Introduction
Melanoma, which is derived from melanocytes, is the
most aggressive type of skin cancer (1). According to epidemiological studies,
the incidence and mortality rates of patients with melanoma are
continuing to increase in the United States (2). Despite improvements in the diagnosis
and treatment of melanoma, the prognosis of patients with advanced
melanoma remains unfavorable, with a 3-year survival rate of only
15% (3,4). Therefore, it is critical to define the
molecular mechanisms of action underlying melanoma growth and
metastasis, with the aim to develop novel therapeutic
strategies.
ZW10 interactor (Zwint) is a kinetochore protein
found in higher eukaryotes, which plays an important role in the
mitotic checkpoint (5,6). The interaction between Zwint and ZW10
mediates stable ZW10 kinetochore residency at prometaphase
kinetochores, which is indispensable for mitotic checkpoint arrest
(7,8). Given that the mitotic checkpoint is a
primary mechanism ensuring high fidelity of segregation during
mitosis, abnormalities often lead to the development of tumors
(9). Therefore, it may be
hypothesized that an abnormal expression or structure of Zwint may
be associated with carcinogenesis. Previous studies have reported
that Zwint is overexpressed in various types of tumors and is
associated with a poor prognosis (10-12).
In addition, it has been demonstrated that Zwint may act as an
oncogene (11,13). However, whether Zwint is associated
with melanoma progression and its specific molecular mechanism of
action remain unclear.
The aim of the present study was to investigate
whether Zwint regulates melanoma growth and metastasis and whether
its effects are mediated by promoting c-Myc expression, in order to
determine whether Zwint may serve as a novel target for the
treatment of melanoma.
Materials and methods
Data Download
Microarray data set (GSE46517) (14) was downloaded from Gene Expression
Omnibus (ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46517). The
expressing level of Zwint in 31 primary melanoma samples and 9 nevi
tissues were from GSE46517 (GPL96 Affymetrix Human Genome U133A
Array).
Cell culture
The human melanoma cell lines A375, A2058
(amelanotic melanoma) and 451Lu (metastatic melanoma) were
purchased from GeneChem, Inc. A375 and A2058 were maintained in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS (Invitrogen; Thermo Fisher Scientific, Inc.). 451Lu cell lines
were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Invitrogen; Thermo
Fisher Scientific, Inc.). The immortalized normal human epidermal
melanocyte cell line, PIG1 (a gift from Dr Caroline Le Poole,
Loyola University Chicago, USA), was cultured in Medium 254
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with
Human Melanocyte Growth Supplement (Invitrogen; Thermo Fisher
Scientific, Inc.) and 5% FBS (Invitrogen; Thermo Fisher Scientific,
Inc.).
Plasmids, short hairpin (sh)RNA and
transfection
The lenti-plasmid vector system CV224
(Ubi-MCS-SV40-Cherry-IRES-puromycin) and lenti-shRNA vector system
GV115 (hU6-MCS-CMV-EGFP) were constructed, packed and purified by
GeneChem, Inc. A second generation system was used for lentivirus
construction. Zwint targeting shRNA-1
(5'-CCGGCAGAGAATCTTCCAGATGATACTCGAGTATCATCTGGAAGATTCTCTGTTT TTG-3')
or shRNA-2
(5'-CCGGCTACAACCTGCTGGAGATGTACTCGAGTACATCTCCAGCAGGTTGTAGTTTTTG-3')
and control shRNA
(5'-CCGGCCTTCTCCGAACGTGTCACGTCTCGAGTAAATGCTGGTACTCAAATGGTTTTT-3')
were designed with a hairpin and inserted into the GV115 vector.
c-Myc cDNA, MMP-2 cDNA and Slug cDNA were cloned into CV224 vector
and empty vector was used as negative control. Then, 293T cells
(Cell Bank of the Chinese Academy of Sciences) were seeded into a
10 cm dish (5x106 cells) and transfected using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
vectors were transfected in the following proportions: GV/CV vector
plasmid: pHelper1.0 vector plasmid: Phelper 2.0 vector plasmid; 20
µg: 15 µg: 10 µg for 48 h at 37˚C in an incubator before the 293T
lentiviral supernatant was harvested by centrifugation at 4,000 x g
for 10 min at room temperature. Phelper 1.0 vector contains the
gag gene of HIV virus, encoding the main structural proteins
of the virus; the pol gene, encoding a virus-specific
enzyme; the rev gene encodes regulatory factors that
regulate the expression of gag and pol genes. Phelper
2.0 vector contains the herpes simplex virus-derived VSV-G gene,
which provides the envelope protein needed for virus packaging. The
lentiviral supernatant was concentrated and purified for further
concentration detection, and 10 MOI was used for the next
transfection. A375 cells were seeded into a six-well plate at a
density of 1x105 cells/well and were infected with the
lenti-shRNA vectors alone or co-infected with lenti-plasmid-Cherry
vectors for 24 h. After that, culture medium containing the virus
was replaced with fresh culture medium. Cells were collected 72 h
after infection for further analysis. The expression levels of the
reporter gene Cherry or GFP were observed using fluorescence
microscopy (Olympus Corporation; magnification, x100). If the
fluorescence rate was >70% and the cells were considered
healthy, subsequent experiments were performed.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA extracted from the melanoma cells was
isolated using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and then reverse transcribed with
Superscript II Reverse Transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. RT-qPCR
experiments were performed using the SYBR-Green Premix Ex Taq kit
(Takara Biotechnology, Co., Ltd.) according to the manufacturer's
protocol. BIO-RAD MJ Mini Opticon Real-Time PCR System was used to
determine the relative mRNA levels. The following thermocycling
conditions were used: Initial denaturation at 95˚C for 30 sec, 40
cycles at 95˚C for 5 sec, 60˚C for 30 sec and 72˚C for 20 sec.
Threshold cycle values were used to calculate the fold-change in
the transcript levels by using the 2-ΔΔCq method
(15). The relative mRNA expression
levels were normalized to the GAPDH gene. The following primer
sequences were used: Zwint forward, 5'-CACGTAGAGGCCATCAAAATTGG-3'
and reverse, 5'-CGGAGTTGTGTCCGTTTCCT-3; GAPDH forward,
5'-TGACTTCAACAGCGACACCCA-3' and reverse,
5'-CACCCTGTTGCTGTAGCCAAA-3'; MMP-2 forward,
5'-GATACCCCTTTGACGGTAAGGA-3' and reverse,
5'-CCTTCTCCCAAGGTCCATAGC-3'; Slug forward,
5'-GCCAAACTACAGCGAACTGG-3' and reverse, 5'-ATTGGGTAGCTGGGCGTGGA-3';
c-Myc forward, 5'-TGGTGCTCCATGAGGAGACA-3'; and reverse,
5'-TGTGAGGAGGTTTGCTGTGG-3'.
Western blot analysis
Western blot analysis was performed as previously
described (16). Melanoma cells
were washed with phosphate buffered saline (PBS) and lysed using
RIPA lysis buffer (Xi'an Runde Biotechnology Co., Ltd.) and
phenylmethanesulfonyl fluoride (protease inhibitor mix;
Sigma-Aldrich; Merck KGaA). Cell lysates were cleared by
centrifugation for 15 min at 13,000 x g at 4˚C. Total protein was
quantified using a bicinchoninic acid assay kit (Beyotime Institute
of Biotechnology) and 20 µg protein/lane was separated by 10%
SDS-PAGE and blotted onto a nitrocellulose membrane. Following
blocking in a solution of 5% non-fat dry milk diluted in
tris-buffered saline (TBS; pH 7.4) containing 0.05% Tween-20 at
room temperature for 1 h, the membranes were washed and incubated
with primary antibodies overnight at 4˚C. After washing, the
membranes were incubated with horseradish peroxidase-conjugated
secondary antibodies for 1 h at room temperature. Bound antibodies
were detected using a chemiluminescence detection kit (KPL, Inc.).
Primary antibodies against Zwint (1:1,000; cat. no. ab71982), c-Myc
(1:1,000; cat. no. ab32072), Twist (1:1,000; cat. no. ab50887),
p-mTOR (1:2,000; cat. no. ab109268), N-Cadherin (1:1,000; cat. no.
ab18203), Fibronectin (1:500; cat. no. ab32419), and GAPDH
(1:2,000; cat. no. ab37168) were purchased from Abcam. Antibodies
against Slug (1:1,000; cat. no. 9585), MMP-2 (1:500; cat. no.
13132), p-AKT (1:1,000; cat. no. 4060), Snail (1:2,000; cat. no.
3879), AKT (1:2,000; cat. no. 4691), β-catenin (1:2,000; cat. no.
8480), NF-κB p65 (1:2,000; cat. no. 8242), Vimentin (1:1,000; cat.
no.3932), p-NF-κB p65 (1:1,000; cat. no. 3033), p-p38 (1:1,000;
cat. no. 4631), MMP-9 (1:500; cat. no. 13667), p38 (1:2,000; cat.
no. 8690), p-β-catenin (1:1,000; cat. no. 2009), mTOR (1:500; cat.
no. 2972), ERK (1:2,000; cat. no. 9102), E-Cadherin (1:2,000; cat.
no. 9107) and p-ERK (1:2,000; cat. no. 4376) were purchased from
Cell Signaling Technology, Inc. Secondary antibodies including
anti-rabbit IgG (1:10,000; cat. no. AS1107) or anti-mouse IgG
(1:10,000; cat. no. AS1106) were purchased from ASPEN Biotechnology
CO., LTD.
Cell proliferation assay
Following transduction, A375 cells from each group
were trypsinized and counted. Cells were seeded into 96-well plates
at a density of 2x103 cells/well in quintuplicate and
incubated at 37˚C for 1, 2, 3, 4 and 5 days. Cell numbers were
counted over the 5 continuous days. Fluorescence microscopy was
used to assess the cell counts using Celigo Imaging Cytometer
(Nexcelom Bioscience LLC; magnification, x100). Cell proliferation
was also assessed using MTT assay (Beijing Dingguo Changsheng
Biotechnology Co., Ltd.). Following transduction, cells were
incubated with 20 µl MTT at 37˚C for 4 h. Following MTT incubation,
the purple formazan crystals were dissolved using DMSO and cell
proliferation was subsequently analyzed at a wavelength of 490 nm
using a microplate reader (infinite M2009PR; Tecan Group,
Ltd.).
Transwell assay
Following transduction, A375 cells were trypsinized
and resuspended in serum-free DMEM at a density of 1x105
cells/ml. Cell suspensions (100 µl) were plated in the upper
chambers (pore size, 8 µm; Corning, Inc.), while 600 µl DMEM
supplemented with 30% FBS was plated in the lower chambers.
Following incubation at 37˚C for 24 h, the migratory cells were
fixed with 4% paraformaldehyde for 30 min at room temperature and
subsequently stained with 4 g/l crystal violet for 10 min at room
temperature. Stained cells were counted in nine randomly selected
fields using a light microscope (Olympus Corporation;
magnification, x100).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism v5.00 software (GraphPad Software, Inc.). All experiments
were performed in triplicate and data are presented as the mean ±
SD. Unpaired student's t-test was used to compare differences
between two groups. One-way ANOVA followed by Tukey's post-hoc test
was used for multiple comparisons. P<0.05 was considered to
indicate a statistically significant difference.
Results
Zwint expression in melanoma
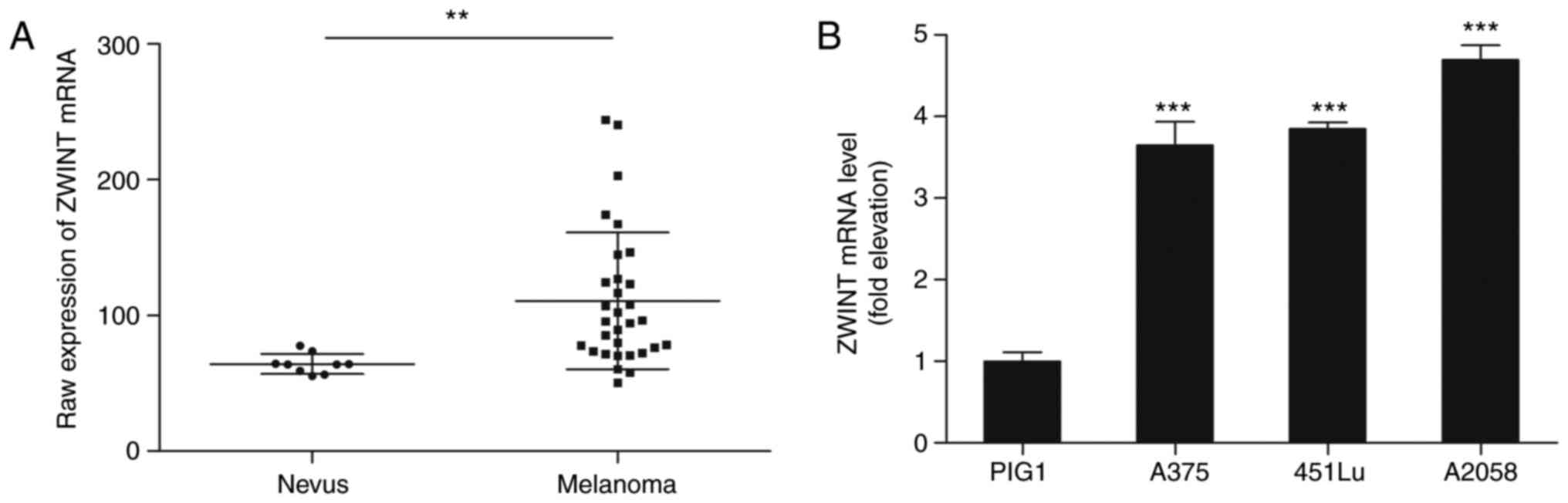
To determine the role of Zwint in melanoma, the
GSE46517 dataset was downloaded from the Gene Expression Omnibus
database. The analysis demonstrated that Zwint expression was
significantly upregulated in primary melanoma lesions compared with
nevus samples (P=0.00178; Fig. 1A).
Subsequently, the mRNA level of Zwint was examined in various
melanoma cell lines (A375, 451Lu and A2058) as well as in the
immortalized normal human epidermal melanocyte cell line PIG1. The
mRNA level of Zwint in all melanoma cell lines was increased
compared with that in the PIG1 cells (Fig. 1B).
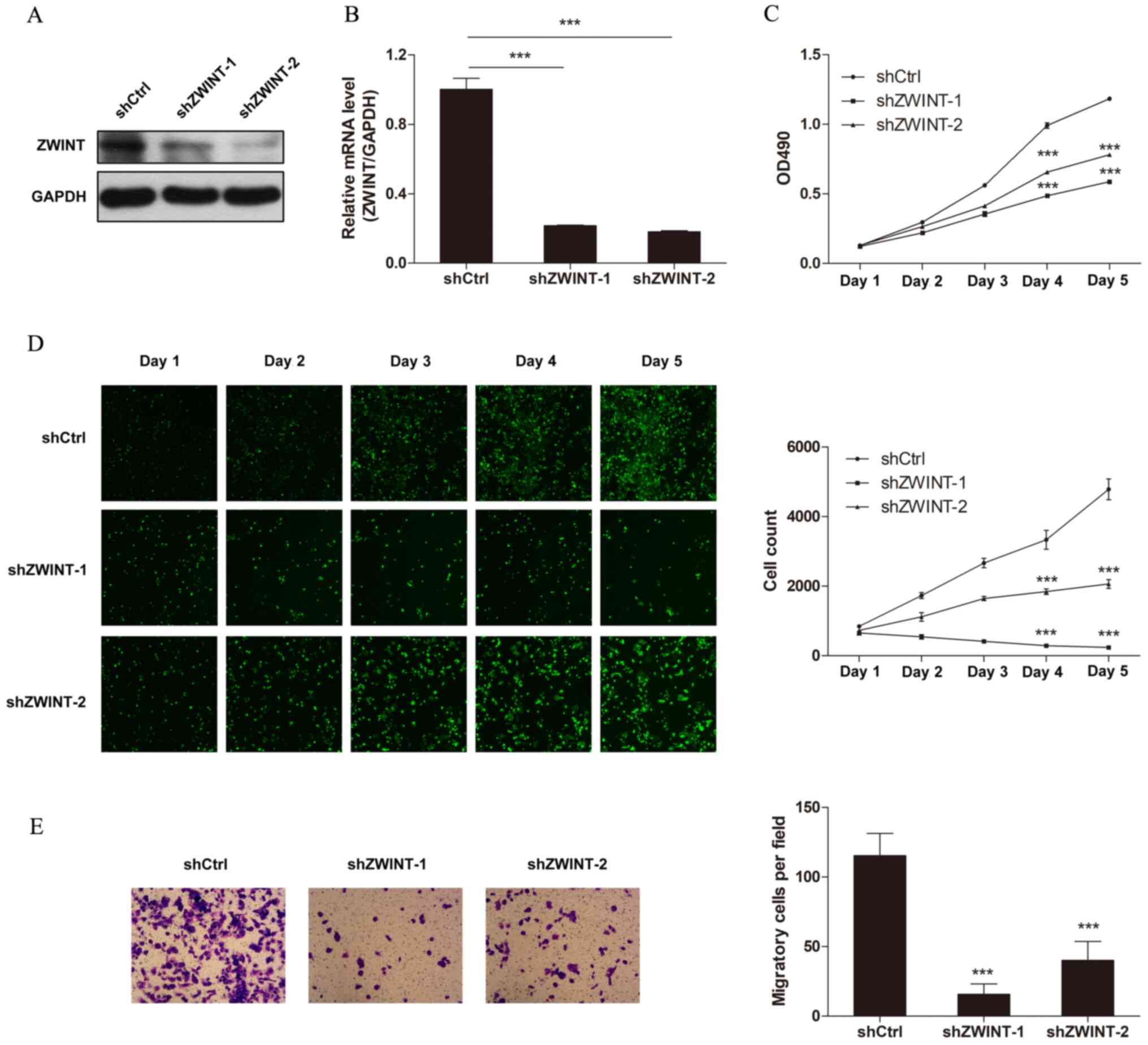
Zwint knockdown suppresses the
proliferation and migration of melanoma cells
To investigate the effect of Zwint knockdown on the
malignant growth potential, A375 cells were transfected with two
different shRNAs targeting Zwint. Western blot and RT-qPCR analyses
demonstrated that Zwint expression significantly decreased
following transfection with both shRNAs compared with the control
shRNA (Fig. 2A and B). The proliferation of melanoma cells was
assessed via cell counting and MTT assay. The results demonstrated
that Zwint knockdown notably decreased cell proliferation (Fig. 2C and D). The ability of Zwint to modify the
motility of melanoma cells was also assessed. The results of the
Transwell migration assay demonstrated that A375 cells transfected
with either Zwint shRNA-1 or shRNA-2 migrated at a slower rate
compared with the control cells (Fig.
2E). Notably, the inhibitory effect on cell proliferation and
migration was more pronounced in A375 cells transfected with Zwint
shRNA-1 compared with Zwint shRNA-2. Thus, Zwint shRNA-1 was
selected for subsequent experiments.
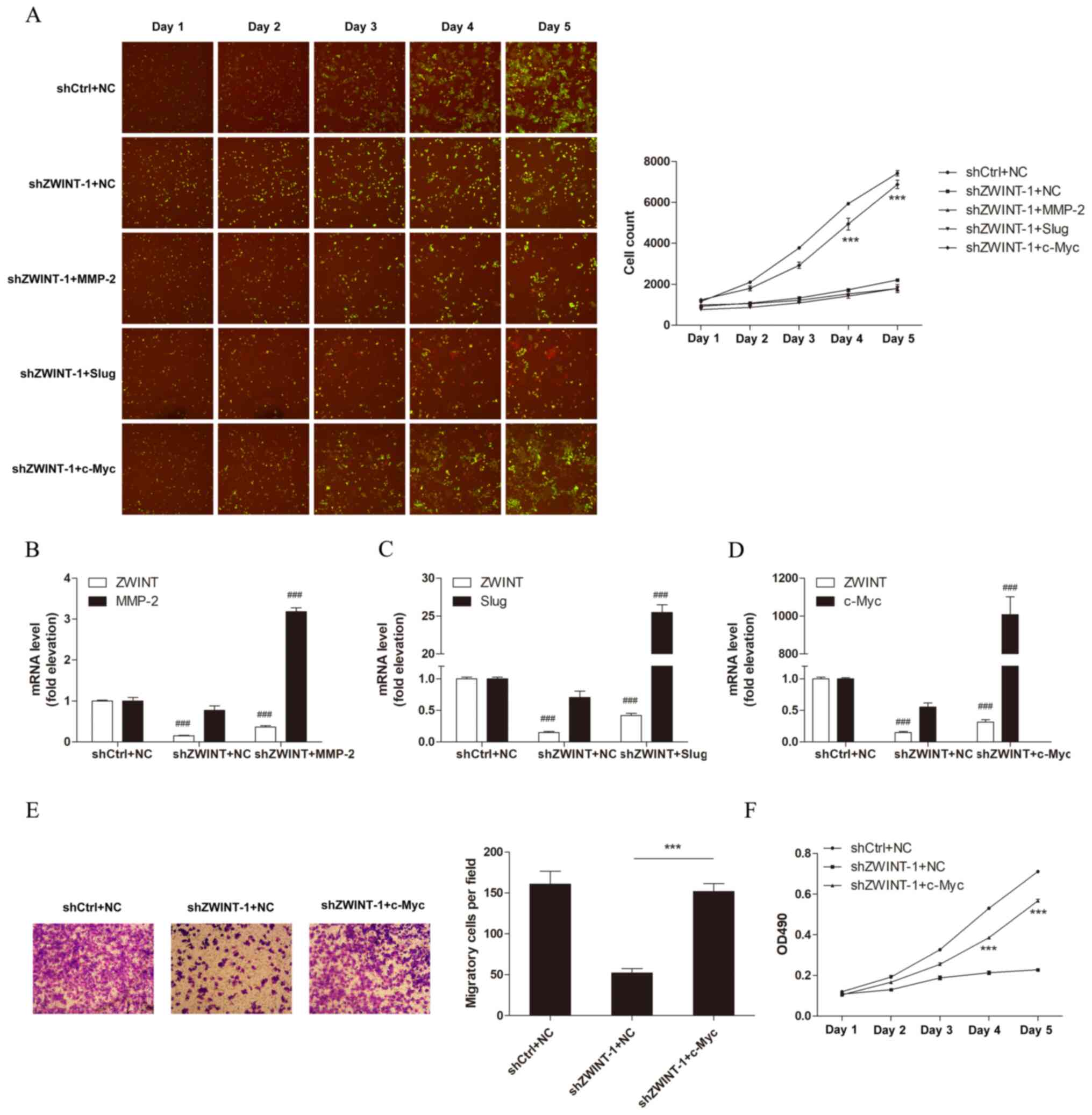
Rescue of c-Myc in Zwint knocked-down
cells restores cell proliferation and migration
Given the notable effects of Zwint knockdown on
melanoma cells, the signaling pathways involved in tumor growth and
migration were assessed via western blot analysis. The results
demonstrated that the expression levels of c-Myc, MMP-2, Slug,
mTOR, phosphorylated (p)-mTOR, p-p38 and fibronectin were decreased
following Zwint knockdown, while the expression levels of
E-cadherin and MMP-9 were increased. However, the expression levels
of ERK, p-ERK, N-cadherin, Snail, AKT, p-AKT, β-catenin,
p-β-catenin, p38, NF-κB p65, p- NF-κB p65, Twist and Vimentin did
not change following Zwint knockdown. (Fig. 3A-H). To identify which gene may
serve a crucial role in mediating the function of Zwint, MMP-2,
Slug and c-Myc were overexpressed to investigate their effects on
cell proliferation and migration following Zwint knockdown. The
overexpression efficiency of MMP-2, Slug and c-Myc vectors was
verified by western blotting (Fig.
S1). Furthermore, Zwint knockdown or MMP-2, Slug and c-Myc
overexpression efficiency was verified by RT-qPCR (Fig. 4B-D). The results of the cell
counting tests demonstrated that c-Myc overexpression promoted cell
proliferation in Zwint depleted cells compared with cells
transfected with the empty vector (Fig.
4A), suggesting that c-Myc may be a downstream effector of
Zwint. Furthermore, the results of the MTT and Transwell migration
assays demonstrated that c-Myc overexpression in Zwint depleted
cells restored melanoma cell migration and proliferation (Fig. 4E and F).
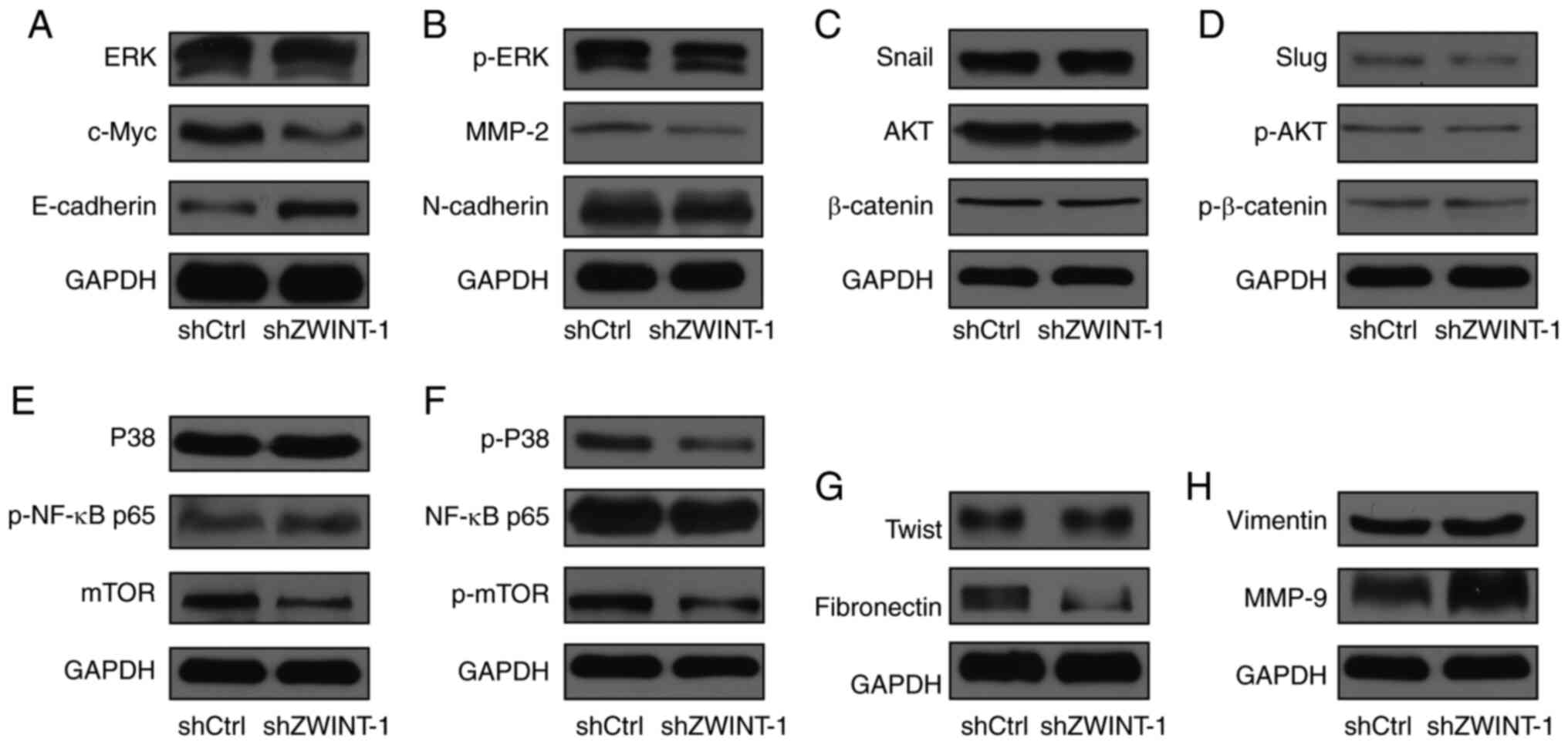 | Figure 3Screening for the downstream effectors
regulated by Zwint. (A-H) Western blot analysis was performed to
detect the protein expression changes of (A) ERK, c-Myc and
E-cadherin; (B) p-ERK, MMP-2 and N-cadherin; (C) Snail, AKT and
β-catenin; (D) Slug, p-AKT and p-β-catenin; (E) p38, p-NF-κB and
mTOR; (F) p-p38, NF-κB and p-mTOR; (G) Twist and fibronectin; and
(H) vimentin and MMP-9, following Zwint knockdown in A375 melanoma
cells. p, phosphorylated; sh, short hairpin; Ctrl, control; ZWINT,
ZW10-interactor. |
Discussion
The present study aimed to investigate the
expression and function of Zwint in melanoma and elucidate its
molecular mechanism. The results demonstrated that Zwint was highly
expressed in melanoma tissues and cells. In addition, Zwint
depletion suppressed melanoma cell proliferation and migration.
Consistent with these results, previous studies have demonstrated
that Zwint is highly expressed in different types of cancer and
exerts an oncogenic effect. For example, Zhou et al
(17) reported that Zwint is
upregulated in breast cancer tissues compared with adjacent normal
tissue and can promote breast cancer cell proliferation.
Furthermore, Peng et al (13) demonstrated that Zwint is
overexpressed in lung adenocarcinoma compared with adjacent normal
tissue, and Zwint knockdown suppresses tumor malignant behaviors,
such as proliferation, migration, invasion and colony formation.
Upregulated Zwint expression has also been observed in human
hepatocellular carcinoma, ovarian cancer and breast cancer compared
with normal adjacent tissue and has been associated with a poor
prognosis (11,12,17).
Consistent with previous findings, the results of the present study
suggested that Zwint acts as an oncogene and may be a potential
therapeutic target in melanoma. However, the molecular mechanism
underlying its carcinogenic effects remains to be fully
elucidated.
To further investigate the molecular mechanism of
Zwint in melanoma, western blot analysis was performed to screen
downstream molecules associated with cell proliferation and
migration, following Zwint knockdown. These downstream candidates
included key molecules in proliferation-associated MAPK and
PI3K-AKT signaling pathways (18,19),
such as ERK, p-ERK, p38, p-p38, c-Myc, AKT, p-AKT, NF-κB p65,
p-NF-κB p65, mTOR and p-mTOR. The candidates also included
molecules associated with cell adhesion and extracellular matrix
that promote tumor migration (20)
such as Twist, Fibronectin, Snail, β-catenin, p-β-catenin, Slug,
Vimentin, MMP-2, MMP-9, E-Cadherin and N-Cadherin. The results
demonstrated that the expression levels of c-Myc, MMP-2, Slug,
mTOR, p-mTOR, p-p38 and fibronectin were decreased following Zwint
knockdown, while the expression levels of E-cadherin and MMP-9 were
increased. Furthermore, subsequent experiments indicated that
overexpression of c-Myc rescued the effects of Zwint depletion on
melanoma cell proliferation and migration. Taken together, these
results suggested that Zwint acts as an oncogene in melanoma by
regulating c-Myc expression. However, whether other molecules
regulated by Zwint also play a role in this process remains to be
investigated. Zwint serves an important role in the spindle
assembly checkpoint (7,8), which suggests that it may play a role
in the pathogenesis of tumors. In addition to its role in the
mitotic spindle checkpoint, increasing evidence has suggested that
Zwint may participate in other signaling pathways. For instance,
Zwint expression was indicated to be associated with the expression
levels of PCNA, Cdc25C, cyclin B1 and CDK1(11). Li et al (10) reported that high Zwint expression
was closely associated with cell cycle-related molecules and
pathways, such as Myc targets V1/2, DNA repair, mitotic spindle,
G2M checkpoint and E2F targets. In addition, Peng et al
(13) demonstrated that Zwint
knockdown affected the expression of molecules associated with TNF
signaling and other pathways. However, these potential molecular
mechanisms of action of Zwint have not been confirmed by subsequent
experiments. To the best of our knowledge, the present study was
the first to demonstrate that Zwint promotes the malignant
phenotype of melanoma by regulating c-Myc expression, which also
supports previous findings with regards to the association between
Zwint expression and the Myc pathway in bioinformatics analysis of
gene expression profiles in breast cancer (10).
Dysregulation of the c-Myc oncogene has been
observed in different types of tumor, such as liver and colon
cancer, as well as Burkitt's lymphoma (21) and is known to contribute to the
development of melanoma and other types of human cancer (22-24).
The molecular mechanism regulating c-Myc expression is intricate.
In addition to gene aberrances, such as genetic amplifications,
insertions and translocations (25-27),
a variety of factors can affect c-Myc expression. For example,
Jiang et al (28) reported
that TIP30 can repress estrogen receptor-α-mediated c-Myc
transcription. Furthermore, Sachdeva et al (29) demonstrated that microRNA-145 can
inhibit c-Myc expression at the post-transcriptional level. c-Myc
expression is also regulated by acetylation and ubiquitination
(30-32).
However, the specific molecular mechanism underlying Zwint
regulation of c-Myc expression remains unclear and requires further
study. Given that c-Myc is considered ‘undruggable’ at the protein
level (33), Zwint, as a regulator
of c-Myc, may be of value as a novel target for the treatment of
melanoma.
In conclusion, the results of the present study
demonstrated that Zwint expression was upregulated in melanoma and
promoted tumor progression. Furthermore, depletion of Zwint was
indicated to impair melanoma cell proliferation and migration,
potentially by downregulating c-Myc expression. Taken together,
these results provided novel insights into the molecular mechanism
of melanoma development and suggested that Zwint may constitute a
promising target for melanoma therapy.
Supplementary Material
Western blot analyses demonstrating
overexpression efficiency following transfection of A375 melanoma
cells with MMP-2, Slug or c-Myc plasmids. NC, negative
control.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81802727) and the
Fundamental Research Fund for Central Universities (grant no.
xzy012019098).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
KM and RG conceived the experiments. KM, JZ and LW
performed most of the experiments. KM and RG confirm the
authenticity of all the raw data. XM and WL helped analyze the
data. All authors discussed the results and RG drafted the initial
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Coricovac D, Dehelean C, Moaca EA, Pinzaru
I, Bratu T, Navolan D and Boruga O: Cutaneous melanoma-a long road
from experimental models to clinical outcome: A review. Int J Mol
Sci. 19(1566)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rozeman EA, Dekker T, Haanen J and Blank
CU: Advanced melanoma: Current treatment options, biomarkers, and
future perspectives. Am J Clin Dermatol. 19:303–317.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Eggermont AM, Spatz A and Robert C:
Cutaneous melanoma. Lancet. 383:816–827. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Woo SD, Yeop YS, Chung WJ, Cho DH, Kim JS
and Su OJ: Zwint-1 is required for spindle assembly checkpoint
function and kinetochore-microtubule attachment during oocyte
meiosis. Sci Rep. 5(15431)2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Starr DA, Saffery R, Li Z, Simpson AE,
Choo KH, Yen TJ and Goldberg ML: HZwint-1, a novel human
kinetochore component that interacts with HZW10. J Cell Sci.
113:1939–1950. 2000.PubMed/NCBI
|
|
7
|
Vos LJ, Famulski JK and Chan GK: hZwint-1
bridges the inner and outer kinetochore: identification of the
kinetochore localization domain and the hZw10-interaction domain.
Biochem J. 436:157–168. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Famulski JK, Vos L, Sun X and Chan G:
Stable hZW10 kinetochore residency, mediated by hZwint-1
interaction, is essential for the mitotic checkpoint. J Cell Biol.
180:507–520. 2008.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Faesen AC, Thanasoula M, Maffini S, Breit
C, Müller F, van Gerwen S, Bange T and Musacchio A: Basis of
catalytic assembly of the mitotic checkpoint complex. Nature.
542:498–502. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li HN, Zheng WH, Du YY, Wang G, Dong ML,
Yang ZF and Li XR: ZW10 interacting kinetochore protein may serve
as a prognostic biomarker for human breast cancer: An integrated
bioinformatics analysis. Oncol Lett. 19:2163–2174. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ying H, Xu Z, Chen M, Zhou S, Liang X and
Cai X: Overexpression of Zwint predicts poor prognosis and promotes
the proliferation of hepatocellular carcinoma by regulating
cell-cycle-related proteins. Onco Targets Ther. 11:689–702.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xu Z, Zhou Y, Cao Y, Dinh TL, Wan J and
Zhao M: Identification of candidate biomarkers and analysis of
prognostic values in ovarian cancer by integrated bioinformatics
analysis. Med Oncol. 33(130)2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Peng F, Li Q, Niu SQ, Shen GP, Luo Y, Chen
M and Bao Y: ZWINT is the next potential target for lung cancer
therapy. J Cancer Res Clin Oncol. 145:661–673. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kabbarah O, Nogueira C, Feng B, Nazarian
RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C,
et al: Integrative genome comparison of primary and metastatic
melanomas. PLoS One. 5(e10770)2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ge R, Liu L, Dai W, Zhang W, Yang Y, Wang
H, Shi Q, Guo S, Yi X, Wang G, et al: Xeroderma pigmentosum group A
promotes autophagy to facilitate cisplatin resistance in melanoma
cells through the activation of PARP1. J Invest Dermatol.
136:1219–1228. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou G, Shen M and Zhang Z: ZW10 binding
factor (ZWINT), a direct target of Mir-204, predicts poor survival
and promotes proliferation in breast cancer. Med Sci Monit.
26(e921659)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hoxhaj G and Manning BD: The PI3K-AKT
network at the interface of oncogenic signalling and cancer
metabolism. Nat Rev Cancer. 20:74–88. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Multhaupt HA, Leitinger B, Gullberg D and
Couchman JR: Extracellular matrix component signaling in cancer.
Adv Drug Deliv Rev. 97:28–40. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hsieh AL, Walton ZE, Altman BJ, Stine ZE
and Dang CV: MYC and metabolism on the path to cancer. Semin Cell
Dev Biol. 43:11–21. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mannava S, Grachtchouk V, Wheeler LJ, Im
M, Zhuang D, Slavina EG, Mathews CK, Shewach DS and Nikiforov MA:
Direct role of nucleotide metabolism in C-MYC-dependent
proliferation of melanoma cells. Cell Cycle. 7:2392–2400.
2008.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Zhuang D, Mannava S, Grachtchouk V, Tang
WH, Patil S, Wawrzyniak JA, Berman AE, Giordano TJ, Prochownik EV,
Soengas MS and Nikiforov MA: C-MYC overexpression is required for
continuous suppression of oncogene-induced senescence in melanoma
cells. Oncogene. 27:6623–6634. 2008.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Beroukhim R, Mermel CH, Porter D, Wei G,
Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J,
Urashima M, et al: The landscape of somatic copy-number alteration
across human cancers. Nature. 463:899–905. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Niitsu N, Okamoto M, Miura I and Hirano M:
Clinical features and prognosis of de novo diffuse large B-cell
lymphoma with t(14;18) and 8q24/c-MYC translocations. Leukemia.
23:777–783. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rajan L, Broussard D, Lozano M, Lee CG,
Kozak CA and Dudley JP: The c-myc locus is a common integration
site in type B retrovirus-induced T-cell lymphomas. J Virol.
74:2466–2471. 2000.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jiang C, Ito M, Piening V, Bruck K, Roeder
RG and Xiao H: TIP30 interacts with an estrogen receptor
alpha-interacting coactivator CIA and regulates c-myc
transcription. J Biol Chem. 279:27781–27789. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V,
Kumar S, Elble R, Watabe K and Mo YY: p53 represses c-Myc through
induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S
A. 106:3207–3212. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wu H, Yang TY, Li Y, Ye WL, Liu F, He XS,
Wang JR, Gan WJ, Li XM, Zhang S, et al: Tumor necrosis factor
receptor-associated factor 6 promotes hepatocarcinogenesis by
interacting with histone deacetylase 3 to enhance c-Myc gene
expression and protein stability. Hepatology. 71:148–163.
2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Pan J, Deng Q, Jiang C, Wang X, Niu T, Li
H, Chen T, Jin J, Pan W, Cai X, et al: USP37 directly
deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene.
34:3957–3967. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Patel JH, Du Y, Ard PG, Phillips C,
Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane
WS, et al: The c-MYC oncoprotein is a substrate of the
acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol.
24:10826–10834. 2004.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Dang CV, Reddy EP, Shokat KM and Soucek L:
Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer.
17:502–508. 2017.PubMed/NCBI View Article : Google Scholar
|