Introduction
Successful embryo implantation is a key step in
pregnancy and is a complicated physiological process that consists
of various steps, including blastocyst hatching, invasion,
migration, attachment and placentation (1,2).
During this period, the uterus undergoes several alterations to
establish an optimal environment for embryonic growth. A complex
regulatory network of molecular interactions exists at the
fetomaternal interface (3). IFN-τ
is produced during the early embryo implantation, especially in
ruminants, such as cattle, sheep and deer, and serves a key role in
the maternal recognition of pregnancy (4-6).
Previous studies have indicated that IFN-τ upregulated the
expression of bovine leukocyte antigen (BoLA)-I, which is the
equivalent of the major histocompatibility complex class I (MHC-I)
antigen in bovines, and may modulate immune responses and
contribute to fetomaternal tolerance in dairy cattle (7-10).
Our previous has revealed that IFN-τ stimulation activated a wide
variety of microRNAs (miRNAs/miRs) in BEECs (11). Whether the upregulation of BoLA-I
expression is associated with miRNAs is still unknown. Moreover,
the underlying mechanisms of the contributions of IFN-τ to embryo
implantation also remain unclear.
miRNAs are a class of small, noncoding,
single-stranded RNAs comprising 22-25 nucleotides. miRNAs have been
suggested to negatively regulate gene expression by targeting mRNAs
to interfere with post-transcriptional protein translation
(12). Because of their ability to
silence genes, miRNAs can modulate a variety of physiological and
pathological processes, including cellular proliferation, apoptosis
and immune responses (13-15).
A previous study has indicated that miRNAs regulated the molecules
involved in peri-implantation and pregnancy, such as let-7a and
miR-320(16). The functional study
of miRNAs may be helpful in revealing the molecular pathways
associated with the embryo implantation process.
Preliminary deep sequencing data indicated that
bta-miRNA-204 was downregulated in bovine endometrial epithelial
cells (bEECs) following IFN-τ treatment (11). Gene Ontology and Kyoto Encyclopedia
of Genes and Genomes pathway analyses indicated the target genes of
bta-miRNA-204, whose function analysis found that those predicted
genes were enriched in graft-vs.-host disease and allograft
rejection, including BoLA-I (17,18) In
this research, we will reveal the inner association between
bta-miRNA-204 and BoLA-IIn the present study, bioinformatics
algorithms were used to predict which genes may be regulated by
bta-miRNA-204. Of note, BoLA was indicated to be a target gene of
bta-miRNA-204. BoLA gene, which is also known as BoLA-A, belongs to
the classic BoLA-I family, and encodes the heavy chain of BoLA
class I molecules (19). As BoLA-I
serves a crucial role in the regulation of immunosuppression and
implantation during early embryonic development (20), the present study investigated
whether IFN-τ-mediated regulation of bta-miRNA-204 contributes to
effective embryo implantation.
Programmed cell death receptor 1 (PD-1) binds to the
programmed death-ligand 1 or 2 (PD-L1/PD-L2) to form a
costimulatory signal that negatively regulates T cell immunity.
PD-L1 and PD-L2 are expressed by antigen presenting cells (APCs)
(21). On the other hand, APCs also
express MHC molecules that can activate T cells by interacting with
the T cell receptor (TCR) (22,23).
Tumor cells frequently upregulate the expression of PD-L1 or PD-L2
to facilitate their escape from the immune system. Although there
have been numerous studies about the PD-1/PD-L signaling pathway in
tumor immune escape, this pathway has not been well described in
pregnant immune tolerance (24). As
the expression of PD-L1 and PD-L2 is regulated by several factors
and IFN-τ can stimulate bEECs to produce MHC, whether it can also
stimulate bEECs to express PD-L1 and PD-L2 is still unknown
(10,25,26).
Moreover, there may be a connection between MHC and PD1 ligands
based on the costimulatory signaling pathway of T cells (27), but few reports exist on this domain,
therefore their relationship should be further examined. The main
purpose of the present study was to preliminary explore whether
IFN-τ could stimulate bEECs to produce both MHC and PD-L1 and
PD-L2.
Materials and methods
Reagents
Recombinant ovine IFN-τ was purchased from Creative
Bioarray. FBS was purchased from SAFC Biosciences Pty Ltd. Bovine
leukocyte antigen (HLA) class I (Thermo Fisher Scientific, Inc.;
cat. no. MA5-28477), Actin Monoclonal Antibody (Thermo Fisher
Scientific, Inc.; cat. no. MA1-744) HRP-conjugated goat anti-rabbit
antibody and the primary antibody anti-cytokeratin-18 (CK-18; cat.
no. MA5-12104) were provided by Abcam. Cell Counting Kit-8 (CCK-8)
was purchased from Dojindo Molecular Technologies, Inc.
LightCycler® FastStart DNA Master PLUS SYBR Green kit
was purchased from Roche Applied Science. The microRNA and U6 small
nuclear RNA normalization reverse transcription-quantitative PCR
(RT-qPCR) kit was purchased from Shanghai GenePharma Co., Ltd. The
bta-miR-204 mimic (bta-miR-204 agomir) and an inhibitor
(bta-miR-204 antagomir), as well as were three BoLA small
interfering (si)RNAs and a negative control (NC) siRNA were
synthesized by Shanghai GenePharma Co., Ltd. The FastDigest
XhoI and NotI, Lipofectamine® 2000 and
Lipofectamine RNAiMAX kits were obtained from Thermo Fisher
Scientific, Inc. Dual-Luciferase Reporter Assay System and
psi-CHECK™-2 plasmid were obtained from Promega Corporation. The
sequences of all primers were synthesized by Shanghai GenePharma
Co., Ltd. and are listed in Table
I. The sequences of the agomirs and antagomirs are presented in
Table II. All other chemicals were
reagent grade.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| A, Primers for
3'-UTR cloning |
|---|
| Name | Sequence
(5'-3') |
|---|
| BoLA 3'-UTR-F |
ATCTCGAGATGACATCGAGTGGCCAGAG |
| BoLA 3'-UTR-R |
GAGCGGCCGCAGGCGATTGGATTTGTCGGC |
| Mut-BoLA
3'-UTR-F |
ATACTCGAGCGAAAGCATGCGTCGTACCT |
| Mut-BoLA
3'-UTR-R |
ATGCGGCCGGCCGAAAGTTCCTTTGTGGG |
| B, Primers for
reverse transcription-quantitative PCR |
| Name | Sequence
(5'-3') |
| β-actin-F |
TGGACTTCGAGCAGGAGAT |
| β-actin-R |
CGTCACACTTCATGATGGAA |
| IFN-τ-F |
TGAACAGACTCTCTCCTCATCCC |
| IFN-τ-R |
TGGTTGATGAAGAGAGGGCTCT |
| BoLA-F |
CTCACACCGTCCAAGAGATG |
| BoLA-R |
CTCGTTCAGGGCGATGTAAT |
| RT-bta-miR-204 |
CTCAACTGGTGTCGTGGAGTCGG
CAATTCAGTTGAGAGGCATAG |
| bta-miR-204-F |
CGTGGACTTCCCTTTGTCA |
| bta-miR-204-R |
CTCAACTGGTGTCGTGGA |
| PD-L1-F |
TTGGTCATCCCAGAACCATATC |
| PD-L1-R |
CCTTCCAGGGTACCTTTATTCC |
| PD-L2-F |
CTACAAGTACCTGACGCTGAAA |
| PD-L2-R |
CAACGATGAGGGAGAGAATGAA |
| U6-F |
CTCGCTTCGGCAGCACATATACT |
| U6-R |
ACGCTTCACGAATTTGCGTGTC |
 | Table IISequences of agomirs and
antagomirs. |
Table II
Sequences of agomirs and
antagomirs.
| Name | Sequence
(5'-3') |
|---|
| bta-miR-204
agomir |
UUCCCUUUGUCAUCCUAUGCCU
GCAUAGGAUGACAAAGGGAAUU |
| bta-miR-204 agomir
NC |
UUCUCCGAACGUGUCACGUTT
ACGUGACACGUUCGGAGAATT |
| bta-miR-204
antagomir |
AGGCAUAGGAUGACAAAGGGAA |
| bta-miR-204
antagomir NC |
CAGUACUUUUGUGUAGUACAA |
Animals and experimental groups
All experimental procedures involving animals and
their care conformed to the Guide for the Care and Use of
Laboratory Animals of National Veterinary Research of China. The
present study was approved by the Huazhong Agricultural University
Animal Care and Use Committee (Wuhan, China; approval no.
20171354CA).
The pregnant cow (n=3) and nonpregnant female dairy
cattle (n=5) were obtained from the Animal Experimental Center of
Huazhong Agricultural University. All cattle were between 16 and 24
months old, and weighed between 350 and 390 kg. They were
acclimatized for one week (24±1˚C, relative humidity of 60-65%, 12
h light/dark cycle with ad libitum supply of food and water.
The body temperature and food intake of each cow was recorded every
day. The dairy cattle were anesthetized with an intravenous
injection of 40 mg/kg sodium pentobarbital to minimize suffering
(28). The endometrial epithelium
(for cell culture) and endometrial tissues of nonpregnant cows
(n=5) were collected before ovulation. Pregnancy was induced by
artificial insemination, and the day of insemination was designated
day 0 of pregnancy (10,29). The endometrial tissues of cattle in
early pregnancy were collected during implantation (day 9-25; n=3).
An intravenous injection of ≥100 mg/kg sodium pentobarbital was
used for euthanasia. All samples were obtained within 30 min after
exsanguination and immediately transported to the laboratory on
ice.
Primary bovine endometrial epithelial
cell (bEEC) culture and identification
The collected caruncular endometrial epithelium was
mixed with adequate 1% collagenase I (10-15 ml), cut into 1x1 mm
pieces and incubated for 1 h in a sealed container in a
thermostatic shaker at 37˚C and 88 revolutions/minute. Collagenase
I was neutralized with FBS (1-1.5 ml) after the incubation period,
and the tissue pieces were placed in a culture dish (35 mm) in
0.5-1 cm intervals. The culture dish was incubated in 5%
CO2 at 37˚C for 3 h. After the cells adhered to the
dish, the bEECs were cultured in DMEM/F12 (Thermo Fisher
Scientific, Inc.; cat. no. 21041033) supplemented with 15% FBS, 2
mM L-glutamine, 50 U/ml penicillin, 50 U/ml streptomycin, 100 U/ml
gentamicin and 10 ng/ml EGF, and maintained in a 5% CO2
humidified incubator at 37˚C. The nutrient solution was replaced
after 8 h, and thereafter it was replaced every 12 h. Following 48
h, the tissue block was removed, and the medium was replaced every
48 h. The cells were transferred (via trypsinization at room
temperature for 20 sec) to 6-well plates on coverslips and analyzed
for the expression of the epithelial-specific marker CK-18. Cells
were grown to approximately 70% confluence, fixed with 4%
paraformaldehyde for 15 min at room temperature and washed three
times with PBS. The cells were blocked with 10% normal goat serum
at room temperature for 30 min and incubated with CK-18 primary
antibody (diluted 1:100) overnight at 4˚C. The fluorescently
labeled DyLight 594 (diluted 1:1,000) secondary antibody was
incubated for 45 min at room temperature. DAPI (300 nM) was used to
stain the cell nuclei for 1-5 min at room temperature. Fluorescent
images were captured using laser scanning confocal microscopy
(magnification, x50 and x100) and analyzed using ImageJ (https://imagej.nih.gov/ij/, National Institutes of
Health; ImageJ bundled with 64-bit Java 1.8.0_172). The sixth or
seventh generation of bEECs was used for subsequent
experiments.
bta-miR-204 target analysis
The bioinformatics database TargetScan 7.2
(http://www.targetscan.org/) and
RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/)
were used to search for the target genes of bta-miR-204. The
duplexes and the minimum free energy (mFE) between bta-miR-204 and
the 3'-untraslated regions (3'-UTRs) of the potential targets were
analyzed by RNA hybridization (30).
Plasmid construction
To construct the wild-type and mutant BoLA 3'-UTR
luciferase reporter plasmids, the full length of the BoLA 3'-UTR or
fragments covering the putative bta-miR-204 binding site were
amplified by RT-qPCR using cDNA resulting from extractions from
dairy cattle uterine tissues. The amplified products were subcloned
into the XhoI and NotI sites of the psi-CHECK-2
vector. Mutagenesis of the seed region (the bta-miR-204 target
site) was achieved by PCR (Shanghai GenePharma Co., Ltd.), using
the 3'-UTR plasmid as the template. The following thermocycling
conditions were used: 94˚C for 2 min; followed by 35 cycles of 94˚C
for 20 sec; 56˚C for 20 sec and 72˚C for 150 sec; followed by 72˚C
for 5 min and 15˚C for 1 min. The amplified products were digested
using the DpnI restriction enzyme. E. coli DH5α cells
were transformed with the plasmids, and colonies were grown on
Luria-Bertani plates containing ampicillin at 37˚C for 16-18 h. The
wild-type and mutant sequences were confirmed by enzyme digestion
and sequencing. The recombinant wild-type and mutant plasmids were
named Luc-BoLA (3'UTR) and Luc-BoLA (3'UTR)-Mut, respectively.
Dual-luciferase reporter assay
The dual-luciferase reporter assay included two
reporters. One was Renilla luciferase, and the other was
firefly luciferase in pmirGLO vector (Promega Corporation), which
contained the examined 3'-UTR sequence. For the luciferase reporter
assay, bEECs were plated in a 6-well plate to a density of 20-30%,
1 day before transfection. On the second day, 200 ng luciferase
reporter plasmid and 10 pmol bta-miR-204 agomir, agomir NC,
antagomir and antagomir NC were also transfected into bEECs using
Lipofectamine 2000. Luc-BoLA (3'UTR) was transfected into bEECs
without the indicated agomirs/antagomirs as the control. The cells
were collected at 24 h post transfection, and dual-luciferase
activity assays were performed using Dual-Luciferase Reporter Assay
System according to the manufacturer's instructions. The luciferase
activity was detected using a Lumat LB 9507 Ultra-Sensitive Tube
Luminometer (Titertek-Berthold). The firefly luciferase activity of
each sample was normalized to the Renilla luciferase
activity. At least three independent repeats were performed for all
the aforementioned transfection experiments.
siRNA design and cell
transfection
The selection of siRNAs was based on the
characterization of siRNAs in a previous study (31). According to the sequence
characteristics of siRNA, the general design principle and previous
design experience, several siRNAs were designed using siRNA online
design tools. Three siRNA sequences were established to target
cattle BoLA mRNA (BoLA siRNA1-3; Table III). To identify the most
effective siRNA, 100 pmol (the amount recommended in the
transfection protocol) of each of these siRNAs were used for
transfection into primary bEECs using Lipofectamine RNAiMAX.
RT-qPCR was used to evaluate the efficacy of the siRNAs in
downregulating BoLA expression in the cells. The BoLA siRNA that
exhibited the best inhibitory effect on BoLA mRNA expression was
used in subsequent experiments. Primary bEECs were seeded into a
6-well plate with 30-40% cell density. The bEECs were treated with
IFN-τ (200 ng/ml dissolved in DMSO). The blank group was treated
with the same amount of DMSO. In addition, the other experimental
groups were transfected with 100 pmol BoLA siRNA or NC siRNA at
37˚C for 6 h and subsequently treated with 200 ng/ml IFN-τ at 37˚C
for another 12 h immediately after removal of transfection
media.
 | Table IIISequences of siRNA
oligonucleotides. |
Table III
Sequences of siRNA
oligonucleotides.
| Name | Sequence
(5'-3') |
|---|
| BoLA siRNA1 |
GCUCAAGUCACCAAGCACATT
UGUGCUUGGUGACUUGAGCTT |
| BoLA siRNA2 |
GCAUCAUUGUUGGACUGGUTT
ACCAGUCCAACAAUGAUGCTT |
| BoLA siRNA3 |
GUGUCUCUCAUGGUUCCUATT
UAGGAACCAUGAGAGACACTT |
| NC siRNA |
UUCUCCGAACGUGUCACGUTT
ACGUGACACGUUCGGAGAATT |
Cell treatment and cell proliferation
assays
Primary bEECs were seeded into a 6-well plate with
30-40% cell density. The bEECs were treated with 200 ng/ml IFN-τ
(11). The blank group was treated
with the same amount of DMSO. Furthermore, the other experimental
groups were transfected with 100 pmol (in accordance with the
transfection protocol) bta-miR-204 agomir, agomir NC, antagomir or
antagomir NC using Lipofectamine 2000 at 37˚C for 6 h, as
aforementioned. The siRNA groups were transfected with 100 pmol
BoLA siRNA or NC siRNA. CCK-8 was used to examine cell
proliferation according to the manufacturer's protocol at 6, 12, 24
and 48 h post-treatment. The cells were treated with 1 ml DMEM/F-12
with CCK-8 reagent (1:10 v/v) and incubated for 1 h. The absorbance
of each well was measured at 450 nm using a microplate reader
(Bio-Rad Laboratories, Inc.) to estimate the cell number.
RT-qPCR
Total RNA from primary bEECs was isolated using
TRIzol® Reagent (cat. no. 15596018; Thermo Fisher
Scientific, Inc.) and converted into cDNA (30˚C for 10 min followed
by 42˚C for 30 min) with the PrimeScript 1st strand cDNA Synthesis
Kit according to the manufacturer's instructions (Takara Bio,
Inc.). Primer Premier 5 software (Premier Biosoft International)
was used to design the specific primers (Table I). qPCR was performed on a StepOne
Real-time PCR System (Thermo Fisher Scientific, Inc.) with the
LightCycler® FastStart DNA Master PLUS SYBR Green mix in
a 25-µl reaction. U6 was used as the housekeeping gene. The PCR
producer for the bta-miR-204 reverse transcription reaction was
95˚C for 5 min; 40 cycles of 95˚C for 10 sec and 60˚C for 30 sec;
95˚C for 15 sec, 60˚C for 60 sec, 95˚C for 15 sec. The producer for
the PCR reaction was 95˚C 3 min; followed by 95˚C for 12 sec and
62˚C for 40 sec. Each sample was assayed in triplicate. The results
(fold changes) were quantified using the
2-ΔΔCq method (32).
Western blot analysis
Total protein from primary bECCs and tissues was
extracted by RIPA Lysis and Extraction Buffer (cat. no. 8990,
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The protein concentration was determined using a BCA
Protein Assay kit. Samples with equal amounts of protein (50 µg)
were separated using 10% SDS-PAGE and transferred to a PVDF
membrane, which was blocked in 5% skimmed milk in 0.4% TBS-Tween-20
(TBST) at room temperature for 2 h. The membrane was incubated with
the primary antibody (1:500 dilution) at 4˚C overnight. Following
washing with TBST, the membrane was incubated with the secondary
antibody (1:1,500 dilution) at room temperature for 2 h. Protein
expression was detected using the ECL Plus Western Blotting
Detection system (Hangzhou Bioer Co., Ltd) and analyzed using
ImageQuant LAS 4000 mini software (Cytiva) according to the
ImageQuant LAS 4000 User Manual 28-9607-42 AC. β-actin was used as
a loading control.
Statistical analysis
Data are presented as the mean ± SEM (n=3).
Statistical analyses were performed using Microsoft Excel 2016
(Microsoft Corporation) and GraphPad Prism 6 (GraphPad Software,
Inc.). Comparisons among all groups were performed with one-way
ANOVA followed by Tukey's multiple comparisons test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of bta-miR-204, IFN-τ and
BoLA in the endometrial tissues of dairy cattle
A previous study using deep sequencing indicated
that IFN-τ (200 ng/ml) decreased the expression of bta-miRNA-204 in
bEECs (10). To evaluate whether
pregnancy affects the expression of IFN-τ, bta-miR-204 and BoLA,
their levels in pregnant cattle were compared with the respective
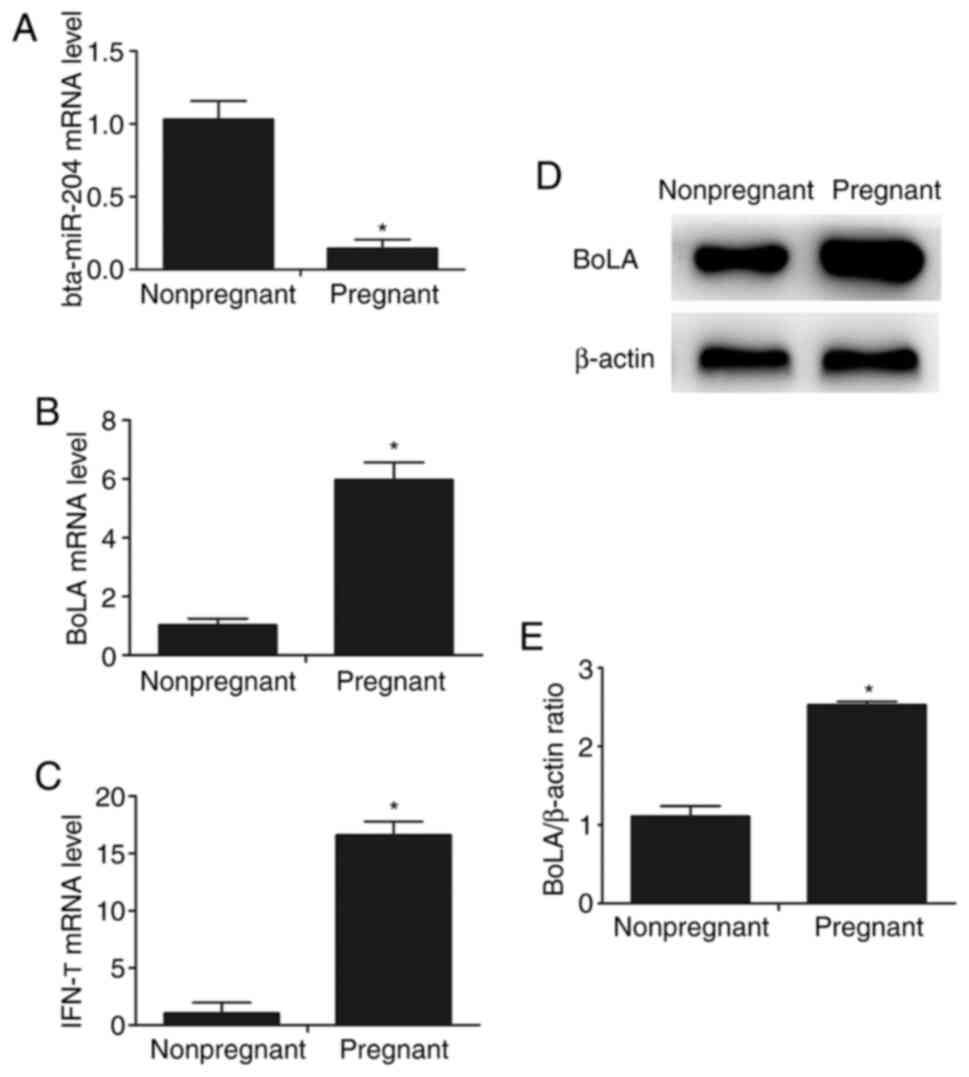
levels in nonpregnant cattle. The results revealed that the mRNA
expression level of bta-miR-204 was significantly decreased in
cattle during early pregnancy, but the expression level of BoLA in
early pregnant cattle was increased compared with that in
nonpregnant cattle (Fig. 1A and
B). Moreover, the mRNA level of
IFN-τ was significantly higher in the endometrium of early pregnant
cattle compared with that in nonpregnant cattle (Fig. 1C). Moreover, the protein expression
level of BoLA was indicated to be increased in cattle during early
pregnancy using western blot analysis (Fig. 1D and E).
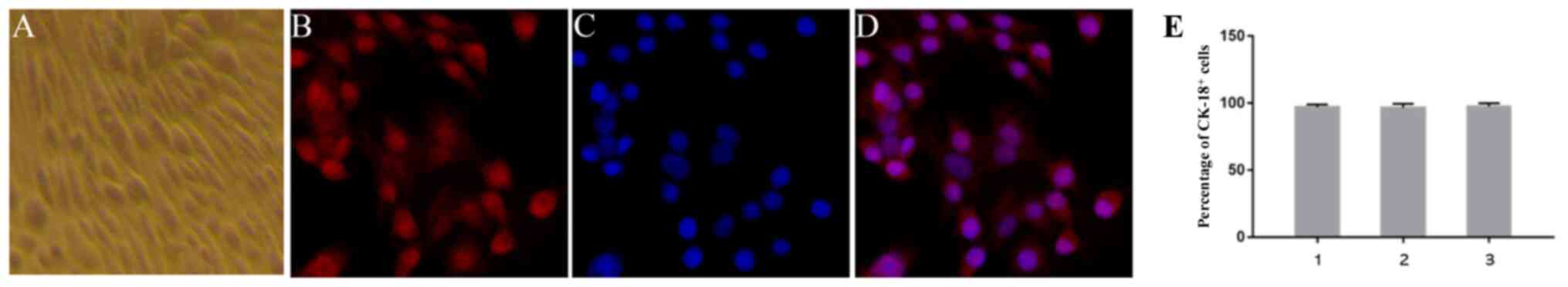
Identification of primary bEECs
Immunofluorescent studies were performed according
to the aforementioned procedure. Primary bEECs were positive for
the epithelial-specific marker CK-18. The proportion of
CK-18+ cells was >95%, as determined using confocal
microscopy (Fig. 2).
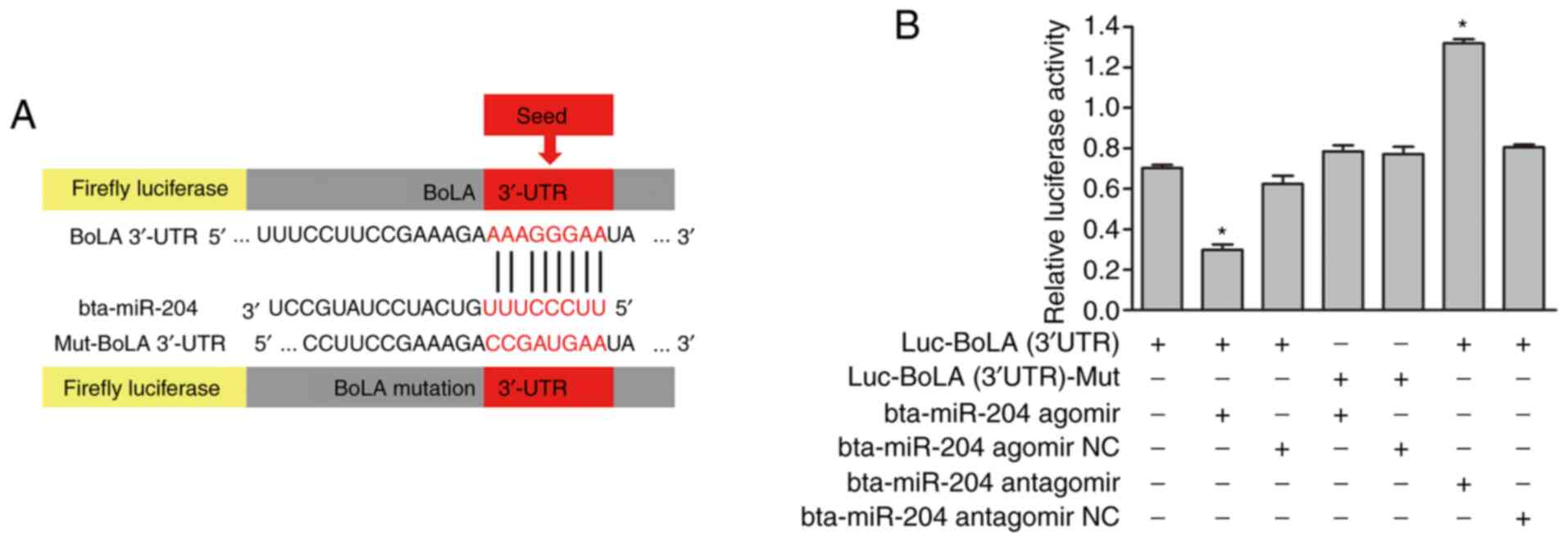
Prediction of the target gene of
bta-miR-204
As miRNAs are well documented to exert their
function by affecting the expression of their target gene(s)
(33), the present study attempted
to identify the direct target of bta-miR-204 associated with IFN-τ
treatment. Data collection and analysis revealed that BoLA was a
potential target gene of bta-miR-204. The predicted target site is
a 1437-1462 base sequence of BoLA mRNA, and the seed region of
bta-miR-204 is a 23-30 base sequence (Fig. 3A). The mFE between the BoLA 3'-UTR
and bta-miR-204 was calculated with RNAhybrid software (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/).
The mFE was ~16.9 kcal/mol, which indicated high stability of the
duplex.
BoLA is a direct target of
bta-miR-204
Luciferase reporter assays were performed to further
investigate whether bta-miR-204 directly targets the 3'-UTR of BoLA
in bEECs. The target sequences of the wild-type and mutant 3'-UTR
of BoLA were cloned separately into a luciferase reporter vector to
generate Luc-BoLA (3'UTR) and Luc-BoLA (3'UTR)-Mut, respectively
(Fig. 3A). The luciferase activity
was inhibited by the binding of bta-miR-204 to the 3'-UTR of BoLA.
Co-transfection of Luc-BoLA (3'UTR) with bta-miR-204 agomir and
bta-miR-204 antagomir in bEECs resulted in a significant alteration
in the luciferase activity compared with that in the control
groups, which were transfected with Luc-BoLA (3'UTR) only or
co-transfected with Luc-BoLA (3'UTR) and bta-miR-204 agomir NC or
Luc-BoLA (3'UTR) and bta-miR-204 antagomir NC. BoLA 3'-UTR
luciferase activity was inhibited by bta-miR-204 agomir, which
mimics bta-miR-204, while it was increased following transfection
with bta-miR-204 antagomir, which is an inhibitor of bta-miR-204.
Furthermore, the luciferase activity of BoLA (3'UTR)-Mut was
unaffected following transfection with bta-miR-204 agomir compared
with the transfection with Luc-BoLA (3'UTR) only or the
co-transfection of Luc-BoLA (3'UTR)-Mut and bta-miR-204 agomir NC.
This result indicated that the mutated seed sequence of BoLA
(3'-UTR) and bta-miR-204 agomir did not bind to each other to
affect the luciferase activity (Fig.
3B). The results demonstrated that bta-miR-204 negatively
regulated BoLA expression by directly binding to its complementary
sequence in the 3'-UTR of BoLA in a sequence-specific manner.
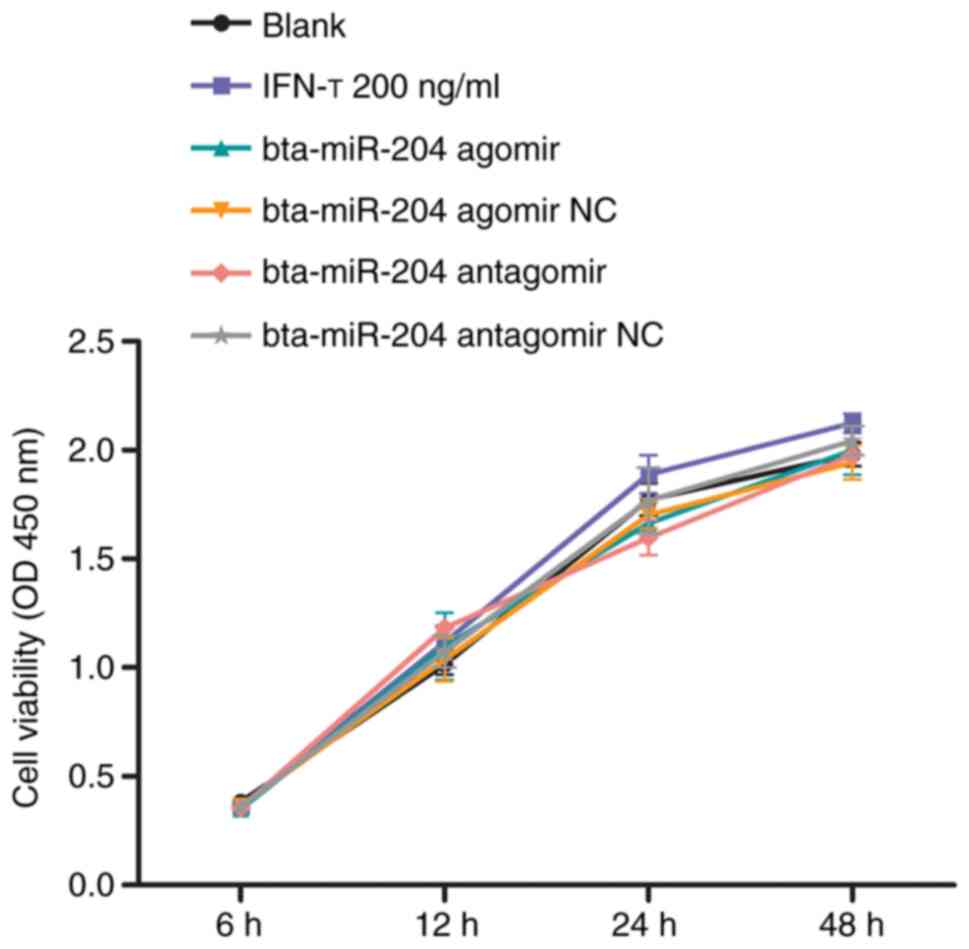
Effects of IFN-τ, bta-miR-204 and BoLA
siRNAs on cell proliferation
CCK-8 cell proliferation assays were performed to
investigate whether the proliferative capacity of bEEC was affected
by IFN-τ, bta-miR-204 agomir/NC, bta-miR-204 antagomir/NC and BoLA
siRNAs/NC. The results indicated that little difference existed
between the IFN-τ-treated group and the blank group. Similarly,
little difference was observed between the transfection groups and
the blank group, respectively (Fig.
4). Therefore, IFN-τ, bta-miR-204 agomir, and bta-miR-204
antagomir exhibited little effect on the proliferation of primary
bEECs compared with the control groups.
IFN-τ positively regulates BoLA
expression
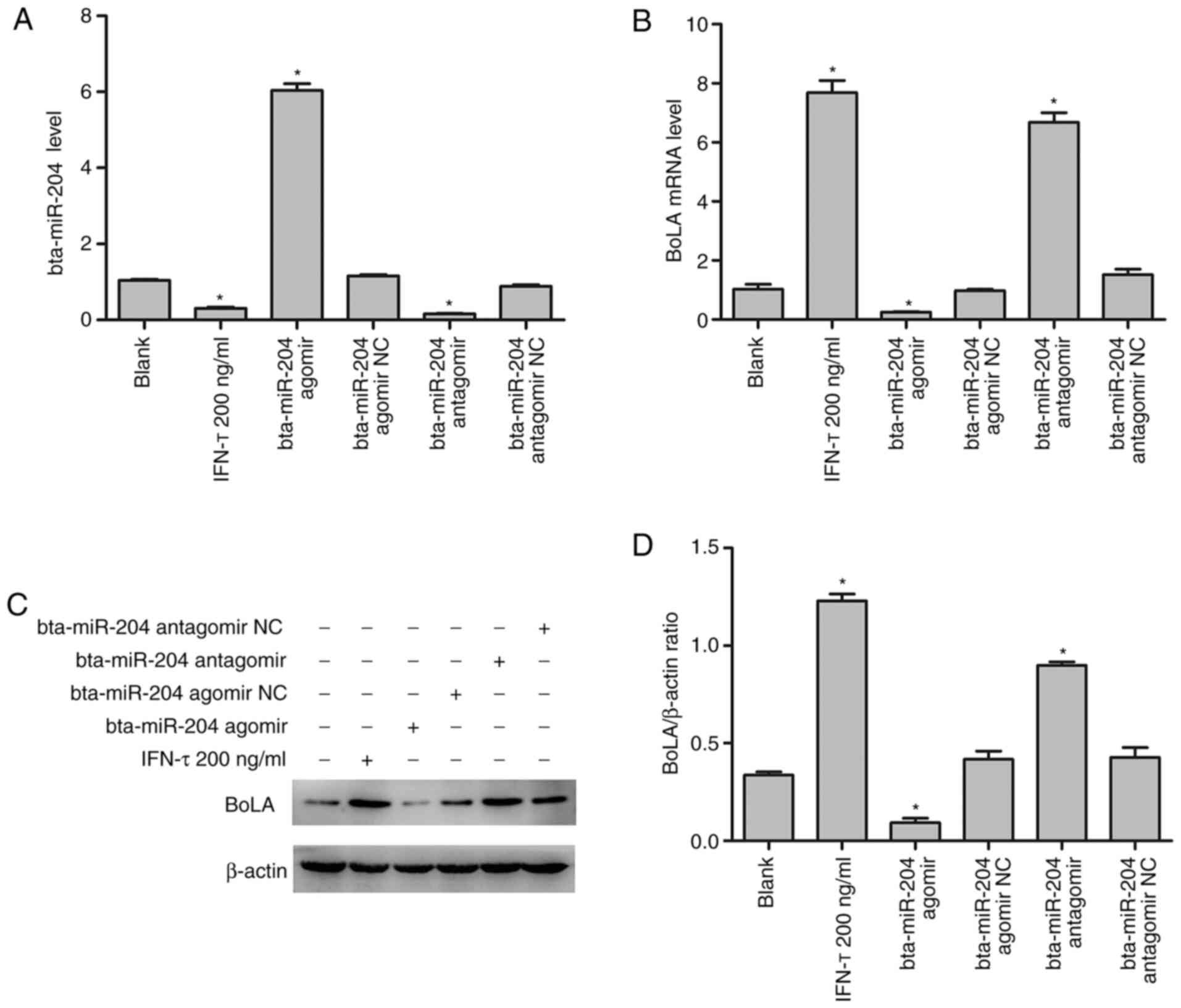
To determine the roles of IFN-τ and bta-miR-204 in
the regulation of BoLA expression, treatment of bEECs with IFN-τ
and suppression or overexpression of bta-miR-204 was performed.
After transient transfection with bta-miR-204 agomir, bta-miR-204
overexpression was detected in bEECs by qPCR (Fig. 5A). Furthermore, bta-miR-204 agomir
NC, bta-miR-204 antagomir and bta-miR-204 antagomir NC were also
overexpressed in bEECs via transient transfection. The statistical
analysis demonstrated that compared with the blank group, the
expression level of bta-miR-204 was downregulated both after IFN-τ
treatment and in the bta-miR-204 antagomir group (Fig. 5A). In addition, compared with the NC
group the mRNA (Fig. 5B) and
protein (Fig. 5C) expression of
BoLA were upregulated as a consequence of the decreased expression
of bta-miR-204 While, mRNA and protein expression levels of BoLA
were downregulated in the bta-miR-204 groups. Collectively, these
results indicated that IFN-τ upregulated BoLA expression by
negatively regulating bta-miR-204 expression in bEECs.
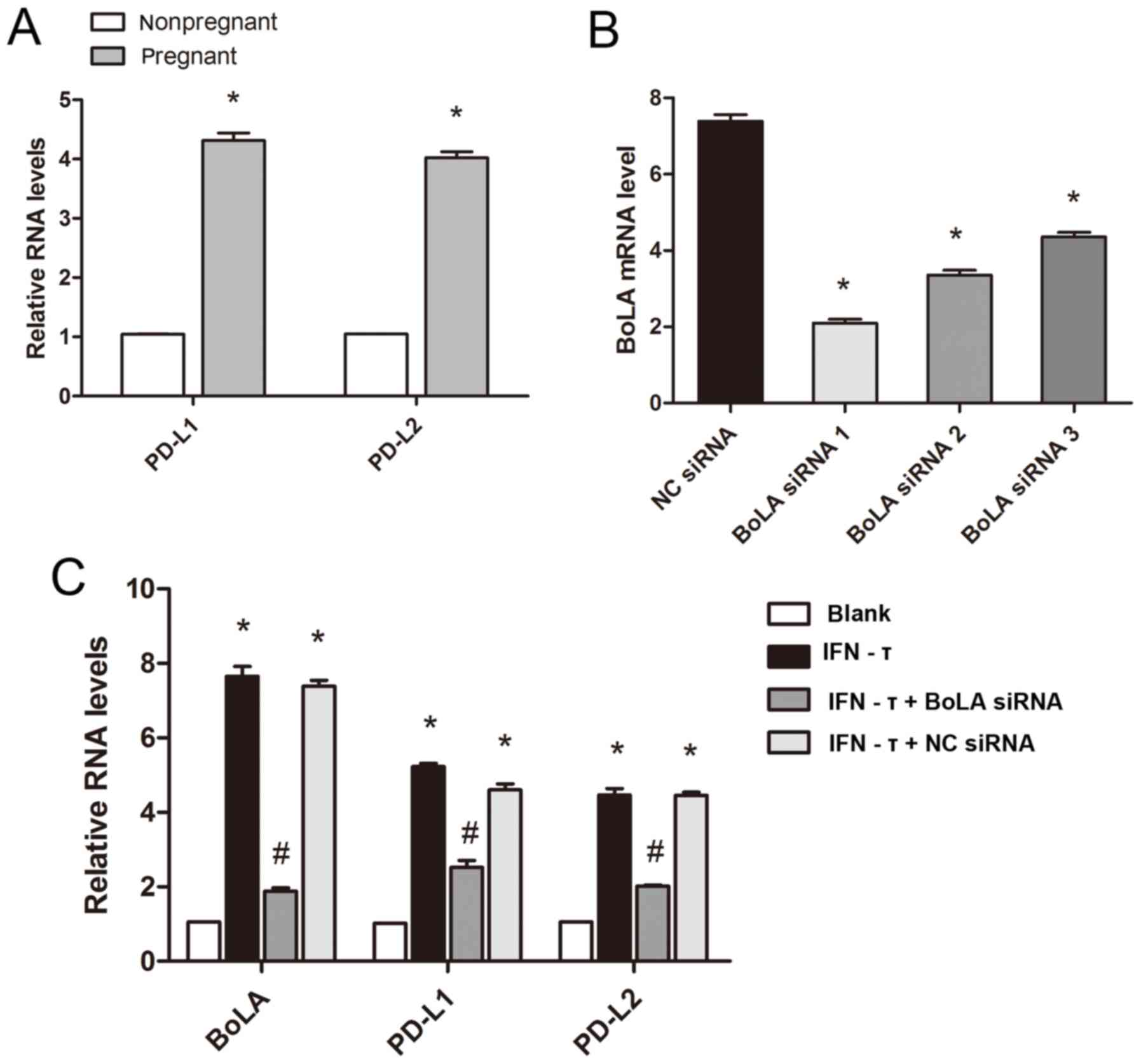
Expression of PD-L1 and PD-L2 in
IFN-τ-treated bEECs and endometrial tissues of dairy cattle
A previous study has reported that the expression
level of MHC-I was positively associated with that of PD-L1 and
PD-L2(34). Uterine tissues of
pregnant or nonpregnant cattle and BoLA siRNA-transfected bEECs
were used in this experiment to evaluate the effect of IFN-τ and
BoLA on the PD-L1 and PD-L2 expression level. The results revealed
that the mRNA expression of PD-L1 and PD-L2 in cattle during early
pregnancy was significantly increased compared with that in
nonpregnant cattle (Fig. 6A).
RT-qPCR was used to evaluate the efficacy of the siRNAs in
downregulating BoLA expression in bEECs. The results indicated that
BoLA siRNA 1 exhibited the best inhibitory effect on BoLA mRNA
expression, therefore it was used in subsequent experiments
(Fig. 6B). In accordance with the
in vivo results, the mRNA expression level of PD-L1 and
PD-L2 in the bEECs was increased following IFN-τ treatment compared
with that in the blank group. However, the expression level of
PD-L1 and PD-L2 in the BoLA siRNA-transfected groups decreased
compared with that in the IFN-τ treatment group (Fig. 6C). These results indicated that
IFN-τ may induce PD-L1 and PD-L2 transcription via regulating BoLA
expression.
Discussion
Embryo implantation is an important step for the
establishment of normal pregnancy. In ruminants and humans, the
interaction between the trophoblast cells and the cells of the
apical surface of the luminal epithelium indicates implantation
(35). The difference between
ruminants and humans in controlling embryo implantation is that, in
addition to estrogen and progesterone, ruminants also secrete IFN-τ
to regulate the expression of various cytokines and transcription
factors (36,37).
Previous studies have identified an important
function of miRNAs in regulating potential gene expression during
implantation in a range of species, such as humans, mice and swine
(38-42).
For instance, miR-200a was reported to regulate
progesterone-progesterone receptor signaling via negatively
regulating the expression of progesterone receptor and
20-hydroxysteroid dehydrogenase to influence endometrial
receptivity and embryo implantation (43). miR-29a was indicated to regulate the
expression of proapoptotic and antiapoptotic factors, thereby
serving an important role during embryo implantation (44). Moreover, miR-148a and miR-152 were
revealed to regulate the expression of HLA-G to affect the
acceptance of the fetus (45,46).
In the present study, IFN-τ was indicated to reduce
the expression of bta-miR-204 in bEECs. miR-204 is highly conserved
in humans, rabbits, rats, mice and other vertebrates, and it has
been revealed to be one of the most commonly altered miRNA in
tumors (47-49).
Previous studies have indicated that the expression of miR-204 was
downregulated in tumor tissues and cells (50-52).
This finding suggested that low-level miR-204 via the negative
regulation of its target genes may aid the tumor to escape the
immune system or proliferate, migrate and invade the tissues.
Previous studies have demonstrated that there were a
number of similarities between embryo implantation and tumor
invasion and metastasis, such as pathophysiological processes, gene
expression, angiogenesis and immune escape (53-55).
Based on these similarities, an analogy may be drawn between
pregnancy and cancer in terms of immune tolerance (35,56).
To further verify whether bta-miR-204 serves an important role in
regulating embryo implantation as in regulating tumors, it was
firstly observed that the mRNA and protein expression of BoLA were
significantly increased in the endometrial tissues of pregnant
dairy cattle compared with nonpregnant cattle and in bEECs treated
with IFN-τ compared with control cells. These results were in
accordance with the results of previous studies, which demonstrated
the important and beneficial role of MHC-I in the establishment of
pregnancy (57-59).
Subsequently, using dual-luciferase reporter assay, BoLA was
verified to be the direct target gene of the bta-miR-204.
Although previous research has revealed MHC-I as a
key factor in regulating embryonic development, the specific
molecular mechanisms and genes involved are not well understood
(60). Several embryonic
development-associated genes were analyzed to explore the possible
mechanisms in embryo development (61). PD-L1 and PD-L2 are two ligands known
to bind to PD-1 and have been associated with the regulation of
tolerance and autoimmunity (62,63).
PD-L1 and PD-L2 mRNAs have been detected in a variety of tissues,
including the heart, lungs, placenta and tumor tissues (64,65).
In the present study, the mRNA levels of PD-L1, PD-L2, BoLA and
IFN-τ were indicated to be significantly increased in the
endometrial tissues of pregnant dairy cattle compared with those in
nonpregnant cattle. Aust et al (34) reported that a large number of MHC-I
and MHC-II genes were positively associated with PD-L1 expression
levels in tumor cells. To further elucidate the relationship
between BoLA and PD-L1 or PD-L2, experiments were conducted using
bEECs. The results revealed that PD-L1 and PD-L2 mRNA expression
level increased following IFN-τ treatment, whereas it decreased
after transfection with BoLA siRNA. These results indicated that
IFN-τ upregulated PD-L1 and PD-L2 transcription, potentially via
regulating BoLA expression. These data may provide a basis for
explaining the diversity in immune escape mechanisms in pregnant
dairy cattle during embryo implantation. A common immune escape
mechanism of tumor cells is the downregulation of HLA-I (66). Another common immune escape
mechanism in tumors and transplants is the activation of the
PD/PD-L negative costimulatory pathway, which alters the balance
between pathogenic and regulatory T cells (21,67,68).
In particular, PD-L1 expression is crucial for the maintenance of
tolerance at the utero-placental interface (69).
In the present study, the results revealed that
IFN-τ may induce BoLA expression due to its negative regulation of
bta-miR-204. In addition, as the expression level of BoLA was
induced by IFN-τ, the mRNA level of PD-L1 and PD-L2 was also
increased, and it was positively associated with the expression
level of BoLA. These results further indicated the immune escape
mechanism of IFN-τ in regulating implantation during early
pregnancy in dairy cattle. However, the use of a single animal
model is a limitation to the present study, and although the method
presented was accurate, different species of animals should be
tested in vivo or in vitro.
To conclude, on the basis of the experimental
results, the current study indicated that IFN-τ increased the
expression of BoLA by reducing the expression of bta-miR-204, and
IFN-τ may induce the transcription of PD-L1 and PD-L2 by inhibiting
bta-miR-204 to upregulate the expression of BoLA, thereby affecting
the immune microenvironment of the maternal-fetal interface and
promoting fetal immune escape.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by National Natural
Science Foundation of China (grant nos. 31472254 and 31772816).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW and GD designed the study and participated in the
data analysis and interpretation, manuscript drafting and critical
revision. XW, NY and XM performed the experiments and contributed
to data acquisition. TY, YY, JY and AS provided reagents and
contributed to data analysis and interpretation and to critical
revision of the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Huazhong
Agricultural University Animal Care and Use Committee (Wuhan,
China; approval no. 20171354CA).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Spencer TE, Johnson GA, Bazer FW and
Burghardt RC: Fetal-maternal interactions during the establishment
of pregnancy in ruminants. Soc Reprod Fertil Suppl. 64:379–396.
2007.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Spencer TE and Bazer FW: Uterine and
placental factors regulating conceptus growth in domestic animals.
J Anim Sci. 82 (Suppl):E4–E13. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Saito S: Cytokine network at the
feto-maternal interface. J Reprod Immunol. 47:87–103.
2000.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Roberts RM: Interferon-tau, a type 1
interferon involved in maternal recognition of pregnancy. Cytokine
Growth Factor Rev. 18:403–408. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bazer FW, Spencer TE and Ott TL:
Interferon tau: A novel pregnancy recognition signal. Am J Reprod
Immunol. 37(412)1997.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Demmers KJ, Derecka K and Flint A:
Trophoblast interferon and pregnancy. Reproduction. 121:41–49.
2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yao GD, Shu YM, Shi SL, Peng ZF, Song WY,
Jin HX and Sun YP: Expression and potential roles of HLA-G in human
spermatogenesis and early embryonic development. PLoS One.
9(e92889)2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ozato K, Wan YJ and Orrison BM: Mouse
major histocompatibility class I gene expression begins at
midsomite stage and is inducible in earlier-stage embryos by
interferon. Proc Natl Acad Sci USA. 82:2427–2431. 1985.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Talbot NC, Powell AM, Ocón OM, Caperna TJ,
Camp M, Garrett WM and Ealy AD: U.S. Department of Agriculture,
Agricultural Research Service. Comparison of the interferon-tau
expression from primary trophectoderm outgrowths derived from IVP,
NT, and parthenogenote bovine blastocysts. Mol Reprod Dev.
75:299–308. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhu Z, Li B, Wu Y, Wang X and Deng G:
Interferon-τ increases BoLA-I for implantation during early
pregnancy in dairy cows. Oncotarget. 8:95095–95107. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wu H, Tao Z, Ma X, Jiang K, Zhao G, Qiu C
and Deng G: Specific microRNA library of IFN-τ, on bovine
endometrial epithelial cells. Oncotarget. 8:61487–61498.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Skommer J, Rana I, Marques FZ, Zhu W, Du Z
and Charchar FJ: Small molecules, big effects: The role of
microRNAs in regulation of cardiomyocyte death. Cell Death Dis.
5(e1325)2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sun E and Shi Y: MicroRNAs: Small
molecules with big roles in neurodevelopment and diseases. Exp
Neurol. 268:46–53. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Smyth LA, Boardman DA, Tung SL, Lechler R
and Lombardi G: MicroRNAs affect dendritic cell function and
phenotype. Immunology. 144:197–205. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bidarimath M, Khalaj K, Wessels JM and
Tayade C: MicroRNAs, immune cells and pregnancy. Cell Mol Immunol.
11:538–547. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ye ZH, Wen DY, Cai XY, Liang L, Wu PR, Qin
H, Yang H, He Y and Chen G: The protective value of miR-204-5p for
prognosis and its potential gene network in various malignancies: A
comprehensive exploration based on RNA-seq high-throughput data and
bioinformatics. Oncotarget. 8:104960–104980. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jacob RJ and Cramer R: PIGOK: Linking
protein identity to gene ontology and function. J Proteome Res.
5:3429–3432. 2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bo W, Xiao-Li HE, Yong-Sheng W, Xue-Jiao
W, Yong Z and Yue-Mao Z: BoLA-I gene expression in early
development of bovine SCNT embryo. Chin J Veterinary: ence,
2015.
|
|
20
|
Loustau M, Wiendl H, Ferrone S and
Carosella ED: HLA-G 2012 conference: The 15-year milestone update.
Tissue Antigens. 81:127–136. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Keir ME, Butte MJ, Freeman GJ and Sharpe
AH: PD-1 and its ligands in tolerance and immunity. Annu Rev
Immunol. 26:677–704. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mincheva-Nilsson L and Baranov V:
Placenta-derived exosomes and syncytiotrophoblast microparticles
and their role in human reproduction: Immune modulation for
pregnancy success. Am J Reprod Immunol. 72:440–457. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bardhan K, Anagnostou T and Boussiotis VA:
The PD1: PD-L1/2 pathway from discovery to clinical implementation.
Front Immunol. 7(550)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Okazaki T and Honjo T: PD-1 and PD-1
ligands: From discovery to clinical application. Int Immunol.
19:813–824. 2007.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Eppihimer MJ, Gunn J, Freeman GJ,
Greenfield EA, Chernova T, Erickson J and Leonard JP: Expression
and regulation of the PD-L1 immunoinhibitory molecule on
microvascular endothelial cells. Microcirculation. 9:133–145.
2002.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen J, Feng Y, Lu L, Wang H, Dai L, Li Y
and Zhang P: Interferon-γ-induced PD-L1 surface expression on human
oral squamous carcinoma via PKD2 signal pathway. Immunobiology.
217:385–393. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ashizawa T, Iizuka A, Nonomura C, Kondou
R, Maeda C, Miyata H, Sugino T, Mitsuya K, Hayashi N, Nakasu Y, et
al: Antitumor effect of programmed death-1 (PD-1) blockade in
humanized the NOG-MHC double knockout mouse. Clin Cancer Res.
23:149–158. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Blank C, Metzner M, Lorch A and Klee W:
Euthanasia of cattle: A clinical comparison of T 61 and
pentobarbital (Eutha 77). Berl Munch Tierarztl Wochenschr.
123:96–102. 2010.PubMed/NCBI(In German).
|
|
29
|
Mishra B, Kizaki K, Koshi K, Ushizawa K,
Takahashi T, Hosoe M, Sato T, Ito A and Hashizume K: Expression of
extracellular matrix metalloproteinase inducer (EMMPRIN) and its
expected roles in the bovine endometrium during gestation. Domest
Anim Endocrinol. 42:63–73. 2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Rehmsmeier M, Steffen P, Höchsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Elbashir SM, Lendeckel W and Tuschl T: RNA
interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev.
15:188–200. 2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531.
2004.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Aust S, Felix S, Auer K, Bachmayr-Heyda A,
Kenner L, Dekan S, Meier SM, Gerner C, Grimm C and Pils D: Absence
of PD-L1 on tumor cells is associated with reduced MHC I expression
and PD-L1 expression increases in recurrent serous ovarian cancer.
Sci Rep. 7(42929)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang S, Lin H, Kong S, Wang S, Wang H,
Wang H and Armant DR: Physiological and molecular determinants of
embryo implantation. Mol Aspects Med. 34:939–980. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bazer FW, Ying W, Wang X, Dunlap KA, Zhou
B, Johnson GA and Wu G: The many faces of interferon tau. Amino
Acids. 47:449–460. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Shirasuna K, Matsumoto H, Matsuyama S,
Kimura K, Bollwein H and Miyamoto A: Possible role of interferon
tau on the bovine corpus luteum and neutrophils during the early
pregnancy. Reproduction. 150:217–225. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Galliano D and Pellicer A: MicroRNA and
implantation. Fertil Steril. 101:1531–1544. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu W, Niu Z, Li Q, Pang RT, Chiu PC and
Yeung WS: MicroRNA and embryo implantation. Am J Reprod Immunol.
75:263–271. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Su L, Liu R, Cheng W, Zhu M, Li X, Zhao S
and Yu M: Expression patterns of microRNAs in porcine endometrium
and their potential roles in embryo implantation and placentation.
PLoS One. 9(e87867)2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Song Y, An X, Zhang L, Fu M, Peng J, Han
P, Hou J, Zhou Z and Cao B: Identification and profiling of
microRNAs in goat endometrium during embryo implantation. PLoS One.
10(e0122202)2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Ponsuksili S, Tesfaye D, Schellander K,
Hoelker M, Hadlich F, Schwerin M and Wimmers K: Differential
expression of miRNAs and their target mRNAs in endometria prior to
maternal recognition of pregnancy associates with endometrial
receptivity for in vivo- and in vitro-produced bovine
embryos. Biol Reprod. 91(135)2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Haraguchi H, Saito-fujita T, Hirota Y,
Egashira M, Matsumoto L, Matsuo M, Hiraoka T, Koga K, Yamauchi N,
Fukayama M, et al: MicroRNA-200a locally attenuates progesterone
signaling in the cervix, preventing embryo implantation. Mol
Endocrinol. 28:1108–1117. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Xia HF, Jin XH, Cao ZF, Hu Y and Ma X:
MicroRNA expression and regulation in the uterus during embryo
implantation in rat. FEBS J. 281:1872–1891. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Rebmann V, da Silva Nardi F, Wagner B and
Horn PA: HLA-G as a tolerogenic molecule in transplantation and
pregnancy. J Immunol Res. 2014(297073)2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Manaster I, Goldman-Wohl D, Greenfield C,
Nachmani D, Tsukerman P, Hamani Y, Yagel S and Mandelboim O:
MiRNA-mediated control of HLA-G expression and function. PLoS One.
7(e33395)2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Mohammed CP, Rhee H, Phee BK, Kim K, Kim
HJ, Lee H, Park JH, Jung JH, Kim JY, Kim HC, et al: MiR-204
downregulates EphB2 in aging mouse hippocampal neurons. Aging Cell.
15:380–388. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li T, Pan H and Li R: The dual regulatory
role of miR-204 in cancer. Tumour Biol. 37:11667–11677.
2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Xia Z, Liu F, Zhang J and Liu L: Decreased
expression of MiRNA-204-5p contributes to glioma progression and
promotes glioma cell growth, migration and invasion. PLoS One.
10(e0132399)2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Shi Y, Huang J, Zhou J, Liu Y, Fu X, Li Y,
Yin G and Wen J: MicroRNA-204 inhibits proliferation, migration,
invasion and epithelial-mesenchymal transition in osteosarcoma
cells via targeting Sirtuin 1. Oncol Rep. 34:399–406.
2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Yin JJ, Liang B and Zhan XR: MicroRNA-204
inhibits cell proliferation in T-cell acute lymphoblastic leukemia
by down-regulating SOX4. Int J Clin Exp Pathol. 8:9189–9195.
2015.PubMed/NCBI
|
|
52
|
Liu L, Wang J, Li X, Ma J, Shi C, Zhu H,
Xi Q, Zhang J, Zhao X and Gu M: MiR-204-5p suppresses cell
proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma.
Biochem Biophys Res Commun. 457:621–626. 2015.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ferretti C, Bruni L, Dangles-Marie V,
Pecking AP and Bellet D: Molecular circuits shared by placental and
cancer cells, and their implications in the proliferative, invasive
and migratory capacities of trophoblasts. Hum Reprod Update.
13:121–141. 2007.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Holtan SG, Creedon DJ, Haluska P and
Markovic SN: Cancer and pregnancy: Parallels in growth, invasion,
and immune modulation and implications for cancer therapeutic
agents. Mayo Clin Proc. 84:985–1000. 2009.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Piechowski J: Trophoblastic implantation,
a model of tumor and metastasis implantation. Bull Cancer.
102:806–813. 2015.PubMed/NCBI View Article : Google Scholar : (In French).
|
|
56
|
Chen T, Darrassejèze G, Bergot AS, Courau
T, Churlaud G, Valdivia K, Strominger JL, Ruocco MG, Chaouat G and
Klatzmann D: Self-specific memory regulatory T cells protect
embryos at implantation in mice. J Immunol. 191:2273–2281.
2013.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Thompson RN, Mcmillon R, Napier A and
Wekesa KS: Pregnancy block by MHC class I peptides is mediated via
the production of inositol 1,4,5-trisphosphate in the mouse
vomeronasal organ. J Exp Biol. 210:1406–1412. 2007.PubMed/NCBI View Article : Google Scholar
|
|
58
|
O'Gorman GM, Al Naib A, Ellis SA, Mamo S,
O'Doherty AM, Lonergan P and Fair T: Regulation of a Bovine
nonclassical major histocompatibility complex class I gene
promoter1. Biol Reprod. 83:296–306. 2010.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Al Naib A, Mamo S, O'Gorman GM, Lonergan
P, Swales A and Fair T: Regulation of non-classical major
histocompatability complex class I mRNA expression in bovine
embryos. J Reprod Immunol. 91:31–40. 2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Cheng Y, King NJ and Kesson AM: Major
histocompatibility complex class I (MHC-I) induction by West Nile
Virus: Involvement of 2 signaling pathways in MHC-I Up-Regulation.
J Infect Dis. 189:658–668. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
61
|
Townson DH: Immune cell-endothelial cell
interactions in the bovine corpus luteum. Integr Comp Biol.
46:1055–1059. 2006.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Liu H, Bakthavatsalam R, Meng Z, Li Z, Li
W, Perkins JD and Reyes J: PD-L1 signal on liver dendritic cells is
critical for Foxp3+CD4+CD25+ Treg
and liver tolerance induction in mice. Transplant Proc.
45:1853–1855. 2013.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Liang SC, Latchman YE, Buhlmann JE,
Tomczak MF, Horwitz BH, Freeman GJ and Sharpe AH: Regulation of
PD-1, PD-L1, and PD-L2 expression during normal and autoimmune
responses. Eur J Immunol. 33:2706–2716. 2003.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Kim MY, Koh J, Kim S, Go H, Jeon YK and
Chung DH: Clinicopathological analysis of PD-L1 and PD-L2
expression in pulmonary squamous cell carcinoma: Comparison with
tumor-infiltrating T cells and the status of oncogenic drivers.
Lung Cancer. 88:24–33. 2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Yang H, Bueso-ramos C, Dinardo C, Estecio
MR, Davanlou M, Geng QR, Fang Z, Nguyen M, Pierce S, Wei Y, et al:
Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic
syndromes is enhanced by treatment with hypomethylating agents.
Leukemia. 28:1280–1288. 2014.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Garcia-Lora A, Algarra I and Garrido F:
MHC class I antigens, immune surveillance, and tumor immune escape.
J Cell Physiol. 195:346–355. 2003.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Francisco LM, Sage PT and Sharpe AH: The
PD-1 pathway in tolerance and autoimmunity. Immunol Rev.
236:219–242. 2010.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Francisco LM, Salinas VH, Brown KE,
Vanguri VK, Freeman GJ, Kuchroo VK and Sharpe AH: PD-L1 regulates
the development, maintenance, and function of induced regulatory T
cells. J Exp Med. 206:3015–3029. 2009.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Guleria I, Khosroshahi A, Ansari MJ,
Habicht A, Azuma M, Yagita H, Noelle RJ, Coyle A, Mellor AL, Khoury
SJ and Sayegh MH: A critical role for the programmed death ligand 1
in fetomaternal tolerance. J Exp Med. 202:231–237. 2005.PubMed/NCBI View Article : Google Scholar
|