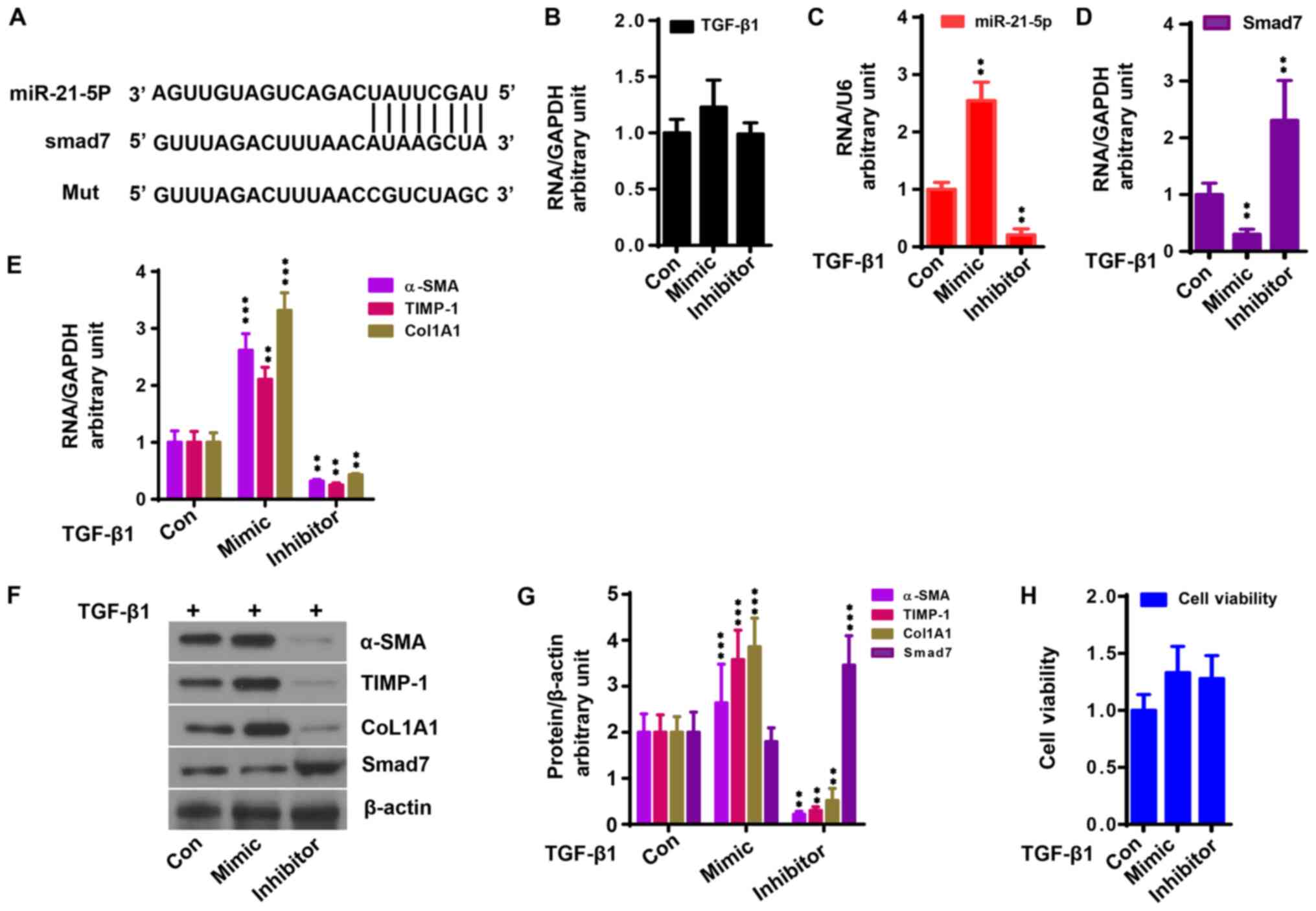 | Figure 3miR-21-5p upregulates TGF-β1
activation in LX2 cells. Control, mimic and inhibitor of miR-21-5p
were transfected into LX2 cells with TGF-β1 (10 ng/ml) stimulation.
(A) miR-21-5p directly binds to the Smad7 3'-untranslated region.
(B) miR-21-5p mimic had no effect on TGF-β1 mRNA expression.
miR-21-5p mimic (C) enhanced miR-21-5p expression and (D) reduced
Smad7 mRNA expression induced by TGF-β1 based on qPCR analysis. (E)
miR-21-5p mimic enhanced α-SMA, TIMP-1 and CoL1A1 mRNA expression
in LX2 cells induced by TGF-β1 based on qPCR analysis, which was
reduced by miR-21-5p inhibitor (P<0.05). (F) Protein expression
of Smad7, α-SMA, TIMP-1 and CoL1A1 in LX2 cells. (G) Relative
protein expression of Smad7, α-SMA, TIMP-1 and CoL1A1 in LX2 cells
to β-actin. (H) miR-21-5p mimic and inhibitor had no effect on cell
viability (P>0.05). **P<0.01 and
***P<0.001 vs. control. SMA, smooth muscle actin;
CoL1A1, collagen type 1 α 1; TIMP, tissue inhibitor of
metalloproteinase; qPCR, quantitative PCR; miR, microRNA; Con,
control. |
HBV replication contributes to liver
fibrosis by upregulating TGF-β1/miR-21-5p
To investigate the mechanism underlying HBV-induced
liver fibrosis, the interaction between HBV-infected NTCP-Huh7.5.1
cells and LX2 cells was observed in a co-culture model (Fig. 4A) as previously described (27). LX2 cells were transfected with
antagomiR-21 or miR-21 mimic prior to co-culture. As a result,
HBV-infected NTCP-Huh7.5.1 cell co-culture significantly increased
the mRNA and protein expression of CoL1A1 in LX2 cells compared
with the control group, which was enhanced by miR-21 mimic and
reduced by antagomiR-21 compared with HBV-infected NTCP-Huh7.5.1
cell co-culture (Fig. 4C, E and F;
P<0.05). As expected, miR-21-5p expression exhibited a similar
trend, while Smad7 expression exhibited an opposite trend compared
with CoL1A1 (Fig. 4D-F; P<0.05).
LX2 cell viability was not affected by miR-21 mimic, antagomiR-21
or HBV transfection (Fig. 4H).
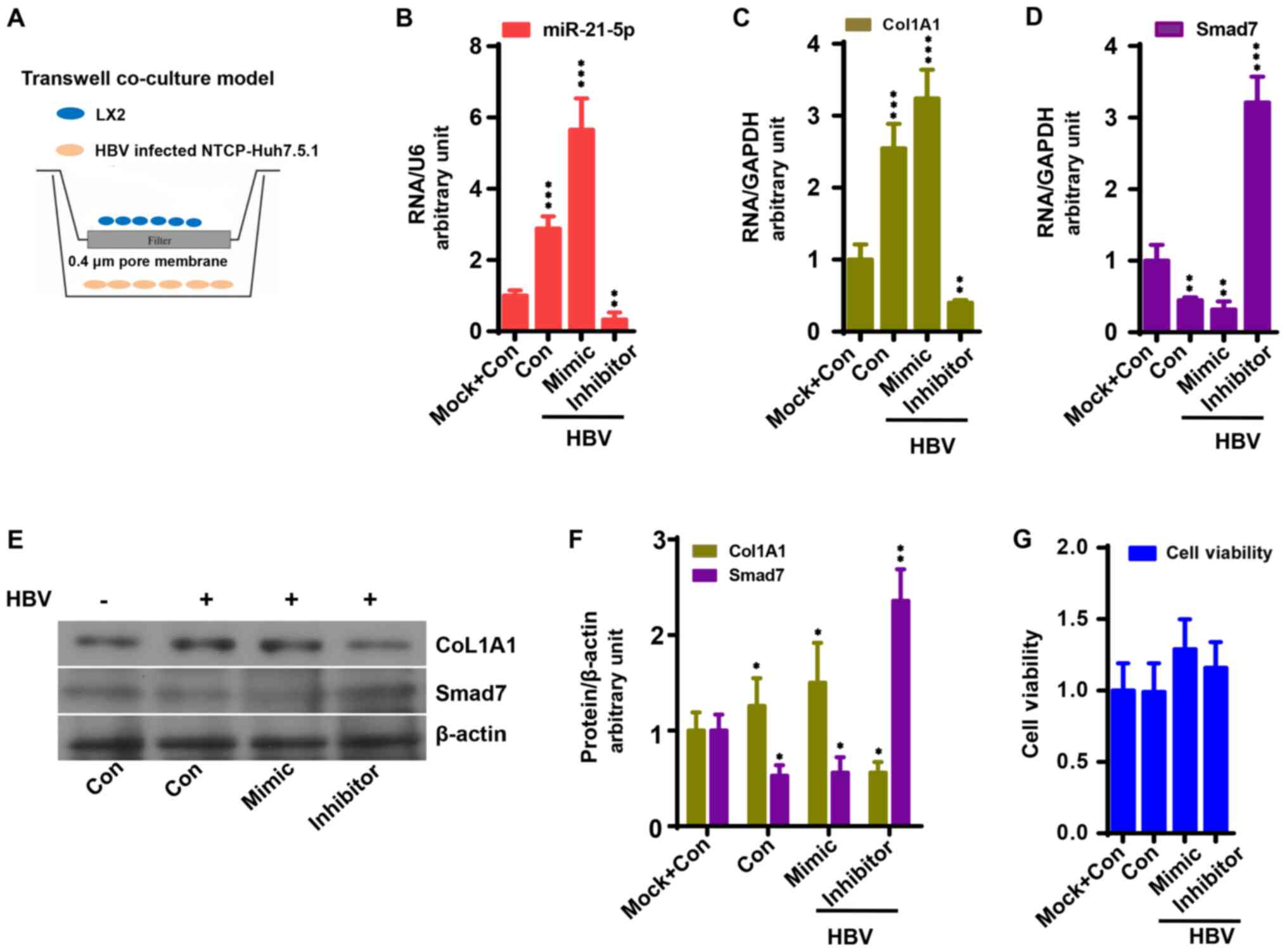 | Figure 4miR-21-5p accelerates the activation
of LX2 cells co-cultured with HBV-infected NTCP-Huh7.5.1 cells. (A)
Co-culture system. miR-21-5p mimic enhanced (B) miR-21-5p and (C)
CoL1A1 mRNA expression in LX2 cells co-cultured with HBV-infected
NTCP-Huh7.5.1 cells, which was reduced by miR-21-5p inhibitor
(P<0.05). (D) miR-21-5p mimic reduced Smad7 mRNA expression in
LX2 cells co-cultured with HBV-infected NTCP-Huh7.5.1 cells based
on quantitative PCR analysis, which was increased by miR-21-5p
inhibitor (P<0.05). (E) Protein expression of Smad7 and CoL1A1
in LX2 cells. (F) Relative protein expression of Smad7, α-SMA,
TIMP-1 and CoL1A1 in LX2 cells to β-actin. (G) miR-21-5p mimic and
inhibitor had no effect on cell viability (P>0.05).
*P<0.05, **P<0.01 and
***P<0.001 vs. control. HBV, hepatitis B virus; NTCP,
sodium taurocholate co-transporting polypeptide; CoL1A1, collagen
type 1 α 1; Con, control; miR, microRNA. |
General and clinical data of patients
with HBV
The present study included 29 patients with HBV
infection to assess the effect of HBV infection on miR-21; 5
patients without HBV infection were enrolled as controls. The
demographic, biological and virological data are shown in Table II. The data demonstrated that
patients with HBV infection had higher serum levels of ALT and AST
as well as HBV DNA levels, compared with the control group
(P<0.05). Furthermore, the liver fibrosis score of HBV-infected
patients was higher compared with the control group
(P<0.05).
 | Table IIClinical, biochemical and laboratory
data of patients. |
Table II
Clinical, biochemical and laboratory
data of patients.
| Characteristic | Control (n=5) | Patients
(n=29) | P-value |
|---|
| Sex (M/F) | 4/1 | 13/16 | >0.05 |
| Age (years) | 38.2±19.4 | 42.0±12.3 | >0.05 |
| Liver fibrosis
(S:1/2/3/4) | 4/1/0/0 | 19/4/4/2 | <0.05 |
| ALT (IU/L) | 13.4±3.4 | 24.0±23.4 | <0.05 |
| AST (IU/L) | 16.0±2.3 | 25.0±10.1 | <0.05 |
| HBV DNA (log
copy/ml) | 0 | 4.0±2.3 | <0.05 |
IHC staining and liver fibrosis
assessment
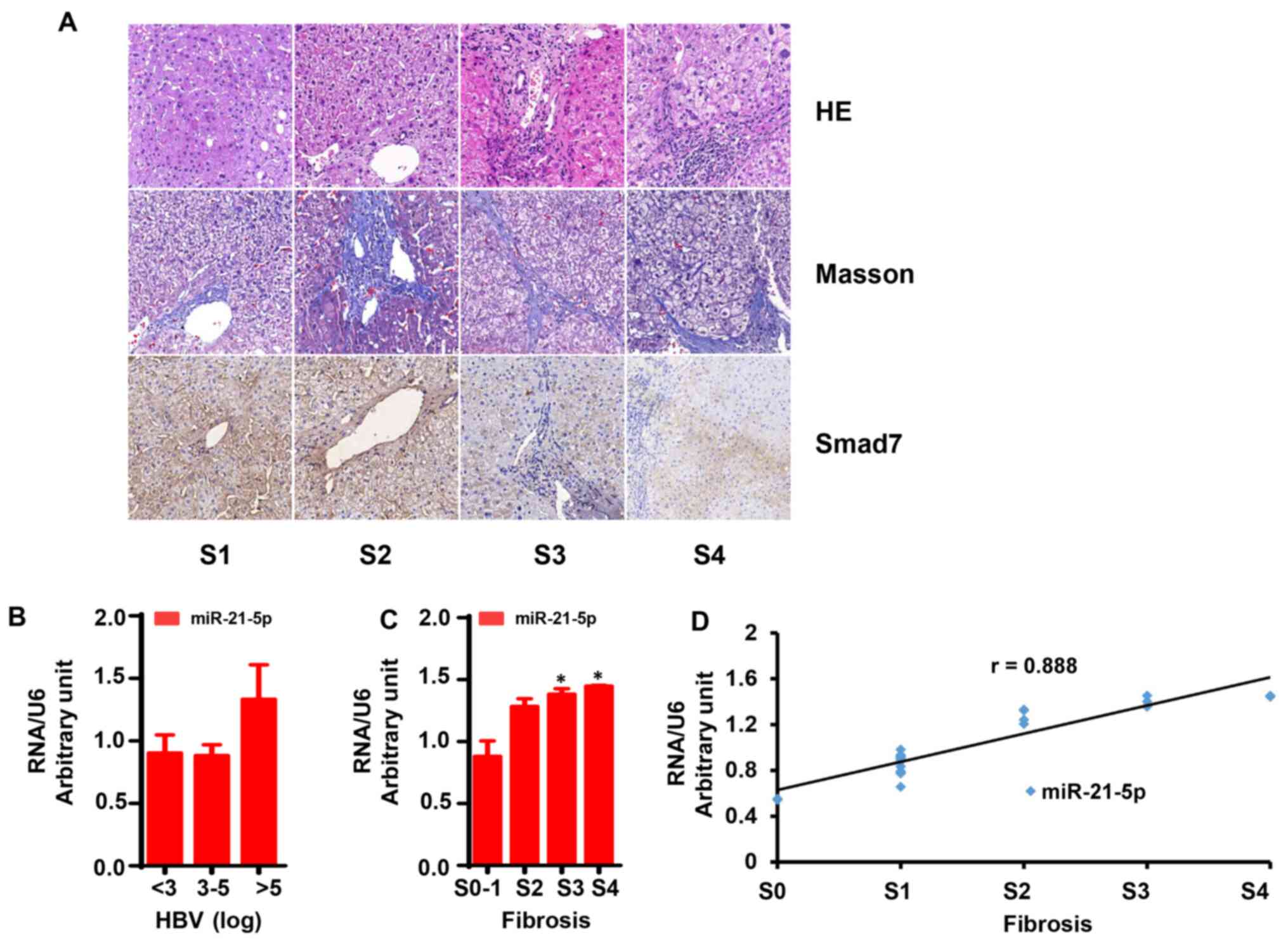
The liver fibrosis score was evaluated using HE and
Masson's staining. As shown in Fig.
5A, liver tissue samples with no fibrosis (S0-1)
exhibited normal lobular architecture with central veins and
radiating hepatic cords. Liver tissue samples with mild fibrosis
(S2) exhibited disordered architecture and bridging
fibrosis. In liver tissue samples with severe fibrosis
(S3-4), normal structure was not visible, while a large
number of pseudolobules was readily identifiable.
IHC staining was also used to assess Smad7 protein
expression in liver tissue. As shown in Fig. 5A, Smad7 expression was significantly
lower in liver tissues with severe fibrosis compared with mild
fibrosis (P<0.05), while mild fibrosis was associated with lower
Smad7 expression compared with liver tissue with no fibrosis
(P<0.05).
mRNA expression of miR-21-5p and
fibrosis genes in liver tissues
The mRNA expression of miR-21-5p in liver tissue was
detected by qPCR. The results demonstrated that liver tissues with
mild fibrosis (S2) had low mRNA levels of miR-21-5p
compared with severe fibrosis (S3-4), and its levels
were even lower in liver tissue with no fibrosis (S0-1)
(P<0.05).
Patients in the present study were divided into two
series as follows: Series 1, HBV DNA levels (log copies/ml) ≤3, 3-5
and ≥5 and series 2, liver fibrosis score S0-1,
S2, S3 and S4. As shown in
Fig. 5B, miR-21 levels exhibited no
significant difference between patients with HBV DNA >5 log and
those with HBV DNA <5 log. Of note, the miR-21 levels in
patients with severe fibrosis was significantly higher compared
with patients with mild fibrosis (Fig.
5C; P<0.05).
Correlation between miR-21-5p and
liver fibrosis
Correlation analysis revealed that miR-21-5p
exhibited a strong positive correlation with liver fibrosis score
(Fig. 5D; r=0.888; P<0.05).
However, there was no significant correlation between miR-21-5p and
HBV DNA levels (r=-0.158; P>0.05).
Discussion
There is a consensus that liver fibrosis is the
healing response of the liver to chronic injuries, including
hepatitis, immune compounds and drugs (28). Generally, hepatitis B remains a
major health concern worldwide, infecting ~350 million patients
(29). HBV also ranks first among
the causes of liver fibrosis in China, infecting ~7% of the entire
population (30). At present,
although anti-HBV treatment has limited success, chronic HBV
infection gradually leads to the development of liver fibrosis and
even liver cancer (31,32).
HBV causes liver damage by engaging hepatocytes,
macrophages and HSCs in a complicated process that remains poorly
understood (33-35).
The main obstacle in HBV research is the lack of cell models of HBV
infection. The present study condensed HBV stock from the HepAD38
cell line, a derivative of hepatoblastoma cells. HepAD38 can stably
produce HBV particles, and have been widely used in scientific
research (25). However, this cell
line could not stimulate the identification and entry of HBV
particles (25,36). NTCP has been demonstrated to be the
receptor for HBV (25,36,37).
The present study constructed a NTCP-Huh7.5.1 cell line by
transfecting NTCP into Huh7.5.1 cells, a derivative of
hepatocellular carcinoma cells. Using this cell line, the presents
study was able to stimulate the natural process of HBV infection.
As a result, HBV levels in the supernatant of HBV-infected
NTCP-Huh7.5.1 cells remained stable at 1x104 copies/ml,
indicating that NTCP-Huh7.5.1 cells supported HBV infection
stably.
Among the various cytokines implicated in liver
fibrosis, TGF-β1 has been proven to be the most important (38). It was demonstrated that TGF-β1
initiates signaling by binding to its receptors on the surface of
LX2 cells, resulting in LX2 activation and ECM production (39). Interestingly, it was previously
suggested that TGF-β1 can be secreted by hepatocytes and
macrophages, in turn activating HSCs (40). Accumulating evidence indicated that
TGF-β1 mainly transduces signals through Smad (41,42).
We have also reported that Smad assisted β-catenin transport to the
nucleus of HSCs, initiating fibrosis gene expression (43). We also demonstrated that HBV
infection promoted TGF-β1 production in hepatocellular carcinoma
cells (26). The present study
demonstrated that HBV-infected NTCP-Huh7.5.1 cells enhanced TGF-β1
production by 2.0-fold compared with NTCP-Huh7.5.1 cells with mock
infection.
miRNAs are a group of non-coding RNAs that play
important roles in regulating the expression of genes involved in
chronic hepatitis B, liver fibrosis and liver cancer (44-46).
Wang et al (21) reported
that miR-21-5p was a key mediator in the TGF-β1/Smad pathway
through inhibiting Smad7 expression in the process of spinal
fibrosis by targeting the AUAAGCUA sequence in the Smad7 3'-UTR,
which was consistent with other reports (46). To the best of our knowledge, no
study has revealed the interaction between miR-21-5p and fibrosis
genes such as α-SAM, TIMP-1 and Col1A1. The present study sought to
investigate the role of miR-21-5p in the TGF-β1/Smad pathway in
HBV-induced liver fibrosis. As expected, TGF-β1 stimulation
upregulated miR-21-5p expression in LX2 cells by 2.1-fold compared
with the control group, and increased the expression of liver
fibrosis genes, such as α-SAM, TIMP-1 and CoL1A1. Moreover, the
TGF-β1-induced liver fibrosis gene expression in LX2 cells was
enhanced by miR-21-5p expression and inhibited by miR-21-5p
knockdown, indicating that miR-21-5p acts as the key mediator of
TGF-β1-induced liver fibrosis. The present study also examined the
interaction between miR-21-5p and Smad7 and observed that
inhibition of miR-21-5p overturned the inhibitory effects of TGF-β1
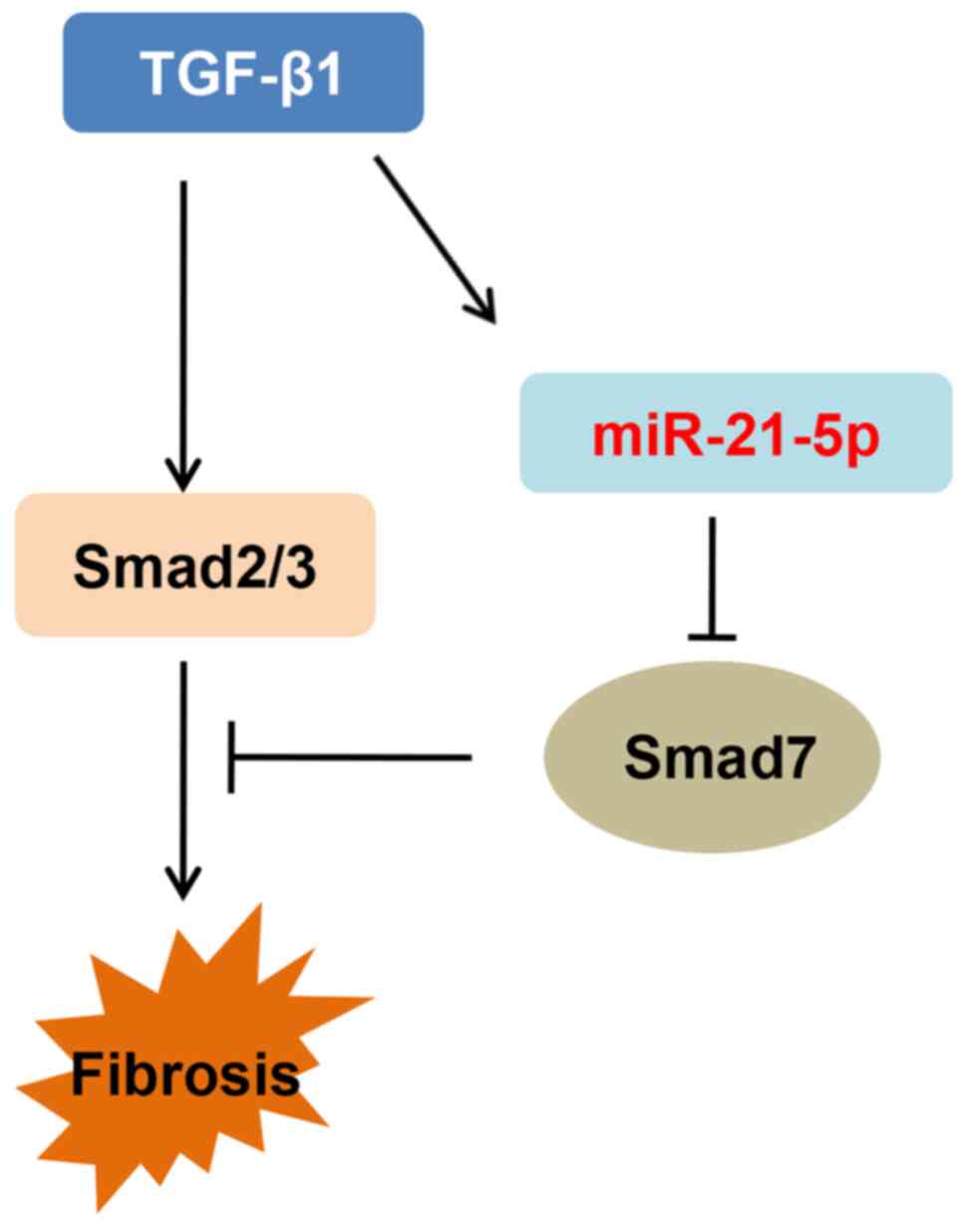
on Smad7. These results suggested that miR-21-5p accelerates liver
fibrosis by inhibiting Smad7 activation (Fig 6).
Liver fibrosis is characterized by excessive
deposition of ECM in the liver, with ensuing destruction of the
liver architecture (28). HSCs are
the main sources of ECM, and their activation has been considered
as a key event in liver fibrosis (40). In this process, cytokines released
from injured hepatocytes initiates HSC activation (41). In the present study, hepatocyte-HSC
interaction was stimulated using a co-culture system, as previously
reported (27). It was demonstrated
that co-culture with HBV-infected NTCP-Huh7.5.1 cells significantly
enhanced CoL1A1 expression in LX2 cells compared with mock
infection, which was reduced by miR-21-5p overexpression,
suggesting that miR-21-5p inhibited HBV-induced liver fibrosis.
In addition, the correlation between miR-21-5p and
liver fibrosis was examined. Results from 29 patients indicated
that liver miR-21-5p expression exhibited a strong positive
correlation with liver fibrosis (r=0.888; P<0.05). Furthermore,
miR-21-5p levels were evaluated according to the HBV DNA levels,
and it was observed that liver miR-21-5p levels did not differ
significantly between patients with HBV DNA >5 log and those
with HBV levels <5 log. There was no significant positive
correlation between miR-21-5p and HBV DNA levels (r=-0.158).
In conclusion, the results of the present study
revealed that HBV leads to liver fibrosis by regulating the
TGF-β1/miR-21-5p pathway. Therefore, miR-21-5p may be a novel
target for the treatment of HBV-induced liver fibrosis.
Acknowledgements
Not applicable.
Funding
The study was supported by the Natural Science
Foundation of Anhui Province (grant no. 1508085MH172) and the
Fundamental Research Funds for the Central Universities (grant no.
WK9110000048).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WL, XY and XC performed the experiments and drafted
the manuscript. MY participated in the liver biopsy and specimen
storage. ZZ participated in the liver inflammation and fibrosis
assessment. CZ was involved in research design, literature review
and data examination. ZW participated in the statistical analysis.
All authors read and approved final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Anhui Provincial Hospital (approval no.
2019-ky026).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang K, Shi Z, Zhang M, Dong X, Zheng L,
Li G, Han X, Yao Z, Han T and Hong W: Silencing lncRNA Lfar1
alleviates the classical activation and pyoptosis of macrophage in
hepatic fibrosis. Cell Death Dis. 11(132)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Su QD, Zhang S, Wang F, Liu H, Zhang GM,
Zheng H, Qiu F, Sun XJ, Liang XF, Bi SL, et al: Epidemiological
distribution of hepatitis B virus genotypes in 1-29-year-olds in
the mainland of China. Vaccine. 38:8238–8246. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mohd-Ismail NK, Lim Z, Gunaratne J and Tan
YJ: Mapping the interactions of HBV cccDNA with host factors. Int J
Mol Sci. 20(4276)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sun Y, Wu X, Zhou J, Meng T, Wang B, Chen
S, Liu H, Wang T, Zhao X, Wu S, et al: Persistent low level of
hepatitis B virus promotes fibrosis progression during therapy.
Clin Gastroenterol Hepatol. 18:2582–2591.e6. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Dai Y, Che F, Jiang X, Cui D, Zhou H, Xu
X, Sun C and Cheng J: Clinical characteristics and association
analysis of persistent low-level HBsAg expression in a physical
examination population with HBV infection. Exp Ther Med. 19:19–32.
2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kim J, Kang W, Kang SH, Park SH, Kim JY,
Yang S, Ha SY and Paik YH: Proline-rich tyrosine kinase 2 mediates
transforming growth factor-beta-induced hepatic stellate cell
activation and liver fibrosis. Sci Rep. 10(21018)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Clayton SW, Ban GI, Liu C and Serra R:
Canonical and noncanonical TGF-β signaling regulate fibrous tissue
differentiation in the axial skeleton. Sci Rep.
10(21364)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chang CJ, Lin CF, Lee CH, Chuang HC, Shih
FC, Wan SW, Tai C and Chen CL: Overcoming interferon (IFN)-γ
resistance ameliorates transforming growth factor (TGF)-β-mediated
lung fibroblast-to-myofibroblast transition and bleomycin-induced
pulmonary fibrosis. Biochem Pharmacol. 183(114356)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Derynck R, Turley SJ and Akhurst RJ: TGFβ
biology in cancer progression and immunotherapy. Nat Rev Clin
Oncol. 18:9–34. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kori M and Arga KY: Pathways involved in
viral oncogenesis: New perspectives from virus-host protein
interactomics. Biochim Biophys Acta Mol Basis Dis.
1866(165885)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ho CH, Chang TT and Chien RN: Telbivudine
on IgG-associated hypergammaglobulinemia and TGF-β1 hyperactivity
in hepatitis B virus-related liver cirrhosis. PLoS One.
14(e0225482)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xu Y, Sun X, Zhang R, Cao T, Cai SY, Boyer
JL, Zhang X, Li D and Huang Y: A positive feedback loop of TET3 and
TGF-β1 promotes liver fibrosis. Cell Rep. 30:1310–1318.e5.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kim JY, Kim KM, Yang JH, Cho SS, Kim SJ,
Park SJ, Ahn SG, Lee GH, Yang JW, Lim SC, et al: Induction of E6AP
by microRNA-302c dysregulation inhibits TGF-β-dependent
fibrogenesis in hepatic stellate cells. Sci Rep.
10(444)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang Y, Li J, Wang S, Yang F, Zhou Y, Liu
Y, Zhu W and Shi X: HBx-associated long non-coding RNA activated by
TGF-β promotes cell invasion and migration by inducing autophagy in
primary liver cancer. Int J Oncol. 56:337–347. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li MH, Chen QQ, Zhang L, Lu HH, Sun FF,
Zeng Z, Lu Y, Yi W and Xie Y: Association of cytokines with
hepatitis B virus and its antigen. J Med Virol: Jul 14, 2020 (Epub
ahead of print).
|
|
16
|
Martin N, Ziegler DV, Parent R and Bernard
D: Hepatic stellate cell senescence in liver tumorigenesis.
Hepatology: Sep 15, 2020 (Epub ahead of print).
|
|
17
|
Zhang J, Jiang N, Ping J and Xu L:
TGF-β1-induced autophagy activates hepatic stellate cells via the
ERK and JNK signaling pathways. Int J Mol Med: Nov 3, 2020 (Epub
ahead of print).
|
|
18
|
Wang X, He Y, Mackowiak B and Gao B:
MicroRNAs as regulators, biomarkers and therapeutic targets in
liver diseases. Gut: Oct 30, 2020 (Epub ahead of print).
|
|
19
|
Ezhilarasan D: MicroRNA interplay between
hepatic stellate cell quiescence and activation. Eur J Pharmacol.
885(173507)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Salimi S, Noorbakhsh F, Faghihzadeh S,
Ghaffarpour S and Ghazanfari T: Expression of miR-15b-5p,
miR-21-5p, and SMAD7 in lung tissue of sulfur mustard-exposed
individuals with long-term pulmonary complications. Iran J Allergy
Asthma Immunol. 18:332–339. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang W, Liu R, Su Y, Li H, Xie W and Ning
B: MicroRNA-21-5p mediates TGF-β-regulated fibrogenic activation of
spinal fibroblasts and the formation of fibrotic scars after spinal
cord injury. Int J Biol Sci. 14:178–188. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chinese Society of Hepatology, Chinese
Medical Association; Chinese Society of Infectious Diseases. The
guideline of prevention and treatment for chronic hepatitis B: A
2019 update. Chin J Infect Dis. 37:711–736. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhu CL, Li WT, Li Y and Gao RT: Serum
levels of tissue inhibitor of metalloproteinase-1 are correlated
with liver fibrosis in patients with chronic hepatitis B. J Dig
Dis. 13:558–563. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bedossa P and Poynard T: An algorithm for
the grading of activity in chronic hepatitis C The METAVIR
cooperative study group. Hepatology. 24:289–293. 1996.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Duan X, Li S, Holmes JA, Tu Z, Li Y, Cai
D, Liu X, Li W, Yang C, Jiao B, et al: MicroRNA 130a regulates both
hepatitis C virus and hepatitis B virus replication through a
central metabolic pathway. J Virol. 92:e02009–17. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li W, Yu X, Zhu C, Wang Z, Zhao Z, Li Y
and Zhang Y: Notum attenuates HBV-related liver fibrosis through
inhibiting Wnt 5a mediated non-canonical pathways. Biol Res.
52(10)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kisseleva T and Brenner D: Molecular and
cellular mechanisms of liver fibrosis and its regression. Nat Rev
Gastroenterol Hepatol: Oct30, 2020 (Epub ahead of print).
|
|
29
|
Liu L, Zhu J, Yang J, Li X, Yuan J, Wu J
and Liu Z: GP73 facilitates hepatitis B virus replication by
repressing the NF-κB signaling pathway. J Med Virol, February 20,
2020 (Online ahead of print).
|
|
30
|
Wang T, Dai Y, Lu W, Zhou H, Chen Y, Xu X,
Sun C and Cheng J: An epidemiological survey of HBV infection and
low-level HBsAg in military camps in eastern China. Medicine
(Baltimore). 97(e12201)2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li Z, Hu Y, Wang H, Wang M, Gu X, Ping Y,
Zeng Q, Li H, Yan J and Yu Z: Predictors for the progression of
hepatic cirrhosis to hepatocellular carcinoma under long-term
antiviral therapy. Eur J Gastroenterol Hepatol. 32:447–453.
2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yotsuyanagi H, Takano T, Tanaka M, Amano
K, Imamura M, Ogawa K, Yasunaka T, Yasui Y, Hayashi K, Tanaka Y, et
al: Hepatitis B virus-related hepatocellular carcinoma in young
adults: Efficacy of nationwide selective vaccination. Hepatol Res.
50:182–189. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Duriez M, Mandouri Y, Lekbaby B, Wang H,
Schnuriger A, Redelsperger F, Guerrera CI, Lefevre M, Fauveau V,
Ahodantin J, et al: Alternative splicing of hepatitis B virus: A
novel virus/host interaction altering liver immunity. J Hepatol.
67:687–699. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yuan L, Jiang J, Liu X, Zhang Y, Zhang L,
Xin J, Wu K, Li X, Cao J, Guo X, et al: HBV infection-induced liver
cirrhosis development in dual-humanised mice with human bone
mesenchymal stem cell transplantation. Gut. 68:2044–2056.
2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bility MT, Cheng L, Zhang Z, Luan Y, Li F,
Chi L, Zhang L, Tu Z, Gao Y, Fu Y, et al: Hepatitis B virus
infection and immunopathogenesis in a humanized mouse model:
Induction of human-specific liver fibrosis and M2-like macrophages.
PLoS Pathog. 10(e1004032)2014.PubMed/NCBI View Article : Google Scholar : Yan H, Zhong G, Xu
G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H et al:
Sodium taurocholate cotransporting polypeptide is a functional
receptor for human hepatitis B and D virus. Elife 1: e00049,
2012.
|
|
36
|
Donkers JM, Appelman MD and van de Graaf
SFJ: Mechanistic insights into the inhibition of NTCP by myrcludex
B. JHEP Rep. 1:278–285. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tian F, Liu Y, Gao J, Yang N, Shang X, Lv
J, Ba D, Zhou X, Zhang C and Ma X: Study on the association between
TGF-β1 and liver fibrosis in patients with hepatic cystic
echinococcosis. Exp Ther Med. 19:1275–1280. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tao L, Xue D, Shen D, Ma W, Zhang J, Wang
X, Zhang W, Wu L, Pan K, Yang Y, et al: MicroRNA-942 mediates
hepatic stellate cell activation by regulating BAMBI expression in
human liver fibrosis. Arch Toxicol. 92:2935–2946. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Dewidar B, Meyer C, Dooley S and
Meindl-Beinker AN: TGF-β in hepatic stellate cell activation and
liver fibrogenesis-updated 2019. Cells. 8(1419)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Frangogiannis N: Transforming growth
factor-β in tissue fibrosis. J Exp Med.
217(e20190103)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Marvin DL, Heijboer R, Ten Dijke P and
Ritsma L: TGF-β signaling in liver metastasis. Clin Transl Med.
10(e160)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li W, Zhu C, Chen X, Li Y, Gao R and Wu Q:
Pokeweed antiviral protein down-regulates Wnt/β-catenin signalling
to attenuate liver fibrogenesis in vitro and in vivo. Dig Liver
Dis. 43:559–566. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Sagnelli E, Potenza N, Onorato L, Sagnelli
C, Coppola N and Russo A: Micro-RNAs in hepatitis B virus-related
chronic liver diseases and hepatocellular carcinoma. World J
Hepatol. 10:558–570. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhi SC, Chen SZ, Li YY, Li JJ, Zheng YH
and Yu FX: Rosiglitazone inhibits activation of hepatic stellate
cells via up-regulating Micro-RNA-124-3p to alleviate hepatic
fibrosis. Dig Dis Sci. 64:1560–1570. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sendi H, Mehrab-Mohseni M, Russo MW,
Steuerwald N, Jacobs C, Clemens MG and Bonkovsky HL: Baseline
hepatic levels of miR-29b and claudin are respectively associated
with the stage of fibrosis and HCV RNA in hepatitis C. Clin Exp
Gastroenterol Hepatol. 1(105)2019.PubMed/NCBI
|
|
46
|
Li Q, Li B, Li Q, Wei S, He Z, Huang X,
Wang L, Xia Y, Xu Z, Li Z, et al: Exosomal miR-21-5p derived from
gastric cancer promotes peritoneal metastasis via
mesothelial-to-mesenchymal transition. Cell Death Dis.
9(854)2018.PubMed/NCBI View Article : Google Scholar
|