Introduction
Diabetes mellitus (DM) is a group of clinical
syndromes characterized by disorders of glucose metabolism caused
by genetic and environmental factors; these syndromes are divided
into type 1 DM (T1DM), T2DM, other special types and gestational
diabetes. T1DM is caused by the destruction of islet β cells and
absolute insulin deficiency, while T2DM is characterized by
insufficient insulin secretion and insulin resistance. The current
diagnostic criteria for DM primarily include the World Health
Organization-recommended 75-g oral glucose tolerance test and the
American Diabetes Association-recommended level of glycated
hemoglobin A1c (HbA1c) (1).
HbA1c is a product of the Hb in red blood cells
(RBCs) that binds to glucose in serum, is formed by the
nonenzymatic attachment of glucose (known as glycation) to Hb and
normally reflects the ambient glucose concentration of the previous
2 to 3 months, which is the lifespan of an RBC, suggesting that
HbA1c is a convenient measure of long-term blood glucose
concentrations. In addition, HbA1c is not affected by several
factors, such as time, fasting and exercise. Given these
characteristics, the measurement of HbA1c is among the most
important laboratory medical advances in diabetes care in decades
and this measurement is now an accepted diagnostic test for T2DM
and is used for monitoring glycemic control in patients with DM
(2).
However, it has been indicated that numerous factors
may influence HbA1c levels, including variability in erythrocyte
life span and erythropoiesis (3).
In addition, severe hypertriglyceridemia and chronic alcoholism may
interfere with the measurement of HbA1c (3). Previous studies suggested that Hb
variants may also affect the detection of HbA1c (1,4,5). In
the present study, the HbA1c levels in a patient diagnosed with
T2DM were observed to be in the normal range, which is inconsistent
with the clinical diagnosis; this discrepancy was investigated and
discussed in the present study.
Case report
A 55-year-old female patient presented at the
Department of Endocrinology and Metabolism of the Affiliated
Hospital of Qingdao University (Qingdao, China) in Laoshan District
of Qingdao on January 16, 2019 and requested assistance to manage
her T2DM. She was diagnosed with T2DM 12 months previously based on
fasting blood glucose (FBG) levels of 11 mmol/l. Prescribed
metformin (500 mg) was given for 12 months and the FBG level
decreased to 7 mmol/l, while the postprandial blood glucose was 8-9
mmol/l. Selected laboratory test data for the patient are presented
in Table I. The results suggested
that the proportion of HbA1c obtained for this patient during a
routine examination using the Variant II (Bio-Rad Laboratories,
Inc.) was 3.8%, which was in the normal range and inconsistent with
the clinical diagnosis. Subsequently, to further confirm whether
the values obtained at our laboratory were correct, the levels of
HbA1c, FBG and glycated albumin were further measured at another
clinical laboratory at the headquarters of the Affiliated Hospital
of Qingdao University (Qingdao, China) in Shinan District of
Qingdao 3 days later. The resulting laboratory test data are
provided in Table II. These tests
provided a proportion of HbA1c of 3.7%, which was also measured
during a routine examination using the Variant II (Bio-Rad
Laboratories, Inc.); this result was slightly below the normal
range and was still inconsistent with the clinical diagnosis.
 | Table ISummary of selected laboratory test
data measured at the Affiliated Hospital of Qingdao University in
Laoshan District of Qingdao. |
Table I
Summary of selected laboratory test
data measured at the Affiliated Hospital of Qingdao University in
Laoshan District of Qingdao.
| Parameter | Patient's value | Reference range |
|---|
| Hb, g/l | 142.0 | 115.0-150.0 |
| MCH, pg | 30.6 | 27.0-34.0 |
| MCHC, g/l | 349.0 | 316.0-354.0 |
| MCV, fl | 87.7 | 82.0-100.0 |
| HCT, % | 40.7 | 35.0-45.0 |
| RBC,
x1012/l | 4.64 | 3.8-5.1 |
| RDW, % | 11.4a | 11.6-16.5 |
| PLT,
x109/l | 249 | 125-350 |
| WBC,
x109/l | 6.42 | 3.5-9.5 |
| FBG, mmol/l | 7.63a | 3.90-6.16 |
| HbA1c, % | 3.8 | 3.6-6.0 |
| ALT, U/l | 16.0 | 7.0-40.0 |
| AST, U/l | 17.0 | 13.0-35.0 |
| TG, mmol/l | 1.34 | 0.30-1.92 |
| TC, mmol/l | 4.29 | 2.32-5.62 |
| HDL, mmol/l | 1.31 | 0.80-2.35 |
| LDL, mmol/l | 2.57 | 1.90-3.12 |
| BUN, mmol/l | 6.14 | 2.6-7.5 |
| Cre, µmol/l | 88.0 | 31.0-132.0 |
| UA, µmol/l | 280.0 | 89.2-339.0 |
| GA, % | 21.7a | 10.4-15.7 |
 | Table IISummary of selected laboratory test
data measured at the headquarters of the Affiliated Hospital of
Qingdao University in the Shinan District of Qingdao. |
Table II
Summary of selected laboratory test
data measured at the headquarters of the Affiliated Hospital of
Qingdao University in the Shinan District of Qingdao.
| Parameter | Patient's value | Reference range |
|---|
| FBG, mmol/l | 7.40a | 3.90-6.16 |
| HbA1c, % | 3.7 | 4.3-6.3 |
| GA, % | 18.1 | 10.4-15.7 |
Next, genomic DNA was extracted from the patient's
blood using the Ezup Column Blood Genomic DNA Purification kit
(Sangon Biotech Co., Ltd.) and genomic DNA was used to amplify HbA
genes with specific forward and reverse primers by targeting the
HbA α1/2- and β-chain genes (HbA-α1 forward,
5'-CCACCACCAAGACCTACT-3' and reverse, 5'-TCACAGAAGCCAGGAACT-3';
HbA-α2 forward, 5'-CCACCACCAAGACCTACT-3' and reverse,
5'-TCACAGAAGCCAGGAACT-3'; HbA-β 1-forward,
5'-ACTCCTAAGCCAGTGCCAGA-3' and 1-reverse,
5'-AGATCCCCAAAGGACTCAAAG-3'; HbA-β 2-forward,
5'-TGAGGAGAAGTCTGCCGTTAC-3' and 2-reverse,
5'-AAAACGATCCTGAGACTTCCAC-3'; HbA-β 3-forward,
5'-TGATAATTTCTGGGTTAAGGCAA-3' and HbA-β 3-reverse,
5'-TAACCTCCAAATCAAGCCTCTAC-3'). PCR was performed in 50 µl of
solution containing 1-2 µl DNA (20-50 ng/µl), 5 µl Taq buffer
(10X), 2 µl dNTPs (10 mM), 2.5 U Taq DNA polymerase (Sangon Biotech
Co., Ltd.) and 2 µl of each of the two primers (10 µM).
ddH2O was then added to increase the reaction volume to
50 µl. The following thermocycling conditions were used: 95˚C for 5
min, followed by 35 cycles at 94˚C for 30 sec, 30 sec at 55-60˚C,
and 50 sec at 72˚C, finishing with 8 min at 72˚C. The PCR products
were detected by gel electrophoresis. In addition, the HbA genes
were analyzed using Sanger sequencing. The PCR products were
purified using the SanPrep Column PCR Product Purification kit
(Sangon Biotech Co., Ltd.). Sequencing PCR was subsequently
performed in 20 µl of solution containing 10 ng purified PCR
products, 4 µl BigDye (2.5X; Thermo Fisher Scientific, Inc.), 2 µl
BigDye Seq Buffer (5X; Thermo Fisher Scientific, Inc.) and 1 µl
sequencing primers (3.2 pmol/µl), with 12 µl ddH2O added
to increase the reaction volume to 20 µl. The thermocycling
conditions were as follows: 1 min at 96˚C, followed by 25 cycles of
10 sec at 96˚C, 5 sec at 50˚C and 4 min at 60˚C, finishing at 4˚C.
The sequencing PCR products were purified using the following
method: 2 µl EDTA (125 mM) and 2 µl NaAc (3 M) were added per well
to a 96-well plate, after which 50 µl alcohol (100%) was added and
the solution was mixed and incubated for 15 min at room
temperature. The plate was centrifuged at 3,000 x g at 4˚C for 30
min, after which the plate was inverted. A subsequent
centrifugation was performed at 4˚C and the power was turned off as
soon as the speed was up to 185 x g. Next, 70 µl alcohol (70%) was
added to the plate and samples were centrifuged at 3,000 x g at 4˚C
for 15 min. After the plate was inverted, samples were further
centrifuged at 4˚C and the power was turned off as soon as the
speed was up to 185 x g. Once the alcohol had evaporated at room
temperature, 10 µl Hi-Di Formamide (Sangon Biotech Co., Ltd.) was
added to dissolve the DNA. The DNA was subsequently denaturized at
95˚C for 4 min and 4˚C for 4 min in the PCR instrument. Samples
were then sequenced on an ABI 3730XL DNA analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The Sanger sequencing
results were then analyzed using SeqMan DNASTAR Lasergene 11.0
software (DNASTAR Inc.). The results of the Sanger sequencing
indicated that the patient's HbA α1/2-chains had no gene mutations
(data not shown). However, gene sequencing of the HbA β-chain PCR
products suggested that the patient had two HbA β-chain gene
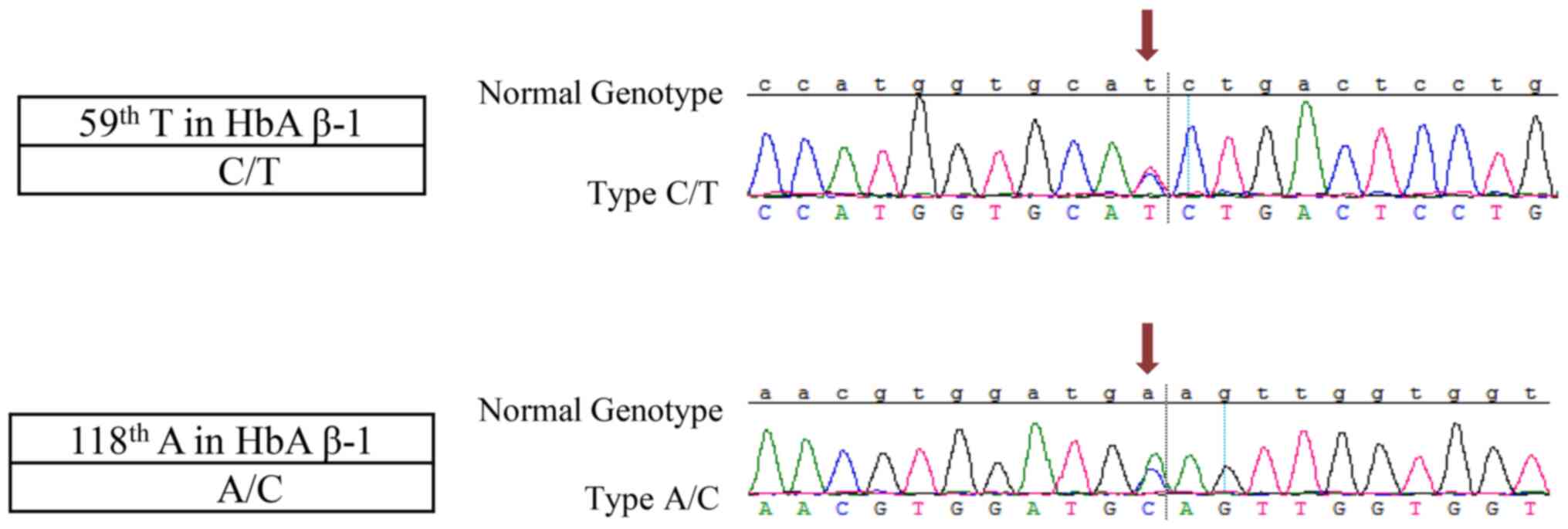
mutations (Fig. 1). The first
mutation was a C/T point mutation that occurred at point 59, which
comprises CAT>CAC at codon 2 in the HbA β-chain and is named
HBB:c.9T>C variant described in the human hemoglobin variant
(HbVar) database (http://globin.bx.psu.edu/hbvar). The next mutation was
an A/T point mutation that occurred at point 118, which comprises
GAA>GCA at codon 22 in HbA β-chain and is named the Hb
G-Coushatta variant, also known as the Hb G Hsin Chu variant, Hb
G-Saskatoon variant or Hb G-Taegu variant described in HbVar. As
ion-exchange high-performance liquid chromatography (HPLC) for
HbA1c quantification is based on charge differences and is known to
be susceptible to interference from Hb variants (6). The HBB:c.9T>C variant is a silent
mutation, resulting in no change in the charge of the patient's HbA
β-chain; thus, the HBB:c.9T>C variant has no effect on the HbA1c
level when detected by ion-exchange HPLC. However, previous studies
have indicated that the Hb G-Coushatta variant usually leads to
underestimation of HbA1c (7-9).
HbA variants constitute a well-known cause of analytical
interference in HbA1c measurements (10). Thus, it is most likely that the Hb
G-Coushatta variant leads to HbA1c levels that are clearly
inconsistent with the clinical manifestations.
Discussion
The measurement of HbA1c levels is an accepted
diagnostic test for T2DM. The interindividual variation in HbA1c,
defined as the glycation gap, is not attributable to mean blood
glucose values and has been indicated to be associated with
diabetic nephropathy (3,11). Several studies have described low
HbA1c levels in patients with Hb variants (9,12,13).
According to the laboratory test data obtained for the patient of
the present study, numerous factors that may lead to low HbA1c
levels were excluded, except for Hb variants.
In the present study, to determine why the HbA1c
level was clearly inconsistent with the clinical manifestations,
the HbA1c sequence of this patient was analyzed using Sanger
sequencing and the results suggested that the patient's HbA β-chain
had two gene mutations. To confirm the Hb variants of the patient,
the mutations were searched in HbVar. According to HbVar, the first
gene mutation is named the HBB:c.9T>C variant, which is a silent
mutation that results in no change in the charge of the patient's
HbA β-chain and the data from the ClinVar Miner website (https://clinvarminer.genetics.utah.edu)
also indicated that the HBB:c.9T>C variant is considered to be
benign and is not related to any pathological conditions; In
addition, ion-exchange HPLC for HbA1c quantification are based on
charge differences (6), thus, the
HBB:c.9T>C variant has no effect on the HbA1c level when
detected by ion-exchange HPLC. The second gene mutation is named
the Hb G-Coushatta variant, which has been observed to occur in
certain populations, such as native Americans, Chinese, Koreans,
Japanese, Turks and Algerians. Previous studies have indicated that
this variant usually leads to underestimation of HbA1c and HbA
variants constitute a well-known cause of analytical interference
in the measurement of HbA1c levels (7-10).
One of these studies suggested that the Hb G-Coushatta variant
affected the determination of HbA1c levels, with the ion-exchange
HPLC method measuring an HbA1c level that was 45% lower than that
obtained by the immunoturbidimetric assay method (9). Thus, it may be concluded that the Hb
G-Coushatta variant is the most likely cause of the HbA1c measured
by HPLC appearing normal, which was clearly inconsistent with the
clinical manifestations.
An unusually low level of HbA1c or discordance
between blood sugar and HbA1c values should alert clinicians to the
possibility of Hb variants. Since the effect of a particular Hb
variant on HbA1c results is frequently method-dependent (e.g.,
ion-exchange HPLC methods are frequently affected by the presence
of Hb variants), repeat analysis should ideally be performed using
an alternative method based on a different analytical principle
from the initial assay (14); these
alternative methods may include electrophoresis, immunoassays and
enzymatic analysis. Among the methods, the principle of
electrophoresis is based on the fact that colloidal particles with
different electric charges under certain conditions move through an
electric field by electrostatic attraction, which may identify
abnormal Hb (12). In the future,
when the HbA1c levels of patients are clearly inconsistent with the
clinical manifestations, electrophoresis should be used to measure
HbA1c levels, which may help with the monitoring of HbA1c levels
when HbA variants are present in those patients.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by grant from the Key R&D
Projects of Shandong Province (grant no. 2019GSF108108).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available in a public curated database named figshare
(https://figshare.com/articles/figure/HbA_mutations_in_a_T2DM_patient/12922502;
DOI: 10.6084/m9.figshare.12922502). In addition, the .sqd file
contains the high-throughput sequencing data.
Authors' contributions
JW analyzed and interpreted of the data, wrote the
initial draft and revised the manuscript. YW, MH and QW contributed
to the acquisition and analysis of the data. WL and LY contributed
to evaluation and interpretation of the data. MH and QW made
substantial contributions to the conception and design of the
current study. JW and QW confirmed the authenticity of all the raw
data. JW and QW agreed to be accountable for all aspects of the
work. All authors critically reviewed the manuscript and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Hospital of Qingdao University
(Qingdao, China). The patient provided written informed
consent.
Patient consent for publication
The patient had provided written informed consent
regarding the publication of the case details.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
International Expert Committee.
International expert committee report on the role of the A1C assay
in the diagnosis of diabetes. Diabetes Care. 32:1327–1334.
2009.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Standards of medical care in
diabetes-2016: Summary of revisions. Diabetes Care. 39 (Suppl
1):S4–S5. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gallagher EJ, Le Roith D and Bloomgarden
Z: Review of hemoglobin A(1c) in the management of diabetes. J
Diabetes. 1:9–17. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Little RR, Rohlfing CL, Hanson S, Connolly
S, Higgins T, Weykamp CW, D'Costa M, Luzzi V, Owen WE and Roberts
WL: Effects of hemoglobin (Hb) E and HbD traits on measurements of
glycated Hb (HbA1c) by 23 methods. Clin Chem. 54:1277–1282.
2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Dessi M, Pieri M, Pignalosa S, Martino FG
and Zenobi R: Performances of capillary electrophoresis and HPLC
methods in HbA1c determination: Diagnostic accuracy in HbS and
HbD-Iran variants' presence. J Clin Lab Anal. 29:57–60.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lorenzo-Medina M, De-La-Iglesia S, Ropero
P, Nogueira-Salgueiro P and Santana-Benitez J: Effects of
hemoglobin variants on hemoglobin a1c values measured using a
high-performance liquid chromatography method. J Diabetes Sci
Technol. 8:1168–1176. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Al-Hakeim HK and Abdulzahra MS:
Correlation between glycated hemoglobin and homa indices in type 2
diabetes mellitus: Prediction of beta-cell function from glycated
hemoglobin. J Med Biochem. 34:191–199. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ogawa K, Bando T, Ogawa M, Miyazaki A,
Nakanishi T and Shimizu A: Hemoglobin variant HbG-coushatta
(beta-22 Glu->Ala) found by dissociation of blood glucose from
values of HbA1C measured by HPLC. Intern Med. 42:781–787.
2003.PubMed/NCBI
|
|
9
|
Kurtoğlu AU, Eren E, Aslan V, Erkal Ö,
Kurtoğlu E and Yilmaz N: Heterozygote hemoglobin G-coushatta as the
cause of a falsely decreased hemoglobin A1C in an Ion-exchange HPLC
method. J Med Biochem. 36:270–273. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rohlfing C, Hanson S, Weykamp C, Siebelder
C, Higgins T, Molinaro R, Yip PM and Little RR: Effects of
hemoglobin C, D, E and S traits on measurements of hemoglobin A1c
by twelve methods. Clin Chim Acta. 455:80–83. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Rodriguez-Segade S, Rodriguez J,
Cabezas-Agricola JM, Casanueva FF and Camiña F: Progression of
nephropathy in type 2 diabetes: The glycation gap is a significant
predictor after adjustment for glycohemoglobin (Hb A1c). Clin Chem.
57:264–271. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Schnedl WJ, Reisinger EC, Katzensteiner S,
Lipp RW, Schreiber F, Hopmeier P and Krejs GJ: Haemoglobin O Padova
and falsely low haemoglobin A1c in a patient with type I diabetes.
J Clin Pathol. 50:434–435. 1997.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Estey MP, Rodriguez-Capote K, Adelowokan T
and Higgins T: Hemoglobin Hirose: A rare beta chain variant causing
falsely low HbA1c by HPLC. Clin Biochem. 49:498–501.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xu A, Sun J, Li J, Chen W, Zheng R, Han Z
and Ji L: Hb I: A alpha-globin chain variant causing unexpected
HbA1c results. J Clin Lab Anal. 33(e22671)2019.PubMed/NCBI View Article : Google Scholar
|