Introduction
Osteoporosis is a bone disease characterized by
decreased bone mass, bone microstructure degeneration or
destruction and an increased fracture rate (1). Bone marrow-derived mesenchymal stem
cells (BMSCs) can differentiate into a variety of cells, such as
adipocytes, chondrocytes and nerve cells. However, their
differentiation into cell types other than osteoblasts may lead to
a decrease in the number of bone cells, eventually resulting in
osteoporosis (2). Previous studies
have shown that exogenous mesenchymal stem cell (MSC)
transplantation can restore the impaired function of BMSCs, thereby
promoting osteoblast formation and increasing bone regeneration
(3,4). Mo et al (5) also reported that a compound extracted
from Phyllanthus amarus promotes the osteogenic
differentiation of BMSCs and increases bone mass by activating the
Wnt/β-catenin signaling pathway. In addition, Luo et al
(6) found that runt-related
transcription factor (RUNX) 1 regulates the osteogenic
differentiation of BMSCs by inhibiting adipogenesis-related
pathways. Therefore, BMSCs are beneficial for osteoblast
regeneration and bone remodeling, and are expected to be utilized
for the treatment of bone-related diseases such as osteoporosis and
osteoarthritis (7).
Factors that affect the differentiation of BMSCs
include physical (cell shape and external mechanical forces),
chemical (dexamethasone and insulin are required for adipogenic
differentiation), biological (mineral deposition and cell
proliferation) and age (senescence) (8,9).
Osteocalcin is a vitamin K-dependent bone protein that is
synthesized in the human body by osteoblasts (10). In particular, γ-glutamate
carboxylase can catalyze the carboxylation of three glutamates at
positions 17, 21 and 24 in the molecular structure of osteocalcin
(11). According to whether these
three sites are fully carboxylated, osteocalcin can be divided into
two subtypes: γ-Carboxyl osteocalcin and incomplete
carboxylated/uncarboxylated osteocalcin (unOC) (11). Previous studies have shown that
osteocalcin-knockout in mice can affect bone mineralization
(12,13). Therefore, osteocalcin is
hypothesized to affect osteoblast and osteoclast activity to
regulate mineralization (14,15). A
previous study indicated that unOC can promote the differentiation
of BMSCs into osteoblasts (16).
However, further study is required to identify the specific
mechanism by which unOC regulates the osteogenic differentiation of
MSCs.
Studies have shown that sirtuin 1 (SIRT1) is
associated with the differentiation of MSCs (17,18).
SIRT1 is a nicotinamide adenine dinucleotide
(NAD+)-dependent lysine deacetylase. It activates the
Wnt signaling pathway and promotes MSC osteogenic differentiation
by the deacetylation of secreted frizzled-related protein 1
(sFRP1), sFRP2 and disheveled-binding antagonist of β catenin 1
(19,20). SIRT1 positively regulates RUNX2, a
key transcription factor associated with osteoblasts, to activate
the transcription of BMSC osteogenic differentiation (21). SIRT1 can also deacetylate peroxisome
proliferator-activated receptor γ (PPARγ), inhibiting PPARγ
activity and attenuating lipogenesis to enhance osteogenic
differentiation (22). Therefore,
it was hypothesized that unOC can promote the differentiation of
BMSCs through SIRT1, thereby promoting osteogenic formation.
5'AMP-activated protein kinase (AMPK) is a protein
kinase closely associated with glucose metabolism (23). Increasing evidence suggests that
SIRT1 and AMPK can interact to regulate the osteogenic and
adipogenic differentiation of MSCs (24). AMPK can increase SIRT1 activity by
promoting NAD+ biosynthesis, where SIRT1 can also
upregulate AMPK activity through the liver kinase B1 (LKB1)-AMPK
axis (25). A previous study
reported that protein kinase A (PKA) regulates SIRT1 expression by
activating serine/threonine kinase LKB1(26). Based on these studies, it was
hypothesized that unOC regulated BMSC differentiation into
osteoblasts via the PKA-AMPK-SIRT1 axis.
The present study aimed to further explore the
mechanism and the signaling pathways associated with the promotion
of BMSC osteogenic differentiation by unOC and to uncover novel
avenues for the treatment of osteoporosis.
Materials and methods
Preparation of unOC
unOC was prepared as previously described by Liu and
Yang (16) with slight
modifications. The method used to lyse the bacteria in the present
study was to freeze the bacteria at -80˚C before using an
ultrasonic cell disruptor. In brief, the plasmid pet30a (cat no.
JX210976.1 GI:392880915; Biovector Science Lab, Inc.; http://www.biovector.net/product/1040481.html)
containing the mouse unOC recombinant gene was previously
constructed in the laboratory (27). It was made by amplifying the mouse
osteocalcin gene (accession no. NM_007541) sequences by PCR based
on NCBI database (The source of the template: Mouse bone tissue),
cloning into the pet-30a vector to obtain the pet-30a-OC
recombinant plasmid. It was introduced into Escherichia
coli, and bacteria populations containing the plasmid vector
with a resistance gene were selected with kanamycin antibiotic
(1:1,000; cat. no. K1010; Beijing Lablead Biotechnology Co., Ltd.).
The selected populations were cultured in liquid phase to the log
phase at 37˚C for 8 h and induced to express recombinant
osteocalcin with 1 mM isopropyl β-d-1-thiogalactopyranoside at
28.5˚C for 3 h. Protein (mouse unOC with a six-histidine tag)
extraction and purification processes were performed in accordance
with the previous description (28). The obtained osteocalcin was
isolated, purified by binding to a recombinant protein in a Ni
Sepharose™ 6 Fast Flow column (cat. no. 17531801; Cytiva), and
eluted with 150 mM of imidazole (cat. no. II0070; Beijing Solarbio
Science & Technology Co., Ltd.). The purified protein was
dialyzed and concentrated. The molecular weight of the protein was
determined using 10% SDS-PAGE (the amount of protein loaded per
lane: 20 µg) and Coomassie blue staining (Coomassie Blue Fast
Staining Solution; cat. no. P0017; Beyotime Institute of
Biotechnology) was used. Experimental conditions for Coomassie
staining: 50 ml deionized water was added to the protein gel and
heated at 100˚C for 3 min, before being shaken on a horizontal
shaker at 37˚C for 5 min. the gels were then incubated in ~20 ml
Coomassie Blue Fast Staining Solution at 100˚C for 3 min and
incubated on a shaker at a temperature of 37˚C to decolorize.
Protein bands were observed at room temperature after 2 h. Western
blot analysis was used for protein detection.
Cell culture and differentiation
In total, 12 4-week-old male C57BL/6 mice
(certificate no. SCXK 2016-0006) were purchased from Charles River
Laboratories, Inc. During the research process, all animal
experiments were conducted in accordance with the standards in
University of Chinese Academy of Sciences Institutional Committee
for the Use and Care of Animals. The mice (weight range, 16-18 g)
were used for each experiment. Housing conditions are 25˚C,
relative air humidity: 50-60%. Mice had free access to food and
water and were on a 12-h light/dark cycle. All experimental
protocols for the present study were approved by The Institutional
Animal Care and Use Committee of the University of Chinese Academy
of Sciences (Beijing, China). Isolation and culture of mouse BMSCs
were performed as described previously by Cai et al
(29). Briefly, mice were
sacrificed through cervical dislocation, the tibia and femur were
separated and the surrounding muscle tissue was removed. The
removed bone tissue was placed in PBS with 100 U/ml penicillin and
100 mg/l streptomycin (the double antibody, and both ends of the
tibia and femur were removed. Bone marrow was flushed out using
serum-free α-Minimal Essential Medium (α-MEM; Gibco; Thermo Fisher
Scientific, Inc.). The collected media were centrifuged at 278 x g
for 10 min at 37˚C, resuspended in α-MEM supplemented with 10% FBS
(cat. no. S601P-500; Sera Pro), 100 U/ml penicillin and 100 mg/l
streptomycin. Cells were incubated at 37˚C in 5% CO2 for
72 h, and half of the growing medium was replaced every 12 h. Fresh
medium was added and was replaced every 2 days thereafter. BMSCs
were then expanded for a maximum of four passages, trypsinized and
seeded at a density of ~1x106 cells per well in six-well
plates, followed by adipogenic or osteogenic differentiation
analysis.
To induce BMSCs to differentiate into osteoblasts,
cells were cultured in α-MEM medium and osteogenic differentiation
inducer (ODI) containing 50 mg/ml L-ascorbic acid (cat. no.
A4544-25G), 10 mM β-glyceryl phosphate (cat. no. 50020) and 100 nM
dexamethasone (cat. no. D1756) (all Sigma-Aldrich; Merck KGaA) was
used. Control cells were cultured in α-MEM medium containing 10%
FBS and 1% penicillin/streptomycin only. unOC-treated cells were
cultured in osteogenic differentiation medium containing 3 ng/ml of
unOC. BMSCs were divided into four groups: Normal control group
(NG), unOC-treated group, NG + ODI group and unOC + ODI group. All
cells were incubated at 37˚C and 5% CO2 for 3 weeks.
The third-generation BMSCs were used to detect the
adipogenic differentiation potential of the cells. The composition
of the adipogenic differentiation inducer (ADI) was as follows: 1
mmol/l Dexamethasone, 0.5 mmol/l 3-isobutyl-1-methylxanthine, 10
mg/l insulin and 0.2 mmol/l indomethacin (all Sigma-Aldrich; Merck
KGaA). The BMSCs with ADI were placed in a CO2
constant-temperature incubator, the cells were cultured at 37˚C and
5% CO2 for 3 weeks, and the medium was changed every
day.
Alizarin red staining and oil red O
staining
After the BMSCs were cultured under different
osteoblast differentiation conditions for 3 weeks, cells were
washed twice with PBS, fixed with 10% neutral formaldehyde at 37˚C
for 30 min, and washed twice with PBS before subjected to staining
with Alizarin Red S (cat. no. G3281; Beijing Solarbio Science &
Technology Co., Ltd.) at 37˚C for 30 min. Optical microscope (Leica
Microsystems GmbH) was used to capture high-resolution images of
the stained calcium nodules (light, x40 magnification). In order to
quantify the calcium content in each sample, Alizarin Red was
eluted with 10% (wt/vol) cetylpyridinium chloride (cat. no. C9890;
Beijing Solarbio Science & Technology Co., Ltd.), and the
optical density of the eluate was determined using an automatic
microplate reader (BioTek China) at a wavelength of 540 nm.
After culturing with ADI for 3 weeks, the lipid
droplets in the cells were measured using Oil red O (ORO) staining.
The ORO dye was prepared by slowly adding 0.1 g Oil red O
(Sigma-Aldrich; Merck KGaA) to 100 ml 60% isopropanol while mixing
the solution continuously, followed by heating for 5 min at 60˚C,
filtering and diluting with deionized water before use (Oil red O
saturated liquid: Deionized water, 3:2). The treated cells were
fixed with 10% paraformaldehyde (Biosharp Life Sciences) at 37˚C
for 30 min. The excess liquid was removed and 0.5% ORO dye solution
was added for 20 min at 37˚C. Before microscopic examination, a
small amount of 75% ethanol was used to wash out excess dye if
necessary. Lipid droplet images were obtained using a
high-resolution optical microscope (Leica Microsystems GmbH)
(light, x40 magnification). The dye was extracted with 100%
isopropanol and the absorbance was measured at 500 nm with an
automatic microplate reader (BioTek China).
Cell transfection
Transfection was performed when mouse BMSCs were
passaged to the third generation and 60-80% of the adherent cells
were confluent. Small interfering RNA (siRNA) targeting SIRT1
(forward 5'-CCCUCAAGCCAUGUUUGAUTT-3' and reverse
5'-AUCAAACAUGGCUUGAGGGTT-3') and negative control (NC; forward
5'-UUCUCCGAACGUGUCACGUTT-3' and reverse
5'-ACGUGACACGUUCGGAGAATT-3') siRNA were purchased from JTS
Scientific (30). The overall
transfection procedure was in accordance with the recommendations
of the manufacturer. Lipofectamine® 3000 (cat. no.
L3000015; Invitrogen; Thermo Fisher Scientific, Inc.) was used to
transfect 20 nM of the siRNA and the NC siRNA into the cells at
37˚C. The cells were continuously transfected in medium with or
without unOC until they were used for experiments. Finally, protein
samples or RNA were collected for further experiments. The time
interval between transfection and subsequent experiments was 48
h.
RNA preparation and reverse
transcription-quantitative (RT-q)PCR analysis
Total cellular RNA was extracted using 1 ml
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) per 10-cm plate. Total RNA was reverse transcribed into cDNA
using TransScript® One-Step gDNA Removal and cDNA
Synthesis SuperMix (cat. no. A T311-03; Beijing Transgen Biotech
Co., Ltd.) and TransStart® Top Green qPCR SuperMix (+Dye
II) for qPCR (cat. no. AQ132-24; Beijing Transgen Biotech Co.,
Ltd.). Full temperature protocol for RT: The product after cDNA
synthesis was incubated at 42˚C for 15 min, and then heated at 85˚C
for 5 sec to inactivate excess reagents. The thermocycling
conditions were 95˚C for 5 min, then 40 cycles of 94˚C for 30 sec,
56˚C for 30 sec and 72˚C for 30 sec. Gapdh served as the
internal standard. The expression levels of target genes were
normalized to the expression of Gapdh mRNA. The following
primers were used to determine the expression levels of proteins
associated with osteogenic differentiation: Fatty acid-binding
protein 4 (Fabp4) forward 5'-AAGTGGGAGTGGGCTTTG-3', and
reverse 5'-GTCGTCTGCGGTGATTTC-3'; fatty acid synthase (Fas)
forward 5'-TGCTTGCTGGCTCACAGTTAAGAG-3', and reverse
5'-TCAGGTTGGCATGGTTGACAGC-3'; osterix (Osx) forward
5'-CTAGTTCCTATGCTCCGACC-3', and reverse 5'-TCATCACATCATCATCGTG-3';
alkaline phosphatase (Alp) forward
5'-CAAAGGCTTCTTCTTGCTGGT-3', and reverse
5'-AAGGGCTTCTTGTCCGTGTC-3'; and Gapdh forward
5'-GGCATTGCTCTCAATGACAA-3', and reverse
5'-TGTGAGGGAGATGCTCAGTG-3'.
Immunoprecipitation
The cell culture medium was aspirated and washed
three times with ice-cold 4˚C PBS. Afterward, RIPA lysis buffer
(cat. no. R0020; Beijing Solarbio Science & Technology Co.,
Ltd.) was added to lyse the cells. Detection of protein
concentration: BCA Protein Assay kit (Beyotime Institute of
Biotechnology). A total of 200 µg protein samples and 20 µg protein
A+G Agarose beads (cat. no. P2012; Beyotime Institute of
Biotechnology) were used. The mixture was shaken slowly at 4˚C,
then centrifuged at 1,000 x g for 5 min and the supernatant was
collected for subsequent immunoprecipitation experiments. In total,
2 µg the primary antibody, including anti-RUNX2 (cat. no. 12556;
Cell Signaling Technology, Inc.), anti-PPARγ (cat. no. 2443; Cell
Signaling Technology, Inc.), anti-acetylated-lysine (cat. no. 9441;
Cell Signaling Technology, Inc.) was added and shaken slowly
overnight at 4˚C. Afterwards, completely resuspended 40 µg Protein
A + G Agarose was added and the mixture was slowly shaken at 4˚C
for 3 h. Centrifuge at 278 x g for 5 min at 4˚C for washing. After
the last wash, the supernatant was removed, 5X SDS-PAGE
electrophoresis loading buffer was added, and the resulting pellet
was resuspended and vortexed. The sample was centrifuged to the
bottom of the tube by instantaneous high-speed centrifugation.
After 5 min incubation in a 100˚C water bath, some or all samples
were collected for western blotting.
Western blotting
BMSCs were washed three times with ice-cold PBS, and
lysed with RIPA (cat. no. R0010; Beijing Solarbio Science &
Technology Co., Ltd.) buffer. Lysate was crushed with an ultrasonic
cell disruptor and the supernatant was reserved. Conditions used
for cell were 300 W sound waves, duration of 3 min with, 3-sec
intervals at 0˚C. The protein concentrations were determined using
a Beyotime BCA Protein Assay kit (Beyotime Institute of
Biotechnology). Afterward, 20 µg cell lysate (each electrophoresis
lane) was separated using a 10% SDS-PAGE gel and transferred to a
PVDF membrane. After blocking with 5% milk at 37˚C for 2 h, the
membrane was incubated with primary antibodies (1:1,1000) at 4˚C
for 12 h. The membrane was washed three times using Tris-buffered
saline with 0.1% Tween-20 (TBST) on a shaker for 10 min, and then
incubated with secondary antibody at 37˚C for 1 h. After washing
three times with TBST, 10 min each time, bands were detected with
Immobilon Western Chemilum HRP Substrate (EMD Millipore). The
band's optical density value was quantified using Image Lab
Software for PC Version 6.1 (Bio-Rad Laboratories, Inc.). The
protein quantity was calculated as the optical density value of the
protein measured/the optical density value of an internal
reference.
Antibodies used in western blot analysis included
the following: Anti-β-actin (cat. no. CPA9066; Cohesion
Biosciences, Ltd.), anti-SIRT1 (cat. no. 3931), anti-RUNX2 (cat.
no. 12556), anti-PPARγ (cat. no. 2443), anti-CCAAT-enhancer-binding
protein alpha (C/EBPα; cat. no. 2295), anti-acetylated-lysine (cat.
no. 9441), anti-PKA (cat. no. 4782), anti-phosphorylated (p)-PKA
(cat. no. 4781), anti-AMPK (cat. no. 5831) and anti-p-AMPK (cat.
no. 2535) (all Cell Signaling Technology, Inc.). The secondary
antibody used was Goat anti-Rabbit IgG (H+L)-HRP (cat. no.
S0101-100; Beijing Lamblide Trading Co., Ltd.) at a dilution of
1:5,000.
Signaling pathway inhibition
Cells were pretreated with or without 10 µM H89 (a
potent inhibitor of PKA; cat. no.T6250) (31) or 10 µM Dorsomorphin (an effective
inhibitor of AMPK; cat. no. T1977; both TargetMol) (32) for 1 h, then cultured for 48 h with
or without unOC.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Statistical analyses for two groups were performed using the
two-tailed paired Student's t-test. Examination of more than two
groups was conducted using one-way ANOVA using SPSS software
(version 19.0; IBM Corp.). Bonferroni's correction method was
performed as a post hoc test after one-way ANOVA. P<0.05 was
considered to indicate a statistically significant difference.
Results
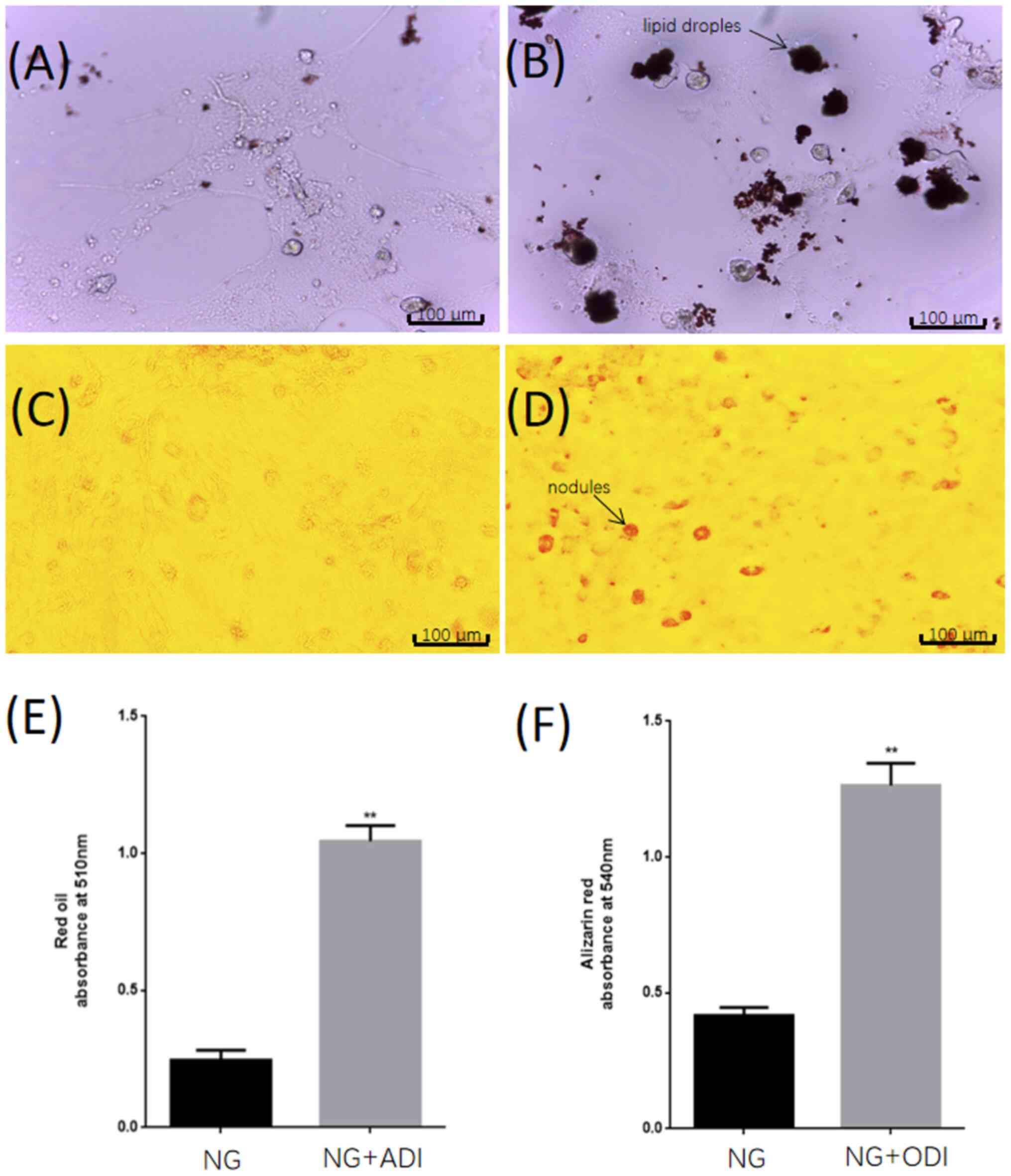
ODI induces osteogenic differentiation
of BMSCs
First, the osteogenic differentiation potential of
BMSCs was determined. In contrast to undifferentiated BMSCs,
differentiated osteoblasts accumulate a large amount of
extracellular calcium deposits (mineralization). This process is
accompanied by the formation of bone nodules (33). Osteoblast-mediated mineralization is
therefore indicative of the formation of bone mass and can be
specifically detected using Alizarin Red S (34,35).
Confluent fourth passage cells were cultured in medium containing
ODI and control cells were cultured in the same medium without ODI.
After 3 weeks, cells were stained with Alizarin Red to gauge the
degree of osteogenic differentiation. The ODI-treated group was
observed to have more calcium nodules compared with the control
group (Fig. 1C and D). In addition, the NG + ADI group showed
more lipid droplets than the NG group (Fig. 1A and B). The quantitative results were
statistically significant, indicating that BMSCs had the potential
for osteogenic differentiation and adipogenic differentiation
(Fig. 1E and F).
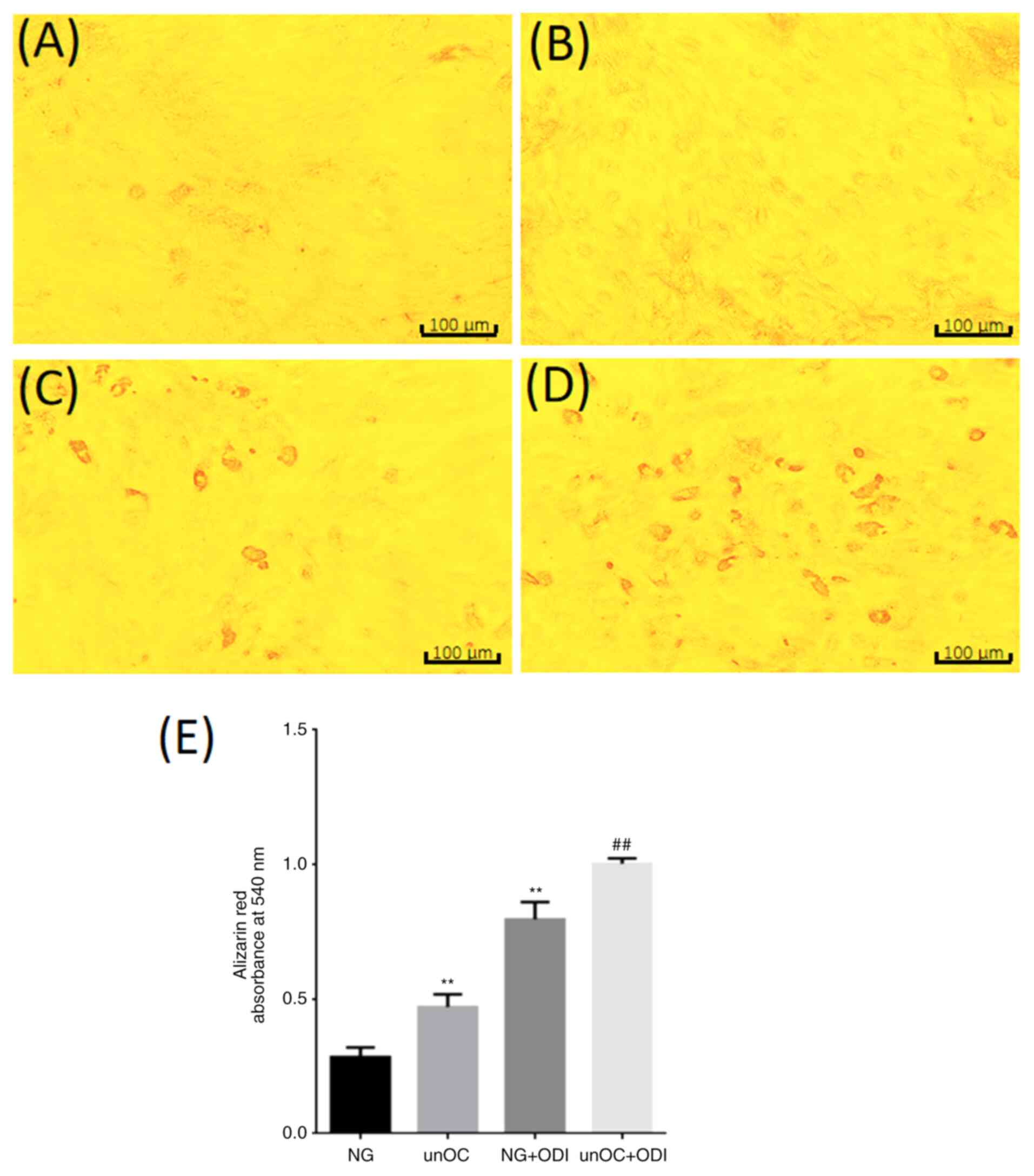
unOC promotes osteogenic
differentiation of BMSCs and inhibits adipogenic differentiation
through SIRT1 deacetylation of RUNX2 and PPARγ unOC promotes
osteogenic differentiation of BMSCs
To determine the effect of unOC on osteogenic
differentiation, the BMSCs were divided into four groups: NG, unOC,
NG + ODI And unOC + ODI. Alizarin Red staining was performed after
3 weeks of culture. It was found that the unOC group had more
calcium nodules compared with the NG group (Fig. 2A and B). Similar results were observed in the NG
+ ODI group, indicating that unOC can function in the same manner
as ODI to promote the differentiation of BMSCs into osteoblasts
(Fig. 2C). In addition, the number
of calcium nodules in the unOC + ODI treatment group was higher
compared with that in the NG + ODI group (Fig. 2D). Quantification of Alizarin
Red-positive areas indicated that significant increase in unOC and
unOC + ODI groups compared with that in the NG group (Fig. 2E), suggesting that unOC can promote
and facilitate the differentiation of mouse BMSCs into
osteoblasts.
unOC promotes osteogenic
differentiation of BMSCs and inhibits adipogenic differentiation
through SIRT1
To assess whether unOC-induced osteogenic
differentiation of BMSCs was mediated by SIRT1, transient
transfection analysis was performed using SIRT1 siRNA and RT-qPCR
analysis was used to determine that expression levels of two
differentiation-related factors, ALP and OSX. Cells were divided
into four groups: NG + NC, unOC Treatment + NC group, SIRT1 siRNA
group and SIRT1 siRNA and unOC treatment group (unOC + SIRT1).
RT-qPCR analysis showed that the expression levels of Sirt1,
Alp and Osx significantly increased in the unOC + NC
treatment group compared with the NG + NC group (Fig. 3A-C). In contrast, the expression
levels of Alp and Osx significantly decreased in the
SIRT1 siRNA transient transfected group compared with the NG + NC
group. Moreover, their expression could not be recovered even when
unOC was applied, suggesting that unOC promotes the osteogenic
differentiation of mouse BMSCs through SIRT1 and that unOC cannot
counteract the effects of SIRT1 siRNA.
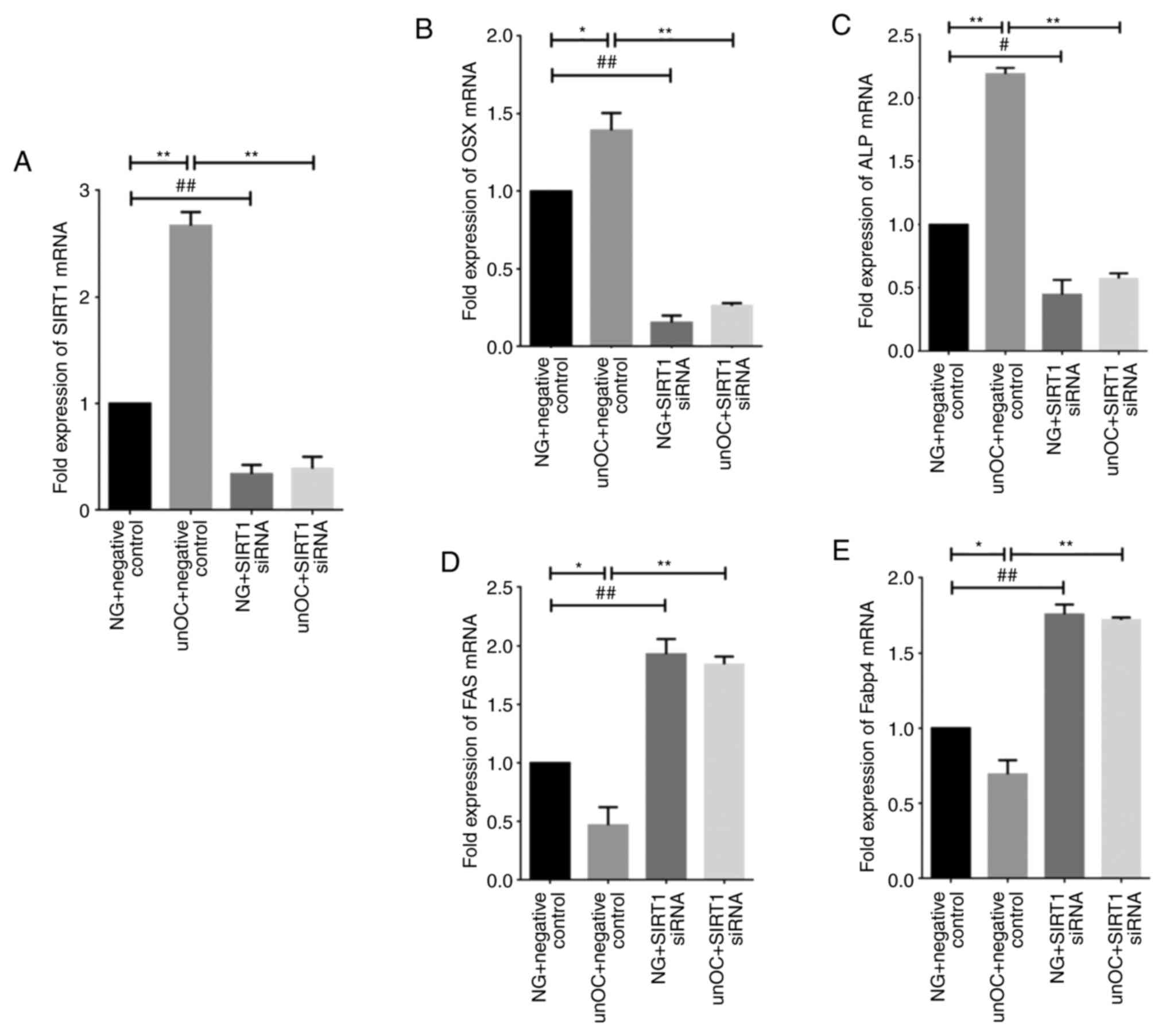 | Figure 3unOC promotes osteogenic
differentiation and inhibits adipogenic differentiation of BMSCs
through SIRT1. RT-qPCR analysis of total cellular RNA from four
cell culture groups: NG + NC Group, unOC + NC group, SIRT1 siRNA
group and unOC + SIRT1 siRNA treatment group. (A) mRNA expression
levels of SIRT1 in the different groups. RT-qPCR was used to
determine the mRNA expression levels of (B) OSX, (C) ALP, (D) FAS
and (E) Fabp4. Experiments were repeated at least three times.
*P<0.05, **P<0.01 vs. unOC + NC;
#P<0.05, ##P<0.01 vs. NG + SIRT1 siRNA.
unOC, uncarboxylated osteocalcin; BMSCs, bone marrow-derived
mesenchymal stem cells; SIRT1, sirtuin 1; RT-qPCR, reverse
transcription-quantitative PCR; NG, normal group; NC, negative
control; si-small interfering; FAS, fatty acid synthase; Fabp4,
fatty acid-binding protein 4; Osx, osterix; ALP, alkaline
phosphatase. |
To determine whether unOC inhibits adipogenic
differentiation through SIRT1, two notable factors associated with
adipogenic differentiation were examined, FAS and FABP4. It was
observed that Fas and Fabp4 expression levels
decreased in the unOC treatment group, indicating that unOC
inhibited the adipogenesis of mouse BMSCs (Fig. 3D and E). The expression levels of Fas and
Fabp4 significantly increased in the SIRT1 siRNA-transfected
group compared with those in the NG + NC group. After knocking down
SIRT1, this increase was not reversible even after reintroducing
unOC, indicating that the inhibition of the adipogenic
differentiation by unOC was mediated by SIRT1.
unOC regulates BMSC differentiation
through SIRT1 deacetylation of RUNX2 and PPARγ
To determine whether the ability of unOC to regulate
osteogenic differentiation was mediated through the acetylation
function of SIRT1, SIRT1's downstream targets, RUNX2 and PPARr,
were examined. Cells were divided into four groups: NG + NC, unOC +
NC, SIRT1 siRNA And SIRT1 siRNA + unOC. Western blot analysis was
performed in the total cell lysates for each group. It was found
that unOC treatment significantly reduced the acetylation levels of
PPARγ and RUNX2 compared with those in the NG + NC group. By
contrast, their acetylation levels increased after SIRT1-knockdown
(Fig. 4A, B, D and
E). The acetylation states of PPARγ
and RUNX2 have opposite effects on their activity and the PPARγ
activity is enhanced in the acetylation state (22,36).
Therefore, SIRT1 can reduce the activity of PPARγ and inhibit
adipogenic differentiation. The results of the present study
suggested that unOC deacetylated PPARγ through SIRT1 and
downregulated the activity of PPARγ, thereby inhibiting the
differentiation of BMSCs into adipocytes.
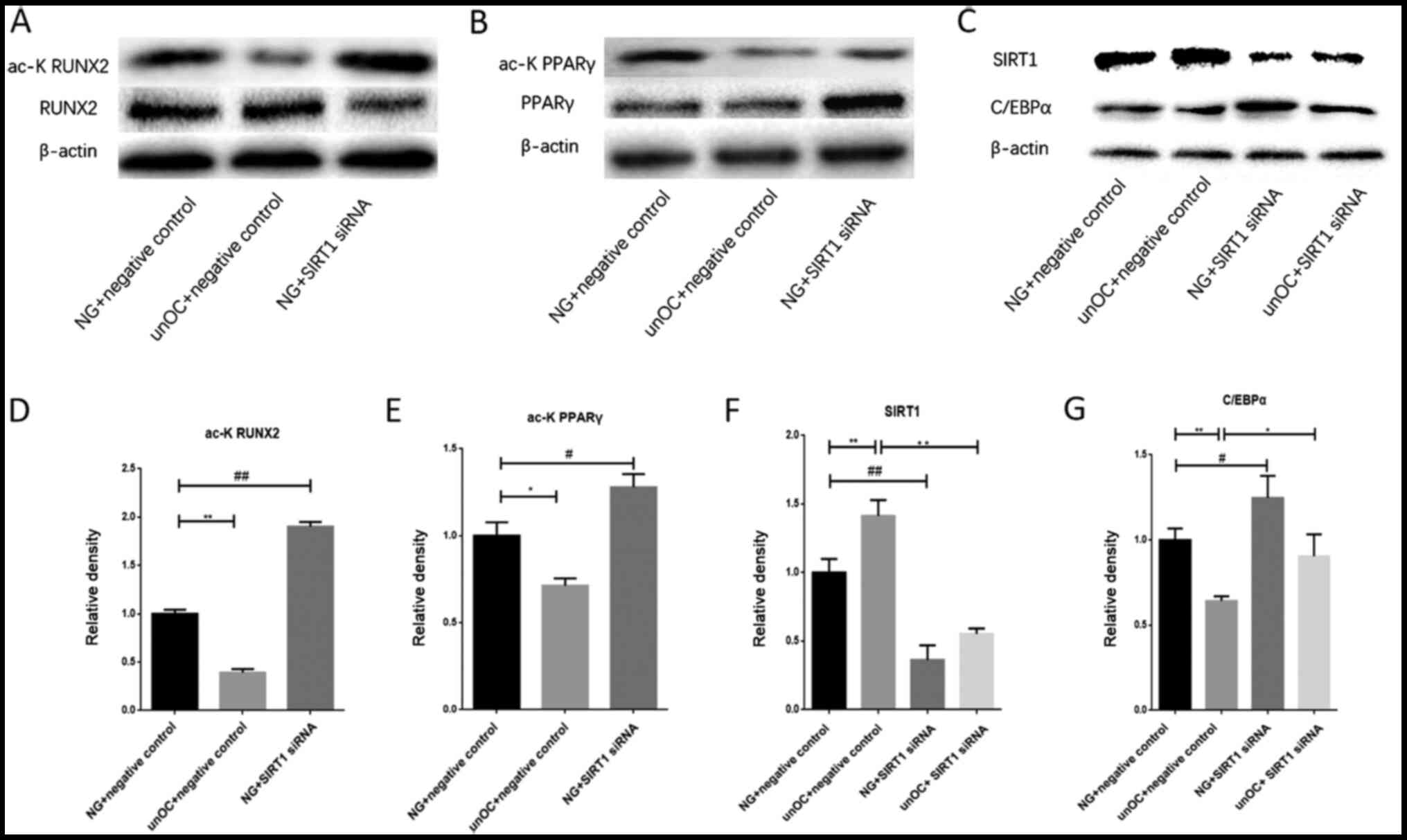 | Figure 4unOC regulates the differentiation of
BMSCs through SIRT1 deacetylation of RUNX2 and PPARγ. Western blot
analysis of the total and acetylated (A) RUNX2 and (B) PPARγ. (C)
Western blot analysis of SIRT1 and C/EBPa protein expression levels
in the cell lysates of the four treatment groups after transfection
with the lentiviral vector or vector control. Ratios of acetylated
protein:total protein of (D) RUNX2 and (E) PPARγ. Gray value
analysis of (F) SIRT1 and (G) C/EBPα. Experiments were repeated at
least three times. *P<0.05, **P<0.01
vs. unOC + NC; #P<0.05, ##P<0.01 vs. NG
+ SIRT1 siRNA. unOC, uncarboxylated osteocalcin; BMSCs, bone
marrow-derived mesenchymal stem cells; SIRT1, sirtuin 1; RUNX2,
runt-related transcription factor 2; PPARγ, peroxisome
proliferator-activated receptor γ; C/EBPa, CCAAT-enhancer-binding
protein α; NG, normal group; NC, negative control; si-small
interfering. C/EBPα, CCAAT-enhancer-binding protein a. |
To further confirm that unOC inhibited the
adipogenic differentiation of BMSCs by upregulating SIRT1, the
adipogenic differentiation-related factor, C/EBPα, was analyzed and
its expression was examined in four groups of cells: NG + NC, unOC
+ NC, SIRT1 siRNA And SIRT1 siRNA + unOC. It was found that
SIRT1-knockdown resulted in marked increases in the C/EBPα protein
abundance (Fig. 4C) and mRNA
expression levels (Fig. 4G) as
compared with those in the control (NG + NC) group. unOC + NC
treatment significantly decreased the expression level of C/EBPα
compared with the NG + NC group and the SIRT1 siRNA group (Fig. 4C and G). There was no significant difference in
C/EBPα expression between the unOC + SIRT1 siRNA treatment group
and the SIRT1 siRNA group. The protein expression levels of SIRT1
markedly increased in the unOC + NC treatment group compared with
those in the NG + NC group (Fig. 4C
and F). By contrast, the expression
levels of SIRT1 were markedly decreased in the SIRT1 siRNA group
compared with those in the NG + NC group. There was no significant
difference between the unOC + SIRT1 siRNA treatment group and the
NG + SIRT1 siRNA group. Collectively, the data suggested that the
regulation of SIRT1 by unOC in BMSCs inhibited adipogenic
differentiation and, thus, promoted osteogenic differentiation.
unOC promotes osteogenic
differentiation of BMSCs through PKA-AMPK-SIRT1 unOC can upregulate
the expression levels of p-PKA, p-AMPK and SIRT1
As PKA and AMPK regulate SIRT1 in cell
differentiation (37,38), the PKA-AMPK pathway was examined to
find if it was required for unOC to promote osteogenic
differentiation in BMSCs. The PKA inhibitor, H89, was used to
detect the signaling pathway through which unOC regulated the
differentiation of mouse BMSCs. It was found that unOC treatment
significantly increased the protein level of p-PKA, p-AMPK and
SIRT1 in BMSCs compared with their expression in the NG group
(Fig. 5A-D). In contrast, the
increased expression levels were completely abolished and even
reversed by H89, while the expression levels of PKA and AMPK
remained the same, indicating that unOC positively regulated the
PKA-AMPK-SIRT1 pathway by upregulating p-PKA and p-AMPK.
Furthermore, simultaneous treatment with H89 and unOC restored the
expression of p-PKA, p-AMPK and SIRT1 (Fig. 5). Collectively, the data indicated
that unOC functioned upstream of the PKA-AMPK-SIRT1 pathway,
thereby promoting the expression levels of p-AMPK and SIRT1.
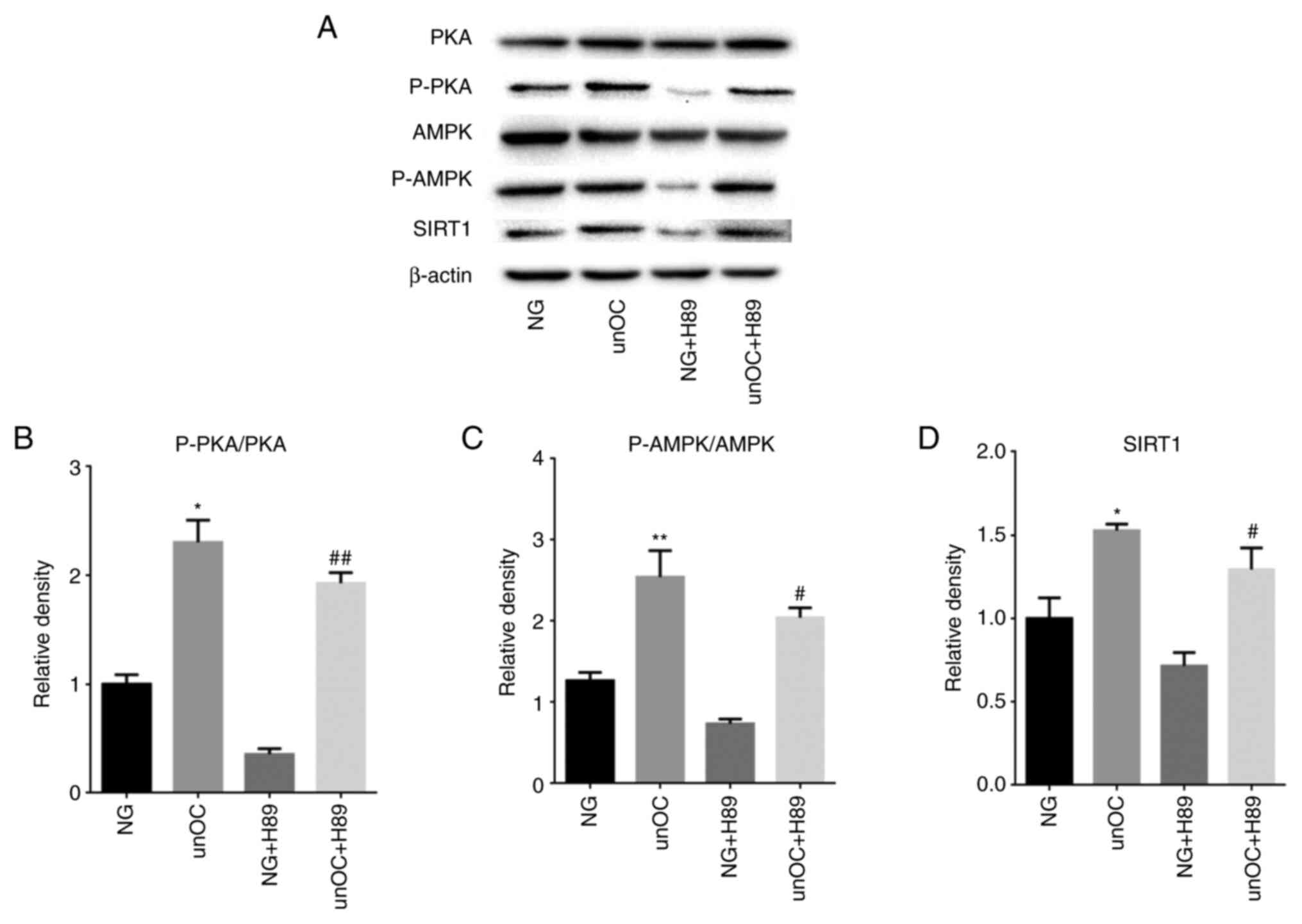 | Figure 5unOC upregulates the expression of
p-PKA and p-AMPK and SIRT1. Cells were divided into the NG,
unOC-treated, PKA inhibitor H89 and PKA inhibitor H89 + unOC
groups. (A) Western blot analysis to detect the levels of total
PKA, total AMPK, p-AMPK, p-PKA and SIRT1. p-protein was normalized
with the corresponding total protein, and the ratio was calculated
for (B) PKA and (C) AMPK. (D) Quantified SIRT1 protein levels.
Experiments were repeated at least three times.
*P<0.05, **P<0.01 vs. NG;
#P<0.05, ##P<0.01 vs. NG + H89. unOC,
uncarboxylated osteocalcin; BMSCs, bone marrow-derived mesenchymal
stem cells; SIRT1, sirtuin 1; NG, normal group; p-phosphorylated;
PKA, protein kinase A; AMPK, AMP-activated protein kinase. |
unOC regulates the differentiation of
BMSCs through the PKA-AMPK-SIRT1 pathway
To study the unOC-regulated PKA signaling pathways
involved in the differentiation of mouse BMSCs, the AMPK inhibitor,
Dorsomorphin, was examined using western blot analysis. Compared
with the NG group, the levels of p-AMPK, p-PKA and SIRT1 were
significantly increased in the unOC group, but there was no obvious
trend in total PKA (Fig. 6). This
change also appeared in the comparison between the NG +
Dorsomorphin group and the unOC + Dorsomorphin group (Fig. 6). However, in the NG + Dorsomorphin
group compared with the NG group, or the unOC + Dorsomorphin group
compared with the unOC group, there was no statistical difference
between P-PKA and total PKA. This is related to Dorsomorphin only
inhibiting AMPK, but also indicates that AMPK is a downstream
regulator of PKA (Fig. 6). After
treating cells with Dorsomorphin, the phosphorylation of AMPK and
SIRT1 in the NG + Dorsomorphin decreased compared with that in the
NG group, but that of PKA did not change significantly (Fig. 6B-D). In addition, after adding
Dorsomorphin and unOC to the BMSCs, both p-AMPK and SIRT1
expression levels were restored to similar levels of the NG group.
These results indicated that unOC upregulated the expression of
PKA, increased the phosphorylation level of the downstream target
AMPK and promoted the expression of SIRT1. Through the
aforementioned experiments, it was concluded that unOC promoted the
osteogenic differentiation of mouse BMSCs through the
PKA-AMPK-SIRT1 pathway.
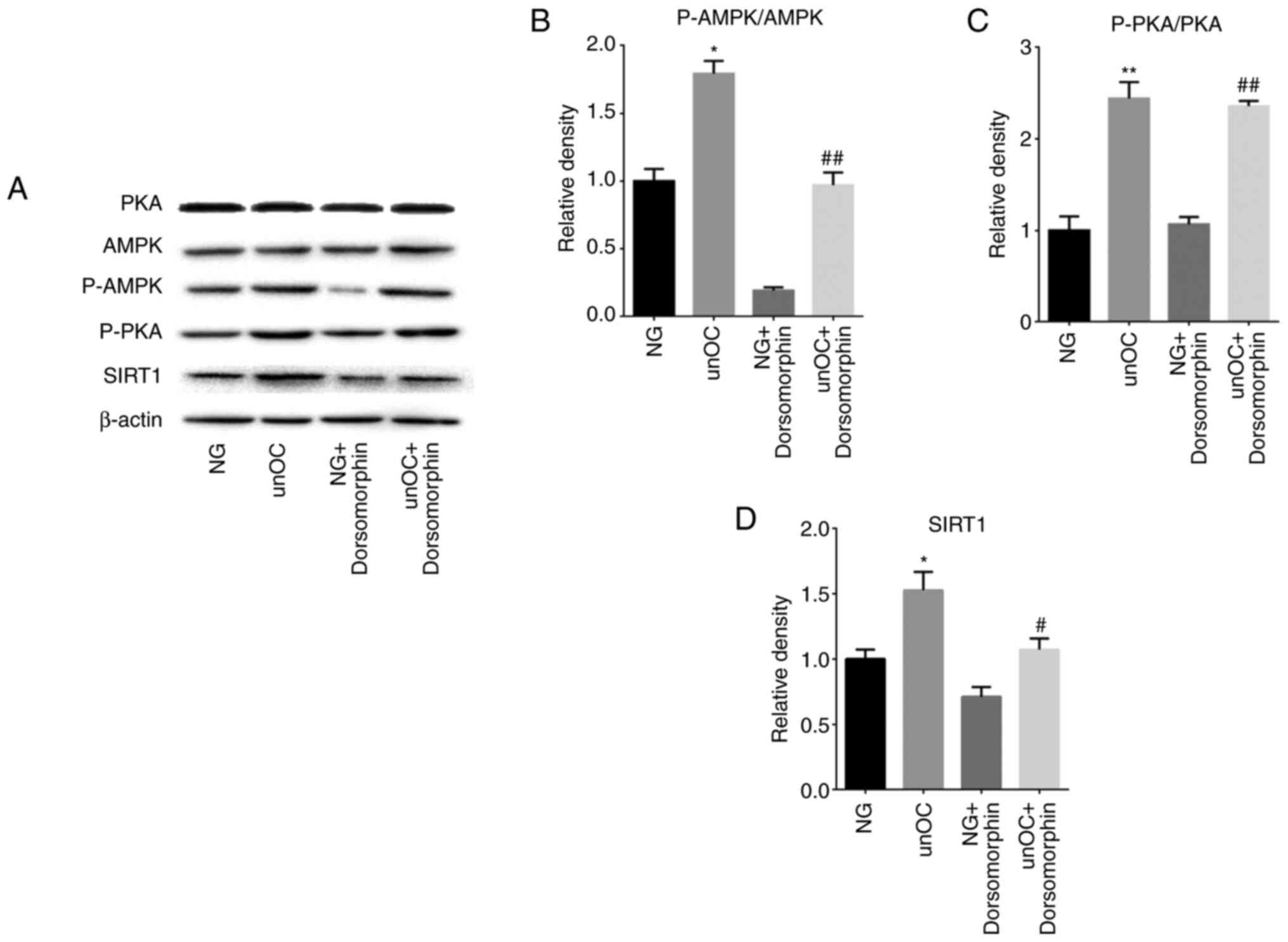 | Figure 6unOC regulates mesenchymal stem cell
differentiation through the PKA-AMPK-SIRT1 axis. Cells were divided
into: NG, unOC-Treated, AMPK inhibitor Dorsomorphin and
Dorsomorphin and unOC groups. (A) Western blot analysis to detect
total AMPK, total PKA, p-PKA, p-AMPK and SIRT1 protein expression
levels after 2 days. P-protein was normalized to the corresponding
total protein, and the ratio was calculated for (B) AMPK and (C)
PKA. (D) Quantified SIRT1 protein levels. Experiments were repeated
at least three times. *P<0.05, **P<0.01
vs. NG; #P<0.05, ##P<0.01 vs. NG +
Dorsomorphin. unOC, uncarboxylated osteocalcin; BMSCs, bone
marrow-derived mesenchymal stem cells; SIRT1, sirtuin 1; NG, normal
group; p-phosphorylated; PKA, protein kinase A; AMPK, AMP-activated
protein kinase. |
Discussion
Previous studies have shown that the differentiation
of BMSCs plays a noteworthy role in osteoporosis treatment as a
therapeutic application (39-41).
BMSCs can differentiate into osteoblasts, which can serve important
functions in the synthesizing bone matrix (42). Osteoblasts, as the key cells for
mediating bone formation, are embedded into the bone tissue after
the mineralization process to eventually become bone cells and form
are one of the most important components of the bone (43). Studies have shown that osteoporosis
is related to the adipogenic differentiation and aging of BMSCs
(40,44). At the same time, promoting the
osteogenic differentiation of BMSCs has predictable help in
treating or reducing osteoporosis (45,46).
Although the regulation and shift of the cell differentiation of
BMSCs to osteoblasts rather than adipocytes is needed for the
treatment of osteoporosis, the molecular mechanism responsible for
this regulation of BMSCs has not been defined (39). Our previous study showed that unOC
can regulate the differentiation shift of BMSCs to osteoblasts
(16), suggesting its role as a
potential treatment candidate for osteoporosis. The current study
provided insights into the underlying mechanisms by identifying the
downstream unOC regulatory pathway and the critical factor,
SIRT1.
SIRT1, a core member of the sirtuin family of
proteins, is an NAD+-dependent deacetylase. The main
function of SIRT1 is to deacetylate histones and non-histone
proteins in an organism (47). It
affects cell proliferation, differentiation, metabolism, autophagy
(48), cancer (49), inflammation, diabetes (50), osteoporosis (51) and other diseases closely associated
with the health and aging of the body (52). It also plays an important role in
regulating the differentiation of BMSCs. BMSC-specific
SIRT1-knockout mice display reduced cell proliferation and
accelerated cell senescence (53).
In contrast, the overexpression of SIRT1 delays senescence in
BMSCs, which extends the cell life span in vitro, and the
cells do not lose the potential for osteogenic and adipogenic
differentiation (53). These
findings uncovered potential therapeutic applications of BMSCs in
tissue engineering (54). SIRT1
also prevents senescence in human BMSCs (53), suggesting a regulatory role for
SIRT1 in the proliferation and differentiation of MSCs in humans.
The present study found that the expression of SIRT1 is positively
regulated by unOC in BMSCs, suggesting that SIRT1 is likely to play
a role in mediating the effect of unOC on BMSC differentiation.
The dynamic balance between bone formation and bone
resorption is the basis of bone remodeling and homeostasis
(55). In addition to regulating
the differentiation of MSCs (56),
SIRT1 is also involved in the regulation of bone metabolism in the
body (57,58). It maintains the normal bone
remodeling process by regulating factors that are associated with
bone metabolism, such as SOX9, PPARγ, NF-κB and RUNX2. SIRT1
deacetylates SOX9 and PPARγ, enhances SOX9 activity and reduces
PPARγ activity, thereby regulating the differentiation of MSCs into
chondrocytes and inhibiting their differentiation into adipocytes
(22). SIRT1 inhibits NF-κB
signaling to maintain normal skeletal remodeling (59).
SIRT1 is known to play a notable role in bone
metabolism by promoting osteoblast differentiation, inhibiting
osteoclast formation and reducing osteolysis (57,59).
First, SIRT1 promotes osteogenic differentiation and inhibits
adipogenic differentiation in human dental pulp stem cells by
regulating the Wnt/β-catenin signaling pathway (20,60).
Second, SIRT1 is a positive regulator of the osteoblast
transcription factor, RUNX2. RUNX2 is one of the key osteogenic
cytokines (61,62). RUNX2-deficient mice exhibit a
complete absence of mature osteoblasts and obvious defects in bone
mineralization (63). Zainabadi
et al (21) found that
SIRT1-deficient mice had lower expression levels of RUNX2
downstream targets, including osteopontin and OSX, and decreased
osteogenic differentiation. In contrast, MSCs treated with SIRT1
agonists show decreased RUNX2 acetylation, increased expression
levels of RUNX2 targets and enhanced osteoblast differentiation.
Thirdly, SIRT1 inhibits the osteoclast process by regulating the
NF-κB signaling pathway. NF-κB is a type of nuclear transcription
factor associated with cellular immunity, apoptosis and
differentiation. The activation of NF-κB promotes the receptor
activator of nuclear factor-κΒ ligand-induced osteoclast formation
(64). SIRT1 and NF-κB have a
mutual inhibitory relationship. The SIRT1 activator, resveratrol,
activates the expression of NF-κB inhibitory protein α.
Furthermore, SIRT1 directly inhibits NF-κB signaling by
deacetylating the p65 subunit of the NF-κB complex and inhibits its
accumulation in the nucleus (65).
Thus, the inhibition of NF-κB signaling reduces the formation of
osteoclasts and adipocytes and increases the number of osteoblasts,
which further reduces bone resorption, increases bone formation,
restores bone mass and corrects bone metabolism imbalance (51). In contrast, NF-κB downregulates
SIRT1 activity through decreased intracellular NAD+
levels (66). Thus, both NF-κB and
SIRT1 restrict each other and participate in the regulation of
osteogenic differentiation and bone metabolism.
In summary, SIRT1 has potential for restoring bone
metabolism and treating osteoporosis by regulating the osteogenic
differentiation of MSCs. The findings of the present study are
consistent with previous reports that SIRT1 deacetylates RUNX2 and
PPARγ and regulates their activity (67). unOC treatment was also found to
upregulate SIRT1 expression, which lead to increased deacetylation
of RUNX2 and PPARγ. According to a previous study, the activity of
RUNX2 was enhanced after deacetylation whereas the activity of
deacetylated PPARγ was reduced, thereby promoting the
differentiation of BMSCs into osteoblasts and inhibiting their
differentiation into adipocytes (21,22).
unOC promotes BMSC differentiation and inhibits
adipogenesis through SIRT1, which may serve as a novel target for
osteoporosis treatment (68).
Studies have shown that unOC has a wide range of biological
functions. It is involved in vital life processes, such as
differentiation, metabolism and cognition (69-73).
unOC also plays an active role in the regulation of bone metabolism
(74), cardiovascular disease
(75) and melanoma (76).
Multiple lines of evidence indicate that unOC is
involved in BMSC differentiation. Studies have shown that mineral
maturation and total hydroxyapatite content are reduced as a result
of decreased levels of the osteocalcin gene in BMSCs (77-79).
This suggests that osteocalcin can be induced by promoting bone
differentiation (80) and is
conducive to the recovery of bone metabolism (81). Our previous studies have shown that
unOC stimulates the differentiation of pre-bone MC3T3-E1 cells
(27) and BMSCs (16) into bone cells. The present study
demonstrated that unOC promoted osteogenic differentiation and
inhibited adipogenic differentiation in BMSCs via the
PKA-AMPK-SIRT1 signaling pathway as the PKA inhibitor, H89,
abolished the upregulation of SIRT1, p-PKA and AMPK via unOC.
Moreover, treatment with Dorsomorphin, an AMPK inhibitor, reversed
the effect of unOC on SIRT1 and AMPK, but not on PKA. These
findings suggested that unOC functioned via the PKA-AMPK-SIRT1
pathway in regulating BMSC differentiation. However, Dorsomorphin
is also an inhibitor to bone morphogenetic protein (BMP), which is
known to be involved in adipogenesis (82,83).
Therefore, it is possible that BMP also plays a role in mediating
the function of unOC on BMSC differentiation. Further studies are
required to confirm this theory.
There are some limitations in the present study.
First of all, the cells used in the present study were extracted
from the femur of C57 mice, which are different from the BMSCs of
humans (84,85), Especially the osteoarthritis of
patients with osteoarthritis will show higher levels of osteocalcin
and type I collagen (86). In
addition, used 3rd to 5th generations of BMSCs were used from
4-week-old mice, where it has been reported that the pathological
characteristics of patients with osteoporosis are significantly
different in the early, middle and late stages (87). Therefore, future research would need
to involve the comparison of the treatment effects of unOC on
osteoporosis at different pathological stages.
The present study provided several lines of evidence
that SIRT1 was a key target of unOC-mediated regulation of BMSC
differentiation into osteoblasts. Firstly, unOC directly shifted
the differentiation potential via SIRT1. SIRT1 was required for
unOC-mediated upregulation of pro-osteogenic differentiation
factors, ALP and OSX, and unOC-induced downregulation of
pro-adipogenic differentiation factors, FAS and Fabp4. Secondly,
unOC shifted the differentiation potential of BMSCs via SIRT1 by
inducing the deacetylation of the downstream targets, RUNX2 and
PPARγ. Finally, unOC upregulated SIRT1 expression through the
PKA-AMPK pathway. Collectively, these data indicated that unOC has
the potential to promote the differentiation of BMSCs into
osteoblasts or tissue regeneration, suggesting its potential in
clinical practice, especially in the treatment of osteoporosis.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from The
Knowledge Innovation Program of The Chinese Academy of Sciences
(grant nos. KSCX2-EW-J-29 and Y129015EA2) and The College of Life
Sciences, University of Chinese Academy of Sciences (grant no.
KJRH2015-006).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LG designed and performed the majority of the
investigation and performed data analysis. FZG and JHY provided
assistance in designing animal experiments. LYM, JHY and FZG
contributed to the interpretation of the data and analyses. All
authors have read and approved the final manuscript. LG and FZG
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by The Institutional
Animal Care and Use Committee of The University of Chinese Academy
of Sciences (Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Peterson JA: Osteoporosis overview.
Geriatr Nurs. 22:17–21. 2001.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang C, Meng H, Wang X, Zhao C, Peng J and
Wang Y: Differentiation of bone marrow mesenchymal stem cells in
osteoblasts and adipocytes and its role in treatment of
osteoporosis. Med Sci Monit. 22:226–233. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kiernan J, Davies JE and Stanford WL:
Concise review: Musculoskeletal stem cells to treat age-related
osteoporosis. Stem Cells Transl Med. 6:1930–1939. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Fu X, Liu G, Halim A, Ju Y, Luo Q and Song
AG: Mesenchymal stem cell migration and tissue repair. Cells.
8(784)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mo J, Yang R, Li F, He B, Zhang X, Zhao Y,
Shen Z and Chen P: Geraniin promotes osteogenic differentiation of
bone marrow mesenchymal stem cells (BMSCs) via activating
β-catenin: A comparative study between BMSCs from normal and
osteoporotic rats. J Nat Med. 73:262–272. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Luo Y, Zhang Y, Miao G, Zhang Y, Liu Y and
Huang Y: Runx1 regulates osteogenic differentiation of BMSCs by
inhibiting adipogenesis through Wnt/β-catenin pathway. Arch Oral
Biol. 97:176–184. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ganguly P, El-Jawhari JJ, Giannoudis PV,
Burska AN, Ponchel F and Jones EA: Age-related changes in bone
marrow mesenchymal stromal cells: A potential impact on
osteoporosis and osteoarthritis development. Cell Transplant.
26:1520–1529. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Baker N, Boyette LB and Tuan RS:
Characterization of bone marrow-derived mesenchymal stem cells in
aging. Bone. 70:37–47. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen Q, Shou P, Zheng C, Jiang M, Cao G,
Yang Q, Cao J, Xie N, Velletri T, Zhang X, et al: Fate decision of
mesenchymal stem cells: Adipocytes or osteoblasts? Cell Death
Differ. 23:1128–1139. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ducy P: The role of osteocalcin in the
endocrine cross-talk between bone remodelling and energy
metabolism. Diabetologia. 54:1291–1297. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zoch ML, Clemens TL and Riddle RC: New
insights into the biology of osteocalcin. Bone. 82:42–49.
2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Iwamoto J, Takeda T and Sato Y: Effects of
vitamin K2 on osteoporosis. Curr Pharm Des. 10:2557–2576.
2004.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Berezovska O, Yildirim G, Budell WC,
Yagerman S, Pidhaynyy B, Bastien C, van der Meulen MCH and Dowd TL:
Osteocalcin affects bone mineral and mechanical properties in
female mice. Bone. 128(115031)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Neve A, Corrado A and Cantatore FP:
Osteocalcin: Skeletal and extra-skeletal effects. J Cell Physiol.
228:1149–1153. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Le VD and Marcil V: Osteocalcin and
glucose metabolism: Assessment of human studies. Med Sci (Paris).
33:417–422. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu Z and Yang J: Uncarboxylated
osteocalcin promotes osteogenic differentiation of mouse bone
marrow-derived mesenchymal stem cells by activating the
Erk-Smad/β-catenin signalling pathways. Cell Biochem Funct.
38:87–96. 2020.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Choi SM, Lee KM, Ryu SB, Park YJ, Hwang
YG, Baek D, Choi Y, Park KH, Park KD and Lee JW: Enhanced articular
cartilage regeneration with SIRT1-activated MSCs using
gelatin-based hydrogel. Cell Death Dis. 9(866)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen Y, Zhou F, Liu H, Li J, Che H, Shen J
and Luo E: SIRT1, a promising regulator of bone homeostasis. Life
Sci. 269(119041)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Simic P, Zainabadi K, Bell E, Sykes DB,
Saez B, Lotinun S, Baron R, Scadden D, Schipani E and Guarente L:
SIRT1 regulates differentiation of mesenchymal stem cells by
deacetylating β-catenin. EMBO Mol Med. 5:430–440. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhou Y, Song T and Peng J, Zhou Z, Wei H,
Zhou R, Jiang S and Peng J: SIRT1 suppresses adipogenesis by
activating Wnt/β-catenin signaling in vivo and in vitro.
Oncotarget. 7:77707–77720. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zainabadi K, Liu CJ and Guarente L: SIRT1
is a positive regulator of the master osteoblast transcription
factor, RUNX2. PLoS One. 12(e0178520)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Qu P, Wang L, Min Y, McKennett L, Keller
JR and Lin PC: Vav1 regulates mesenchymal stem cell differentiation
decision between adipocyte and chondrocyte via Sirt1. Stem Cells.
34:1934–1946. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ha J, Guan KL and Kim J: AMPK and
autophagy in glucose/glycogen metabolism. Mol Aspects Med.
46:46–62. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang Y, Chen G, Yan J, Chen X, He F, Zhu
C, Zhang J, Lin J, Pan G, Yu J, et al: Upregulation of SIRT1 by
kartogenin enhances antioxidant functions and promotes osteogenesis
in human mesenchymal stem cells. Oxid Med Cell Longev.
2018(1368142)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chen H, Liu X, Chen H, Cao J, Zhang L, Hu
X and Wang J: Role of SIRT1 and AMPK in mesenchymal stem cells
differentiation. Ageing Res Rev. 13:55–64. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shelly M, Cancedda L, Heilshorn S, Sumbre
G and Poo MM: LKB1/STRAD promotes axon initiation during neuronal
polarization. Cell. 129:565–577. 2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu J and Yang J: Uncarboxylated
osteocalcin inhibits high glucose-induced ROS production and
stimulates osteoblastic differentiation by preventing the
activation of PI3K/Akt in MC3T3-E1 cells. Int J Mol Med.
37:173–181. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kim JH, Park S, Kim HW and Jang JH:
Recombinant expression of mouse osteocalcin protein in Escherichia
coli. Biotechnol Lett. 29:1631–1635. 2007.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cai Y, Liu T, Fang F, Xiong C and Shen S:
Comparisons of mouse mesenchymal stem cells in primary adherent
culture of compact bone fragments and whole bone marrow. Stem Cells
Int. 2015(708906)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liu G, Bi Y, Shen B, Yang H, Zhang Y, Wang
X, Liu H, Lu Y, Liao J, Chen X and Chu Y: SIRT1 limits the function
and fate of myeloid-derived suppressor cells in tumors by
orchestrating HIF-1α-dependent glycolysis. Cancer Res. 74:727–737.
2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Amini E, Nassireslami E, Payandemehr B,
Khodagholi F, Foolad F, Khalaj S, Hamedani MP, Azimi L and
Sharifzadeh M: Paradoxical role of PKA inhibitor on
amyloidβ-induced memory deficit. Physiol Behav. 149:76–85.
2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liu X, Chhipa RR, Nakano I and Dasgupta B:
The AMPK inhibitor compound C is a potent AMPK-independent
antiglioma agent. Mol Cancer Ther. 13:596–605. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hasegawa T: Ultrastructure and biological
function of matrix vesicles in bone mineralization. Histochem Cell
Biol. 149:289–304. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ishikane S, Ikushima E, Igawa K, Tomooka K
and Takahashi-Yanaga F: Differentiation-inducing factor-1
potentiates adipogenic differentiation and attenuates the
osteogenic differentiation of bone marrow-derived mesenchymal stem
cells. Biochim Biophys Acta Mol Cell Res.
1868(118909)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li D, Zhang R, Zhu W, Xue Y, Zhang Y,
Huang Q, Liu M and Liu Y: S100A16 inhibits osteogenesis but
stimulates adipogenesis. Mol Biol Rep. 40:3465–3473.
2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Picard F, Kurtev M, Chung N, Topark-Ngarm
A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW and
Guarente L: Sirt1 promotes fat mobilization in white adipocytes by
repressing PPAR-gamma. Nature. 429:771–776. 2004.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Huang Y, Zhu X, Chen K, Lang H, Zhang Y,
Hou P, Ran L, Zhou M, Zheng J, Yi L, et al: Resveratrol prevents
sarcopenic obesity by reversing mitochondrial dysfunction and
oxidative stress via the PKA/LKB1/AMPK pathway. Aging.
11:2217–2240. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen Y, Zhang LS, Ren JL, Zhang YR, Wu N,
Jia MZ, Yu YR, Ning ZP, Tang CS and Qi YF:
Intermedin1-53 attenuates aging-associated vascular
calcification in rats by upregulating sirtuin 1. Aging.
12:5651–5674. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hu L, Yin C, Zhao F, Ali A, Ma J and Qian
A: Mesenchymal stem cells: Cell fate decision to osteoblast or
adipocyte and application in osteoporosis treatment. Int J Mol Sci.
19(360)2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Qadir A, Liang S, Wu Z, Chen Z, Hu L and
Qian A: Senile osteoporosis: The involvement of differentiation and
senescence of bone marrow stromal cells. Int J Mol Sci.
21(349)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yang X, Yang J, Lei P and Wen T: LncRNA
MALAT1 shuttled by bone marrow-derived mesenchymal stem
cells-secreted exosomes alleviates osteoporosis through mediating
microRNA-34c/SATB2 axis. Aging. 11:8777–8791. 2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Blair HC, Larrouture QC, Li Y, Lin H,
Beer-Stoltz D, Liu L, Tuan RS, Robinson LJ, Schlesinger PH and
Nelson DJ: Osteoblast differentiation and bone matrix formation in
vivo and in vitro. Tissue Eng Part B Rev. 23:268–280.
2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Florencio-Silva R, Sasso GR, Sasso-Cerri
E, Simões MJ and Cerri PS: Biology of bone tissue: Structure,
function, and factors that influence bone cells. Biomed Res Int.
2015(421746)2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Li JY, Wei X, Sun Q, Zhao XQ, Zheng CY,
Bai CX, Du J, Zhang Z, Zhu LG and Jia YS: MicroRNA-449b-5p promotes
the progression of osteoporosis by inhibiting osteogenic
differentiation of BMSCs via targeting Satb2. Eur Rev Med Pharmacol
Sci. 23:6394–6403. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Li M, Xie Z, Li J, Lin J, Zheng G, Liu W,
Tang S, Cen S, Ye G, Li Z, et al: GAS5 protects against
osteoporosis by targeting UPF1/SMAD7 axis in osteoblast
differentiation. Elife. 9(e59079)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhou J, Nie H, Liu P, Wang Z, Yao B and
Yang L: Down-regulation of miR-339 promotes differentiation of
BMSCs and alleviates osteoporosis by targeting DLX5. Eur Rev Med
Pharmacol Sci. 23:29–36. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ding RB, Bao J and Deng CX: Emerging roles
of SIRT1 in fatty liver diseases. Int J Biol Sci. 13:852–867.
2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Imperatore F, Maurizio J, Vargas Aguilar
S, Busch CJ, Favret J, Kowenz-Leutz E, Cathou W, Gentek R, Perrin
P, Leutz A, et al: SIRT1 regulates macrophage self-renewal. EMBO J.
36:2353–2372. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Mu N, Lei Y, Wang Y, Wang Y, Duan Q, Ma G,
Liu X and Su L: Inhibition of SIRT1/2 upregulates HSPA5 acetylation
and induces pro-survival autophagy via ATF4-DDIT4-mTORC1 axis in
human lung cancer cells. Apoptosis. 24:798–811. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Strycharz J, Rygielska Z, Swiderska E,
Drzewoski J, Szemraj J, Szmigiero L and Sliwinska A: SIRT1 as a
therapeutic target in diabetic complications. Curr Med Chem.
25:1002–1035. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wang X, Chen L and Peng W: Protective
effects of resveratrol on osteoporosis via activation of the
SIRT1-NF-κB signaling pathway in rats. Exp Ther Med. 14:5032–5038.
2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chang HC and Guarente L: SIRT1 and other
sirtuins in metabolism. Trends Endocrinol Metab. 25:138–145.
2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Yuan HF, Zhai C, Yan XL, Zhao DD, Wang JX,
Zeng Q, Chen L, Nan X, He LJ, Li ST, et al: SIRT1 is required for
long-term growth of human mesenchymal stem cells. J Mol Med (Berl).
90:389–400. 2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Han X, Liu L, Wang F, Zhao X, Zhao D, Dai
X and Li Y: Reconstruction of tissue-engineered bone with bone
marrow mesenchymal stem cells and partially deproteinized bone in
vitro. Cell Biol Int. 36:1049–1053. 2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chang Y, Yu D, Chu W, Liu Z, Li H and Zhai
Z: LncRNA expression profiles and the negative regulation of
lncRNA-NOMMUT037835.2 in osteoclastogenesis. Bone.
130(115072)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Song J, Li J, Yang F, Ning G, Zhen L, Wu
L, Zheng Y, Zhang Q, Lin D, Xie C and Peng L: Nicotinamide
mononucleotide promotes osteogenesis and reduces adipogenesis by
regulating mesenchymal stromal cells via the SIRT1 pathway in aged
bone marrow. Cell Death Dis. 10(336)2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wang H, Hu Z, Wu J, Mei Y, Zhang Q, Zhang
H, Miao D and Sun W: Sirt1 promotes osteogenic differentiation and
increases alveolar bone mass via Bmi1 activation in mice. J Bone
Miner Res. 34:1169–1181. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Domazetovic V, Marcucci G, Pierucci F,
Bruno G, Di Cesare Mannelli L, Ghelardini C, Brandi ML, Iantomasi
T, Meacci E and Vincenzini MT: Blueberry juice protects osteocytes
and bone precursor cells against oxidative stress partly through
SIRT1. FEBS Open Bio. 9:1082–1096. 2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Edwards JR, Perrien DS, Fleming N, Nyman
JS, Ono K, Connelly L, Moore MM, Lwin ST, Yull FE, Mundy GR and
Elefteriou F: Silent information regulator (Sir)T1 inhibits NF-κB
signaling to maintain normal skeletal remodeling. J Bone Miner Res.
28:960–969. 2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Feng G, Zheng K, Song D, Xu K, Huang D,
Zhang Y, Cao P, Shen S, Zhang J, Feng X and Zhang D: SIRT1 was
involved in TNF-α-promoted osteogenic differentiation of human
DPSCs through Wnt/β-catenin signal. In Vitro Cell Dev Biol Anim.
52:1001–1011. 2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Constanze B, Popper B, Aggarwal BB and
Shakibaei M: Evidence that TNF-β suppresses osteoblast
differentiation of mesenchymal stem cells and resveratrol reverses
it through modulation of NF-κB, Sirt1 and Runx2. Cell Tissue Res.
381:83–98. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Hong W, Wei Z, Qiu Z, Li Z, Fu C, Ye Z and
Xu X: Atorvastatin promotes bone formation in aged apoE(-/-) mice
through the Sirt1-Runx2 axis. J Orthop Surg Res.
15(303)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Komori T, Yagi H, Nomura S, Yamaguchi A,
Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al:
Targeted disruption of Cbfa1 results in a complete lack of bone
formation owing to maturational arrest of osteoblasts. Cell.
89:755–764. 1997.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Lv ZT, Liang S, Chen K, Zhang JM, Cheng P,
Guo JC, Yang Q, Zhou CH, Liao H and Chen AM: FNDC4 inhibits
RANKL-induced osteoclast formation by suppressing NF-κB Activation
and CXCL10 expression. Biomed Res Int. 2018(3936257)2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Kauppinen A, Suuronen T, Ojala J,
Kaarniranta K and Salminen A: Antagonistic crosstalk between NF-κB
and SIRT1 in the regulation of inflammation and metabolic
disorders. Cell Signal. 25:1939–1948. 2013.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Zhou H, Shang L, Li X, Zhang X, Gao G, Guo
C, Chen B, Liu Q, Gong Y and Shao C: Resveratrol augments the
canonical Wnt signaling pathway in promoting osteoblastic
differentiation of multipotent mesenchymal cells. Ex Cell Res.
315:2953–2962. 2009.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Shakibaei M, Shayan P, Busch F, Aldinger
C, Buhrmann C, Lueders C and Mobasheri A: Resveratrol mediated
modulation of Sirt-1/Runx2 promotes osteogenic differentiation of
mesenchymal stem cells: Potential role of Runx2 deacetylation. PLoS
One. 7(e35712)2012.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zainabadi K: Drugs targeting SIRT1, a new
generation of therapeutics for osteoporosis and other bone related
disorders? Pharmacol Resh. 143:97–105. 2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Lieben L, Callewaert F and Bouillon R:
Bone and metabolism: A complex crosstalk. Horm Res. 71 (Suppl
1):S134–S138. 2009.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Confavreux CB: Bone: From a reservoir of
minerals to a regulator of energy metabolism. Kidney Int Suppl.
79:S14–S19. 2011.PubMed/NCBI View Article : Google Scholar
|
|
71
|
De Toni L, De Filippis V, Tescari S,
Ferigo M, Ferlin A, Scattolini V, Avogaro A, Vettor R and Foresta
C: Uncarboxylated osteocalcin stimulates 25-hydroxy vitamin D
production in Leydig cell line through a GPRC6a-dependent pathway.
Endocrinology. 155:4266–4274. 2014.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Gao J, Bai T, Ren L, Ding Y, Zhong X, Wang
H, Guo Y, Li J, Liu Y and Zhang Y: The PLC/PKC/Ras/MEK/Kv channel
pathway is involved in uncarboxylated osteocalcin-regulated insulin
secretion in rats. Peptides. 86:72–79. 2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Guedes JAC, Esteves JV, Morais MR, Zorn TM
and Furuya DT: Osteocalcin improves insulin resistance and
inflammation in obese mice: Participation of white adipose tissue
and bone. Bone. 115:68–82. 2018.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Miyamoto T, Oguma Y, Sato Y, Kobayashi T,
Ito E, Tani M, Miyamoto K, Nishiwaki Y, Ishida H, Otani T, et al:
Elevated creatine kinase and lactic acid dehydrogenase and
decreased osteocalcin and uncarboxylated osteocalcin are associated
with bone stress injuries in young female athletes. Sci Rep.
8(18019)2018.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Kim KM, Lim S, Moon JH, Jin H, Jung KY,
Shin CS, Park KS, Jang HC and Choi SH: Lower uncarboxylated
osteocalcin and higher sclerostin levels are significantly
associated with coronary artery disease. Bone. 83:178–183.
2016.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Hayashi Y, Kawakubo-Yasukochi T, Mizokami
A, Hazekawa M, Yakura T, Naito M, Takeuchi H, Nakamura S and Hirata
M: Uncarboxylated osteocalcin induces antitumor immunity against
mouse melanoma cell growth. J Cancer. 8:2478–2486. 2017.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Tsao YT, Huang YJ, Wu HH, Liu YA, Liu YS
and Lee OK: Osteocalcin mediates biomineralization during
osteogenic maturation in human mesenchymal stromal cells. Int J Mol
Sci. 18(159)2017.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Lee YC, Chan YH, Hsieh SC, Lew WZ and Feng
SW: Comparing the osteogenic potentials and bone regeneration
capacities of bone marrow and dental pulp mesenchymal stem cells in
a rabbit calvarial bone defect model. Int J Mol Sci.
20(5015)2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Zhang J, Zhang W, Dai J, Wang X and Shen
SG: Overexpression of Dlx2 enhances osteogenic differentiation of
BMSCs and MC3T3-E1 cells via direct upregulation of Osteocalcin and
Alp. Int J Oral Sci. 11(12)2019.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Fratzl-Zelman N, Glantschnig H, Rumpler M,
Nader A, Ellinger A and Varga F: The expression of matrix
metalloproteinase-13 and osteocalcin in mouse osteoblasts is
related to osteoblastic differentiation and is modulated by
1,25-dihydroxyvitamin D3 and thyroid hormones. Cell Biol Int.
27:459–468. 2003.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Roach HI: Why does bone matrix contain
non-collagenous proteins? The possible roles of osteocalcin,
osteonectin, osteopontin and bone sialoprotein in bone
mineralisation and resorption. Cell Biol Int. 18:617–628.
1994.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Chung JE, Park JH, Yun JW, Kang YH, Park
BW, Hwang SC, Cho YC, Sung IY, Woo DK and Byun JH: Cultured human
periosteum-derived cells can differentiate into osteoblasts in a
perioxisome proliferator-activated receptor gamma-mediated fashion
via bone morphogenetic protein signaling. Int J Med Sci.
13:806–818. 2016.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Yu PB, Hong CC, Sachidanandan C, Babitt
JL, Deng DY, Hoyng SA, Lin HY, Bloch KD and Peterson RT:
Dorsomorphin inhibits BMP signals required for embryogenesis and
iron metabolism. Nat Chem Biol. 4:33–41. 2008.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Ingersoll MA, Spanbroek R, Lottaz C,
Gautier EL, Frankenberger M, Hoffmann R, Lang R, Haniffa M, Collin
M, Tacke F, et al: Comparison of gene expression profiles between
human and mouse monocyte subsets. Blood. 115:e10–e19.
2010.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Uder C, Brückner S, Winkler S, Tautenhahn
HM and Christ B: Mammalian MSC from selected species: Features and
applications. Cytometry A. 93:32–49. 2018.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Couchourel D, Aubry I, Delalandre A,
Lavigne M, Martel-Pelletier J, Pelletier JP and Lajeunesse D:
Altered mineralization of human osteoarthritic osteoblasts is
attributable to abnormal type I collagen production. Arthritis
Rheum. 60:1438–1450. 2009.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Glaser DL and Kaplan FS: Osteoporosis.
Definition and clinical presentation. Spine (Phila Pa 1976). 22
(Suppl 24):12S–16S. 1997.PubMed/NCBI View Article : Google Scholar
|