Introduction
Degeneration of the intervertebral discs is
characterized by alterations in the morphology of the discs and the
composition of the extracellular matrix, including a reduction in
the number of the intervertebral disc cells (1,2). It
has been reported that when the intervertebral disc degenerates,
the extracellular matrix is reduced, resulting in a decrease in the
water content of the intervertebral disc, thereby resulting in the
degeneration of the intervertebral disc structure (1-4).
Tissue engineering has been increasingly employed in
the medical field as a treatment for degenerative discs, with cell
therapy becoming a potential treatment for intervertebral disc
degeneration (5). Previous studies
have reported that various types of cells, including nucleus
pulposus cells, chondrocytes and mesenchymal stem cells (MSCs), can
be used for cell therapy (6-8).
With further progress, stem cell therapy has been applied in the
clinic, with several studies reporting these results. For example,
Centeno et al (9) reported
that patients with degenerative disc disease (DDD) who are treated
with MSCs to counteract lower back pain with radicular symptoms,
exhibit minor adverse effects and considerable improvements in the
degree of pain, function and overall quality of life during a 6
year follow-up. Li et al (10) investigated the characteristics of
stem cells, including nucleus pulposus-derived stem cells (NPSCs),
annulus fibrosus-derived stem cells (AFSCs) and cartilage
endplate-derived stem cells (CESCs), in human degenerative
intervertebral discs to determine the best stem cell-like
characteristics. No significant differences in cell morphology
among NPSCs, AFSCs and CESCs have been revealed; however AFSCs have
been indicated to exhibit the best stem cell-like characteristics
in the human degenerative intervertebral disc (10).
The present study focused on another type of stem
cell, namely precartilaginous stem cells (PSCs). It has been
indicated that PSCs are localized in the perichondrium surrounding
the epiphysis (11). PSCs are adult
stem cells with a multi-directional differentiation potential,
which have been reported to differentiate into chondrocytes and
osteoblasts to promote the growth of animal limbs (12). PSCs have been isolated and purified
via immunomagnetic bead sorting, and used as seed cells in several
tissue engineering studies (12).
The discovery of PSCs has provided a novel tool for cell
transplantation to repair degenerative intervertebral discs
(13). PSCs have been indicated to
serve an important role in cartilage growth and internalization, as
well as articular cartilage injury repair (14). PSCs are precursor cells of
chondrocytes and exhibit a homology with intervertebral disc
nucleus cells. The alterations in nucleus pulposus cells have been
reported to serve an important role in the degeneration of
intervertebral discs (15). The
nucleus pulposus is an avascular tissue with a limited number of
cells and it is difficult to repair its structure following the
occurrence of a lesion (16).
Therefore, various studies have investigated the use of seed cells
to repair degenerated nucleus pulposus cells (17-19).
The nucleus pulposus cells and chondrocytes of the intervertebral
disc have been reported to exhibit numerous common characteristics,
such as production of collagen II and aggrecan as principal
components of the extracellular matrix (18,19).
Moreover, nucleus pulposus-like cells (NPLCs), which are derived
from the degenerated intervertebral disc, have been indicated to
undergo osteogenesis and promote cartilage repair owing to the fact
that these cells secrete factors (TGF-1, IL-1, TNF, prostaglandin
E2, IL-10, and granulocyte-macrophage colony stimulating factor)
which promote the proliferation and regulate the differentiation of
chondrocytes, as well as promote the synthesis of extracellular
matrix components (20).
Our previous study indicated that PSCs can be
induced to differentiate to NPLCs to repair degenerative
intervertebral discs with unique therapeutic advantages (21). However, the detailed mechanisms of
action behind the differentiation are still unclear. MicroRNAs
(miRNAs/miRs) are a class of small noncoding RNA molecules that
negatively regulate gene expression at the post-transcriptional
level (22). It has been reported
that miRNAs not only regulate several normal physiological
processes but are also associated with the development of DDD
(23). In the present study, miRNA
microarray screening was employed to identify differences in miRNA
expression profiles between TGF-β1-induced rat primary PSCs and
differentiated NPLCs. miR-124-3p was verified as a significantly
differentially expressed miRNA during the differentiation of PSCs
to NPLCs. Moreover, the mechanisms of action through which
miR-124-3p regulates the fate of PSCs and its role in the
differentiation process were also investigated.
Materials and methods
Chemicals, reagents and
antibodies
TGF-β1 was purchased from Selleck Chemicals.
miR-124-3p and negative control (NC/nc) agomirs (cat. no.
miR40000422-4-5) were obtained from Guangzhou RiboBio Co., Ltd.
Primary antibodies against fibroblast growth factor receptor 3
(FGFR-3; cat. no. ab133644), follistatin-related protein 1 (FSTL1;
cat. no. ab232777,), collagen II (cat. no. ab188570), aggrecan
(cat. no. ab3778) and HRP-conjugated secondary antibody GAPDH (cat.
no. ab181602) were obtained from Abcam. FSTL1 small interfering
(si)RNA and siRNAnc sequences were designed, synthesized and
validated by Shanghai GenePharma Co., Ltd. The siRNA sequences used
were as follows: FSTL1, 5'-GCAAAUACUUACGGACUUUU-3'; and siRNAnc,
5'-GCAUCUUGAGAUUUAAUCA-3'. FSTL1 (cat. no. HP100806), TPST2 (cat.
no. HP101143) and enhancer of zeste homolog 2 (EZH2; cat. no.
HP101222) quantitative PCR (qPCR) primers were purchased from Sino
Biological Inc. (Shanghai, China). COS7 and 293 cells were
purchased from the Cell Bank of the Chinese Academy of
Sciences.
Primary PSCs culture
PSCs were generated as previously described
(11). Briefly, the perichondrial
mesenchyme (the rings of LaCroix) were isolated from Sprague Dawley
rats (Trophic Animal Feed High-Tech Co., Ltd.) and subsequently
digested in 0.25% Trypsin Solution (cat. no. 25200072; Thermo
Fisher Scientific, Inc.) supplemented with 0.05% collagenase type I
(Sigma-Aldrich; Merck KGaA) for 3-5 min at 37˚C . The digestion
mixture was resuspended in FBS. Subsequently, the cells were
dispersed and resuspended as a single cell suspension in 0.1 M PBS
with centrifugation speeds of 167.7 x g for 5 min at room
temperature. The cell suspension was incubated with the FGFR-3
antibody (1:500; cat. no. ab133644; Abcam) for 15 min at 4˚C, and
subsequently, an immunomagnetic separation system (Miltenyi Biotec
GmbH) was used to purify the FGFR-3 expressing cells. The isolated
PSCs were cultured in complete DMEM/F12 medium supplemented with
20% FBS (Nanjing KeyGen Biotech Co., Ltd.) at 37˚C with 5%
CO2. The phenotype identification of the rat PSCs was
based on the FGFR-3 protein expression, which was detected via
western blotting. The present study was approved by the
Institutional Animal Care and Use Committee of Wuxi People's
Hospital (approval no. WXPH20190103c0600105). In the current study,
60 rats were used between January and March 2019. Animal health and
behavior were monitored daily. All operations were performed
according to international guidelines for the care and treatment of
experimental animals. The rats were male Sprague-Dawley rats of 5-7
weeks of age weighing 150-200 g. Cages were cleaned and enrichment
items were renewed weekly. The animal room had a controlled 12-h
light/dark cycle (lights on at 6:00 AM), temperature (22±2˚C) and
relative humidity (45-65%). Daily food and water were supplied by
laboratory staff. The rats were maintained in an ordinary housing
facility, which is in accordance with the national standard
‘Laboratory Animal-Requirements of Environment and Housing
Facilities’ (standard no. GB 14925-2010). The animals were
euthanized in accordance with the requirements of the ‘Guidelines
for the Examination of the Scientific Research of Experimental
Animal Welfare’ (standard no. GB/T 35892-2018) using intravenous
administration of pentobarbital (100-150 mg/kg) and death was
verified via examining the animals' heartbeat for complete
cessation and pupil dilation.
PSCs cell differentiation
PSCs were differentiated according to a previously
published protocol (24). Briefly,
PSCs at passage three were trypsinized and transferred to six-well
plates at a concentration of 1x105 cells/ml.
Differentiation of PSCs was induced by culturing the cells in
DMEM/F12 medium supplemented with 10% FBS, 10 ng/ml TGF-β1, 100 nM
dexamethasone, 50 µg/ml L-ascorbic acid 2-phosphate, 100 µg/ml
sodium pyruvate, 40 µg/ml proline and 6.25 mg/l insulin at 37˚C
with 5% CO2 for 7 days.
Cell lines and culturing
conditions
COS-7 and 293 cells were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and maintained
in an incubator at 37˚C and 5% CO2. The medium was
replaced 2-3 times a week. When the cell density reached 70-80%,
cells were digested by 0.25% trypsin (Gibco; Thermo Fisher
Scientific, Inc.) and passaged.
Cell transfection
Small interfering RNA targeting FSTL1 (siFSTL1) was
synthesized by Guangzhou RiboBio Co., Ltd., and a scrambled siRNA
(siNC; Guangzhou RiboBio Co., Ltd.) was used as the negative
control. One day before transfection, cells were digested with
0.25% trypsin (cat. no. C0202; Beyotime Institute of
Biotechnology). Then, 50 µl of Opti-MEM® medium (cat.
no. 31985062; Invitrogen; Thermo Fisher Scientific, Inc.) was mixed
with siFSTL1(cat. no. siB14715103107-1-5; Guangzhou RiboBio Co.,
Ltd.) or siNC. And 3 µl Lipofectamine® 3000 reagent
(cat. no. L3000015; Thermo Fisher Scientific, Inc.) was diluted
with 50 µl OptiMEM. Then, the above two mixtures were mixed. Then
cells were transfected with the mixture according to the
Lipofectamine® 3000 kit (Thermo Fisher Scientific,
Inc.).
miR-124-3p agomir (miR40000422-4-5 Guangzhou RiboBio
Co., Ltd.) and negative control (agomir NC) were synthesized by
Guangzhou RiboBio Co., Ltd. Cells were seeded in six-well plates
(1x105 cells/ml) and transfected with 50 nM miR-124-3p
agomir or 50 nM agomir NC using Lipofectamine® 3000 kit
(Thermo Fisher Scientific, Inc.). The transfection efficiency was
observed by a fluorescence microscope after 24 h.
miRNA microarray analysis
Total RNA from rat primary PSCs and
TGF-β1-differentiated NPLCs was isolated using the
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Following
quantification of total RNA using NanoDrop ND-1000 Ultraviolet
Spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.), a miRNA profiling system (Agilent Technologies, Inc.) was
used to detect the differences in miRNA expression between primary
PSCs and NPLCs. The slides were scanned using a microarray scanner
(GenePix 4100A) and the data were quantified using Feature
Extraction software v10.7 (both from Agilent Technologies, Inc.).
The GeneSpring GX software v12.6 (Agilent Technologies, Inc.) was
additionally used to analyze the miRNA expression data that were
detected using the microarray system.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA of PSCs was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The primers used for the amplification of miR-124-3p (cat.
no. miRA1001726-1-100) and U6 endogenous control (cat. no.
miRAN0002-1-100) were purchased from Guangzhou RiboBio Co., Ltd.
Subsequently, qPCR was performed using the Mir-X™ miRNA RT-qPCR TB
Green® kit according to the manufacturer's instructions
(Takara Biotechnology). Briefly, Total RNA was reverse transcribed
to obtain cDNA. The reverse transcription process of miR-124-3p
used the Mir-X miRNA First-Strand Synthesis Kit (cat. no. 638313;
Takara Biotechnology, Co., Ltd.), the tube was incubated for 1 h at
37˚C, and the reaction terminated at 85˚C for 5 min to inactivate
the enzymes. The PCR reactions were carried out using TB Green
Advantage qPCR Premix (cat. no. 639676; Takara Biotechnology, Co.,
Ltd.) under the following conditions: Denaturation, 95˚C 10 sec;
qPCR x 40 cycles of 95˚C 5 sec and 60˚C 20 sec; dissociation curve,
95˚C 60 sec, 55˚C 30 sec and 95˚C 30 sec. The relative miRNA
expression was calculated using the 2-∆∆Cq method
(25).
Target gene prediction
To predict the potential target genes of
miRNA-124-3p, three bioinformatics analysis tools, TargetScan v7.2
(www.targetscan.org), Pictar (https://pictar.mdc-berlin.de/) and miRanda (http://www.microrna.org/microrna/home.do) were used to
predict its potential target genes.
Luciferase reporter assay
For the luciferase reporter assay, a construct
containing the 3'-untranslated region (UTR) of the FSTL1 mRNA
carrying the putative miRNA-124-3p binding site or a respective
mutant, which was used as a control, were cloned into the pGL3
Luciferase Reporter plasmid (Promega Corporation). Cells
(1x105) were co-transfected with a 50 ng/well reporter
plasmid and 50 nM miR-124-3p agomir or 50 nM agomir NC in a 6-well
plate for 48 h. Firefly and Renilla luciferase activities
were detected using Dual-Luciferase® Reporter Assay
System (cat. no. E1910; Promega Corporation). Briefly, 500 µl of
PLB was added and agitated for 15 min. LAR II (100 µl) and cells
(20 µl) were sequentially added to 96-well plate. Then, 100 µl of
1X Stop & Glo reagent was added to detect the luminescence
intensity of Renilla luciferase.
Western blot analysis
Following miR-124-3p agomir or FTSL1 siRNA
transfection, PSCs were collected and solubilized in RIPA lysis
buffer (cat. no. KGP702; Nanjing KeyGen Biotech Co., Ltd.).
Following centrifugation for 10 min (1,600 x g, 4˚C), the protein
concentration was measured using a BCA assay kit (Sigma-Aldrich;
Merck KGaA). The samples (20 µg/well) were separated on 10%
SDS-PAGE (Nanjing Genscript Biotechnology Co., Ltd.) at 140 V for
50 min. Subsequently, the separated proteins were transferred onto
PVDF membranes at 300 mA for 60 min (Nanjing Genscript
Biotechnology Co., Ltd.). The membranes were subsequently blocked
for 1 h at room temperature with 5% fat-free dried milk diluted in
TBS-0.1% Tween-20, followed by incubation with primary antibodies
against FSTL1 (1:1,000), collagen II (1:1,000), with GAPDH being
used as an internal control (l:80,000), at 4˚C overnight. The
membranes were subsequently incubated with HRP-conjugated secondary
antibodies (anti-rabbit; cat. no. ab6721; 1:10,000; Abcam) for 1 h
at room temperature. The protein bands were detected using an ECL
chemiluminescence kit (Nanjing KeyGen Biotech Co., Ltd., China) and
semi-quantified using ImageQuant TL analysis software v8.1
(Cytiva).
Statistical analysis
All statistical analyses were performed using SPSS
v19 software (IBM Corp.). Each experiment was repeated three times
and data were presented as the mean ± SD. Unpaired Student's t-test
or one-way ANOVA followed by Bonferroni's post hoc test were
applied to identify the statistical significance. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-124-3p is associated with the
differentiation of PSCs to NPLCs
To identify miRNAs that are associated with the
regulation of the differentiation of PSCs to NPLCs, miRNA
microarray technology was used to detect differences in miRNA
expression profiles between TGF-β1-treated rat primary PSCs (day 0)
and differentiated NPLCs (day 15). The efficient induction of PSC
differentiation by TGF-β1 has been previously verified in our
previous study (16). The results
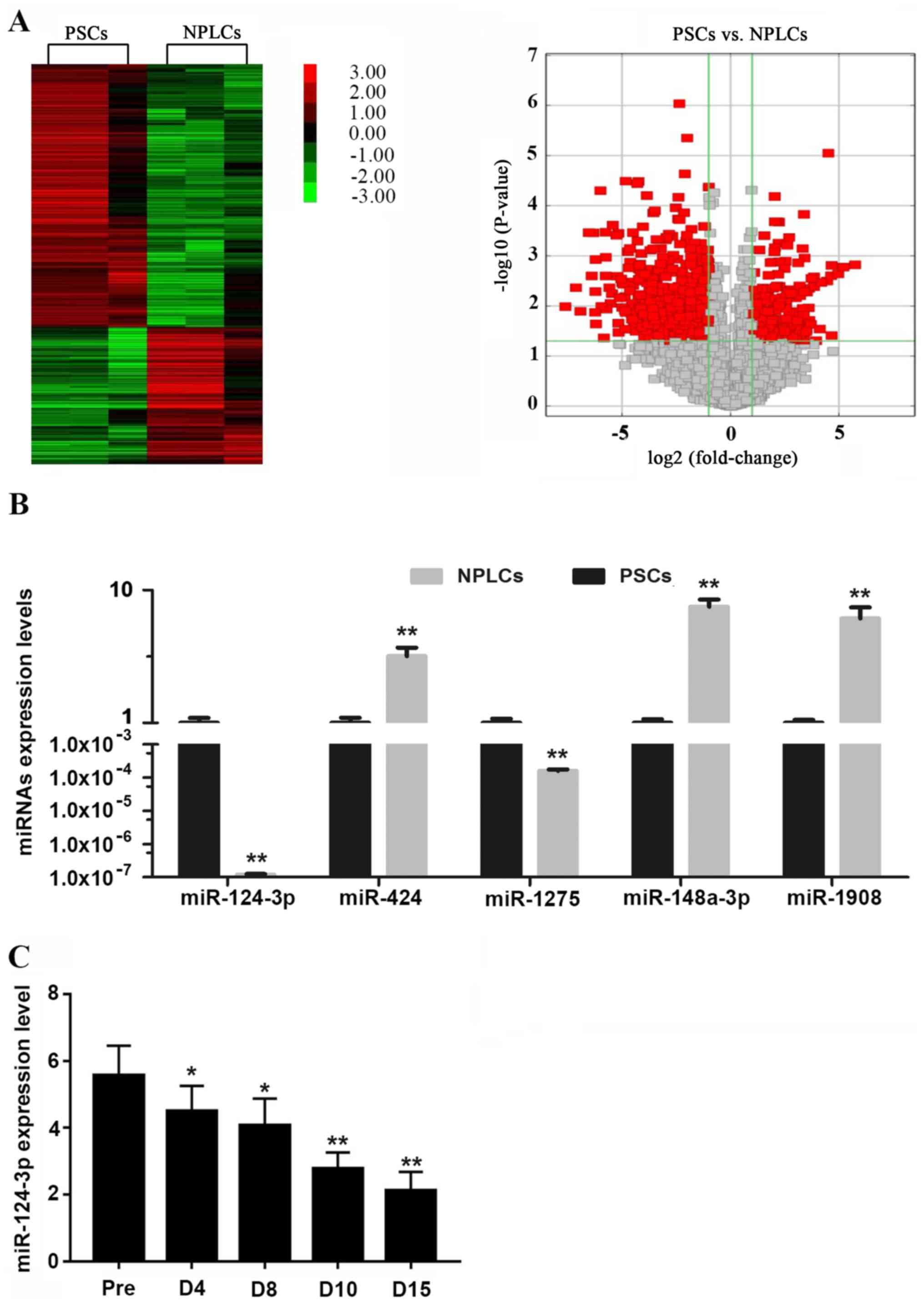
of the present study indicated that when compared with primary
PSCs, 50 miRNAs in the differentiated NPLCs were considerably
upregulated (>3-fold), while 36 miRNAs were downregulated
<3-fold (Fig. 1A). A total of
five miRNAs (miR-124-3p, miR-424, miR-1275, miR-148a-3p and
miR-1908) with a >5-fold expression difference and a small
within group variation were selected for subsequent validation
using RT-qPCR. As indicated in Fig.
1B, the expression tendency of the five miRNAs, was consistent
with the results of the microarray screening. Among them, the
difference in the expression levels of miR-124-3p was the lowest
(Fig. 1B).
The alterations in miR-124-3p expression during the
differentiation of PSCs to NPLCs were subsequently analyzed
(between days 0 to 15). PSCs were induced to differentiate into
NPLCs via treatment with TGF-β1 and the expression levels of
miR-124-3p at various time points of differentiation were detected
using RT-qPCR (days 0, 4, 8, 10 and 15). The results verified that
the expression of miR-124-3p decreased along with the
differentiation of PSCs and exhibited the lowest levels during the
later stages of differentiation (Fig.
1C).
miR-124-3p inhibits the
differentiation of PSCs
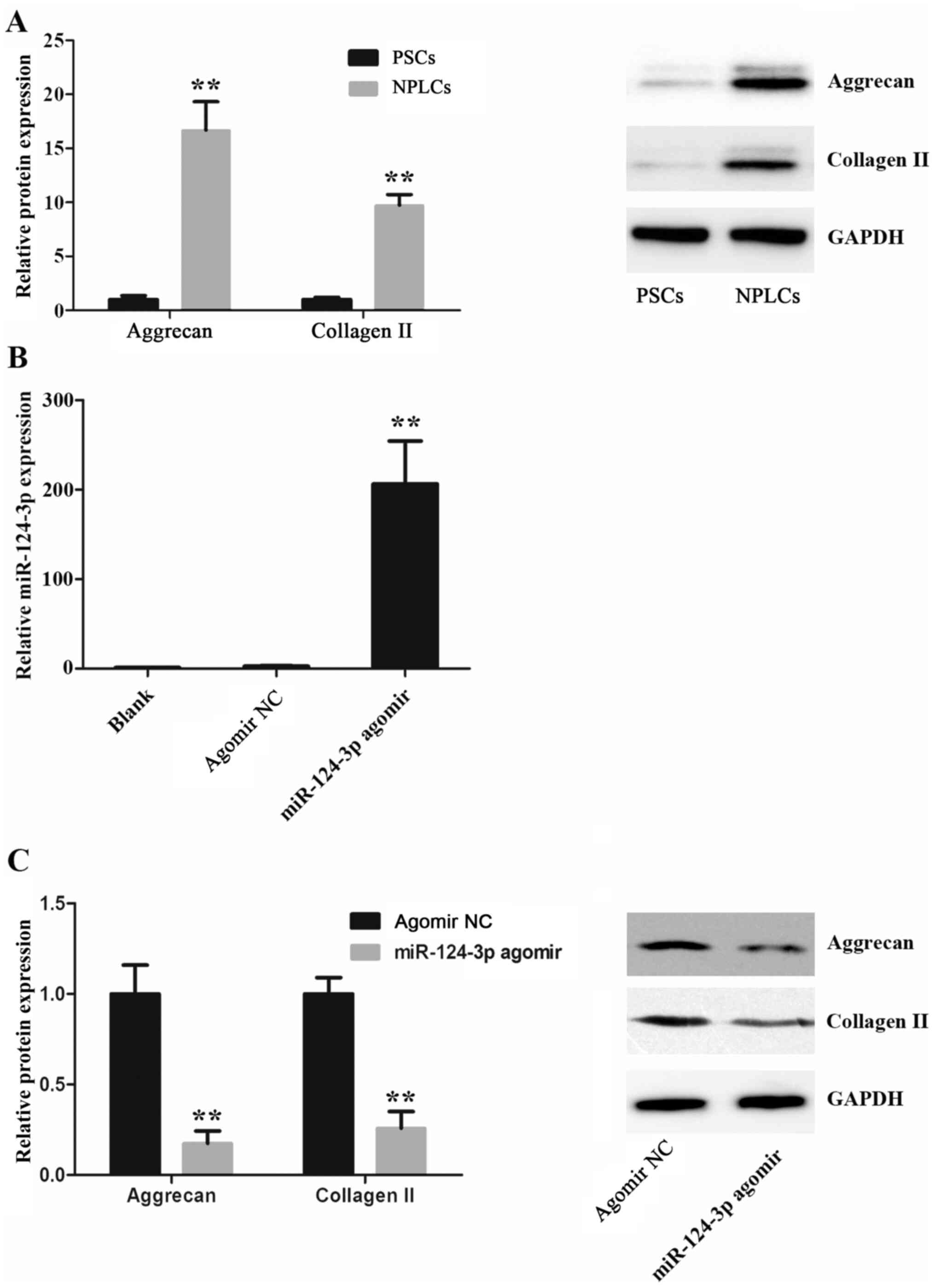
As specific markers for nucleus pulposus cells have
not been identified, collagen II and aggrecan are used to detect
differentiated nucleus pulposus cells (26). Therefore, the expression of collagen
II and aggrecan during the differentiation of PSCs into NPLCs was
detected using western blotting. It was demonstrated that collagen
II and aggrecan protein were expressed at a significantly higher
levels in NPLCs compared with PSCs, which suggested that PSCs were
efficiently differentiated into NPLCs (Fig. 2A). Furthermore, to identify the role
of miR-124-3p in PSCs differentiation, miR-124-3p was overexpressed
in PSCs via transfection with miR-124-3p agomir (Fig. 2B). As presented in Fig. 2C, collagen II and aggrecan protein
expression levels in the miR-124-3p agomir group were significantly
decreased when compared with the NC group on day 18
post-transfection. These results suggested that miR-124-3p
overexpression inhibited the differentiation of PSCs into
NPLCs.
miR-124-3p targets FSTL1 during PSCs
differentiation
Using TargetScan software, 7-9 binding sites were
predicted in the 3'-UTR region of the FSTL1 mRNA. The results
indicated that FSTL1, EZH2 and TPST2 may represent potential target
genes of miR-124-3p (Fig. 3A). The
alterations in the expression levels of the aforementioned three
genes during the differentiation process of PSCs (0, 4, 8 and 15
days) were subsequently determined. As indicated in Fig. 3B, only FSTL1 was demonstrated to be
gradually increased during the differentiation process, in contrast
to the expression levels of miR-124-3p, which were indicated to
decrease during PSCs differentiation, as aforementioned. However,
the expression of EZH2 and TPST2 showed neither gradual increase
nor decrease. Notably, the majority of miRNAs have been indicated
not to affect the mRNA expression of their target gene. However,
certain miRNAs have been reported to result in mRNA degradation of
their target genes (27). In the
present study, it was demonstrated that miR-124-3p overexpression
resulted in decreased FSTL1 mRNA levels in PSCs (Fig. 3C).
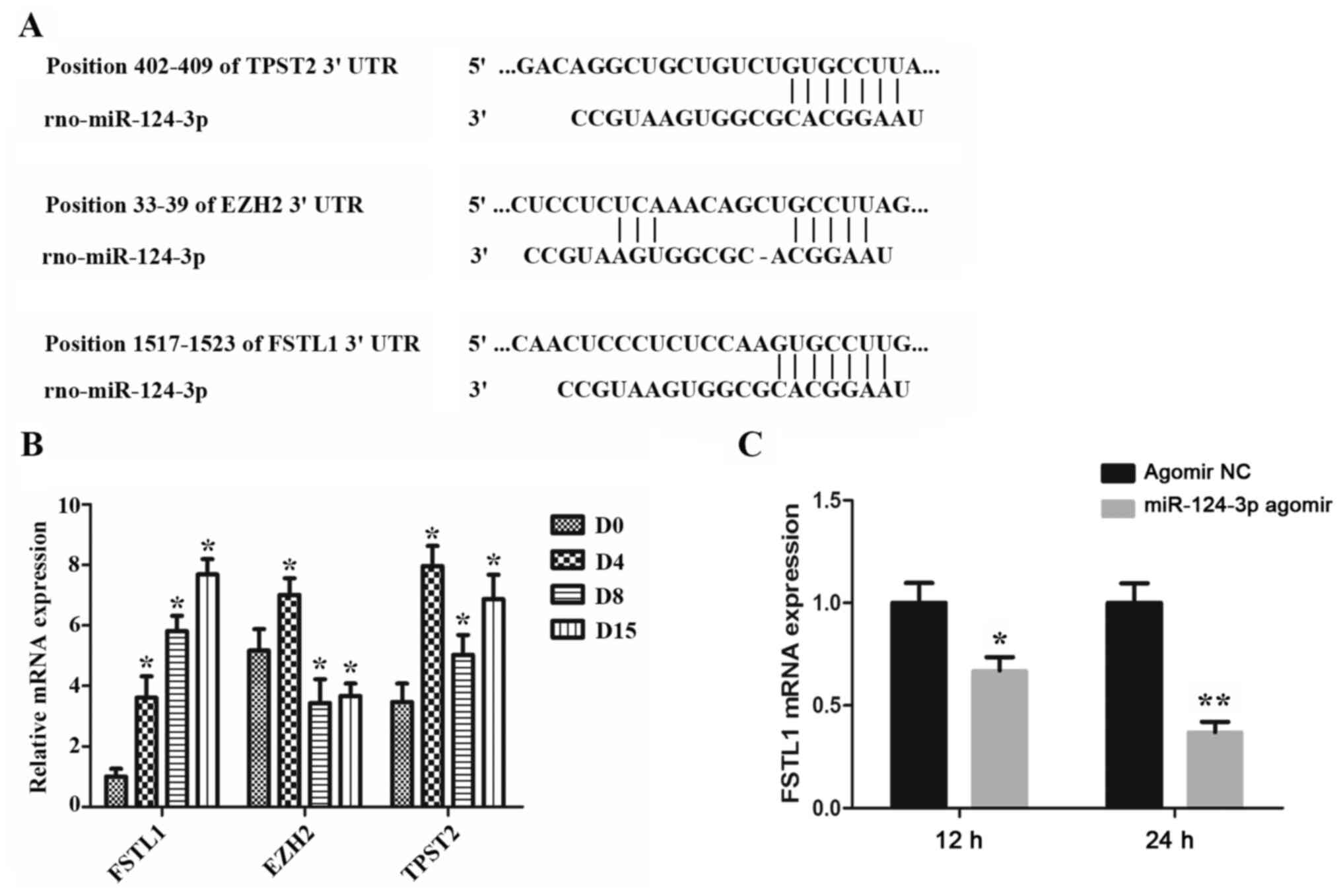 | Figure 3Analysis of the targets of miR-124-3p
during PSCs differentiation. (A) Bioinformatics analysis tools
predicted the potential target genes for miR-124-3p. (B) The
expression of FSTL1, EZH2 and TPST2 during the differentiation of
PSCs to nucleus pulposus-like cells was detected via RT-qPCR.
*P<0.05 vs. D0. (C) PSCs were transfected with
miR-124-3p agomir, followed by the detection of FSTL1 mRNA levels
via RT-qPCR. *P<0.05 and **P<0.01 vs.
agomir NC. miR, microRNA; PSCs, precartilaginous stem cells;
RT-qPCR, reverse transcription-quantitative PCR; D, day; UTR,
untranslated region; FSTL1, follistatin-related protein 1; TPST2,
protein-tyrosine sulfotransferase 2; EZH2, enhancer of zeste
homolog 2; NC, negative control. |
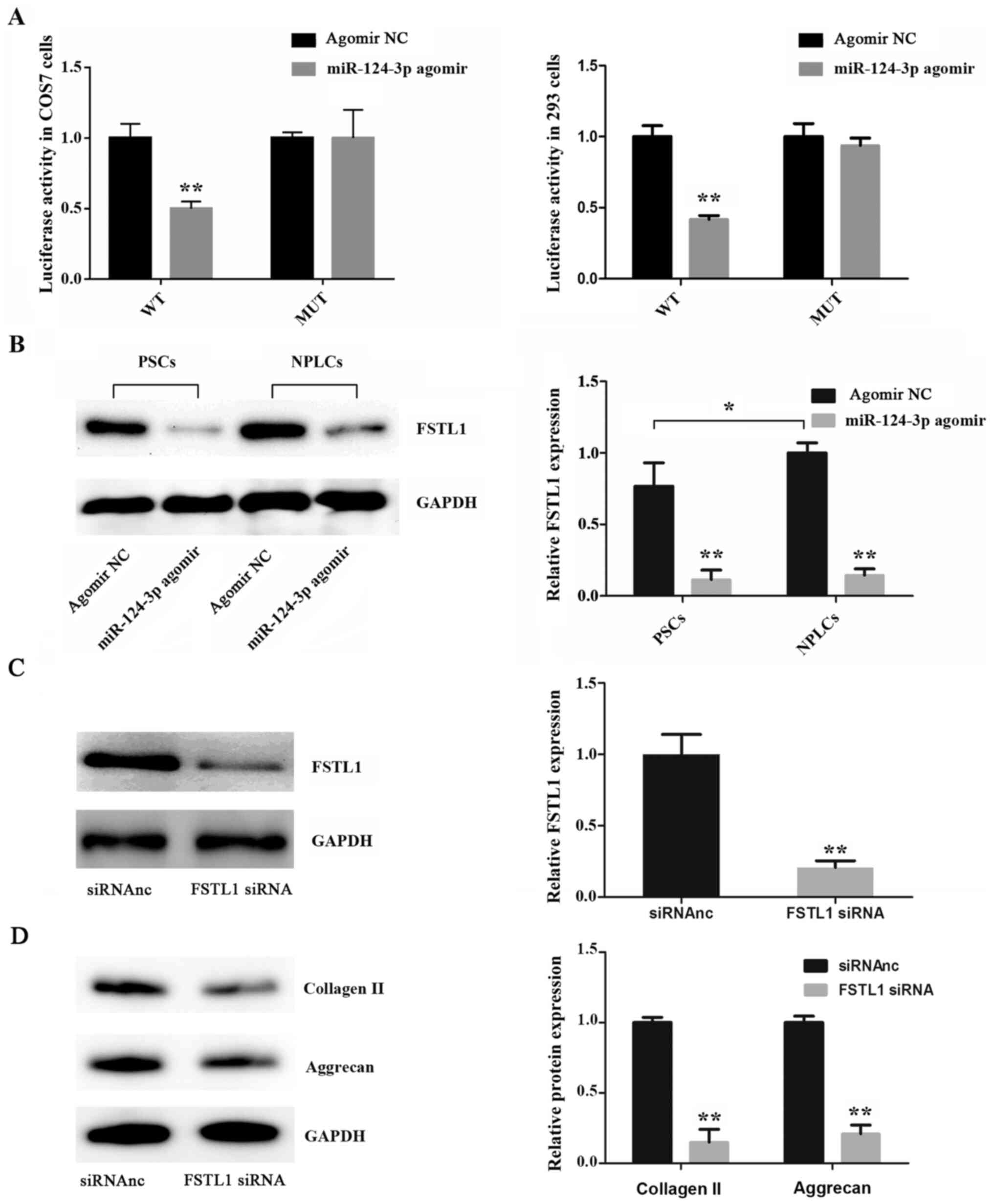
Moreover, in order to verify whether miR-124-3p can
directly bind to and regulate the mRNA expression levels of FSTL1,
a dual-luciferase reporter assay was used in the present study.
Luciferase reporter plasmids containing the predicted GUGCCUU
binding site of the 3'-UTR of FSTL1 and a mutant site (UGCUUGC)
were constructed and co-transfected with miR-124-3p into COS7 and
293 cells to detect the alterations in luciferase activity. As
indicated in Fig. 4A, the results
revealed that miR-124-3p overexpression significantly reduced the
luciferase activity of FSTL1 in the wild-type group when compared
with the mutant group. Furthermore, western blot analysis revealed
that miR-124-3p overexpression resulted in a decrease in FSTL1
expression in both PSCs (day 0) and NPLCs (day 15) compared with
the control group (Fig. 4B).
Subsequently, FSTL1 was knocked down using an FSTL1 siRNA (Fig. 4C) and the expression of collagen II
and aggrecan was determined. The results revealed that FSTL1
knockdown resulted in decreased collagen II and aggrecan protein
expression levels compared with control cells, which indicated that
FSTL1 was associated with the differentiation of PSCs (Fig. 4D).
Discussion
PSC-derived NPLCs may be used to treat
intervertebral discs degeneration, owing to the capacity of NPLCs
to promote tissue remodeling, which has been shown to result in
lower back pain relief (28). The
present study demonstrated that miR-124-3p was associated with the
differentiation of PSCs into NPLCs and that the expression of
miR-124-3p gradually decreased during the differentiation process,
from day 0 to day 15. Previous studies have indicated that
miR-124-3p expression is decreased in Alzheimer's disease (AD) and
several types of tumors, including bladder cancer, hepatocellular
carcinoma and glioma (29-32).
Recently, Zhang et al (33)
reported that miR-124-3p attenuates neuropathic pain induced by
chronic sciatic nerve injury in rats. However, the function of
miR-124-3p in lower back pain, which is induced by DDD, remains
unknown. Therefore, the present study aimed to elucidate the
molecular mechanism of action underlying the association between
miR-124-3p and the differentiation of PSCs into NPLCs, and to
identify the potential target genes which may be involved in this
process.
In AD, Kang et al (29) demonstrated that upregulation of
miR-124-3p expression levels attenuates cell apoptosis and abnormal
hyperphosphorylation of Tau protein in N2a/APP695swe cells. Zo and
Long (30) reported that inhibition
of miR-124-3p expression increases bladder cancer cell
proliferation, migration and invasion, as well as reducing
apoptosis. Furthermore, Luo et al (32) revealed that transfection with
miR-124-3p inhibits the expression of cell cycle and
epithelial-mesenchymal transition regulators, and inhibits the
proliferation, migration and invasion of glioma cells.
To explore the function of miR-124-3p in the
differentiation of PSCs to NPLCs, miR-124-3p was overexpressed in
PSCs using a miR-124-3p agomir. Collagen type II and aggrecan have
been indicated to be produced by nucleus pulposus cells and are
required for their homeostasis (34,35).
The results of the present study revealed that collagen II and
aggrecan levels were increased during the differentiation of PSCs
to NPLCs. However, following transfection of PSCs with miR-124-3p
agomir, the expression of collagen II and aggrecan in NPLCs (day
18) was significantly decreased when compared with the control
group. This suggested that miR-124-3p may influence the
differentiation of PSCs into NPLCs.
However, the mechanism of action through which
miR-124-3p regulates the differentiation of PSCs remained unclear.
Bioinformatics analysis indicated that FSTL1, EZH2 and TPST2 may
represent targets of miR-124-3p during the differentiation of PSCs.
However, the results indicated that only FSTL1 levels increased
gradually during the differentiation of PSCs to NPLCs. Considering
that miRNAs recognize their target genes via complementary base
pairing and subsequently guide the silencing complex to degrade or
repress the translation of the target mRNA according to the degree
of complementarity (27), it was
speculated that FSTL1 may be a potential target gene of miR-124-3p.
FSTL1 was firstly identified in mouse osteoblasts and as TGF-β1 has
been indicated to induce its upregulation, FSTL1 is also known as
TGF-β-stimulated clone-36(36).
FSTL1 is an extracellular matrix protein, which is widely expressed
in all eukaryotic cells and has been associated with cell
differentiation, metabolism, cell proliferation and the immune
response (37-39).
A previous study indicated that FSTL1 also participates in the
regulation of Lumbar disc herniation (40). It was demonstrated that FSTL1
expression is increased during the progression of intervertebral
disc disease and promotes inflammatory reactions in the nucleus
pulposus via the MAPK and NF-κB signaling pathways (40). The present study revealed that FSTL1
was also associated with the differentiation of PSCs to NPLCs, a
process which was indicated to be regulated by miR-124-3p.
Overexpression of miR-124-3p resulted in decreased expression
levels of FSTL1 during the differentiation of PSCs to NPLCs and
dual-luciferase reporter assays identified FSTL1 as a direct target
of miR-124-3p. A previous study reported that PSCs can be induced
to differentiate to NPLCs to repair degenerative intervertebral
discs, which can provide unique therapeutic advantages (24). The present study elucidated the
function and molecular mechanism underlying the role of miR-124-3p
in the differentiation of PSCs to NPLCs. Therefore, it was
hypothesized that miR-124-3p may be a target for the induction of
PSC differentiation and the subsequent repair of degenerated
intervertebral discs.
In conclusion, the present study demonstrated a
novel role for miR-124-3p in the differentiation of PSCs to NPLCs,
and provided evidence that miR-124-3p negatively regulated its
target gene, FSTL1, during PSC differentiation. However, the
limitations of the present study lie in the lack of effective in
vivo experiments to demonstrate the regulatory effect of
miR-124-3p on PSCs. Moreover, whether miR-124-3p can alleviate DDD
remains unclear. Therefore, further investigations into the
function of miR-24-3p in vivo should be an aim of future
studies.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by National Science
Foundation of China (grant no. 81101374).
Availability of data and materials
All data generated and/or analyzed during the
present study are included in the published article.
Authors' contributions
QW designed the current study and acquired funding.
JW, XG, DF, DL and TJ performed the experiments, collected the data
and wrote the manuscript. QW and JW confirmed the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by Institutional
Animal Care and Use Committee of Wuxi People's Hospital (approval
no. WXPH20190103c0600105).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fardon DF, Williams AL, Dohring EJ,
Murtagh FR, Gabriel Rothman SL and Sze GK: Lumbar disc
nomenclature: Version 2.0: Recommendations of the combined task
forces of the North American Spine Society, the American Society of
Spine Radiology, and the American Society of Neuroradiology. Spine
J. 14:2525–2545. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hemanta D, Jiang XX, Feng ZZ, Chen ZX and
Cao YW: Etiology for Degenerative Disc Disease. Chin Med Sci J.
31:185–191. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lu H and Peng L: Efficacy and safety of
Mobi-C cervical artificial disc versus anterior discectomy and
fusion in patients with symptomatic degenerative disc disease: A
meta-analysis. Medicine (Baltimore). 96(e8504)2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Matta A, Karim MZ, Isenman DE and Erwin
WM: Molecular therapy for degenerative disc disease: Clues from
secretome analysis of the notochordal cell-rich nucleus pulposus.
Sci Rep. 7(45623)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liang L, Li X, Li D, Jiang W, Wang H, Chen
J, Sun Z, Zhang N and Zhu Y: The characteristics of stem cells in
human degenerative intervertebral disc. Medicine (Baltimore).
96(e7178)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Guo J, Shao M, Lu F, Jiang J and Xia X:
Role of Sirt1 plays in nucleus pulposus cells and intervertebral
disc degeneration. Spine (Phila Pa 1976). 42:E757–E766.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Vedicherla S and Buckley CT: Cell-based
therapies for intervertebral disc and cartilage regeneration -
Current concepts, parallels, and perspectives. J Orthop Res.
35:8–22. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Skovrlj B, Cunn G, Guzman JZ and Qureshi
SA: Mesenchymal stem cell technology in the treatment of
degenerative disc disease. J Neurosurg Sci. 59:25–35.
2015.PubMed/NCBI
|
|
9
|
Centeno C, Markle J, Dodson E, Stemper I,
Williams CJ, Hyzy M, Ichim T and Freeman M: Treatment of lumbar
degenerative disc disease-associated radicular pain with
culture-expanded autologous mesenchymal stem cells: A pilot study
on safety and efficacy. J Transl Med. 15(197)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li XC, Tang Y, Wu JH, Yang PS, Wang DL and
Ruan DK: Characteristics and potentials of stem cells derived from
human degenerated nucleus pulposus: Potential for regeneration of
the intervertebral disc. BMC Musculoskelet Disord.
18(242)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang S, Chen A, Hu W, Li M, Liao H, Zhu
W, Song D and Guo F: Immunological purification of rat
precartilaginous stem cells and construction of the immortalized
cell strain. Arch Orthop Trauma Surg. 128:1339–1344.
2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li C, Wang Q and Wang JF: Transforming
growth factor-β (TGF-β) induces the expression of
chondrogenesis-related genes through TGF-β receptor II
(TGFRII)-AKT-mTOR signaling in primary cultured mouse
precartilaginous stem cells. Biochem Biophys Res Commun.
450:646–651. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fan MP, Si M, Li BJ, Hu GH, Hou Y, Yang W,
Liu L, Tang B and Nie L: Cell therapy of a knee osteoarthritis rat
model using precartilaginous stem cells. Eur Rev Med Pharmacol Sci.
22:2119–2125. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Richardson SM, Kalamegam G, Pushparaj PN,
Matta C, Memic A, Khademhosseini A, Mobasheri R, Poletti FL,
Hoyland JA and Mobasheri A: Mesenchymal stem cells in regenerative
medicine: Focus on articular cartilage and intervertebral disc
regeneration. Methods. 99:69–80. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Xia K, Gong Z, Zhu J, Yu W, Wang Y, Wang
J, Xu A, Zhou X, Tao H, Li F and Liang C: Differentiation of
pluripotent stem cells into nucleus pulposus progenitor cells for
intervertebral disc regeneration. Curr Stem Cell Res Ther.
14:57–64. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chen P, Ning L, Qiu P, Mo J, Mei S, Xia C,
Zhang J, Lin X and Fan S: Photo-crosslinked gelatin-hyaluronic acid
methacrylate hydrogel-committed nucleus pulposus-like
differentiation of adipose stromal cells for intervertebral disc
repair. J Tissue Eng Regen Med. 13:682–693. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ma K, Chen S, Li Z, Deng X, Huang D, Xiong
L and Shao Z: Mechanisms of endogenous repair failure during
intervertebral disc degeneration. Osteoarthritis Cartilage.
27:41–48. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yang Z, Gao XJ and Zhao X: CDMP1 promotes
type II collagen and aggrecan synthesis of nucleus pulposus cell
via the mediation of ALK6. Eur Rev Med Pharmacol Sci.
24:10975–10983. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Le Maitre CL, Pockert A, Buttle DJ,
Freemont AJ and Hoyland JA: Matrix synthesis and degradation in
human intervertebral disc degeneration. Biochem Soc Trans.
35:652–655. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yang SH, Wu CC, Shih TT, Sun YH and Lin
FH: In vitro study on interaction between human nucleus pulposus
cells and mesenchymal stem cells through paracrine stimulation.
Spine (Phila Pa 1976). 33:1951–1957. 2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang Q, Gu X, Cheng L and Wang J:
Differentiation of immortalized human precartilaginous stem cells
into nucleus pulposus-like cells. Int J Clin Exp Pathol.
8:2816–2822. 2015.PubMed/NCBI
|
|
22
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang C, Wang WJ, Yan YG, Xiang YX, Zhang
J, Tang ZH and Jiang ZS: MicroRNAs: New players in intervertebral
disc degeneration. Clin Chim Acta. 450:333–341. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang Q, Gu X and Wang J: Transforming
growth factor-β1 (TGF-β1) induces mouse precartilaginous stem cell
differentiation through TGFRII-CK1ε-β-catenin signalling. Int J Exp
Pathol. 99:113–120. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou X, Tao Y, Chen E, Wang J, Fang W,
Zhao T, Liang C, Li F and Chen Q: Genipin-cross-linked type II
collagen scaffold promotes the differentiation of adipose-derived
stem cells into nucleus pulposus-like cells. J Biomed Mater Res A.
106:1258–1268. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Vishnoi A and Rani S: miRNA biogenesis and
regulation of diseases: An overview. Methods Mol Biol. 1509:1–10.
2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Weber KT, Jacobsen TD, Maidhof R,
Virojanapa J, Overby C, Bloom O, Quraishi S, Levine M and Chahine
NO: Developments in intervertebral disc disease research:
Pathophysiology, mechanobiology, and therapeutics. Curr Rev
Musculoskelet Med. 8:18–31. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kang Q, Xiang Y, Li D, Liang J, Zhang X,
Zhou F, Qiao M, Nie Y, He Y, Cheng J, et al: miR-124-3p attenuates
hyperphosphorylation of Tau protein-induced apoptosis via
caveolin-1-PI3K/Akt/GSK3β pathway in N2a/APP695swe cells.
Oncotarget. 8:24314–24326. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zo RB and Long Z: miR-124-3p suppresses
bladder cancer by targeting DNA methyltransferase 3B. J Cell
Physiol. 234:464–474. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Long HD, Ma YS, Yang HQ, Xue SB, Liu JB,
Yu F, Lv ZW, Li JY, Xie RT, Chang ZY, et al: Reduced hsa-miR-124-3p
levels are associated with the poor survival of patients with
hepatocellular carcinoma. Mol Biol Rep. 45:2615–2623.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Luo L, Chi H and Ling J: miR-124-3p
suppresses glioma aggressiveness via targeting of Fra-2. Pathol Res
Pract. 214:1825–1834. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang Y, Liu HL, An LJ, Li L, Wei M, Ge DJ
and Su Z: miR-124-3p attenuates neuropathic pain induced by chronic
sciatic nerve injury in rats via targeting EZH2. J Cell Biochem.
120:5747–5755. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tao Y, Zhou X, Liu D, Li H, Liang C, Li F
and Chen Q: Proportion of collagen type II in the extracellular
matrix promotes the differentiation of human adipose-derived
mesenchymal stem cells into nucleus pulposus cells. Biofactors.
42:212–223. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lian C, Gao B, Wu Z, Qiu X, Peng Y, Liang
A, Xu C, Su P and Huang D: Collagen type II is downregulated in the
degenerative nucleus pulposus and contributes to the degeneration
and apoptosis of human nucleus pulposus cells. Mol Med Rep.
16:4730–4736. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shibanuma M, Mashimo J, Mita A, Kuroki T
and Nose K: Cloning from a mouse osteoblastic cell line of a set of
transforming-growth-factor-beta 1-regulated genes, one of which
seems to encode a follistatin-related polypeptide. Eur J Biochem.
217:13–19. 1993.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ogura Y, Ouchi N, Ohashi K, Shibata R,
Kataoka Y, Kambara T, Kito T, Maruyama S, Yuasa D, Matsuo K, et al:
Therapeutic impact of follistatin-like 1 on myocardial ischemic
injury in preclinical models. Circulation. 126:1728–1738.
2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Torres S, Bartolomé RA, Mendes M, Barderas
R, Fernandez-Aceñero MJ, Peláez-García A, Peña C, Lopez-Lucendo M,
Villar-Vázquez R, de Herreros AG, et al: Proteome profiling of
cancer-associated fibroblasts identifies novel proinflammatory
signatures and prognostic markers for colorectal cancer. Clin
Cancer Res. 19:6006–6019. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wilson DC, Marinov AD, Blair HC, Bushnell
DS, Thompson SD, Chaly Y and Hirsch R: Follistatin-like protein 1
is a mesenchyme-derived inflammatory protein and may represent a
biomarker for systemic-onset juvenile rheumatoid arthritis.
Arthritis Rheumatism. 62:2510–2516. 2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Liu Y, Wei J, Zhao Y, Zhang Y, Han Y, Chen
B, Cheng K, Jia J, Nie L and Cheng L: Follistatin-like protein 1
promotes inflammatory reactions in nucleus pulposus cells by
interacting with the MAPK and NFκB signaling pathways. Oncotarget.
8:43023–43034. 2017.PubMed/NCBI View Article : Google Scholar
|