Introduction
Ovarian cancer is a common malignancy of the female
reproductive system with a high mortality rate (1). In the USA, >15,000 patients die of
ovarian cancer and there are 20,000 new diagnosed patients each
year (2). The most common type of
ovarian cancer is epithelial ovarian cancer, also known as ovarian
carcinoma (3). Due to the lack of
effective screening methods, patients are frequently already in the
advanced stages of ovarian cancer at the time of diagnosis, where
extensive metastasis into the abdominal cavity is the main cause of
mortality (4,5). The 5-year survival rate of patients
with ovarian cancer remains only at 60% though progress has been
made in the development of novel chemotherapeutic strategies
(6). Therefore, it is important to
understand the process of ovarian cancer tumorigenesis and
metastasis on a deeper level. In the study of the basic underlying
mechanism underlying ovarian cancer progression, it was previously
found that alterations in the expression profiles of cytokines,
such as VEGF and TNF-α, can be used to reflect the status of
abnormal proliferation or apoptosis in ovarian germinal epithelial
cells, where they can contribute to the occurrence and development
of ovarian cancer cells (7).
B cell receptor associated protein 31 (BAP31 for
protein; BCAP31 for gene) was first identified by Kim et al
(8) in 1994. Due to its selective
binding to membrane immunoglobulin D, it is considered to be a
member of the B cell receptor (9,10).
There are two primary functions of the BAP31 protein: i) To mediate
the transport of numerous types of newly formed proteins from the
endoplasmic reticulum (ER) to the Golgi apparatus as a carrier
molecule (10); and ii) to regulate
apoptosis. In particular, BAP31 is involved in regulating apoptosis
mediated by Bcl-2 and Bcl-XL (11).
The BCAP31 gene is located in the q28 open reading frame of the X
chromosome that is 738-bp in length and encodes 246 amino acids.
The molecular weight of the mature BAP31 protein product is 28 KDa
(9,10). BAP31 is an endoplasmic reticulum
(ER) protein where its N-terminus is located in the ER lumen and
contains three transmembrane domains, which serve to promote
anchoring to the ER (11,12). By contrast, the C-terminus is
located in the cytoplasm, where it mediates protein-protein
interaction and is the area where BAP31 mainly performs its protein
transport and apoptosis regulation functions (11,12).
As a carrier protein, BAP31 serves an important role in apoptosis
(11,13). Compared with those in adjacent
non-cancerous tissues, the expression levels of BAP31 protein were
persistently found to be significantly upregulated in malignant
melanoma, hepatocellular carcinoma or cervical cancer tissues
(14-16).
These previous observations suggested that BAP31 may serve an
important role in the pathogenesis of malignancies. In particular,
it has been previously demonstrated that BAP31 may be an ideal
target for immunotherapy of malignant melanoma (14). The expression levels of BAP31 in
colorectal cancer has also been found to be positively associated
with the degree of liver metastasis. However, the survival rate of
patients with colorectal cancer with lower expression levels of
BAP31 is significantly lower (17).
In addition, BAP31 expression has been revealed to be upregulated
during the progression of cervical cancer into the metastatic
stages (16-18).
However, the expression profile and role of BAP31 in ovarian cancer
remain unknown.
Epithelial-to-mesenchymal transition (EMT) is a
developmental cell program that naturally occurs in a broad range
of tissue types and developmental stages (19). During EMT, epithelial cells
temporarily could lose their cell polarity and exhibit the
characteristics of mesenchymal cells with increased migratory and
invasive phenotypes (20).
Downregulation of E-cadherin expression and the concomitant
upregulation of N-cadherin expression are considered to be key
events during EMT (21).
Downregulation of E-cadherin expression has been found in
esophageal and prostate cancer (22,23).
The internal-to-external signaling mechanism that regulates the
adhesive activity of E-cadherin present on the cell surface is
important in cancer (13,24). Loss of cell adhesion and cell
junctions formed by E-cadherin anchoring allows cells to detach
from the primary tumor, invade surrounding tissues and migrate to
distant tissues (24). Previous
studies have demonstrated the role of E-cadherin and N-cadherin in
non-small cell lung, colorectal and pancreatic cancer and melanoma
invasion and metastasis and how they synergistically contribute to
signaling pathways involved in EMT (21), such as TGF-β (13,25),
MAPK (26) or JAK/STAT (27). Compared with the suppressive role of
E-cadherin on cell migration, N-cadherin enhances the migration and
invasion of tumor cells, irrespective of the expression of
E-cadherin (28). Numerous previous
studies found that increased expression of N-cadherin can enhance
the migratory and invasive capabilities of multiple types of cancer
cells in vivo and in vitro, such as bladder (29) and pancreatic cancer (30).
The transcriptional inhibition of E-cadherin and the
upregulation of N-cadherin i.e. the cadherin switch is considered
to be a marker of EMT (21). This
is mediated by transcription factors (TFs), including the snail
family (Snai1, Snai2/Slug and Snai3/Smuc), zinc finger
E-box-binding homeobox 1 (Zeb)1, Zeb2 proteins, TWIST1 and
TWIST2(31). TWIST is essential for
a number of biological processes, including myogenesis (32), osteogenesis (33) and neurogenesis (34). In addition to its role in normal
physiological development, TWIST has also been found to be
upregulated in a number of cancer types, such as breast and lung
cancer (35,36). In these cancer types, TWIST
expression is associated with poor prognosis, higher grade tumors
and more invasive and metastatic phenotypes (31). Vertebrate animals express two types
of TWIST, which encode two similar proteins that share 90%
homology, named TWIST1 AND TWIST2(37). TWIST1 serves an important role in
cell migration and tissue reorganization during embryo formation
(38). Studies have previously
reported that the Twist1 gene and its expression products are
expressed in a variety of tumor tissues and cells, including
rhabdomyosarcoma, breast cancer, gastric cancer, fibroblastoma,
glioma and pancreatic adenocarcinoma (39-41).
Activated Twist enhances the migration and invasion of cells by
upregulating N-cadherin whilst downregulating E-cadherin (42).
Based on all the aforementioned findings, BAP31 may
serve an important role in the progress of cancer; however, its
function in ovarian cancer is not clear. The present study
investigated the role of BAP31 involved in ovarian cancer cell
invasion and its underlying mechanism. A BAP31 knockdown model was
established, and cell proliferation, migration and invasion ability
was assessed. In addition, downstream genes of BAP31were
investigated to elucidate the underly mechanisms.
Materials and methods
Clinical samples
The present study was approved by the Ethics
Committees of China-Japan Friendship Hospital (approval no.
2020-28-K20; Beijing, China) and written informed consents were
obtained from all participants. Frozen resected surgical tissue
from 10 female patients with ovarian cancer with an average age of
59.1±8.3 years old were enrollment from June 2018 to March 2020
collected from Department of Gynecology and Obstetrics, China-Japan
Friendship Hospital (Beijing, China). All pathological specimens
collected during primary surgery or before neoadjuvant chemotherapy
were analyzed by an expert gynecological pathologist according to
the guidelines of the World Health Organization (WHO) International
Classification of Ovarian Tumors (43). The inclusion and exclusion criteria
are as follows: Inclusion criteria: Patients with complete clinical
data. Exclusion criteria: i) Patients who received chemotherapy or
radiation therapy and ii) patients with missing clinical data. None
of the patients recruited in the present study received
chemotherapy or radiation therapy before the specimens were
collected.
Immunohistochemistry (IHC)
A total of 10 tissue samples from patients with
ovarian cancer were immersed in 4% paraformaldehyde for 4 h at room
temperature and assembled into a tissue microarray (diameter, 2
mm). After cutting to a thickness of 5 µm, paraffin sections were
deparaffinized in xylene for 5 min twice, followed by hydration in
a descending ethanol series (100, 100, 90, 80 and 70%), and rinsed
in distilled water. The paraffin sections were heat-treated with
EDTA antigen retrieval solution (pH, 8.0; cat. no. ZLI-9079;
OriGene Technologies, Inc.) for 20 min at 98˚C Paraffin sections
were incubated in 3% hydrogen peroxide for 10 min at 37˚C for
endogenous peroxidase blocking, washed twice with PBS (pH 7.4; 5
min; room temperature), and then blocked with normal goat serum for
20 min at room temperature (cat. no. ab7481; Abcam). Incubation
with anti-BAP31 (cat. no. ab237485; Abcam; 1:1,000) was performed
overnight at 4˚C. Sections were then incubated with the secondary
antibody for 1 h at room temperature (cat. no. ZB-2301; 1:5,000;
OriGene Technologies, Inc.). Between each step, the sections were
washed three times with PBS (pH 7.4). Coloration with
3,3-diaminobenzidin (1 mg/ml) was performed in the dark for 5 min
at room temperature, followed by counterstaining with hematoxylin
(8 mg/ml) for 5 min at room temperature. Between each staining, the
sections were washed three times with tap water. BAP31-positive
ovarian cancer tissue served as a positive control and tissue
incubated with 10% pre-immune rabbit serum overnight at 4˚C as a
negative control. The stained slides were examined using an Olympus
DP70 light microscope (Olympus Corporation; magnification,
x20).
Cell culture
Human ovarian normal epithelial cell line IOSE80 and
five ovarian cancer cell lines (A2780, Hey-T30, COC1, SKOV3 and
OVCAR3) were obtained from the BeNa Culture Collection; Beijing
Beina Chuanglian Biotechnology Research Institute. Mycoplasma
testing was performed for all cell lines used, and cell lines were
tested and identified using short tandem repeats. All cell lines
were cultured in DMEM supplemented with 10% FBS (ProSpec-Tany
TechnoGene, Ltd.) and 100 mg/ml penicillin/streptomycin
(ProSpec-Tany TechnoGene, Ltd.). Cells were cultured in a
humidified 5% CO2 atmosphere at 37˚C.
Cell transfection
BCAP31 shRNA lentiviral transduction particles and
shRNA negative control (shCtrl) transduction particles were
purchased from Sigma-Aldrich; Merck KGaA. The sequences of shRNA
vectors (pLKO.1; Sigma-Aldrich; Merck KGaA) that targeted BCAP31
were 5'-CCGGCATGGACAAGAAGGAAGAGTACTCGAGTACTCTTCCTTCTTG
TCCATGTTTTTTG-3' [shBCAP31-1, The RNAi Consortium (TRC) no.
TRCN0000179092] and
5'-CCGGCCTATGGCAACACCTTCTTTGCTCGAGCAAAGAAGGTGTTGCCATAGGTTTTTG-3'
(shBCAP31-2, TRC no. TRCN0000242709). The vector shRNA negative
control (shCtrl) was purchased form Sigma-Aldrich
(MISSION® SHC016-1EA; Merck KGaA) and contained shRNA
insert that does not target any known genes from any species. The
BCAP31 shRNA lentiviral plasmid or shRNA negative control plasmid
and pPACK Packaging Plasmid mix (cat. no. K497500; Invitrogen;
Thermo Fisher Scientific, Inc.) were co-transfected into the
healthy digested 293T cells (Type Culture Collection of Chinese
Academy of Sciences, Shanghai, China) using
Lipofectamine® 3000 (cat. no. L3000-015; Invitrogen;
Thermo Fisher Scientific, Inc.) at 37˚C. The medium was replaced at
8 h and cultivation was continued until 48 h. The supernatant of
each culture was collected for centrifugation at 4,000 x g and 4˚C
for 10 min to remove cell debris, followed by further filtration
with A filter (0.45 µm) to obtain concentrated lentivirus solution.
The virus suspension was added to the A2780 and Hey-T30 cells
medium for transfection at a concentration of 1.5x107
international units (IU). The cells were harvested at 2 days
post-transduction for proliferation, migration and invasion assays,
or at 4 days post-transduction for reverse
transcription-quantitative (RT-q)PCR or western blot analysis.
BCAP31 small interfering (si)RNA (siRNA IID.
138059), SNAI1 siRNA (siRNA IID. 107915), SNAI2 siRNA (siRNA IID.
106954), ZEB1 siRNA (siRNA IID. 109651), ZEB2 siRNA (siRNA IID.
108632), TWIST1 siRNA (siRNA ID. 106119), TWIST2 siRNA (siRNA IID.
114998) and control siRNA (siCtrl; cat. no. AM4641) were purchased
from Invitrogen,(silencer®, Thermo Fisher Scientific,
Inc.). Then, 50 nmol/l siRNA sequences and recombinant lentivirus
(BCAP31-shRNA1, BCAP31-shRNA 2 and shRNA-NC) were transfected into
A2780 cells using the Lipofectamine® 3000 reagent (cat.
no. L3000-015; Invitrogen; Thermo Fisher Scientific, Inc.), shBCAP
cell lines and siRNA cell lines were obtained after 48 h.
RNA extracted from A2780 was reverse transcribed to
cDNA using PrimeScript™ RT Master Mix (Perfect Real Time; cat. no.
RR036A; Takara Bio, Inc.) as follows: 25˚C for 2 min, 42˚C for 50
min and 75˚C for 15 min. PrimeSTAR HS DNA Polymerase (cat. no.
R010B; Takara Bio, Inc.) was used for PCR reaction with the primers
listed in Table I. TWIST1 cDNA was
cloned into the pcDNA3.1 vector (Generay Biotech Co., Ltd.) to
construct the expression vector pdTWIST. The successfully
constructed expression vector pdTWIST (5 µg/µl) was transfected
into shBCAP31 cells, and a shBCAP31 + TWIST1 cell line was
obtained. Subsequent in vitro assays were performed 3 days
post-transfection. The empty pcDNA3.1 was transfected into A2780
cells to obtain an OE Ctrl cell line.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene name | Forward, 5'→3' | Reverse, 5'→3' |
|---|
| BAP31 |
GCCACCTTCCTCTACGCAG |
GCCATAGGTCACTACCAACTC |
| CDH1 |
TAACCGATCAGAATGAC |
TTTGTCAGGGACTCAGG |
| CDH2 | ATTGTGGGTG
CGGGGCTTGG |
GGGTGTGGGGCTGCAGATCG |
| VIM |
AGTCCACTGAGTACCGGAGAC | CAT
TTCACGCATCTGGCGTTC |
| Acta2 |
GGGACATCAAGGAGAAACTGTGT |
TCTCTGGGCAGCGGAAAC |
| SNAI1 |
TCTGGTTCTGTGTCCTCTGC |
TTCCCAGTGAGTCTGTCAGC |
| SNAI2 |
TGTCATACCACAACCAGAGA |
CTTGGAGGAGGTGTCAGAT |
| ZEB1 |
AGACTATTCTGATTCCCCAAGTG |
CCTTCTGAGCTAGTGTCTTGTC |
| ZEB2 |
AATGCACAGAGTGTGGCAAGGC |
CTGCTGATGTGCGAACTGTAGG |
| TWIST1 |
GTCCGCAGTCTTACGAGGAG |
CCAGCTTGAGGGTCTGAATC |
| TWIST2 |
TGGACCAAGGCTCTCAGAACA |
ACAGGAGTATGCGGGCAAGA |
| TWIST1* |
CCaagcttAACTCCCAGACACCTCGCGGG |
GCggatccTTCTCTAAATTTTTTATATTTA |
| GAPDH |
ATCACTGCCACCCAGAAGAC |
TTTCTAGACGGCAGGTCAGG |
RNA extraction and RT-qPCR)
RNA was extracted from the cell lines (IOSE80,
A2780, Hey-T30, COC1, SKOV3, OVCAR3, shCtrl, shBCAP31-1,
shBCAP31-2, shBCAP31 + OE WTIST1, OE Ctrl, OE TWIST1, siCtrl,
siBCAP31 and siTWIST1) using QIAzol lysis reagent (Qiagen GmbH) and
purified with a RNeasy mini kit (Qiagen GmbH). RNA was treated with
DNase I (New England BioLabs, Inc.) to remove genomic DNA. RNA
quality and quantity were analyzed using a NanoDrop
spectrophotometer (NanoDrop program 1000 version 3.8.2; NanoDrop
Technologies; Thermo Fisher Scientific, Inc.) and Bioanalyzer
(Agilent, Technologies, Inc.). For RT-qPCR, PrimeScript™ RT Master
Mix (Perfect Real Time; cat. no. RR036A; Takara Bio, Inc.) was used
to reverse transcribe 1,000 ng total RNA to cDNA in a final volume
of 20 µl. RT was performed as follows: 25˚C for 2 min, 42˚C for 50
min and 75˚C for 15 min. qPCR was performed using SYBR Select
Master mix (cat. no. 4472908; Applied Biosystems; Thermo Fisher
Scientific, Inc.). The primers used for BCAP31, cadherin (CDH)1,
CDH2, vimentin (VIM), actin α2 (Acta2), SNAI1, SNAI2, ZEB1, ZEB2,
TWIST1, TWIST2 and GAPDH are listed in Table I. QuantStudio™ 6 Flex Real-Time PCR
system (Thermo Fisher Scientific, Inc.) was used to collect RT-qPCR
data. The RT-qPCR reaction included an initial denaturation step at
95˚C for 10 min, followed by 40 cycles at 93˚C for 15 sec and 60˚C
for 1 min. Each sample was carried out in triplicate, and relative
expression was calculated and normalized using the
2-ΔΔCq method (44)
relative to β-actin.
Transcriptome sequencing
RNA was extracted from shCtrl, shBCAP31-1 and
shBCAP31-2 cells using QIAzol lysis reagent (Qiagen, GmbH) and
subsequently purified with a RNeasy mini kit (Qiagen, GmbH).
Purified RNA was digested with DNase I (New England Biolabs, Inc.)
to remove residual genomic DNA. RNA quality and quantity were
analyzed using a NanoDrop and Bioanalyzer respectively. RNA-seq
library preparation and sequencing were performed at the Beijing
GeneX Health Co., Ltd. Libraries (300 bp) were constructed using a
NuGen® Ovation human FFPE RNA-seq multiplex system kit
(cat. no. 1707160; Ovation® Ultralow System V2; Tecan
Group., Ltd.). Directional mRNA-seq was conducted using the HiSeq
2000 system (Illumina, Inc.) using the single-read 100 cycles
option. The nucleotide length to be sequenced was 300 bp, and
double-end pyrosequencing was performed with the read lengths of
150 bp for each end. The effective concentration of each library
was quantified accurately by qPCR to ensure the quality of the
library, and the library loading concentration was 2.2 pM. The
resulting raw data were transformed into sequenced reads by base
calling. The poor quality reads were filtered by fastq-tools v0.8
(homes.cs.washington.edu/~dcjones/fastq-tools/);
clean reads were mapped to the reference genome by using HISAT2
(ccb.jhu.edu/software/hisat2). DEGseq
version 1.36.1was used to analyze differential expression genes
(45). The differential expression
genes were determined by Log2 fold change >2 and
adjusted P-value <0.05.
Nuclear and cytoplasmic fractionation
of proteins
A nuclear and cytoplasmic protein extraction kit
(Beyotime Institute of Biotechnology) was used to extract cytosolic
and nuclear proteins from shCtrl, shBCAP31 and shBCAP31 + OE TWIST1
cells according to manufacturer's protocol. All isolated fractions
were analyzed by western blotting.
Western blotting
Cell lines (IOSE80, A2780, Hey-T30, COC1, SKOV3,
OVCAR3, shCtrl, shBCAP31-1, shBCAP31-2, siCtrl, shBCAP31 + OE
TWIST, siBCAP31 and siTWIST1) were harvested and processed in lysis
buffer [50 mM Tris-HCl (pH, 7.4), 150 mM NaCl, 1% TritonX-100, 0.2%
SDS, 10 mM β-mercaptoethanol and 5% glycerol] on ice before a BCA
protein assay kit (Nanjing KeyGen Biotech Co., Ltd.) was utilized
to quantify protein concentrations. Cell extracts were boiled for 5
min in loading buffer before equal amounts of protein (6-10 µg)
were separated via 10% SDS-PAGE. Separated protein bands were then
transferred onto polyvinylidene fluoride membranes. The membranes
were blocked in 5% skimmed milk powder in Tris-buffered saline with
0.1%Tween-20 (TBST) for 1 h at 37˚C and subsequently incubated
overnight at 4˚C with primary antibodies against BAP31 (cat. no.
ab237485; 1:3,000), N-cadherin (cat. no. ab76057; 1:1,000),
E-cadherin (cat. no. ab76055; 1:1,000), SNAI1 (cat. no. ab216347;
1:1,000), SNAI2 (cat. no. ab27568; 1:1,000), ZEB1 (cat. no.
ab203829; 1:500), ZEB2 (cat. no. ab138222; 1:1,000), Twist1 (cat.
no. ab175430; 1:1,500), Twist2 (cat. no. ab66031; 1:2,000), VIM
(cat. no. ab137321; 1:2,000), α-smooth muscle actin (α-SMA; cat.
no. ab32575; 1:3,000), β-tubulin (cat. no. ab210797; 1:1,000),
Lamin B1 (cat. no. ab133741; 1:2,000) and β-actin (cat. no.
ab115777; 1:1,000). After washing with TBST three times, the
membrane was incubated with a HRP-conjugated goat anti-rabbit
secondary antibody (cat. no. ab7090; 1:10,000) at room temperature
for 2 h. ECL Substrate kit (cat. no. 36222ES60; Shanghai Yeasen
Biotechnology Co. Ltd.) was used to visualize the bands on the
membrane. The primary and secondary antibodies were purchased from
Abcam. All experiments were independently repeated ≥ three
times.
In vitro proliferation, migration and
invasion assays
Cell lines were used to detect the ability of
proliferation, migration and invasion, including shCtrl,
shBCAP31-1, shBCAP31-1 and shBCAP31 + OE TWIST1 cell lines.
Cell viability
The viability of cells was examined using Cell
Counting Kit-8 (CCK-8) assays (Beijing Solarbio Science &
Technology Co., Ltd.). Cells were seeded at a density of 1,000
cells/well in a 96-well plate and were cultured in humidified 5%
CO2 atmosphere at 37˚C for 24, 48 or 72 h. Before
detection, 10 µl CCK-8 reagent was added into each well and
incubated at 37˚C for an additional 1.5 h at 37˚C. The optical
density value was measured at 450 nm using a microplate reader
(Multimode Reader; PerkinElmer, Inc.). Cell numbers were analyzed
and a growth curve was drawn. The cell count fold-change was
calculated as follows: Cell count fold-change(t) =
OD(t)/OD(t-24). Each experiment was repeated
three times.
Cell migration and invasion
assays
Cell migration and invasion assays were performed in
6.5-mm Transwell inserts (pore size, 8.0 µm; cat. no. 3422;
Corning, Inc.). Cells (2x105) suspended in 100 µl
serum-free DMEM were added into the upper chamber, whilst the lower
chamber was filled with complete DMEM with 10% FBS. For migration
assay, cells were allowed to migrate at 37˚C for 18 h. For the
invasion assay, Transwell inserts were first coated at 37˚C with 20
µl Matrigel (BD Biosciences) mixed with DMEM in a ratio of 1:4 for
30 min and cells were allowed to invade for 24 h at 37˚C. After
removing the non-migratory cells, the membranes were fixed in 20%
methanol at room temperature for 10 min and stained with 100 ng/ml
4,6-diamidino-2-phenylindole dihydrochloride (DAPI) solution
(Thermo Fisher Scientific, Inc.) at room temperature for 15 min.
Microscopy was performed on a Nikon E800 microscope with a 100x
1.40 Plan-Apo objective lens and five fields of view per chamber.
Migratory and invasive cells were imaged using Nikon eclipse Ti2
fluorescent microscope, before cell migration and invasion were
quantified using ImageJ software (1.38, National Institutes of
Health). Each sample was assayed in triplicate.
Co-immunoprecipitation (Co-IP)
assay
Cells were harvested in 1% NP40 lysis buffer [150 mM
NaCl, 50 mM Tris-HCl (pH 7.4) and 1% Nonidet P-40] containing
protease and phosphatase inhibitors (Sangon Biotech Co., Ltd.) and
the supernatant was removed after centrifugation at 1,000 x g for 1
min at 4˚C. The binding of BAP31 (cat. no. ab237485,1:1,000,
Abcam), E-cadherin (cat. no. ab76055; 1:100, Abcam), N-cadherin
(cat. no. ab76057; 1:100, Abcam) or Twist1 (cat. no. ab175430;
1:500, Abcam) antibody to protein G agarose was performed using a
Protein G Immunoprecipitation kit (cat. no. 11719386001;
Sigma-Aldrich; Merck KGaA). The protein A/G agarose slurry (20 µl)
was washed twice with 200 µl PBS buffer and then incubated with 100
µl antibody in PBS (10 µl antibody + 85 µl H2O + 5 µl
20X PBS) at 25˚C for 30 min on a mixer In parallel, 100 µl rabbit
serum (cat. no. ab7487, Abcam) or anti-rabbit IgG peroxidase
secondary antibody (cat. no. ab6721, Abcam) with 2 µg/µl IgG was
prepared as the negative control. The immunoprecipitated products
(isolated by centrifugation at 2,000 x g for 1 min at 4˚C) were
washed with washing buffer five times and eluted with 2X Laemmli
buffer at 100˚C for 10 min. Three independent experiments were
performed. Finally, the eluate and flow-through were separated by
10% SDS-PAGE and analyzed by western blotting.
Statistical analysis
All statistical analyses were performed using the
SPSS 18.0 statistical software package (SPSS, Inc.). All values are
presented as the mean ± standard deviation. All data were
statistically analyzed using one-way analysis of variance (ANOVA)
with a Bonferroni correction. The significance of the differences
between groups were determined by paired Student's t-test (for
comparison between two groups) or one way ANOVA followed by LSD
test. The experiments were performed in triplicate. Data are
presented as the mean ± SD. P<0.05 was considered to indicate a
statistically significant difference.
Results
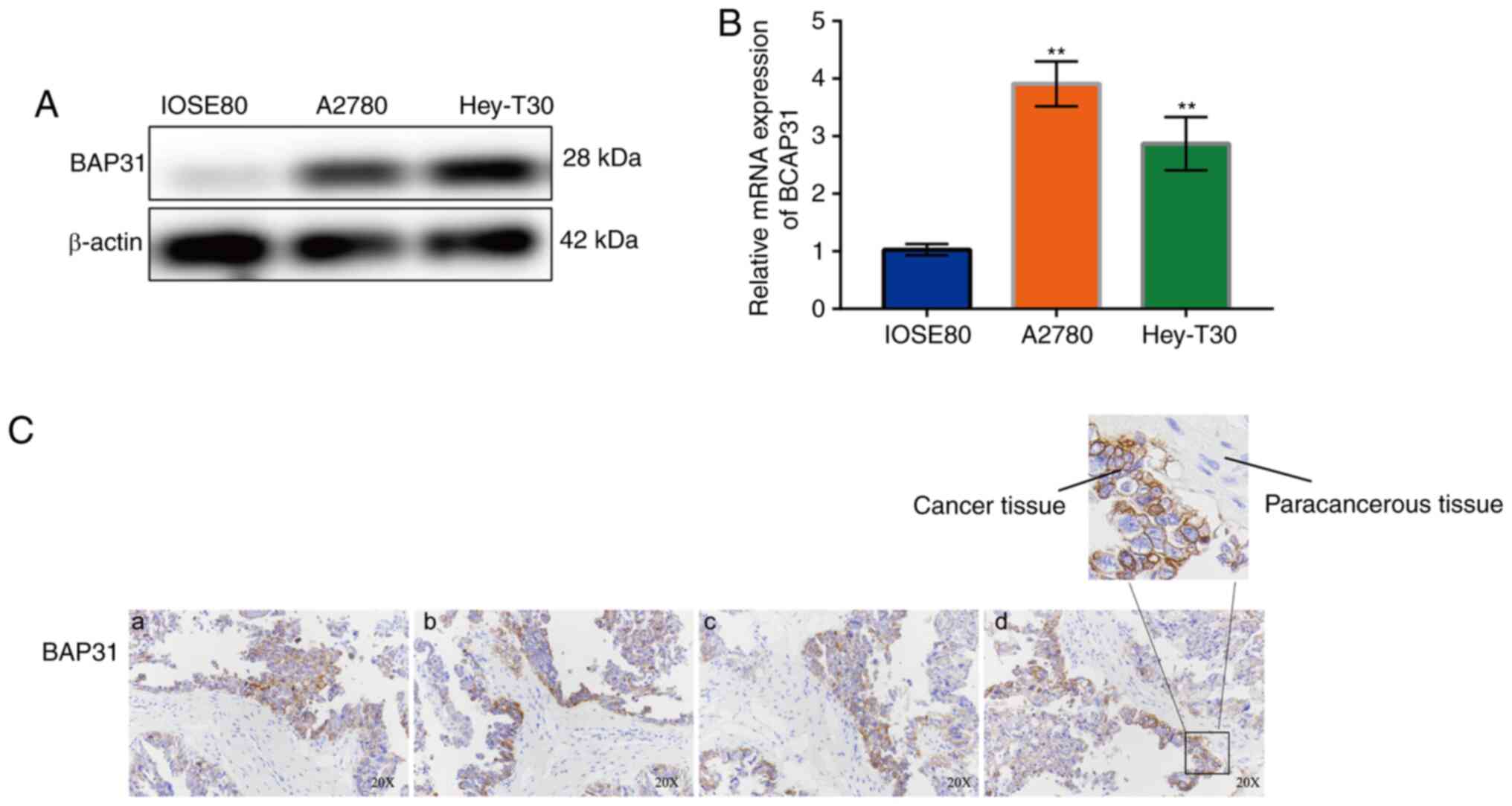
BAP31 expression is upregulated in
ovarian cancer cells
The expression of BAP31 in the human ovarian
epithelial cell line IOSE80 and five ovarian cancer cell lines,
including A2780 and Hey-T30 cells, was measured. The results showed
that the expression of BAP31 in the ovarian cancer cell lines was
markedly higher compared with that in IOSE80 cells on both mRNA and
protein levels (Fig. S1). The
protein expression of BAP31 was observed to be upregulated in
ovarian cancer cells, notably in A2780 (Fig. 1A and B), whilst the mRNA expression of BCAP31 in
the two ovarian cancer cell lines (A2780 and Hey-T30) tested was
significantly higher compared with that observed in IOSE80 cells
(Fig. 1B), particularly for A2780
cells, in which expression levels were nine times higher than those
in IOSE80 cells. An analysis on the relative expression of BAP31 in
cancer and paracancerous tissue from patients with ovarian cancer
was also performed. The results showed that BAP31 was highly
expressed in ovarian cancer tissues compared with that in the
surrounding paracancerous tissues (Fig.
1C).
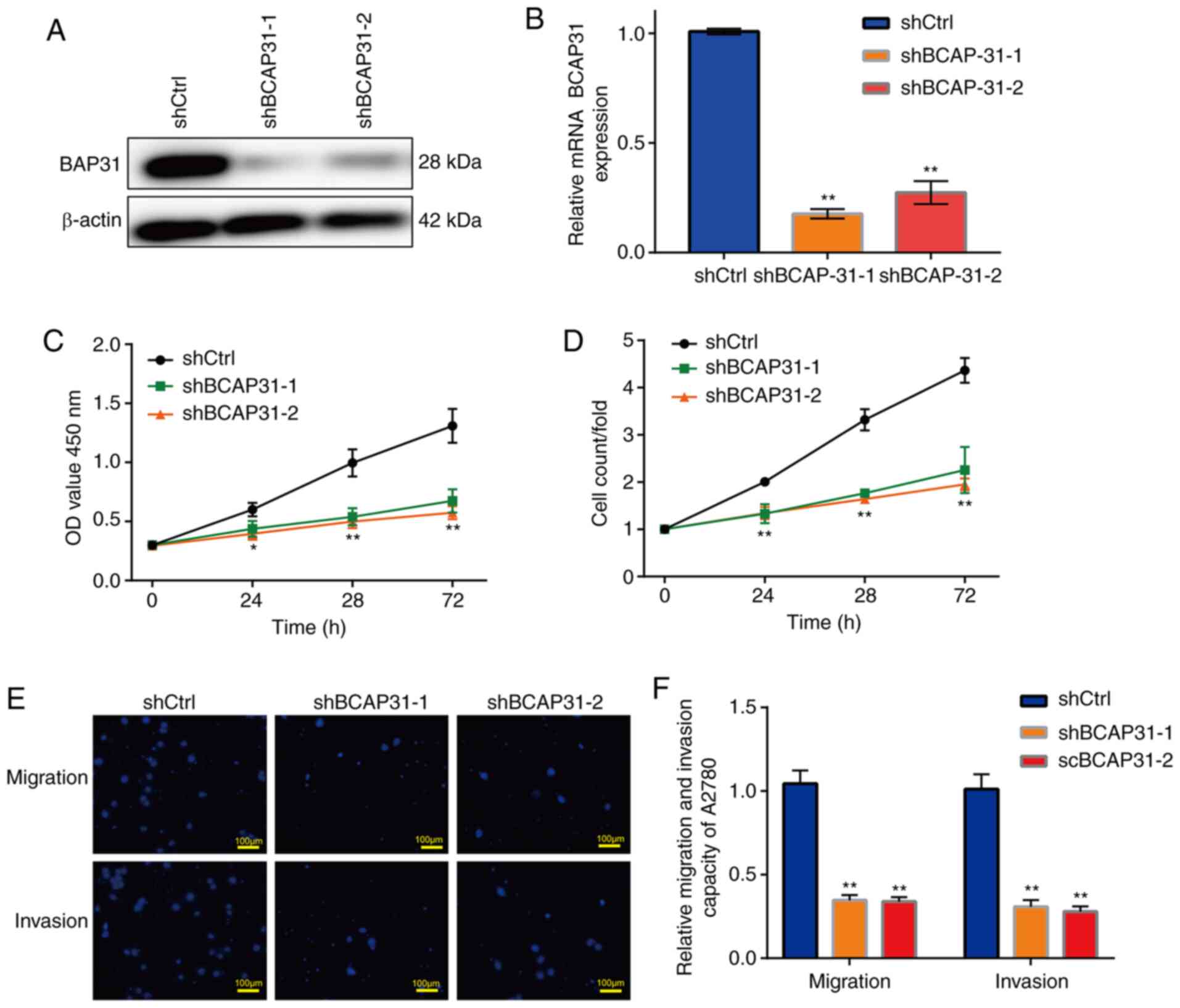
Knockdown of BAP31 inhibits the
viability, migration and invasion of ovarian cancer cells
Two shRNA lentiviral vectors specifically targeting
BCAP31 were constructed, following which A2780 cells were
transfected after viral packaging to obtain the shBCAP31-1 and
shBCAP31-2 cell lines. The protein and mRNA expression of BCAP31
was markedly reduced in the two shBCAP31 groups (Fig. 2A and B), where BCAP31 mRNA expression in the two
shBAP31 groups was significantly lower compared with that observed
in the shCtrl group (Fig. 2B)
according to RT-qPCR analysis. Further examination revealed that
the cell viability, migration and invasion of the shBCAP31 cell
lines were all markedly inhibited. The viability of the two
shBCAP31 A2780 cell lines was significantly reduced compared with
that observed in the shCtrl cells (Fig.
2C and D), whilst the migratory
and invasive abilities were significantly reduced in the two
shBCAP31 A2780 cell lines compared with that in the shCtrl cells
(Fig. 2E and F). Similar trends were also observed in
Hey-T30 cells transfected with shBCAP31-1 and shBCAP31-2 (Fig. S2).
Effects of BAP31 knockdown on the
expression of E-cadherin and N-cadherin
To explore the specific underlying mechanism of
BAP31 on the proliferation, migration and invasion of A2780 cells,
the differentially expressed genes were detected by RNA sequencing
in the shBCAP31-1, shBCAP31-2 and shCtrl groups. Among the 10 most
differentially expressed genes found in the two shBAP31 cell
groups, E-cadherin and N-cadherin were identified (Fig. 3A; Table
SI), which are genes that have important reported roles in EMT
(21). Further verification
revealed that the mRNA (Fig. 3B)
and protein expression (Fig. 3C)
levels of E-cadherin were markedly upregulated in the two shBCAP31
cell lines, whereas N-cadherin expression was markedly
downregulated, compared with those in the shCtrl cells. The
expression levels of vimentin and α-SMA, both of which are
associated with EMT (46), were
next detected. The mRNA (Fig. 3D)
and protein (Fig. 3E) levels were
also markedly downregulated following transfection with shBCAP31-1
and shBCAP31-2 compared with those in shCtrl cells. This suggests
that BAP31 may potentiate the proliferation, migration and invasion
of ovarian cancer cells by regulating EMT.
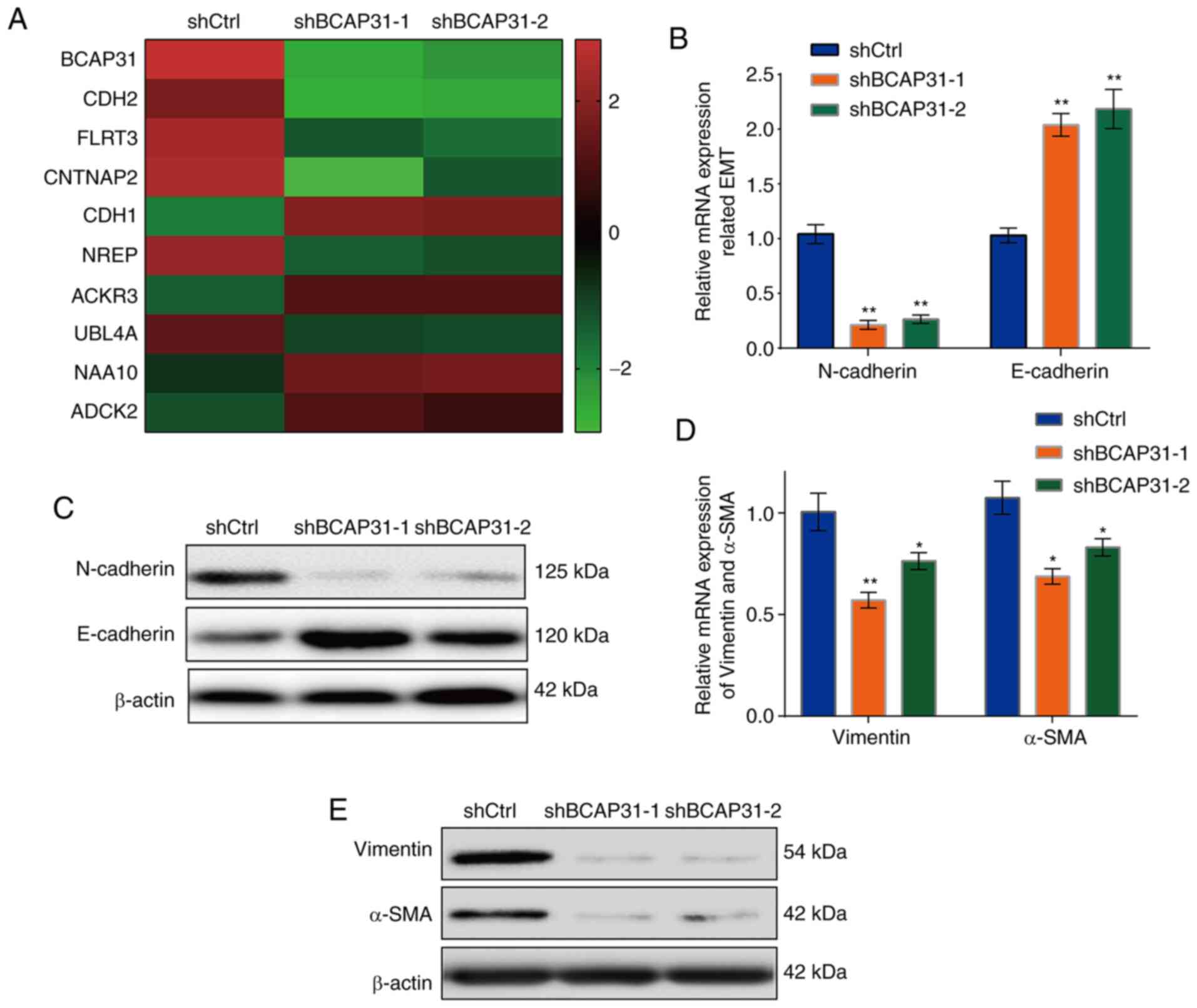 | Figure 3BAP31 regulates the expression of EMT
proteins. (A) Top 10 differentially expressed genes in shCtrl,
shBCAP31-1 and shBCAP31-2 A2780 cells. (B) mRNA and (C) protein
expression of E-cadherin and N-cadherin. (D) mRNA and (E) protein
expression of EMT-related genes vimentin and α-SMA, in shCtrl,
shBCAP31-1 and shBCAP31-2 A2780 cells. *P<0.05 and
**P<0.01 vs. shCtrl. BAP31 or BCAP31, B cell
receptor-associated protein 31; EMT, epithelial-to-mesenchymal
transition; sh, short hairpin RNA; α-SMA, α-smooth muscle actin;
Ctrl, control; CDH, cadherin; FLRT3, fibronectin leucine-rich
transmembrane protein 3; CNTNAP2, contactin-associated protein 2;
NREP, neuronal regeneration-related protein; ACKR3, atypical
chemokine receptor 3; UBL4A, ubiquitin-like 4A; NAA10,
N-α-acetyltransferase 10, NatA catalytic subunit; ADCK2, AarF
domain containing kinase 2. |
BAP31 regulates the expression of
E-cadherin and N-cadherin on a transcriptional level
It was found that the knockdown of BAP31 affected
the relative mRNA expression levels of E-cadherin and N-cadherin
(Fig. 3). No previous studies have
described the DNA-binding domain of BAP31, and there was no
interaction found between BAP31 and E-cadherin or N-cadherin using
Co-IP detection (Fig. 4A). Previous
studies have reported that the TFs, including SNAI1, SNAI2, ZEB1,
ZEB2, TWIST1 and TWIST2, can simultaneously regulate the expression
of E-cadherin and N-cadherin (47).
After the respective siRNAs were used to knock down the expression
levels of these TFs aforementioned in A2780 cells, the mRNA
expression levels of N-cadherin were downregulated by differing
degrees in all siRNAs tested compared with those in cells
transfected with siCtrl (Fig. S3).
By contrast, the mRNA expression levels of E-cadherin were
increased by differing degrees in all siRNAs tested compared with
those in cells transfected with siCtrl, except for those
transfected with siZEB1 (Fig. S3).
The knockdown of BAP31 expression did not affect the mRNA or the
protein expression levels of these TFs (Fig. 4B and C), but altered the nuclear distribution of
TWIST1, specifically markedly reducing its nuclear aggregation
(Fig. 4D). An interaction was
observed between BAP31 and TWIST1 (Fig.
4E). siRNAs were then used to knock down the expression levels
of BAP31 and TWIST1, following which the mRNA and protein
expression levels of E-cadherin and N-cadherin in the TWIST1 siRNA
group exhibited comparable expression as that seen in the siBCAP31
cell line (Fig. 4F and G).
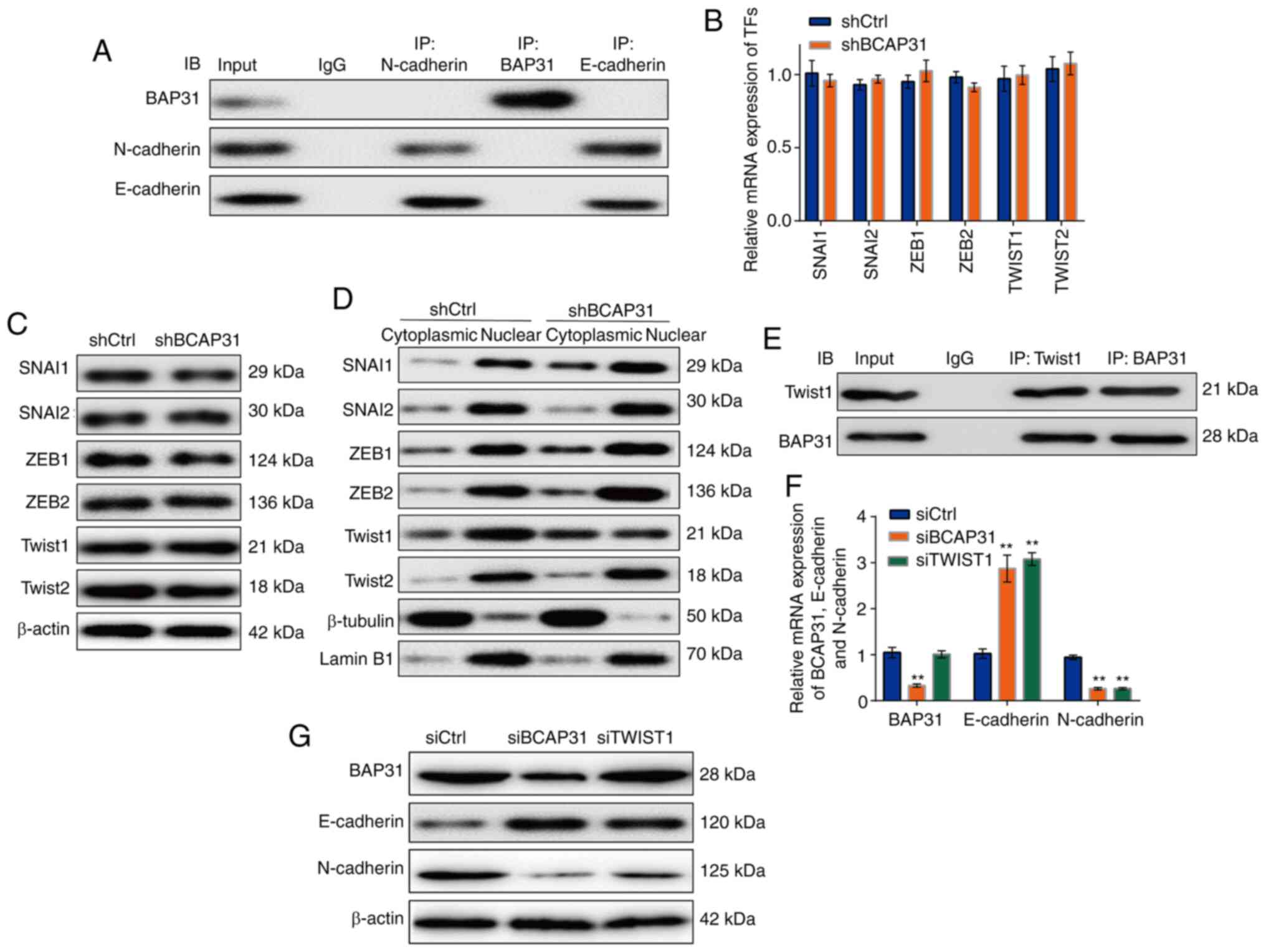 | Figure 4BAP31 regulates the expression of
E-cadherin and N-cadherin at the transcriptional level through
TWIST1. (A) Interaction between BAP31 and E-cadherin or N-cadherin
in A2780 shCtrl cells. (B) mRNA and (C) protein expression of TFs
that may regulate E-cadherin and N-cadherin expression in shCtrl
and shBCAP31 A2780 cells. (D) Protein expression of TFs in the
cytoplasmic and nuclear fractions of shCtrl and shBAP31 A2780
cells. (E) Interaction between BAP31 and TWIST1 in A2780 shCtrl
cells. (F) mRNA and (G) protein expression of N-cadherin and
E-cadherin in A2780 cells transfected with siCtrl, siBCAP31 and
siTWIST1. **P<0.01 vs. siCtrl BAP31 or BCAP31, B cell
receptor-associated protein 31; sh, short hairpin RNA; TF,
transcription factor; si, small interfering RNA; Ctrl, control;
SNAI, snail family; IB, immunoblot; IP, immunoprecipitation; ZEB,
zinc finger E-box-binding homeobox 1; CDH, cadherin. |
Overexpression of TWIST1 in the
shBAP31 cell line can restore cell functions and protein
expression
The expression vector pdTWIST1 (TWIST1 cDNA was
cloned into the pcDNA3.1 vector) was transfected into shBCAP31
A2780 cells to obtain the shBCAP31 + OE TWIST1 A2780 cell line,
before the mRNA expression of BCAP31 and TWIST1 in the A2780 cells
following eight different transfection protocols (shCtrl, siCtrl,
OE Ctrl, shBCAP31, siBCAP31, siTWIST1, OE TWIST1 and shBCAP31 + OE
TWIST1) were analyzed by RT-qPCR. The mRNA expression of BCAP31 was
decreased in the shBCAP31 and shBCAP31 + OE TWIST1 groups compared
with shCtrl group, and which in siBCAP31 group was decreased
compared with siCtrl group. Whilst the mRNA expression of TWIST1
was decreased in siTWIST1 group compared with siCtrl and increased
in the OE TWIST1 and shBCAP31 + OE TWIST1 groups compared with
shCtrl group (Fig. S4). After
TWIST1 was overexpressed in the shBCAP31 A2780 cell line (shBCAP31
+ OE TWIST1), the nuclear and cytoplasmic distribution of TWIST1
was comparable with that found in the shBCAP31 cell line, whilst
the protein levels in the nucleus was markedly higher compared with
those in the shBCAP31 group (Fig.
5A). Cell viability (Fig. 5B),
migration and invasion (Fig. 5C and
D) in the shBCAP31 + OE TWIST1
group were all increased compared with those in the shBCAP31 group,
similar to the level observed in the shCtrl group. Subsequently,
the mRNA and protein expression of N-cadherin and E-cadherin was
explored, where it was found that the mRNA (Fig. 5E) and protein (Fig. 5F) expression levels of N-cadherin in
the shBCAP31 + OE TWIST1 group were markedly higher compared with
those in the shBCAP31 group and comparable with those in the shCtrl
group. By contrast, E-cadherin expression in the shBCAP31 + OE
TWIST1 group exhibited the opposite trend.
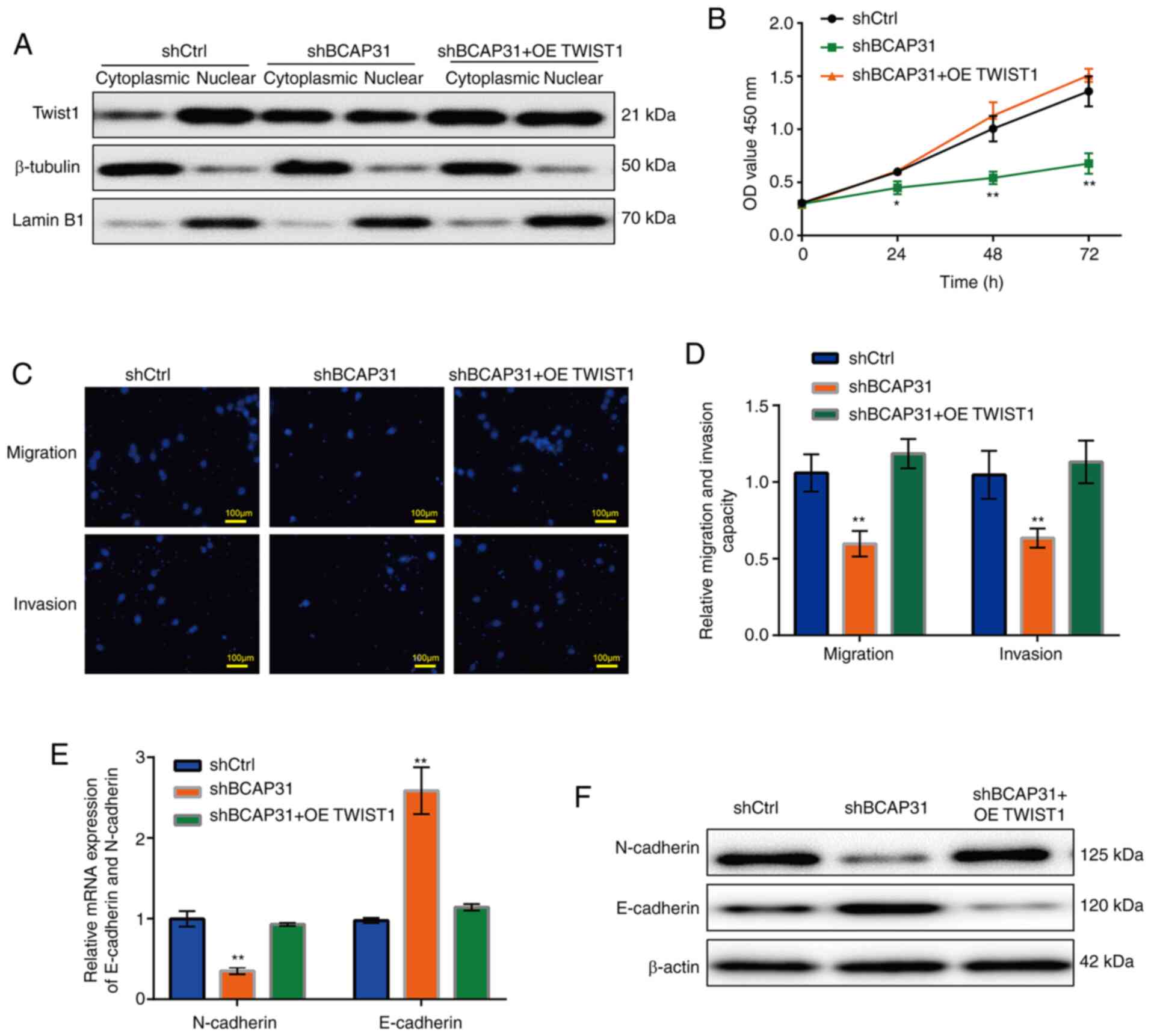 | Figure 5Cell functions and EMT-related
protein expression levels are recovered when TWIST1 was
overexpressed in the shBCAP31 A2780 cells. (A) Protein expression
of TWIST1 in the cytoplasmic and nuclear fractions of shCtrl,
shBCAP31 and shBCAP31 + OE TWIST1 A2780 cells. (B) Growth curve
after transfection with shCtrl, shBCAP31 and shBCAP31 + OE TWIST1.
(C) Representative Transwell assay images of migratory and invasive
shCtrl, shBCAP31 and shBCAP31 + OE TWIST1 A2780 cells. (D) Relative
migratory and invasive abilities of shCtrl, shBAP31 and shBAP31 +
OE TWIST1 A2780 cells. (E) mRNA and (F) protein expression of
N-cadherin and E-cadherin in shCtrl, shBCAP31 and shBCAP31 + OE
TWIST1 A2780 cells. *P<0.05, **P<0.01
vs. shCtrl. BCAP31, B cell receptor-associated protein 31; sh,
short hairpin RNA; OE, overexpression; Ctrl, control; OD, optical
density; CDH cadherin. |
Discussion
BAP31 is a B cell receptor protein that is an
integrated 28-KDa multimer and is evolutionarily conserved and
ubiquitously expressed (8). Studies
have previously shown that in addition to participating in the
activation of B cell receptors, BAP31 can regulate the metabolism
of a number of proteins (10,48-50).
BAP31 can also function as a carrier protein to mediate the
transport of proteins from the ER to the Golgi apparatus for
processing and maturation (11).
BAP31 is located in the ER, where the cleaved
fragment, BAP20, serves an important role in apoptosis (11). BAP20 induces the ER to release
Ca2+, which is taken up by the mitochondria, leading to
mitochondrial fragmentation and subsequent cell death (11). By contrast, the complete BAP31
protein is a direct inhibitor of caspase 8, which initiates
apoptosis (51). BAP31 is highly
expressed in various cancer tissues, including breast, cervical,
liver and colorectal cancer (15-18).
Therefore, the BAP31 protein and mRNA expression profile in ovarian
cancer cells was investigated in the present study.
A previous study demonstrated that BAP31 is a key
gene that promotes the progression of triple negative breast
cancer, where it can interact with epidermal growth factor receptor
(EGFR) to maintain EGFR autophosphorylation and downstream signal
activation (52). In another study,
downregulation of BAP31 expression can inhibit the proliferation
and increase apoptosis of colorectal cancer cells by increasing the
expression of ER-related proteins, including GRP78/BIP, BAX and
PERK/elF2α/ATF4/CHOP (53). BAP31
expression has also been found to be significantly increased in
cervical cancer tissue compared with adjacent normal tissue and
associated highly with poorer clinical outcomes (16,18).
In addition, BAP31 can regulate cervical cancer cell proliferation
by arresting G0/G1 phase cell cycle
progression in cervical cancer cells (18). Depletion of BAP31 can also inhibit
the invasion and migration of cervical cancer cells by decreasing
the expression and uniform distribution in the cel of Drebrin,
myosin phosphatase Rho-interacting protein, calcium-binding protein
SPEC 1A and Nexilin (18). The
expression levels of transforming growth factor-β1, matrix
metalloproteinase (MMP)-2, MMP-9, rho-related protein kinase 1,
α-SMA, vimentin and N-cadherin were found to be significantly lower
in cervical cancer cells following the knockdown of BAP31
expression (16). Furthermore, the
extent intrinsic and extrinsic apoptosis was notably increased in
cells following BAP31 knockdown with the increased levels of
cleaved caspases-8, -9 and -3 in addition to poly (ADP-ribose)
polymerase (16). However, another
study reported that BAP31 can stimulate U2OS osteosarcoma and HeLa
cervical cancer cell death and inhibit autophagy by interacting
with syntaxin 17 to form a protein complex under ER-stress
conditions (49). These previous
findings aforementioned demonstrated that BAP31 can serve different
roles under diverse conditions. To investigate the role of BAP31 in
ovarian cancer in the present study, a shBCAP31 lentiviral vector
was constructed before the ovarian cancer cell lines A2780 and
Hey-T30 were transfected after viral packaging to construct the
shBCAP31 cell line. It was found that the viability of A2780 and
Hey-T30 cells was decreased markedly following the knockdown of
BAP31. The migration and invasion of shBCAP31 cells were also
inhibited compared with those in cells in the shCtrl group. These
results suggest that BAP31 may have an important role in the
migration and invasion of ovarian cancer cells.
To explore the specific mechanism underlying the
effects of BAP31 on ovarian cancer cells, RNA-seq analysis was
performed in the present study to detect the differences in gene
transcription between shBCAP31 and shCtrl cells. E-cadherin and
N-cadherin, two key markers in the EMT process (21), were markedly upregulated and
downregulated following BAP31 knockdown, respectively. This was
also confirmed by RT-qPCR and western blotting. EMT is a process in
which epithelial cells lose their intercellular connections and
acquire a more migratory and aggressive mesenchymal phenotype
(54). However, this process serves
a vital role during normal development in addition to various forms
of tumorigenesis (55). In
addition, EMT has been reported to serve an important role in the
metastasis of a majority of human tumors (20,56),
which can be used as an early indicator of tumor invasion and
metastasis (57). Results from the
present study suggest that the EMT pathway may be manipulated by
altering the expression of BAP31 in ovarian cancer. Cell viability,
migration and invasion of the shBCAP31 ovarian cancer cell lines
were all markedly downregulated, which could be due to inhibition
of the EMT pathway in the shBCAP31 cell line.
Compared with cell lines that express high levels of
N-cadherin, ovarian cancer cell lines with high expression levels
of E-cadherin exhibit poor resistance to cell death, reduced
adhesion to the extracellular matrix and weaker invasiveness
(58). As an indicator of ongoing
EMT, expression of N-cadherin is associated with the development of
various types of tumors, such as prostate (59) and non-small cell lung cancer
(60). During EMT in cancer,
N-cadherin is frequently upregulated, whilst E-cadherin is
downregulated. This cadherin switch is concomitant with enhanced
cell migration and invasion, resulting in the lower survival rate
of patients (61). The present
study showed that BAP31 altered the expression of E-cadherin and
N-cadherin simultaneously in ovarian cancer cells. However, this
was not a result of the direct interaction between BAP31 and E- and
N-cadherin, which was not observed. There was a clear interaction
between E-cadherin and N-cadherin, suggesting that there may be a
direct mutual regulatory mechanism between E-cadherin and
N-cadherin. These results are consistent with previous findings
(21) that demonstrated that
‘cadherin switch’ (N-cadherin is upregulation and E-cadherin
downregulation during the EMT process in cancer), is associated
with enhanced migration and invasion. Since the knockdown of BAP31
affected the expression of E-cadherin and N-cadherin on a
transcriptional level, it was speculated that the expression or
transcriptional activity of TFs that regulate E-cadherin and
N-cadherin expression were influenced by BAP31.
The present study found that the expression of
E-cadherin was upregulated, whereas the expression of N-cadherin
was downregulated, in cells transfected with siTWIST1, which is
consistent with the expression of these two genes analyzed by
RNA-seq in BCAP31 cells. Further analysis revealed an interaction
between BAP31 and TWIST1, where the degree of TWIST1 aggregation in
the nucleus decreased markedly following the knockdown of BAP31
expression. TWIST1 is a highly conserved basic helix-loop-helix TF
that has been previously identified to be a developmental gene and
serves a key role in E-cadherin inhibition and EMT induction
(62). In addition to having an
important role in normal development, TWIST was found to be
upregulated in numerous types of cancer, including breast (35,41),
gastric (40,63) and prostate cancer (64). In vertebrates, two structurally
similar isoforms are expressed, namely TWIST1 and TWIST2(37). TWIST1 contains a glycine-rich
region, which may serve a role in RNA binding, at the N-terminus,
whilst TWIST2 does not (37). In
intestinal gastric cancer, the highly expressed TWIST1 protein
enhances the aggressiveness of gastric cancer cells by
downregulating the expression of E-cadherin and upregulating the
expression of N-cadherin (63). In
the present study, when TWIST1 was overexpressed in the shBCAP31
cells, cell proliferation, migration and invasion were restored to
the levels comparable to those of shCtrl cells. Additionally, the
mRNA and protein expression levels of E-cadherin and N-cadherin
were comparable to those observed in the shCtrl group. These
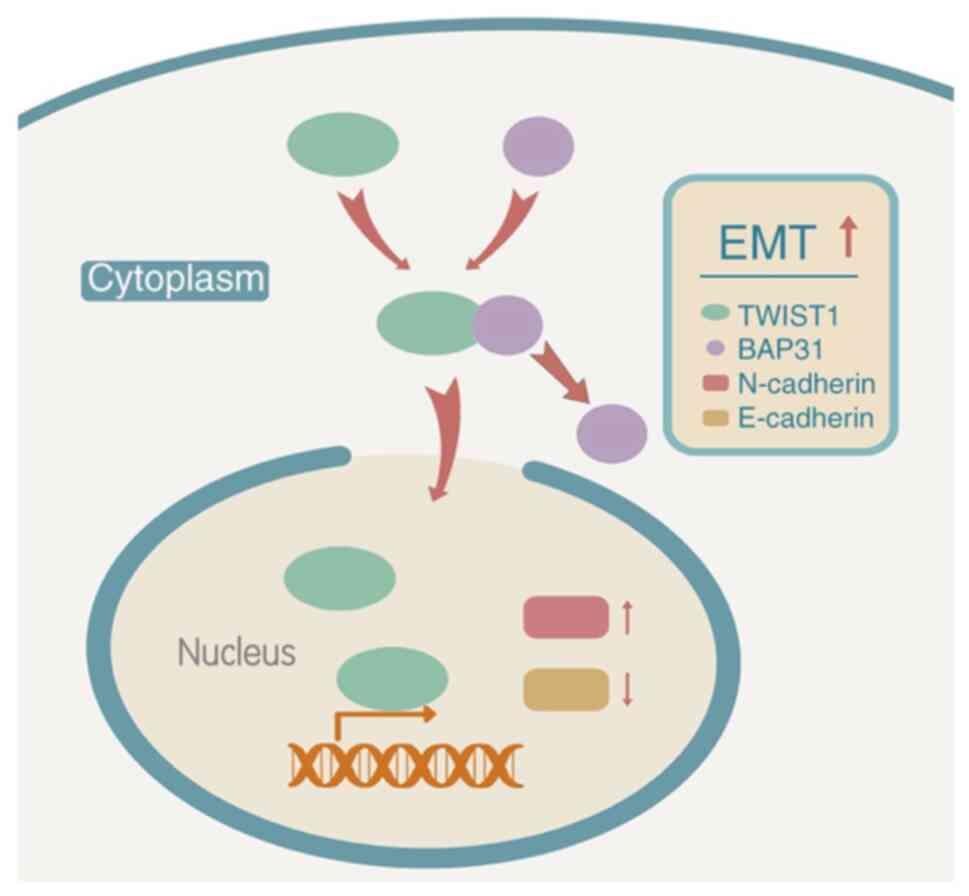
findings suggest that BAP31 may regulate the expression of
E-cadherin and N-cadherin through the subcellular location of
TWIST1, one of the TFs that mediate EMT, which in turn regulates
the proliferation, migration and invasion of ovarian cancer cells
(Fig. 6). Therefore, BAP31 may
promote the EMT pathway by affecting the nuclear and cytoplasmic
distribution of TWIST1 in ovarian cancer cells.
To conclude, results of the present study showed
that BAP31 expression was upregulated in ovarian cancer cells,
suggesting that BAP31 promoted the proliferation, migration and
invasion of ovarian cancer cells. Knockdown of BAP31 was found to
increase the expression of E-cadherin and decrease the expression
of N-cadherin, two markers of the EMT pathway. Concurrently, BAP31
regulated the expression of E-cadherin and N-cadherin on
transcriptional level by affecting the degree of nuclear TWIST1
aggregation, one of the TFs that can regulate the mRNA expression
of E-cadherin and N-cadherin. These findings suggested that BAP31
serve a carcinogenic role in ovarian cancer, which provide
theoretical evidence for the role of BAP31 as a biomarker in
patients with ovarian cancer.
Supplementary Material
BAP31 is overexpressed in ovarian
cancer lines. Protein and mRNA expression of BAP31 in ovarian
cancer cell lines are higher compared with those in IOSE80 cells.
*P<0.05 and **P<0.01 vs. IOSE80 cells.
BAP31, B cell receptor-associated protein 31.
Knockdown of BCAP31 affects the
proliferation, migration and invasion of the ovarian cancer cell
line Hey-T30. (A) Protein and (B) mRNA expression of BAP31 is
downregulated in the shBCAP31-1 and shBCAP31-2 Hey-T30 cells. (C)
Cell count curve of Hey-T30 cells after transfection with shCtrl,
shBCAP31-1 and shBCAP31-2. (D) Representative Transwell assay
images of migratory and invasive shCtrl, shBCAP31-1 and shBCAP31-2
Hey-T30 cells. (E) Relative migratory and invasive activity of
shCtrl, shBCAP31-1 and shBCAP31-2 Hey-T30 cells. Scale bars, 100
μm. **P<0.01 vs. ShCtrl. BAP31 or BCAP31, B cell
receptor-associated protein 31; sh, short hairpin RNA; OD, optical
density.
mRNA expression levels of the various
epithelial-mesenchymal transition-associated TFs, CDH2 and CDH1 in
the cell lines transfected with the various siRNAs. (A) mRNA
expression of TFs in A2780 cells after transfection with siRNA,
which was measured by reverse transcription-quantitative PCR. (B)
mRNA expression of N-cadherin in A2780 cells after transfection
with the six indicated siRNAs. (C) mRNA expression of E-cadherin in
A2780 cells after transfection with the six indicated siRNAs.
*P<0.05 and **P<0.01 vs. siCtrl. TFs,
transcription factor; si, small interfering; Ctrl, control; SNAI,
snail family; ZEB, zinc finger E-box-binding homeobox 1; CDH,
cadherin.
mRNA expression levels of BCAP31 and
TWIST1 after transfection into A2780 cells with the indicated
shRNA, siRNA and plasmids. **P<0.01 vs. shCtrl;
##P<0.01 vs. siCtrl; $$P<0.01 vs. OE
Ctrl. BCAP31, B cell receptor-associated protein 31. Sh, short
hairpin; si, small interfering; OE, overexpression.
Data from RNA-seq analysis.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the National Natural
Science Foundation of China (grant nos. 81372777 and 81372779).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI repository (BioProject ID
PRJNA731548).
Authors' contributions
DF and BL designed the study. HL, DF and BL analyzed
the data. HL wrote the manuscript. JD, ZC, HL and QL performed the
experiments and collected the data. HL and JD confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The use of human tissue was approved by the Ethics
Committee of China-Japan Friendship Hospital (approval no.
2020-28-K20)and written informed consents were obtained from all
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Su Z, Graybill WS and Zhu Y: Detection and
monitoring of ovarian cancer. Clin Chim Acta. 415:341–345.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Narod S: Can advanced-stage ovarian cancer
be cured. Nat Rev Clin Oncol. 13:255–261. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lim D and Oliva E: Precursors and
pathogenesis of ovarian carcinoma. Pathology. 45:229–242.
2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Vang R, Shih IeM and Kurman RJ: Ovarian
low-grade and high-grade serous carcinoma: Pathogenesis,
clinicopathologic and molecular biologic features, and diagnostic
problems. Adv Anat Pathol. 16:267–282. 2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Nelson BH: The impact of T-cell immunity
on ovarian cancer outcomes. Immunol Rev. 222:101–116.
2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kim KM, Adachi T, Nielsen PJ, Terashima M,
Lamers MC, Köhler G and Reth M: Two new proteins preferentially
associated with membrane immunoglobulin D. EMBO J. 13:3793–3800.
1994.PubMed/NCBI
|
|
9
|
Adachi T, Schamel WW, Kim KM, Watanabe T,
Becker B, Nielsen PJ and Reth M: The specificity of association of
the IgD molecule with the accessory proteins BAP31/BAP29 lies in
the IgD transmembrane sequence. EMBO J. 15:1534–1541.
1996.PubMed/NCBI
|
|
10
|
Namba T, Tian F, Chu K, Hwang SY, Yoon KW,
Byun S, Hiraki M, Mandinova A and Lee SW: CDIP1-BAP31 complex
transduces apoptotic signals from endoplasmic reticulum to
mitochondria under endoplasmic reticulum stress. Cell Rep.
5:331–339. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Breckenridge DG, Stojanovic M, Marcellus
RC and Shore GC: Caspase cleavage product of BAP31 induces
mitochondrial fission through endoplasmic reticulum calcium
signals, enhancing cytochrome c release to the cytosol. J
Cell Biol. 160:1115–1127. 2003.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wakana Y, Takai S, Nakajima K, Tani K,
Yamamoto A, Watson P, Stephens DJ, Hauri HP and Tagaya M: Bap31 is
an itinerant protein that moves between the peripheral endoplasmic
reticulum (ER) and a juxtanuclear compartment related to
ER-associated Degradation. Mol Biol Cell. 19:1825–1836.
2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Matroule JY, Carthy CM, Granville DJ,
Jolois O, Hunt DW and Piette J: Mechanism of colon cancer cell
apoptosis mediated by pyropheophorbide-a methylester
photosensitization. Oncogene. 20:4070–4084. 2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yu S, Wang F, Fan L, Wei Y, Li H, Sun Y,
Yang A, Jin B, Song C and Yang K: BAP31, a promising target for the
immunotherapy of malignant melanomas. J Exp Clin Cancer Res.
34(36)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tan N, Liu Q, Liu X, Gong Z, Zeng Y, Pan
G, Xu Q and He S: Low expression of B-cell-associated protein 31 in
human primary hepatocellular carcinoma correlates with poor
prognosis. Histopathology. 68:221–229. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang A, Zhang Y and Cao P: Inhibition of
BAP31 expression inhibits cervical cancer progression by
suppressing metastasis and inducing intrinsic and extrinsic
apoptosis. Biochem Biophys Res Commun. 508:499–506. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ma C, Jin RM, Chen KJ, Hao T, Li BS, Zhao
DH and Jiang H: Low expression of B-Cell-Associated protein 31 is
associated with unfavorable prognosis in human colorectal cancer.
Pathol Res Pract. 214:661–666. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Dang E, Yang S, Song C, Jiang D, Li Z, Fan
W, Sun Y, Tao L, Wang J, Liu T, et al: BAP31, a newly defined
cancer/testis antigen, regulates proliferation, migration, and
invasion to promote cervical cancer progression. Cell Death Dis.
9(791)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Loh CY, Chai JY, Tang TF, Wong WF, Sethi
G, Shanmugam MK, Chong PP and Looi CY: The E-cadherin and
N-cadherin switch in epithelial-to-mesenchymal transition:
Signaling, therapeutic implications, and challenges. Cells.
8(1118)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ling ZQ, Li P, Ge MH, Zhao X, Hu FJ, Fang
XH, Dong ZM and Mao WM: Hypermethylation-modulated down-regulation
of CDH1 expression contributes to the progression of esophageal
cancer. Int J Mol Med. 27:625–635. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Umbas R, Schalken JA, Aalders TW, Carter
BS, Karthaus HF, Schaafsma HE, Debruyne FM and Isaacs WB:
Expression of the cellular adhesion molecule E-cadherin is reduced
or absent in high-grade prostate cancer. Cancer Res. 52:5104–5109.
1992.PubMed/NCBI
|
|
24
|
Petrova YI, Schecterson L and Gumbiner BM:
Roles for E-cadherin cell surface regulation in cancer. Mol Biol
Cell. 27:3233–3244. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fransvea E, Angelotti U, Antonaci S and
Giannelli G: Blocking transforming growth factor-beta up-regulates
E-cadherin and reduces migration and invasion of hepatocellular
carcinoma cells. Hepatology. 47:1557–1566. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tang G, Du R, Tang Z and Kuang Y:
MiRNALet-7a mediates prostate cancer PC-3 cell invasion, migration
by inducing epithelial-mesenchymal transition through CCR7/MAPK
pathway. J Cell Biochem. 119:3725–3731. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
del Valle I, Rudloff S, Carles A, Li Y,
Liszewska E, Vogt R and Kemler R: E-cadherin is required for the
proper activation of the Lifr/Gp130 signaling pathway in mouse
embryonic stem cells. Development. 140:1684–1692. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hazan RB, Qiao R, Keren R, Badano I and
Suyama K: Cadherin switch in tumor progression. Ann N Y Acad Sci.
1014:155–163. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Muramaki M, Miyake H, Terakawa T, Kumano
M, Sakai I and Fujisawa M: Expression profile of E-cadherin and
N-cadherin in non-muscle-invasive bladder cancer as a novel
predictor of intravesical recurrence following transurethral
resection. Urol Oncol. 30:161–166. 2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shintani Y, Hollingsworth MA, Wheelock MJ
and Johnson KR: Collagen I promotes metastasis in pancreatic cancer
by activating c-Jun NH(2)-terminal kinase 1 and up-regulating
N-cadherin expression. Cancer Res. 66:11745–11753. 2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Puisieux A, Brabletz T and Caramel J:
Oncogenic roles of EMT-inducing transcription factors. Nat Cell
Biol. 16:488–494. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hamamori Y, Wu HY, Sartorelli V and Kedes
L: The basic domain of myogenic basic helix-loop-helix (bHLH)
proteins is the novel target for direct inhibition by another bHLH
protein, Twist. Mol Cell Biol. 17:6563–6573. 1997.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lee MS, Lowe GN, Strong DD, Wergedal JE
and Glackin CA: TWIST, a basic helix-loop-helix transcription
factor, can regulate the human osteogenic lineage. J Cell Biochem.
75:566–577. 1999.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Verzi MP, Anderson JP, Dodou E, Kelly KK,
Greene SB, North BJ, Cripps RM and Black BL: N-twist, an
evolutionarily conserved bHLH protein expressed in the developing
CNS, functions as a transcriptional inhibitor. Dev Biol.
249:174–190. 2002.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Mironchik Y, Winnard PT Jr, Vesuna F, Kato
Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van
Diest P, et al: Twist overexpression induces in vivo angiogenesis
and correlates with chromosomal instability in breast cancer.
Cancer Res. 65:10801–10809. 2005.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ansieau S, Bastid J, Doreau A, Morel AP,
Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S,
et al: Induction of EMT by twist proteins as a collateral effect of
tumor-promoting inactivation of premature senescence. Cancer Cell.
14:79–89. 2008.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Rodriguez Y, Gonzalez-Mendez RR and
Cadilla CL: Evolution of the twist subfamily vertebrate proteins:
Discovery of a signature motif and origin of the twist1
glycine-rich motifs in the amino-terminus disordered domain. PLoS
One. 11(e0161029)2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ansieau S, Morel AP, Hinkal G, Bastid J
and Puisieux A: TWISTing an embryonic transcription factor into an
oncoprotein. Oncogene. 29:3173–3184. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Satoh K, Hamada S, Kimura K, Kanno A,
Hirota M, Umino J, Fujibuchi W, Masamune A, Tanaka N, Miura K, et
al: Up-regulation of MSX2 enhances the malignant phenotype and is
associated with twist 1 expression in human pancreatic cancer
cells. Am J Pathol. 172:926–939. 2008.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ru GQ, Wang HJ, Xu WJ and Zhao ZS:
Upregulation of Twist in gastric carcinoma associated with tumor
invasion and poor prognosis. Pathol Oncol Res. 17:341–347.
2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ranganathan S, Krishnan A and
Sivasithambaram ND: Significance of twist and iNOS expression in
human breast carcinoma. Mol Cell Biochem. 412:41–47.
2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Khan MA, Chen HC, Zhang D and Fu J: Twist:
A molecular target in cancer therapeutics. Tumour Biol.
34:2497–2506. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Lu Z and Chen J: Introduction of WHO
classification of tumours of female reproductive organs, fourth
edition. Zhonghua Bing Li Xue Za Zhi. 43:649–650. 2014.PubMed/NCBI(In Chinese).
|
|
44
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang L, Feng Z, Wang X, Wang X and Zhang
X: DEGseq: An R package for identifying differentially expressed
genes from RNA-seq data. Bioinformatics. 26:136–138.
2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Scanlon CS, Van Tubergen EA, Inglehart RC
and D'Silva NJ: Biomarkers of epithelial-mesenchymal transition in
squamous cell carcinoma. J Dent Res. 92:114–121. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Malgulwar PB, Nambirajan A, Pathak P,
Rajeshwari M, Suri V, Sarkar C, Singh M and Sharma MC:
Epithelial-to-mesenchymal transition-related transcription factors
are up-regulated in ependymomas and correlate with a poor
prognosis. Hum Pathol. 82:149–157. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Namba T: BAP31 regulates mitochondrial
function via interaction with Tom40 within ER-mitochondria contact
sites. Sci Adv. 5(eaaw1386)2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Machihara K and Namba T: BAP31 inhibits
cell adaptation to ER stress conditions, negatively regulating
autophagy induction by interaction with STX17. Cells.
8(1350)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Quistgaard EM: BAP31: Physiological
functions and roles in disease. Biochimie. 186:105–129.
2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wilson JD and Barlowe C: Yet1p and Yet3p,
the yeast homologs of BAP29 and BAP31, interact with the
endoplasmic reticulum translocation apparatus and are required for
inositol prototrophy. J Biol Chem. 285:18252–18261. 2010.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Fu W, Sun H, Zhao Y, Chen M, Yang X, Liu Y
and Jin W: BCAP31 drives TNBC development by modulating
ligand-independent EGFR trafficking and spontaneous EGFR
phosphorylation. Theranostics. 9:6468–6484. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Xu K, Han B, Bai Y, Ma XY, Ji ZN, Xiong Y,
Miao SK, Zhang YY and Zhou LM: MiR-451a suppressing BAP31 can
inhibit proliferation and increase apoptosis through inducing ER
stress in colorectal cancer. Cell Death Dis. 10(152)2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
55
|
Shook D and Keller R: Mechanisms,
mechanics and function of epithelial-mesenchymal transitions in
early development. Mech Dev. 120:1351–1383. 2003.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yun SJ and Kim WJ: Role of the
epithelial-mesenchymal transition in bladder cancer: From prognosis
to therapeutic target. Korean J Urol. 54:645–650. 2013.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Franco-Chuaire ML, Magda Carolina SC and
Chuaire-Noack L: Epithelial-mesenchymal transition (EMT):
Principles and clinical impact in cancer therapy. Invest Clin.
54:186–205. 2013.PubMed/NCBI
|
|
58
|
Rosso M, Majem B, Devis L, Lapyckyj L,
Besso MJ, Llauradó M, Abascal MF, Matos ML, Lanau L, Castellví J,
et al: E-cadherin: A determinant molecule associated with ovarian
cancer progression, dissemination and aggressiveness. PLoS One.
12(e0184439)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wang M, Ren D, Guo W, Huang S, Wang Z, Li
Q, Du H, Song L and Peng X: N-cadherin promotes
epithelial-mesenchymal transition and cancer stem cell-like traits
via ErbB signaling in prostate cancer cells. Int J Oncol.
48:595–606. 2016.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Hui L, Zhang S, Dong X, Tian D, Cui Z and
Qiu X: Prognostic significance of twist and N-cadherin expression
in NSCLC. PLoS One. 8(e62171)2013.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Araki K, Shimura T, Suzuki H, Tsutsumi S,
Wada W, Yajima T, Kobayahi T, Kubo N and Kuwano H: E/N-cadherin
switch mediates cancer progression via TGF-β-induced
epithelial-to-mesenchymal transition in extrahepatic
cholangiocarcinoma. Br J Cancer. 105:1885–1893. 2011.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939.
2004.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Rosivatz E, Becker I, Specht K, Fricke E,
Luber B, Busch R, Höfler H and Becker KF: Differential expression
of the epithelial-mesenchymal transition regulators snail, SIP1,
and twist in gastric cancer. Am J Pathol. 161:1881–1891.
2002.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C,
Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, et al: Up-regulation
of TWIST in prostate cancer and its implication as a therapeutic
target. Cancer Res. 65:5153–5162. 2005.PubMed/NCBI View Article : Google Scholar
|