Introduction
Stroke is the second largest cause of death
worldwide (1,2). The current treatment strategies for
ischemic stroke are limited and early reperfusion is primarily
established by intravenous thrombolysis or mechanical thrombectomy.
However, there are no effective clinical drugs that improve patient
prognosis following cerebral tissue infarction (3,4).
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs that control the translation of the target protein through
inhibition of the 3'-untranslated region (UTR) of the target gene
(5-7).
It has been reported that >33% of human genes are regulated by
miRNAs (6). Previous studies have
demonstrated that miRNAs are physiologically and pathologically
involved in ischemic stroke (7,8). Among
them, miR-122 has been reported to be associated with ischemic
stroke. miR-122 is encapsulated in exosomes and secreted into body
fluids, where it is stable in serum and may serve as a promising
biomarker for ischemic stroke (9,10).
miR-122 protects against amyloid-β-induced neuronal injury
(11); however, the molecular
mechanism underlying miR-122-mediated protection against loss of
brain function is not completely understood.
It was initially determined that repressor of RNA
polymerase III transcription MAF1 homolog (Maf1) was highly
expressed in brain tissue with an abundance identified in the
hippocampus and cortex (12).
Subsequently TORC1 in the mTOR pathway was revealed to be involved
in the phosphorylation of Maf1, which led to Maf1 inactivation
(13). Since mTOR protects brain
tissues from ischemic injuries, downregulation of Maf1 may have an
inhibitory effect on the development of ischemic brain injuries
(14,15). In addition,
H2O2 is produced by brain tissue after
ischemic stroke, which can reduce cell function and further
aggravate brain injury (16).
H2O2 can serve as a messenger and promote the
synthesis of the transcription factor Maf1 (17,18).
Therefore, it was hypothesized that Maf1 may serve an important
role in the development and progression of ischemic stroke;
however, the biological function of Maf1 and its relationship with
ischemic stroke are not completely understood.
miR-122 may serve as a potential therapeutic target
for ischemic stroke (19), whereas
Maf1 may aggravate ischemic stroke, but the specific underlying
mechanism is not completely understood (20-23).
Therefore, it was hypothesized that miR-122 may be involved in the
development of ischemic stroke by targeting Maf1. The present study
aimed to investigate the relationship between Maf1 and miR-122 in
the development and progression of ischemic stroke.
Materials and methods
Experimental animals
A total of 60 female C57BL/6 mice (age, 6-8 weeks;
weight, 20-25 g) were purchased from the Experimental Animal Center
of Dalian Medical University. The animals were housed at 23±2˚C
with 55±5% humidity under a 12-h light/dark cycle with free access
to food and water. The present study was approved by the Animal
Care Committee of the Xinhua Hospital affiliated to Dalian
University and performed in accordance with the National Institutes
of Health Guidelines (no. 85-23; revised 1996) for the Care and Use
of Laboratory Animals (24).
Lateral ventricle injection
Mice were randomly divided into six groups (n=6 per
group) for two experimental studies: The effect of miR-122 on
infarct size after stroke (control, miR-122 mimic and miR-122
inhibitor groups) and the effect of miR-122 on Maf1 after stroke
(control, miR-122 mimic and miR-122 inhibitor groups).
Firstly, 3.5 µl miR-122 negative control (NC), mimic
or inhibitor (cat. nos. B04002, B03001 and B02003; Shanghai
GenePharma Co., Ltd.) was mixed with 3.5 µl RNAi-mate reagent (cat.
no. G04002; Shanghai GenePharma Co., Ltd.) at room temperature for
15 min and stored for later use. After the mice were anesthetized
with 400 mg/kg 4% chloral hydrate via intraperitoneal injection,
the mixture (7 µl) was injected into the lateral ventricle (over 20
min) at the coordinate (bregma, -2.5 mm; dorsoventral, 1 mm;
lateral, 1.5 mm). Subsequently, the wound was sutured and the mice
were returned to the cage. At 24 h post-injection, the mouse middle
cerebral artery occlusion (MCAO) model was established.
MCAO model
A total of 60 female C57BL/6 mice were selected for
this model. The sham operated animals received the same operation
as those in the experimental groups except for the coagulation of
the blood vessels. The MCAO model (permanent coagulation of the
distal middle cerebral artery) is highly reproducible and has a
high success rate (the overall mortality is of <5%) (25). Moreover, the relative infarct volume
in relation to brain size corresponds to the majority of human
strokes (25). Mice were
anesthetized with an intraperitoneal injection of 400 mg/kg chloral
hydrate. After aseptic preparation of the surgical site, the skin
between the ear and the eye was cut under a stereomicroscope
(Shanghai Yuyan Instruments Co., Ltd.) using electrocoagulation
forceps (Shanghai Yuyan Instruments Co., Ltd.). Once the temporal
muscle was removed, a drill (Shanghai Yuyan Instruments Co., Ltd.)
with a diameter of 2.5 mm was used to thin out the skull over the
middle cerebral artery. The bone was carefully withdrawn to expose
the middle cerebral artery, which was then coagulated with
electrocoagulation forceps. Subsequently, the wound was sutured and
the animal was maintained at 32˚C for recovery (25). At 24 h post-model induction, 6 mice
in each group were anesthetized with 400 mg/kg chloral hydrate and
sacrificed by decapitation. Brain tissues were harvested and
sectioned into 2-mm slices. Subsequently, the ischemic site was
identified by incubation of the sections with 2% 2,3,5-triphenyl
tetrazolium chloride (cat. no. T8170; Beijing Solarbio Science
& Technology Co., Ltd.) at 37˚C for 15 min in the dark.
Coronal section
Brain tissues were fixed with 4% paraformaldehyde
for 6 h at 4˚C, the surface liquid was removed by blotting and then
the tissues were dehydrated with 15% sucrose solution overnight at
4˚C. Subsequently, the tissues were incubated with 30% sucrose
solution overnight at 4˚C. Coronal sections were prepared into
30-µm sections using a frozen microtome (Leica Microsystems
GmbH).
Nissl stain
Sections were washed twice with PBS for 5 sec and
stained with Nissl staining solution (cat. no. E607316; Sangon
Biotech Co., Ltd.) for 15 min at 25˚C. Subsequently, the sections
were washed twice with PBS for 5 sec and then washed with 95%
ethanol for 5 sec. Stained sections were examined under a Nikon
Eclipse Ti inverted fluorescent microscope with a charge-coupled
device camera at a magnification of x100. Microscopic images were
analyzed with ImageJ (version 1.48; National Institutes of
Health).
Prediction of miR-122 target gene
TargetScan (version 7.2; www.targetscan.org/vert_72), miRWalk (version 3;
mirwalk.umm.uni-heidelberg.de) and
miRDB (version 6.0; mirdb.org) gene prediction software
were used to predict the target genes of miR-122. The intersection
of the results was identified and used to investigate the
association between target genes and ischemic stroke.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cerebral cortex
(n=6 per group) using TRIzol® (cat. no. B610409; Sangon
Biotech Co., Ltd.). Total RNA was reverse transcribed into cDNA
using PrimeScriptTM RT reagent kit (cat. no. RR047A; Takara
Biotechnology Co., Ltd.) at 37˚C for 15 min and 85˚C for 5 sec.
Subsequently, qPCR was performed using SYBR Green Supermix (cat.
no. RR820Q; Takara Biotechnology Co., Ltd.). The following
thermocycling conditions were used for qPCR: Denaturation at 95˚C
for 30 sec; followed by 40 cycles of 95˚C for 10 sec and 60˚C for
30 sec; and cooling to 4˚C. The following primers were used for
qPCR: miR-122 (cat. no. ssD809230039; Sangon Biotech Co., Ltd.);
Maf1 forward, 5'-GATTGCCACCCTCAATGAGTCC-3' and reverse,
5'-CTCCTCATCCACTGCATTCCAC-3' (Sangon Biotech Co., Ltd.); CEL-39
(cat. no. ssD1083145001; Sangon Biotech Co., Ltd.); and actin-β
(ACTB) forward, 5'-GTGCTATGTTGCTCTAGACTTCG-3' and reverse,
5'-ATGCCACAGGATTCCATACC-3' (Sangon Biotech Co., Ltd.). miRNA and
mRNA expression levels were analyzed using the 2-ΔΔCq
method (26) and normalized to the
internal reference genes CEL-39 and ACTB, respectively.
Construction of vector
Maf1-3'UTR primers were designed by Sangon Biotech
Co., Ltd. (forward, 5'-ATCCAGCTCTGACCAATC-3' and reverse,
5'-CCAGGTTCCATCTAAGTCAC-3'). The PCR products were cloned into the
XhoI and NotI sites of the siCheck vector (Invitrogen; Thermo
Fisher Scientific, Inc.) to construct the Maf1-wild-type (WT)-3'UTR
vector. The 3'UTR of Maf1 containing the predicted binding site of
miR-122 was subjected to site-directed mutation (Sangon Biotech
Co., Ltd.) to construct the Maf1-mutant (MUT)-3'UTR vector.
Dual-luciferase reporter assay
Cells (293A) were divided into the following four
groups: i) Maf1-WT-3'UTR + miR-122 NC (Maf1-WT-3'UTR + NC); ii)
Maf1-WT-3'UTR + miR-122 mimic (Maf1-WT-3'UTR + miR-122); iii)
Maf1-MUT-3'UTR vector + miR-122 NC (Maf1-MUT-3'UTR + NC); and iv)
Maf1-MUT-3'UTR vector + miR-122 mimic (Maf1-MUT-3'UTR + miR-122).
293A cells (Dalian University Microanalysis Laboratory) were plated
in 24-well plates at a density of 1x105 cells per well
in DMEM (Thermo Fisher Scientific, Inc.) containing 10% FBS (Thermo
Fisher Scientific, Inc.). Cells were incubated at 37˚C and 5% CO2
and cultured to 90-95% confluence. Subsequently, cells were
co-transfected with 50 nM miRNA and 1 µg vector using
Lipofectamine® 2000 (cat. no. 11668019; Thermo Fisher
Scientific, Inc.). At 48 h post-transfection, luciferase activity
was assessed using a dual-luciferase reporter assay kit (cat. no.
RG027; Beyotime Institute of Biotechnology) and a fluorescence
microplate reader at a wavelength of 560 nm for luciferin and 465
nm for renilla luciferase). Firefly luciferase activity was
normalized to Renilla luciferase activity.
Statistical analysis
Statistical analyses were conducted using SPSS
software (version 22.0; IBM Corp.). The data were analyzed using
unpaired Student's t-test or one-way ANOVA and Tukey's post-hoc
test. Data are expressed as the mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference. All experiments were performed at least six times.
Results
Establishment of the MCAO model
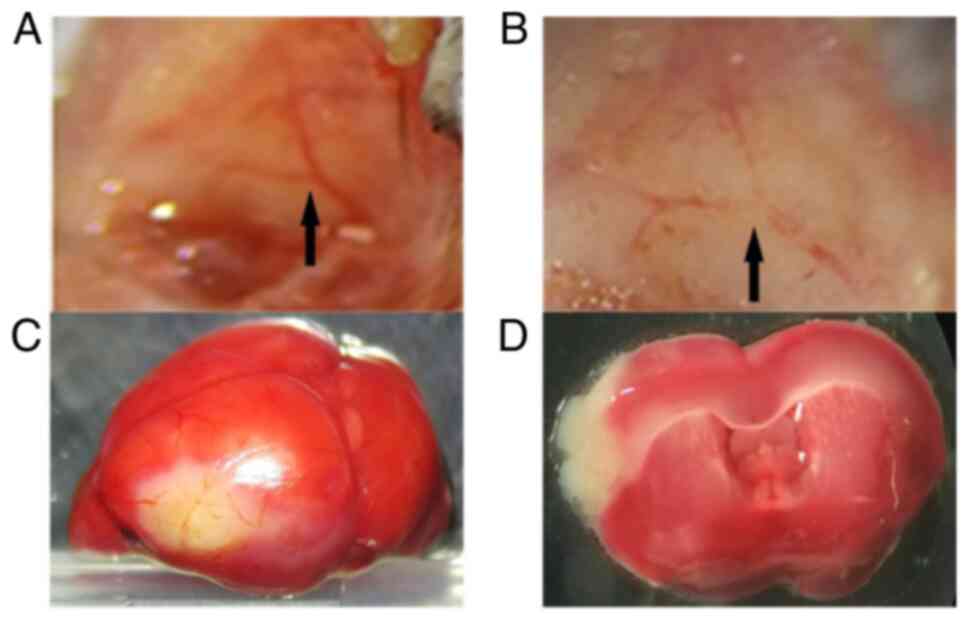
As presented in Fig.
1A, the middle cerebral artery was exposed in a Y shape. After
the middle cerebral artery was coagulated using electrocoagulation
forceps, spontaneous recanalization was not observed (Fig. 1B). After MCAO establishment, the
infarct brain area was white in color, whereas healthy brain
tissues were stained red (Fig. 1C
and D).
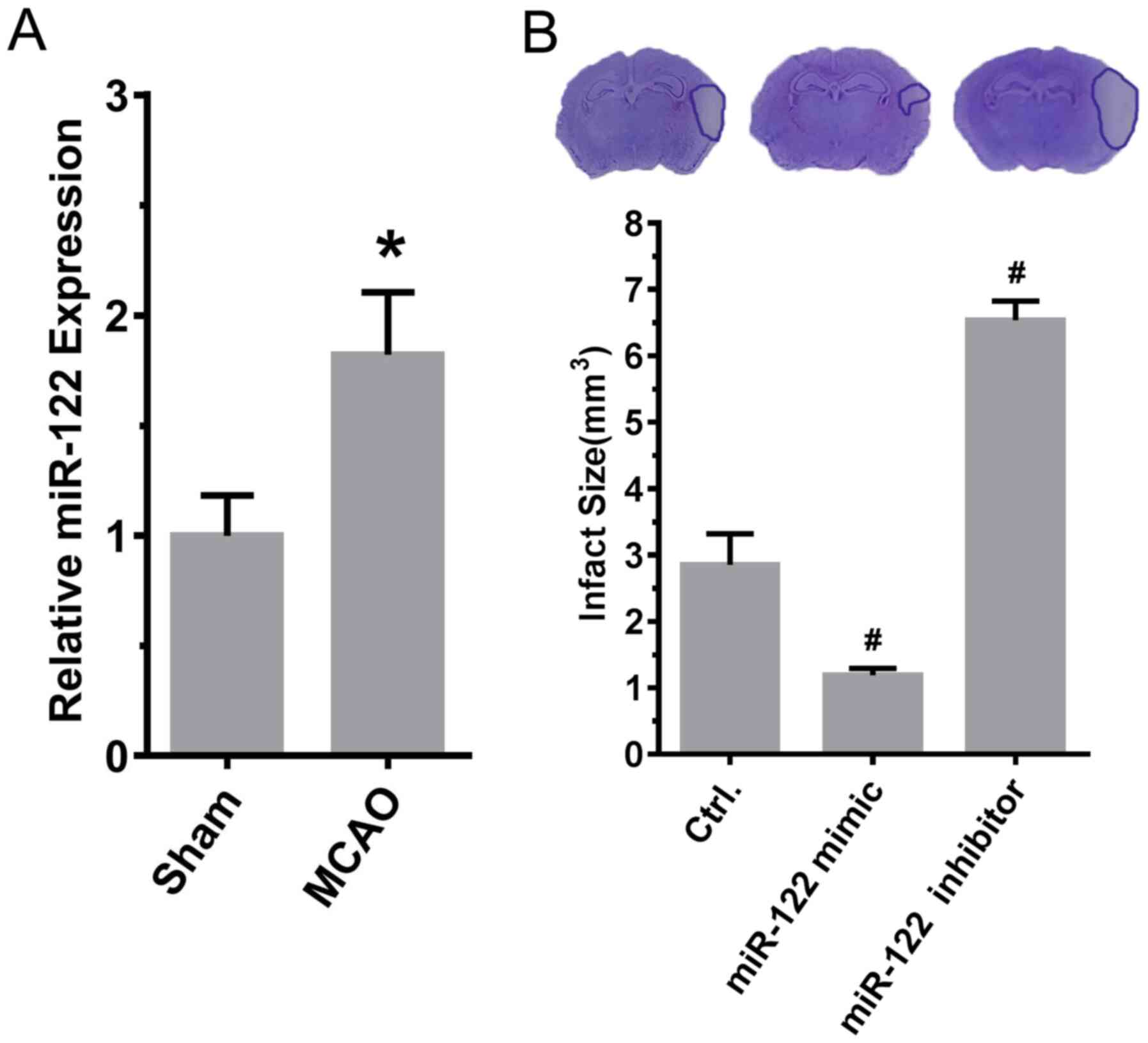
miR-122 decreases the infarct
area
To investigate the relationship between miR-122 and
ischemic stroke, the expression level of miR-122 was examined and
the effect of miR-122 on the infarct area was evaluated. As
presented in Fig. 2A, the
expression of miR-122 (sham, 1.00±0.06; MCAO, 1.82±0.10; P<0.05;
n=6) was significantly increased after MCAO induction in comparison
with sham animals (Fig. 2A).
Compared with MCAO model rats treated with control miRNA, the size
of the area of infarction was significantly reduced in MCAO model
rats that were injected with miR-122 mimic in the lateral ventricle
in comparison with control animals. By contrast, the area of
infarction increased significantly in comparison with the control
following injection of miR-122 inhibitor in the lateral ventricle
(control, 2.85±0.27; mimic, 1.19±0.05; inhibitor 6.53±0.17; mimic
vs. control and inhibitor vs. control, P<0.05; n=6; Fig. 2B).
Predicted target gene of miR-122
To reduce the false positive probability of the
target gene prediction, gene prediction was performed using the
intersection of the prediction results obtained from the three
different types of gene prediction software: TargetScan, miRWalk
and miRtaget2. Maf1 was identified as a target gene of miR-122 in
humans, mice and rats (Table
I).
 | Table IMaf1 is predicted to be a target gene
for miR-122. |
Table I
Maf1 is predicted to be a target gene
for miR-122.
| RNA | Predicted
consequential pairing |
|---|
| mmu-miR-122 | 3'
GUUUGUGGUAACAGUGUGAGGU |
| mouse-Maf1 | 5'
...UCUUAUUGGGCCUGUAA... |
| rat-Maf1 | 5'
...CUUAUUGGGCCUGUACACUCCA... |
| human-Maf1 | 5'
...UUCUACUGGGCCUGCACACUCCA... |
Maf1 is a direct target of
miR-122
After sequence analysis, it was observed that the
3'UTR of Maf1 was a putative target of miR-122 (Fig. 3A). As the 3'UTR of Maf1 mRNA was
predicted to contain a target region of miR-122, a dual-luciferase
reporter assay was conducted to investigate whether miR-122
directly targeted Maf1. 293a cells were transfected with
Maf1-WT-3'UTR + NC as a negative control. Compared with the
negative controls, co-transfection with miR-122 reduced the
luciferase activity of Maf1-WT-3'UTR vector (control, 11.5±0.10;
miR-122, 5.8±0.00; P<0.01; Fig.
3B). However, there was no significant alteration to the
luciferase activity of Maf1-MUT-3'UTR vector following
co-transfection with miR-122 mimics compared with co-transfection
with miR-122 mimics negative control (control, 6.5±0.15; miR-122,
7.5±0.05; P>0.05; Fig. 3B).
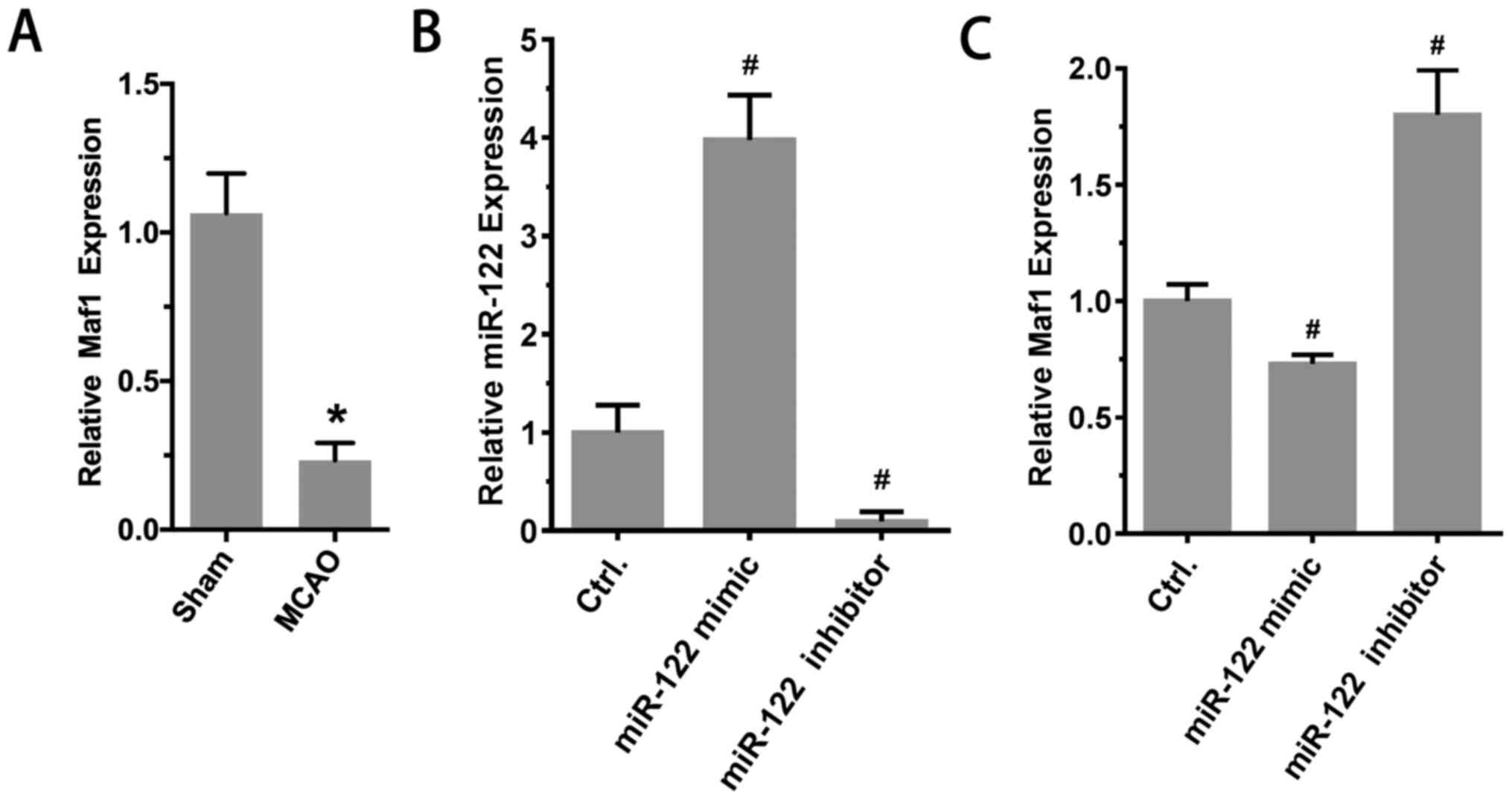
miR-122 inhibits Maf1 expression
Subsequently, whether miR-122 inhibited Maf1
expression in vivo was investigated. The results
demonstrated that compared with the sham group, the expression of
Maf1 in the MCAO group was significantly reduced (sham, 1.00±0.04;
MCAO, 0.21±0.04; P<0.01; n=6; Fig.
4A). Furthermore, RT-qPCR results suggested that the expression
of Maf1 was significantly reduced after injection of miR-122 mimic
in the lateral ventricle. By contrast, the expression of Maf1
increased significantly after the injection of miR-122 inhibitor in
the lateral ventricle. The expression levels of miR-122 were as
follows: 1.00±0.16, 3.98±0.26 and 0.10±0.04 (mimic vs. control and
inhibitor vs. control, P<0.05; n=6; Fig. 4B). Maf1 expression levels were as
follows: 1.00±0.03, 0.73±0.02 and 1.80±0.09 (mimic vs. control and
inhibitor vs. control, P<0.05; n=6; Fig. 4C).
Discussion
It has been reported that there is a close
relationship between miRNA and ischemic stroke (1,27,28),
with the expression of ~20% miRNAs changing during the development
and progression of ischemic stroke (29). By regulating the translation process
of mRNA, miRNAs can affect protein expression, which subsequently
alters biological functions such as axon regeneration, angiogenesis
and inflammatory responses (30-33).
Several studies have indicated that miR-122 can maintain the
caliber and tube shape of blood vessels and reduce neuronal death
in an ischemic stroke model (19,34).
In addition, miR-122 may serve a pivotal immunomodulatory role in
hepatocytes via miR-122-RTKs/STAT3-IRF1-IFNs regulatory circuitry
or target TLR4 (35,36). Nerve cells, blood vessels and
immunity are critical to the prognosis of infarct area in ischemic
stroke (37,38). Therefore, the present study
investigated the effect of miR-122 on the outcome of ischemic
stroke. Compared with the level in the sham group, the expression
of miR-122 increased significantly after MCAO. In addition,
increased miR-122 expression effectively reduced the infarct area.
By contrast, inhibition of miR-122 expression in brain tissue
significantly increased the infarct area, which was consistent with
previous studies (19,39). Therefore, the results indicated that
miR-122 may exert a protective effect against ischemic stroke by
reducing the infarct area.
miRNAs function by regulating the expression of
downstream target genes (1);
therefore, studying the downstream target genes of miR-122 during
the development and progression of ischemic stroke may be
important.
Previous studies have indicated that the Maf1 gene
is associated with oxygen free radicals, such as
H2O2, and the mTOR signaling pathway induced
by ischemia (17-18,20).
It has also been speculated that the Maf1 gene is associated with
ischemic stroke-induced cell death, which further exacerbates brain
damage (21-23).
The present study demonstrated that the 3'UTR of Maf1 mRNA was a
binding site for miR-122 using gene prediction software. Therefore,
Maf1 was investigated as a target gene of miR-122. A
dual-luciferase reporter assay was performed, which verified that
miR-122 bound to the 3'UTR of Maf1. In addition, after MCAO, the
expression of miR-122 was increased in comparison with that in a
sham model, whereas the expression of Maf1 was reduced, indicating
that miR-122 may be negatively associated with Maf1 expression.
Therefore, it was hypothesized that the expression of Maf1 may be
inhibited by miR-122 after MCAO. Increased miR-122 expression
inhibited Maf1 expression and decreased miR-122 expression
increased Maf1 expression in vivo. In summary, the present
study indicated that miR-122 inhibited Maf1 expression after MCAO.
The results of the present study may be beneficial in evaluating
the relationship between Maf1 and ischemic stroke and also
indicated that miR-122 may be able to improve the outcome of
ischemic stroke by regulation of the Maf1 gene. However, the
specific role and regulatory mechanism underlying Maf1 and miR-122
activity during the development of ischemic stroke are not
completely understood and require further investigation. In
contrast to other studies that used male mice, only female mice
were used in the present study. Therefore, potential differences in
findings in experiments using male mice cannot be ruled out. More
studies with male mice may further clarify whether miR-122 promotes
the outcome of acute ischemic stroke by reducing the expression of
Maf1.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Joint Foundation of
Natural Science Foundation of Liaoning Province and Shenyang
National Laboratory for Materials Science (grant no.
2019JH3/30100006), the National Natural Science Foundation of China
(grant no. 21505013), the Liaoning BaiQianWan Talents Program
[grant no. (2019)45] and Dalian Science and Technology Innovation
Funds (grant no. 2018J13SN087).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MW, YB and ML conceived and designed the study, and
MW performed the majority of the experiments. MW, XL and YW
constructed the MCAO model. YW, JC and JS cultured the cells. MW
wrote the manuscript. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
Committee of the Xinhua Hospital affiliated to Dalian University
and performed in accordance with the National Institutes of Health
Guidelines for the Care and Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhou J, Chen L, Chen B, Huang S, Zeng C,
Wu H, Chen C and Long F: Increased serum exosomal miR-134
expression in the acute ischemic stroke patients. BMC Neurol.
18(198)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zheng M, Wang X, Yang J, Ma S, Wei Y and
Liu S: Changes of complement and oxidative stress parameters in
patients with acute cerebral infarction or cerebral hemorrhage and
the clinical significance. Exp Ther Med. 19:703–709.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Catanese L, Tarsia J and Fisher M: Acute
ischemic stroke therapy overview. Circ Res. 120:541–558.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bai Y, Zhang Y, Han B, Yang L, Chen X,
Huang R, Wu F, Chao J, Liu P, Hu G, et al: Circular RNA DLGAP4
ameliorates ischemic stroke outcomes by targeting miR-143 to
regulate endothelial-mesenchymal transition associated with
blood-brain barrier integrity. J Neurosci. 38:32–50.
2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang SW, Liu Z and Shi ZS: Non-Coding RNA
in acute ischemic stroke: Mechanisms, biomarkers and therapeutic
targets. Cell Transplant. 27:1763–1777. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Qi Z, Cai S, Cai J, Chen L, Yao Y, Chen L
and Mao Y: miR-491 regulates glioma cells proliferation by
targeting TRIM28 in vitro. BMC Neurol. 16(248)2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Qi R, Liu H and Liu C, Xu Y and Liu C:
Expression and short-term prognostic value of miR-126 and miR-182
in patients with acute stroke. Exp Ther Med. 19:527–534.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Deng Y, Chen D, Gao F, Lv H, Zhang G, Sun
X, Liu L, Mo D, Ma N, Song L, et al: Exosomes derived from
microRNA-138-5p-overexpressing bone marrow-derived mesenchymal stem
cells confer neuroprotection to astrocytes following ischemic
stroke via inhibition of LCN2. J Biol Eng. 13(71)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Stanzione R, Bianchi F, Cotugno M,
Marchitti S, Forte M, Busceti C, Ryskalin L, Fornai F, Volpe M and
Rubattu S: A decrease of brain MicroRNA-122 level is an early
marker of cerebrovascular disease in the stroke-prone spontaneously
hypertensive rat. Oxid Med Cell Longev.
2017(1206420)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li DB, Liu JL, Wang W, Luo XM, Zhou X, Li
JP, Cao XL, Long XH, Chen JG and Qin C: Plasma exosomal
miRNA-122-5p and miR-300-3p as potential markers for transient
ischaemic attack in rats. Front Aging Neurosci.
10(24)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gu R, Wang L, Tang M, Li SR, Liu R and Hu
X: LncRNA Rpph1 protects amyloid-β induced neuronal injury in
SK-N-SH cells via miR-122/Wnt1 axis. Int J Neurosci. 130:443–453.
2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Smith KR, Oliver PL, Lumb MJ,
Arancibia-Carcamo IL, Revilla-Sanchez R, Brandon NJ, Moss SJ and
Kittler JT: Identification and characterisation of a Maf1/Macoco
protein complex that interacts with GABAA receptors in neurons. Mol
Cell Neurosci. 44:330–341. 2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Shetty M, Noguchi C, Wilson S, Martinez E,
Shiozaki K, Sell C, Mell JC and Noguchi E: Maf1-dependent
transcriptional regulation of tRNAs prevents genomic instability
and is associated with extended lifespan. Aging Cell.
19(e13068)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yamada D, Kawabe K, Tosa I, Tsukamoto S,
Nakazato R, Kou M, Fujikawa K, Nakamura S, Ono M, Oohashi T, et al:
Inhibition of the glutamine transporter SNAT1 confers
neuroprotection in mice by modulating the mTOR-autophagy system.
Commun Biol. 2(346)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ma HX, Hou F, Chen AL, Li TT, Zhu YF and
Zhao QP: Mu-Xiang-You-Fang protects PC12 cells against
OGD/R-induced autophagy via the AMPK/mTOR signaling pathway. J
Ethnopharmacol. 252(112583)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Imai T, Iwata S, Miyo D, Nakamura S,
Shimazawa M and Hara H: A novel free radical scavenger, NSP-116,
ameliorated the brain injury in both ischemic and hemorrhagic
stroke models. J Pharmacol Sci. 141:119–126. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Gunawardena D, Raju R and Münch G:
Hydrogen peroxide mediates pro-inflammatory cell-to-cell signaling:
A new therapeutic target for inflammation. Neural Regen Res.
14:1430–1437. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sies H: Hydrogen peroxide as a central
redox signaling molecule in physiological oxidative stress:
Oxidative eustress. Redox Biol. 11:613–619. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu da Z, Jickling GC, Ander BP, Hull H,
Zhan X, Cox C, Shroff N, Dykstra-Aiello C, Stamova B and Sharp FR:
Elevating microRNA-122 in blood improves outcomes after temporary
middle cerebral artery occlusion in rats. J Cereb Blood Flow Metab.
36:1374–1383. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tsang CK, Chen M, Cheng X, Qi Y, Chen Y,
Das I, Li X, Vallat B, Fu LW, Qian CN, et al: SOD1 phosphorylation
by mTORC1 couples nutrient sensing and redox regulation. Mol Cell.
70:502–515. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu BW, Luo C, Zheng ZG, Xia ZY, Zhang Q,
Ke C, Liu R and Zhao YH: Shengui sansheng san extraction is an
angiogenic switch via regulations of AKT/mTOR, ERK1/2 and Notch1
signal pathways after ischemic stroke. Phytomedicine. 44:20–31.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hou YY, Wang K, Wan WJ, Cheng Y, Pu X and
Ye XF: Resveratrol provides neuroprotection by regulating the
JAK2/STAT3/PI3K/AKT/mTOR pathway after stroke in rats. Genes Dis.
5:245–255. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang SS, Li XX, Wang HY and Zheng XFS:
Beyond regulation of pol III: Role of MAF1 in growth, metabolism,
aging and cancer. Biochim Biophys Acta Gene Regul Mech.
1861:338–343. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
National Research Council (US) Institute
for Laboratory Animal Research: Guide for the Care and Use of
Laboratory Animals. National Academies Press (US), Washington, DC,
1996.
|
|
25
|
Llovera G, Roth S, Plesnila N, Veltkamp R
and Liesz A: Modeling stroke in mice: Permanent coagulation of the
distal middle cerebral artery. J Vis Exp. 31(e51729)2014.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta C(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cheng X, Kan P, Ma Z, Wang Y, Song W,
Huang C and Zhang B: Exploring the potential value of miR-148b-3p,
miR-151b and miR-27b-3p as biomarkers in acute ischemic stroke.
Biosci Rep. 38(BSR20181033)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Si W, Ye S, Ren Z, Liu X, Wu Z, Li Y, Zhou
J, Zhang S, Li Y, Deng R and Chen D: miR-335 promotes stress
granule formation to inhibit apoptosis by targeting ROCK2 in acute
ischemic stroke. Int J Mol Med. 43:1452–1466. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu W, Chen X and Zhang Y: Effects of
microRNA-21 and microRNA-24 inhibitors on neuronal apoptosis in
ischemic stroke. Am J Transl Res. 8:3179–3187. 2016.PubMed/NCBI
|
|
30
|
Shi FP, Wang XH, Zhang HX, Shang MM, Liu
XX, Sun HM and Song YP: MiR-103 regulates the angiogenesis of
ischemic stroke rats by targeting vascular endothelial growth
factor (VEGF). Iran J Basic Med Sci. 21:318–324. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Meng P, Zhu Q, Yang H, Liu D, Lin X, Liu
J, Fan J, Liu X, Su W, Liu L, et al: Leonurine promotes neurite
outgrowth and neurotrophic activity by modulating the GR/SGK1
signaling pathway in cultured PC12 cells. Neuroreport. 30:247–254.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cheng J, Tang JC, Pan MX, Chen SF, Zhao D,
Zhang Y, Liao HB, Zhuang Y, Lei RX, Wang S, et al: l-lysine confers
neuroprotection by suppressing inflammatory response via
microRNA-575/PTEN signaling after mouse intracerebral hemorrhage
injury. Exp Neurol. 327(113214)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Du K, Zhao C, Wang L, Wang Y, Zhang KZ,
Shen XY, Sun HX, Gao W and Lu X: MiR-191 inhibit angiogenesis after
acute ischemic stroke targeting VEZF1. Aging (Albany NY).
11:2762–2786. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Guo D, Ma J, Li T and Yan L: Up-regulation
of miR-122 protects against neuronal cell death in ischemic stroke
through the heat shock protein 70-dependent NF-κB pathway by
targeting FOXO3. Exp Cell Res. 369:34–42. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Shi L, Zheng X, Fan Y, Yang X, Li A and
Qian J: The contribution of miR-122 to the innate immunity by
regulating toll-like receptor 4 in hepatoma cells. BMC
Gastroenterol. 19(130)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Xu H, Xu SJ, Xie SJ, Zhang Y, Yang JH,
Zhang WQ, Zheng MN, Zhou H and Qu LH: MicroRNA-122 supports robust
innate immunity in hepatocytes by targeting the RTKs/STAT3
signaling pathway. Elife. 8(e41159)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hatakeyama M, Ninomiya I and Kanazawa M:
Angiogenesis and neuronal remodeling after ischemic stroke. Neural
Regen Res. 15:16–19. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Jayaraj RL, Azimullah S, Beiram R, Jalal
FY and Rosenberg GA: Neuroinflammation: Friend and foe for ischemic
stroke. J Neuroinflammation. 16(142)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lv B, Cheng X, Sharp FR, Ander BP and Liu
DZ: MicroRNA-122 mimic improves stroke outcomes and indirectly
inhibits NOS2 after middle cerebral artery occlusion in
rats. Front Neurosci. 12(767)2018.PubMed/NCBI View Article : Google Scholar
|