Introduction
Bone homeostasis is maintained by the balance
between the formation of bone by osteoblasts and the resorption of
bone by osteoclasts (1).
Osteoporosis is a systemic skeletal disease that results from the
disruption of bone homeostasis with excessive bone resorption
and/or reduced bone formation (2).
Treatments for osteoporosis include antiresorptive agents to
inhibit bone resorption and anabolic agents to promote bone
formation (3). Although medications
for osteoporosis have been developed and used successfully in
recent years, most of them are antiresorptive drugs. Antiresorptive
drugs including bisphosphonates, estrogen and receptor activator of
NF-κB ligand inhibitors, prevent the loss of bone rather than
restore it (4,5). In addition, parathyroid hormone, the
only US Food and Drug Administration-approved anabolic agent, has
limitations of high cost and invasive modes of administration
(6). Therefore, it is necessary to
explore the mechanisms of osteoblastic differentiation to
facilitate the search for new anabolic agents for the treatment of
osteoporosis.
Leucine-rich repeat-containing G-protein coupled
receptors (LGRs) belong to the G-protein-coupled receptor family,
which transmit extracellular signals into the cytoplasm (7). LGR5 is one of the group B LGR proteins
(LGR4-6), which recognize R-spondin (Rspo) proteins to activate Wnt
signaling (8,9). LGR5 is considered a stem cell marker,
and plays an important role in normal development and cancer. It is
involved in the self-renewal and stem cell development of tissues
including hair follicles, the stomach, small intestine and colon
(10-12).
The genetic deletion of LGR5 in mice results in 100% neonatal
lethality (13). Also, LGR5 has
been shown to promote tumor growth and progression in colorectal
carcinoma (14), basal cell
carcinoma (15), glioblastoma
(16) and neuroblastoma (17). Since its close homologs LGR4 and
LGR6 have been reported to participate in bone formation (18,19),
the role of LGR5 in bone remodeling has also become a topic of
interest. Furthermore, LGR5 is upregulated in Ewing sarcoma, a
malignant bone tumor, and promotes tumor progression through
Wnt/β-catenin signaling (20). A
recent study revealed that mesenchymal stem cells overexpressing
LGR5 promote the healing of fractures through Wnt/ERK signaling
pathways (21). All these previous
findings suggest a potential role of LGR5 in bone remodeling.
However, the effects of LGR5 on osteoblastic differentiation and
the underlying mechanism remain unclear. Thus the present study
aimed to explore the function of LGR5 in osteoblastic
differentiation using the MC3T3-E1 pre-osteoblastic cell line.
Materials and methods
Cell culture
The MC3T3-E1 murine pre-osteoblastic cell line and
C2C12 myoblastic cell line were obtained from the American Type
Culture Collection. MC3T3-E1 cells were cultured in α-minimum
essential medium (HyClone; Cytiva) containing 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). The medium was
supplemented with 1% penicillin/streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C with 5% CO2. C2C12 cells were
cultured in high-glucose DMEM culture medium (HyClone; Cytiva)
containing 10% FBS and 1% penicillin/streptomycin (Gibco; Thermo
Fisher Scientific, Inc.) at 37˚C with 5% CO2. The
culture medium was replaced thrice each week. After the cells had
grown to 70% confluence, the medium was changed to osteogenic media
containing 4 mM β-glycerophosphate and 25 µg/ml ascorbic acid for
the induction of osteoblastic differentiation. Dickkopfs-1 (Dkk-1;
PeproTech, Inc.) was used as a Wnt inhibitor at a concentration of
100 ng/ml and Wnt-3a (PeproTech, Inc.) was used as a Wnt activator
at a concentration of 20 ng/ml. Rspo-2 (PeproTech, Inc.) was added
for osteogenic induction at a concentration of 100 ng/ml. The
duration of treatment is specified in each respective assay
section.
Lentiviral transfection of MC3T3-E1
cells
The lentivirus-based LGR5 overexpression vector
based on green fluorescent protein (GFP)-PURO and short hairpin RNA
(shRNA)-GFP-PURO were purchased from Shanghai GeneChem Co., Ltd.. A
lentivirus-based LGR5 overexpression vector (lv-LGR5) was designed
using the primer sequences of murine LGR5 (GenBank number,
NM_010195.2) cDNA as follows: Sense,
5'-CTTCTCGAGCTACTTCGGGCACCATGGAC-3' and antisense,
5'-GCGGGTACCTTAGAGACATGGGACAAATG-3'. The sequences of the shRNA
targeting LGR5 (sh-LGR5) were as follows: Sense,
5'-GCAACAACAUCAGGUCAAUTT-3' and antisense,
5'-AUUGACCUGAUGUUGUUGCTT-3'. A scrambled shRNA
(TTCTCCGAACGTGTCACGTAA) with no complementary sequences in the
murine genome, was used as a negative control. MC3T3-E1 cells were
seeded in 6-well plates (1.5x104 cells/cm2).
When the cells had grown to 70% confluence, the experimental
lentivirus or negative control lentivirus at a multiplicity of
infection of 20 in 1 ml was used to transfect the cells at 37˚C for
24 h. Post-transfection, the transfection medium was discarded and
replaced by selective medium including 2 µg/ml puromycin
(Sigma-Aldrich; Merck KGaA). After 9 days, the third passage of
stable clones was collected in the following experiments. The
transfection efficiency was evaluated based on the percentage of
GFP-positive cells, and the overexpression or knockdown of LGR5 was
verified using reverse transcription-quantitative PCR (RT-qPCR) and
western blot analysis.
Immunofluorescence (IF)
MC3T3-E1 and C2C12 cells were seeded on coverslips
in 24-well plates (2x104 cells/cm2). After
washing with phosphate-buffered saline (PBS), the cells were fixed
with 4% paraformaldehyde for 20 min at room temperature, and
permeabilized with 0.5% Triton X-100 for another 20 min at room
temperature. The cells were then incubated with primary antibody
against LGR5 (cat. no. ab273092; 1:100) at 4˚C overnight after
blocking with 5% bovine serum albumin (2 h at room temperature).
The next day, cells were incubated with fluorescent-labeled
secondary antibody (cat. no. A0408; 1:200; Beyotime Institute of
Biotechnology) for 1 h at room temperature and treated with
4,6-diamidino-2-phenylindole (cat. no. C1002; 1:1,000; Beyotime
Institute of Biotechnology) for 15 min for nuclear staining at room
temperature. The samples were observed under a confocal
microscope.
Cell viability and apoptosis
assay
For the assessment of cell viability, transfected
cells seeded in 96-well plates (2x104
cells/cm2) were treated with 20 µl/well
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
(MTT; 5 mg/ml). The supernatant was removed after incubation at
37˚C for 2 h. The formazan product in each well was dissolved by
dimethyl sulfoxide with gentle shaking for 5 min. The absorbance
was then detected using a microplate reader (Bio-Rad Laboratories,
Inc.) at 570 nm. Cell viability rates were expressed as fold
changes relative to that of the control group.
Cell apoptosis was detected using a Cell Death
Detection ELISAPLUS kit (Roche Diagnostics) according to
the manufacturer's instructions. Briefly, transfected cells were
lysed within ice-cold lysis buffer for 30 min and then pelleted by
centrifugation for 10 min (200 x g at 4˚C). Mouse monoclonal
antibodies against histones (biotin-labeled) and DNA
(peroxidase-labeled) from the kit were used to bind nucleosomes for
2 h at 15-25˚C, and the antibody-nucleosome complexes were then
bound to the microplate by streptavidin. After washing the
immobilized antibody-histone complexes, the samples were incubated
with 2,2'-azino-di(3-ethylbenzthiazoline-sulfonate). The apoptosis
was colorimetrically determined at 405 nm based on the amount of
mono- and oligonucleosomes in the cytoplasmic fraction of cell
lysates, which was determined using the quantitative sandwich
enzyme immunoassay principle. The results were expressed as the
ratio of absorbance of the treated (apoptotic) sample to that of
the control group.
RT-qPCR analysis
Total cellular RNA was collected using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) at 7 or 14 days after osteogenic induction according to the
manufacturer's instructions. Cells treated with Dkk-1, Wnt-3a and
Rspo-2 were collected at 7 days. The RNA was used to generate cDNA
using a PrimeScript™ RT Reagent kit (Takara Bio, Inc.)
according to the manufacturer's protocol. qPCR was then performed
using SYBR® Premix Ex Taq™ (Takara Bio, Inc.)
and an ABI 7500 Fast Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The thermocycling conditions were
used as follows: Initial denaturation at 95˚C for 30 sec, followed
by 40 cycles at 95˚C for 5 sec and primer annealing and extension
at 60˚C for 30 sec and final extension for 1 min at 72˚C. Each
sample was separately examined in triplicate. The expression values
of the target genes were quantified by the 2-ΔΔCq method
(22) with normalization to
β-actin. The qPCR primers used were as follows: β-actin forward,
5'-TGACAGGATGCAGAAGGAGA-3' and reverse, 5'-CGCTCAGGAGGAGCAATG-3';
LGR5 forward, 5'-TCTTCACCTCCTACCTGGACCT-3' and reverse
5'-GGCGTAGTCTGCTATGTGGTGT-3'; alkaline phosphatase (ALP) forward,
5'-TCGGGACTGGTACTCGGATAAC-3' and reverse,
5'-GTTCAGTGCGGTTCCAGACATAG-3'; osterix (OSX) forward,
5'-GGAGGCACAAAGAAGCCATACGC-3' and reverse
5'-TGCAGGAGAGAGGAGTCCATTG-3'; runt related transcription factor 2
(RUNX2) forward, 5'-GACGAGGCAAGAGTTTCACC-3' and reverse,
5'-GGACCGTCCACTGTCACTTT-3'; collagen type I α1 (COL-1a1) forward,
5'-AAGAAGCACGTCTGGTTGGAG-3' and reverse,
5'-GGTCCATGTAGGCTACGCTGTT-3'; osteocalcin (OCN) forward,
5'-CAAGCAGGGAGGCAATAAGG-3' and reverse, 5'-CGTCACAAGCAGGGTTAAGC-3';
lymphoid enhancer-binding factor 1 (Lef1) forward,
5'-CGCTGATCAATGCCCCAACTTTCCGGAGGA-3' and reverse,
5'-CCGCTCGAGTCAGATGTAGGCAGCTGTCATTCTG-3'; T-cell factor 1 (Tcf1)
forward, 5'-TGCTGTCTATATCCGCAGGAAG-3' and reverse,
5'-CGATCTCTCTGGATTTTATTCTCT-3'; β-catenin forward,
5'-ACGCTGCTCATCCCACTAAT-3' and reverse,
5'-AGTTCCGCGTCATCCTGATA-3'.
Western blot analysis
Cell lysates were collected from cells on days 0, 7
and 14 of osteogenic induction using ice-cold RIPA lysis buffer [50
mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 0.5% sodium
deoxycholate, 0.1% SDS with protease inhibitor cocktail (Roche
Diagnostics)] at 7 or 14 days after osteogenic induction, and total
proteins were extracted using a Protein Extraction kit (Beyotime
Institute of Biotechnology). Nuclear and cytoplasmic proteins were
extracted using a Nucleoprotein Extraction kit (Sangon Biotech Co.,
Ltd.) according to the manufacturer's instructions. Protein content
was quantified using a Bicinchoninic Acid Protein assay kit
(Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Equal amounts of proteins (40 µg/lane)
were loaded onto gels for 8-12% SDS-PAGE and electrotransferred to
polyvinylidene difluoride membranes. The membranes were blocked
with 5% nonfat dry milk diluted in Tris-buffered saline containing
0.05% (v/v) Tween-20 (TBST) for 2 h at room
temperature, and then incubated with primary antibodies at 4˚C
overnight. After washing thrice with TBST, the membranes were
incubated with HRP-conjugated secondary antibody (1:1,000; cat. no.
A0208; Beyotime Institute of Biotechnology) for 1 h at room
temperature. The expression of target proteins was visualized by
the reaction of HRP with a chemiluminescent substrate (EMD
Millipore). GAPDH was used as the control, with the exception of
nuclear protein, where histone H3 was used. The primary antibodies
used in this experiment were as follows: LGR5 (1:1,000; cat. no.
ab273092; Abcam), β-catenin (cat. no. 9562; 1:1,000; Cell Signaling
Technology, Inc.), phosphorylated (inactive) β-catenin (cat. no.
9566; 1:1,000; Cell Signaling Technology, Inc.), GAPDH (cat. no.
8884; 1:2,000, Cell Signaling Technology, Inc.) and histone H3
(cat. no. 4499; 1:5,000; Cell Signaling Technology, Inc.).
ALP staining and Alizarin red
staining
ALP and Alizarin red staining were performed on
MC3T3-E1 cells cultured in osteogenic media for 7 and 14 days,
respectively. For ALP staining, cells fixed with 4%
paraformaldehyde (4˚C for 30 min) were treated with an ALP
substrate mixture from an ALP staining kit (Sigma-Aldrich; Merck
KGaA) in darkness (at room temperature for 30 min). After rinsing
the cells three times with PBS, they were observed under a light
microscope (Leica Microsystems, Inc.). For Alizarin red staining,
cells fixed in ice-cold 70% ethanol (4̊C for 20 min) were stained
with 3% Alizarin red S solution (Sigma-Aldrich; Merck KGaA) for 30
min at room temperature. The cells were washed with
double-distilled water three times, and images of mineralized
nodules were obtained under a light microscope (Leica Microsystems,
Inc.).
TOPflash dual-luciferase reporter
assays
A TOPflash Wnt/b-catenin activity assay was
performed to detect the activation of canonical Wnt signaling.
MC3T3-E1 cells seeded in six-well plates (2x105
cells/well) were transfected with plasmids containing
TOPflash/FOPflash firefly luciferase (500 ng/well; BioVector NTCC,
Inc.) and pRL-SV40-renilla luciferase reporter (20 ng/well;
(BioVector NTCC, Inc.) for 24 h. The medium was then replaced with
osteogenic culture medium containing control lentivirus, lv-LGR5
with or without Wnt-3a or Dkk-1, or Wnt-3a with or without Dkk-1
for another 48 h. The luciferase assay was performed using a
Dual-Luciferase Assay kit (Promega Corporation) according to the
manufacturer's protocol. The luciferase activity was presented as
the ratio of TOPflash/FOPflash, with Renilla luciferase
plasmids as the internal control.
Statistical analysis
All independent experiments were performed at least
three times and the data are presented as the mean ± SD.
Statistical differences were determined by the Student's t-test, or
one-way ANOVA followed by a post hoc Fisher's least significant
difference test or Dunnett's test. Statistical analyses were
performed using SPSS software (version 18.0; SPSS, Inc.). P<0.05
was considered to indicate a statistically significant
difference.
Results
Expression level of LGR5 increases
during the osteoblastic differentiation of MC3T3-E1 cells
To explore whether LGR5 was expressed in MC3T3-E1
cells, IF analysis was first conducted. The results verified the
expression of LGR5 in MC3T3-E1 cells, and demonstrated that LGR5
was mainly distributed in the cytoplasm and membrane (Fig. 1A). The MC3T3-E1 cells were then
induced to undergo osteoblastic differentiation with osteogenic
media for 7 or 14 days. Successful induction was confirmed by the
increased mRNA expression levels of the osteoblast differentiation
markers ALP, OCN and COL-1a1, as well as the transcription factors
RUNX2 and OSX (Fig. 1B).
Osteoblastic differentiation was also confirmed by the results of
ALP staining at day 7 and Alizarin red staining at day 14 during
induction (Fig. 1C). The expression
of LGR5 during osteoblastic differentiation was also detected by
RT-qPCR and western blotting. The mRNA and protein levels of LGR5
increased during differentiation (Fig.
1D and E), suggesting a
potential role of LGR5 in the osteoblastic differentiation of
MC3T3-E1 cells.
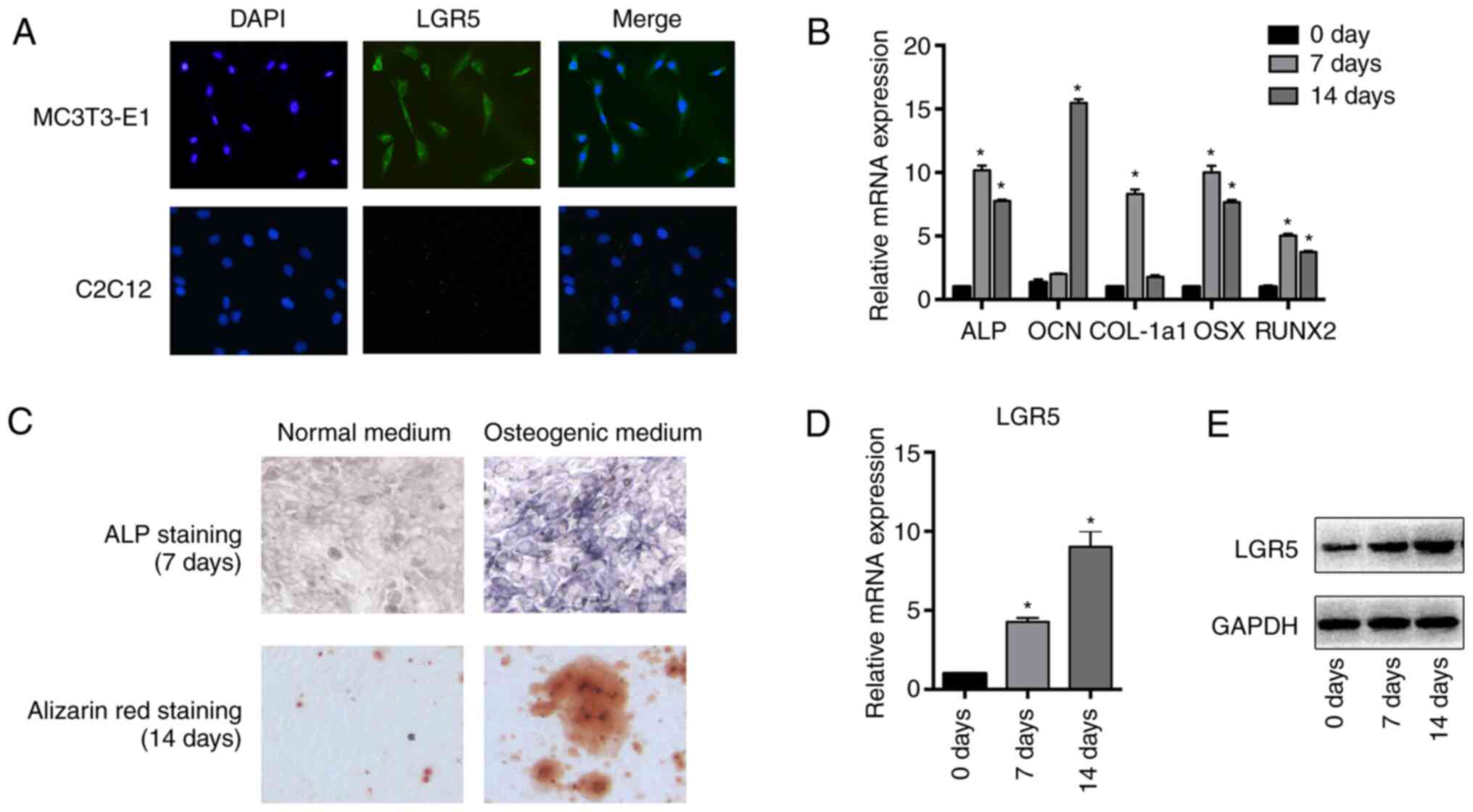 | Figure 1Expression of LGR5 increases during
the osteoblastic differentiation of MC3T3-E1 cells. (A)
Immunofluorescence analysis of LGR5 protein expression in MC3T3-E1
cells. C2C12 mouse myoblast cells were used as control cells
(magnification, x100). (B) MC3T3-E1 cells were induced for 7 or 14
days in osteogenic media, and the mRNA levels of the osteoblast
differentiation markers ALP, COL-a1, OCN, OSX and RUNX2 were
detected by reverse transcription-quantitative PCR analysis. (C)
ALP and Alizarin red staining after osteogenic induction for 7 or
14 days, respectively (magnification, x100). (D) mRNA and (E)
protein expression of LGR5 after osteogenic induction for 7 or 14
days. *P<0.05 vs. 0 day. LGR5, leucine-rich
repeat-containing G-protein coupled receptor; ALP, alkaline
phosphatase; OCN, osteocalcin; COL-1a1, collagen type I α1; OSX,
osterix; Runx2, runt related transcription factor 2. |
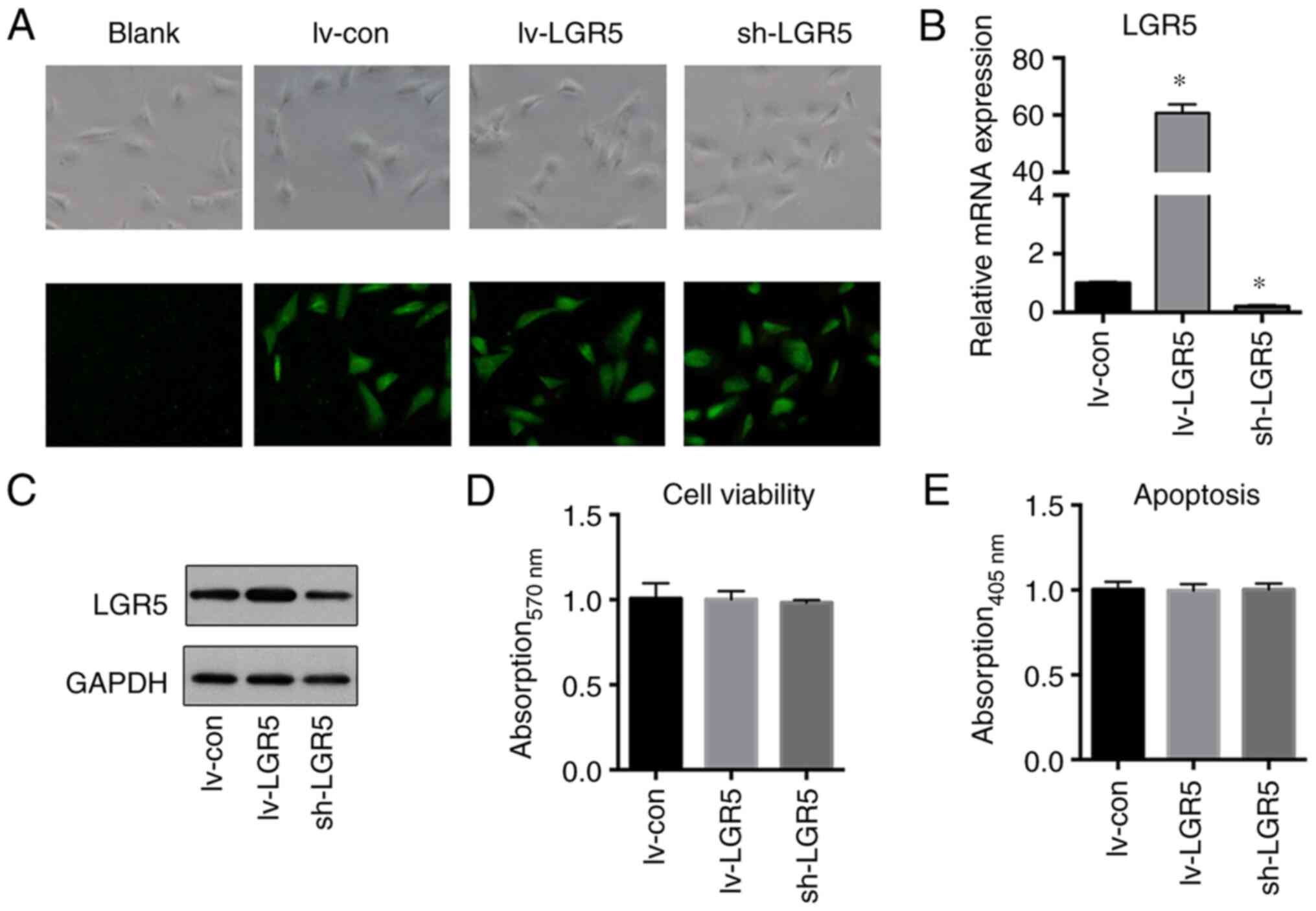
Effects of LGR5 regulation on the
viability and apoptosis of MC3T3-E1 cells
To investigate the effects of LGR5 regulation on
cell viability and apoptosis, LGR5 was overexpressed or knocked
down in MC3T3-E1 cells via lentiviral transfection. Fluorescence
microscopy confirmed high efficiencies of lentivirus transfection
with a large proportion of GFP-positive cells (Fig. 2A). The successful overexpression or
knockdown of LGR5 gene was then verified by RT-qPCR and western
blotting (Fig. 2B and C). Neither overexpression nor knockdown of
the LGR5 gene significantly affected the cell viability or
apoptosis of MC3T3-E1 cells (Fig.
2D and E).
LGR5 enhances the osteoblastic
differentiation of MC3T3-E1 cells
To explore the role of LGR5 in osteoblastic
differentiation, the transfected MC3T3-E1 cells were cultured in
osteogenic media for 7 or 14 days. After 7 days, LGR5
overexpression significantly increased the mRNA levels of ALP and
COL-1a1, which are known as early- and middle-stage osteogenic
differentiation marker genes. The knockdown of LGR5 significantly
inhibited the expression of these mRNAs (Fig. 3A). Concurrently, ALP staining also
suggested that LGR5 played a positive role in the osteoblastic
differentiation of the MC3T3-E1 cells (Fig. 3B). Following 14 days of induction,
the mRNA level of OCN, the late-stage osteogenic gene, was
significantly upregulated by LGR5 overexpression (Fig. 3C). Furthermore, Alizarin red
staining after 14 days also revealed that LGR5 enhanced the
mineralization of MC3T3-E1 cells at the late stage of osteoblastic
differentiation (Fig. 3D). These
results together suggest that LGR5 promoted the osteoblastic
differentiation of MC3T3-E1 cells.
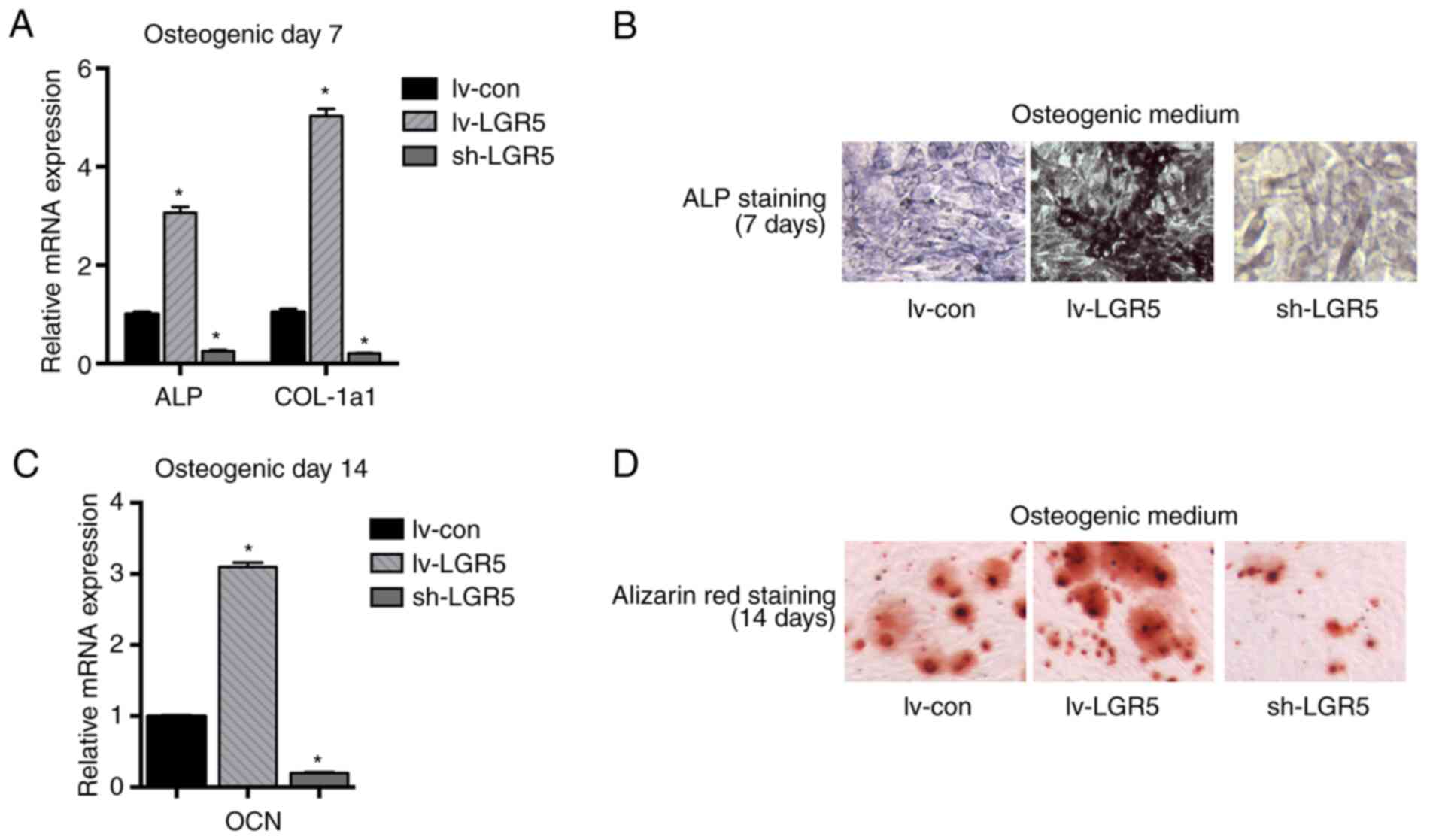 | Figure 3LGR5 promotes the osteogenic
differentiation of MC3T3-E1 cells. (A) mRNA levels of the
osteoblast differentiation markers ALP and COL-1a1 were detected in
MC3T3-E1 cells after osteogenic induction for 7 days. (B) ALP
staining of MC3T3-E1 cells after osteogenic induction for 7 days
(magnification, x100). (C) mRNA levels of the osteoblast
differentiation marker OCN after osteogenic induction for 14 days.
(D) Alizarin red staining of MC3T3-E1 cells after osteogenic
induction for 14 days (magnification, x100). *P<0.05
vs. lv-con. lv-LGR5, lentivirus-based LGR5 overexpression vector;
sh-LGR5, lentivirus-based short hairpin RNA vector targeting LGR5,
lv-con, Lentivirus-based negative shRNA control; LGR5, leucine-rich
repeat-containing G-protein coupled receptor; ALP, alkaline
phosphatase; OCN, osteocalcin; COL-1a1, collagen type I α1. |
LGR5 activates the Wnt signaling
pathway by stabilizing β-catenin
Since LGR5 is a facultative Wnt receptor component
mediating the activation of Wnt signaling (9), and Wnt signaling plays a pivotal role
in osteoblast differentiation (23), whether LGR5 promoted osteoblastic
differentiation through Wnt/β-catenin signaling in MC3T3-E1 cells
was explored. Neither the overexpression nor the knockdown of LGR5
affected the mRNA level of β-catenin (Fig. 4A). The protein level of β-catenin in
transfected MC3T3-E1 cells was then assessed by western blotting.
LGR5 overexpression increased the protein level of β-catenin in the
total cell lysates (Fig. 4B), but
reduced the level of phosphorylated β-catenin in the cytoplasm
(Fig. 4C). In addition, the level
of intranuclear β-catenin was substantially increased by LGR5
overexpression (Fig. 4D). These
results indicate that LGR5 activated Wnt/β-catenin signaling in the
cells by increasing the cytoplasmic stabilization and nuclear
accumulation of β-catenin. Furthermore, LGR5 upregulated the mRNA
levels of Lef1 and Tcf1, two target genes of the Wnt pathway
(Fig. 4E). The TOPflash dual
luciferase activity assay also confirmed the activation of
Wnt/β-catenin signaling in MC3T3-E1 cells with LGR5 overexpression
(Fig. 4F).
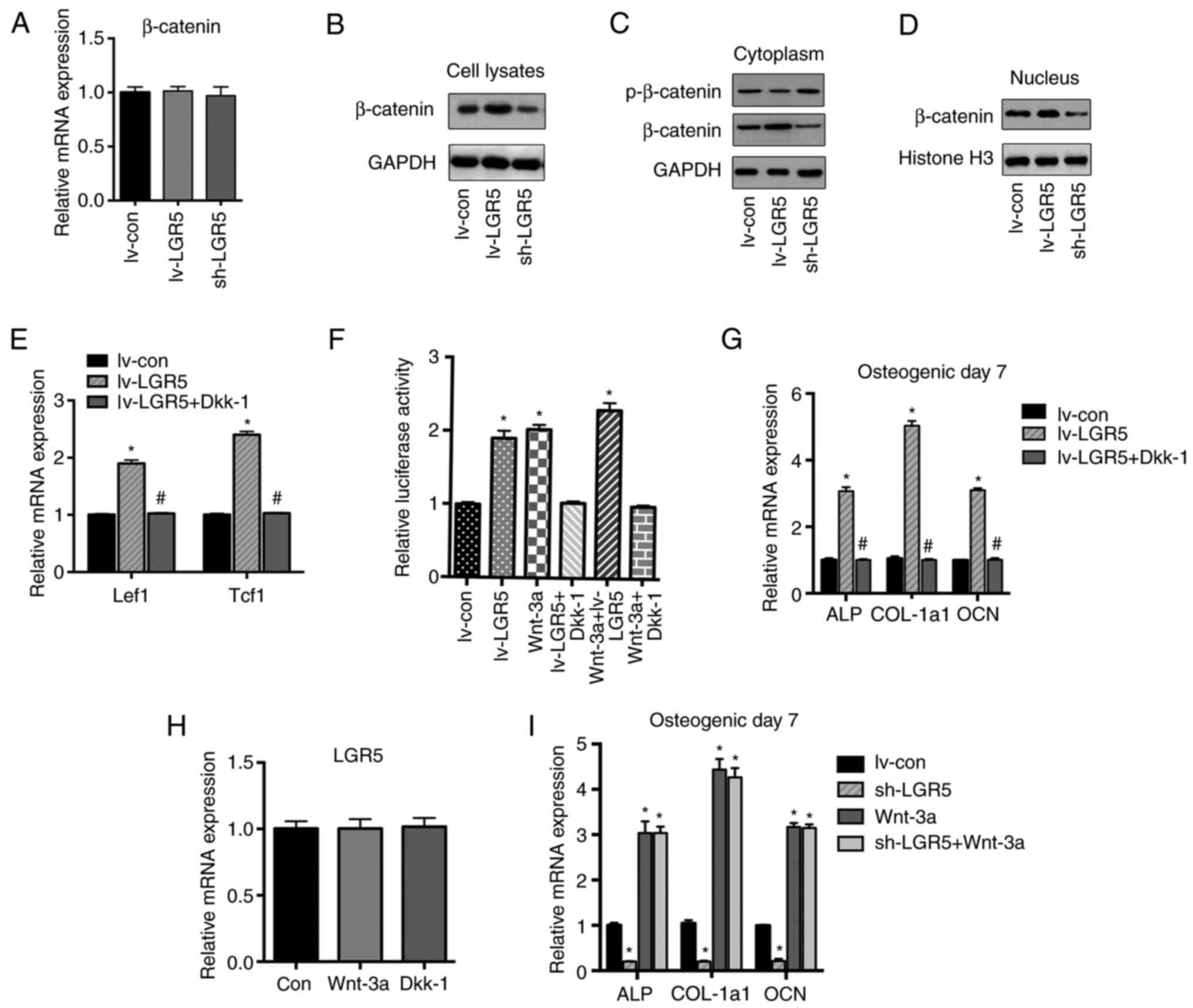 | Figure 4LGR5 activates the Wnt signaling by
stabilizing β-catenin. (A) The mRNA level of β-catenin was detected
in cells transfected with lentiviruses after osteogenic induction
for 7 days. (B) The β-catenin protein level in total cell lysates
was assessed by western blot analysis after the osteogenic
induction of MC3T3-E1 cells for 7 days. (C) p-β-catenin (inactive)
and β-catenin protein levels in the cytoplasm of MC3T3-E1 cells
after osteogenic induction for 7 days. (D) β-catenin protein level
in the nucleus of MC3T3-E1 cells after osteogenic induction for 7
days. (E) Reverse transcription-quantitative PCR analysis of the
Wnt target genes Lef1 and Tcf1 after osteogenic induction for 7
days. (F) TOPflash dual-luciferase activity assay of MC3T3-E1 cells
treated with lentivirus transfection, Dkk-1 and/or Wnt-3a after
induction for 2 days. (G) The mRNA levels of osteogenic marker
genes ALP, COL-a1 and OCN in cells treated with LGR5 overexpression
alone or with Dkk-1 after osteogenic induction for 7 days. (H) The
mRNA level of LGR5 in cells treated with Dkk-1 or Wnt-3a after
osteogenic induction for 7 days. (I) The mRNA levels of osteogenic
marker genes in cells treated with lentivirus transfection and/or
Wnt-3a after osteogenic induction for 7 days. *P<0.05
vs. lv-con. #P<0.05 vs. lv-LGR5. lv-LGR5,
lentivirus-based LGR5 overexpression vector; sh-LGR5,
lentivirus-based short hairpin RNA vector targeting LGR5; lv-con,
lentivirus-based negative shRNA control; con, untreated control;
LGR5, leucine-rich repeat-containing G-protein coupled receptor;
p-, phospho-; Lef1, lymphoid enhancer-binding protein 1; Tcf1,
T-cell factor 1; ALP, alkaline phosphatase; COL-1a1, collagen type
I α1; OCN, osteocalcin; Dkk-1, Dickkopfs-1. |
The Wnt inhibitor Dkk-1 was then added to the cells
to further verify that the potentiating effects of LGR5 on
osteoblastic differentiation were dependent on the Wnt/β-catenin
pathway. The results indicated that LGR5-enhanced osteoblastic
differentiation was significantly abolished by Dkk-1 treatment
(Fig. 4F and G). Thus, it was concluded that LGR5
promoted the osteoblastic differentiation of MC3T3-E1 cells through
activation of the Wnt/β-catenin pathway. Since LGR5 has been
reported to be a target gene of Wnt (9,24-26),
Wnt-3a was added as a Wnt activator, to explore whether LGR5 is
downstream of Wnt/β-catenin signaling. Neither Wnt-3a nor Dkk-1
affected the expression level of LGR5 (Fig. 4H), suggesting that LGR5 was not the
downstream mediator of Wnt/β-catenin signaling in MC3T3-E1 cells.
In addition, Wnt-3a significantly promoted the mRNA levels of the
osteogenic differentiation markers ALP, OCN and COL-1a1, while LGR5
knockdown had no discernible impact on Wnt-3a-induced osteogenesis
(Fig. 4I).
The Rspo family of proteins (Rspo1-4) are agonists
of the canonical Wnt/β-catenin signaling pathway (27). Among the four proteins, Rspo-2 has
the highest affinity for LGR5(23).
Rspo-2 has been identified to be a pivotal protein in embryonic
development (28), tumor growth
(29) and osteoblastogenesis
(30). In the present study, the
effects of Rspo-2 on cells with LGR5 overexpression or knockdown
were explored. The results showed that the knockdown of LGR5
markedly inhibited the Wnt/β-catenin signaling (Fig. 5A and B) and osteogenic differentiation (Fig. 5C and D) induced by Rspo-2. These results
indicate that LGR5 acted as a key receptor for R-spo-2 in the
promotion of osteogenesis.
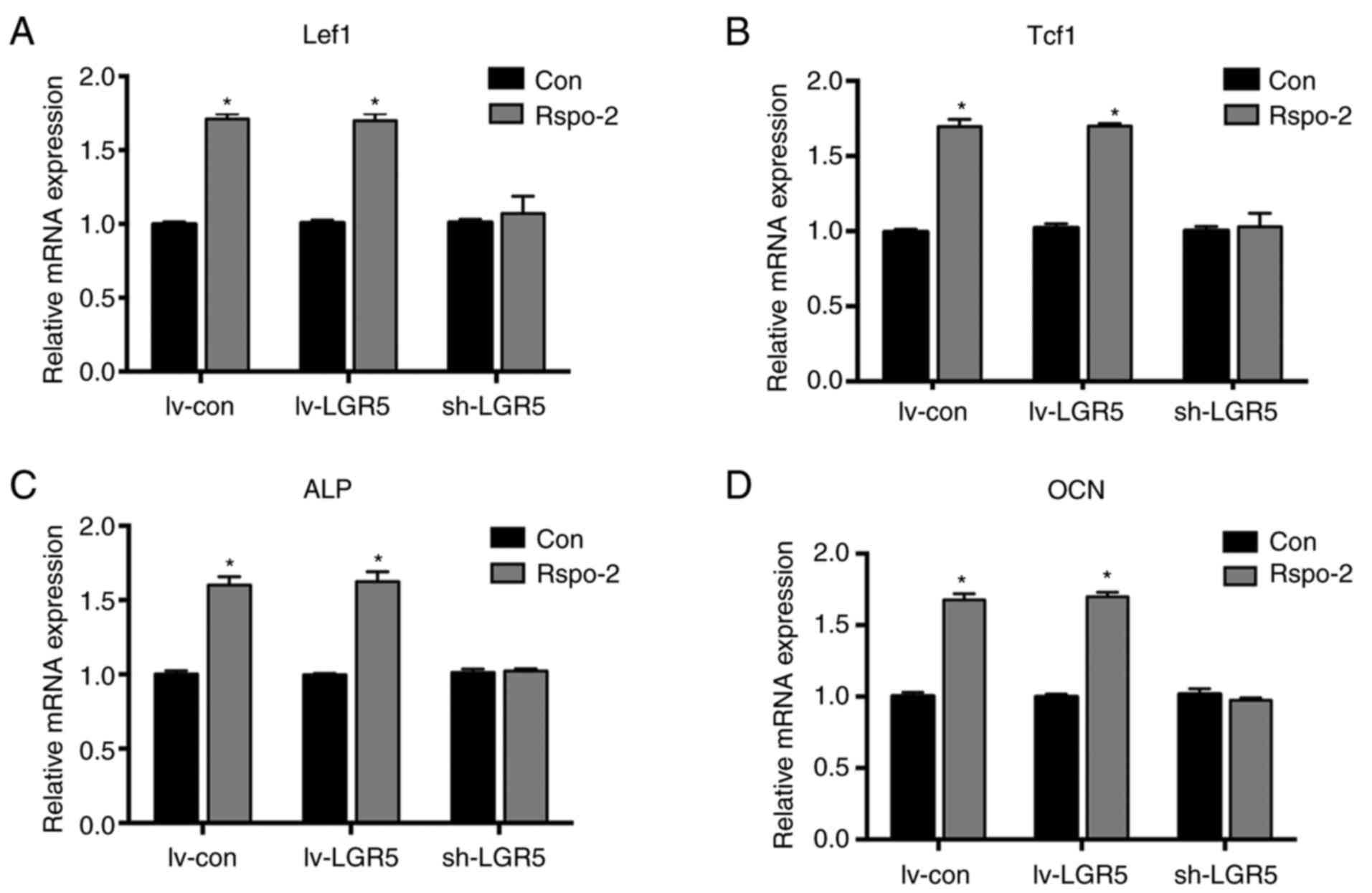 | Figure 5Effects of Rspo-2 on cells with
modulated levels of LGR5. Reverse transcription-quantitative PCR
analysis of the Wnt target genes (A) Lef1 and (B) Tcf1 after
osteogenic induction for 7 days. The mRNA levels of osteogenic
marker genes (C) ALP and (D) OCN in cells with or without Rspo-2
treatment after osteogenic induction for 7 days.
*P<0.05 vs. Con. Con, untreated control; lv-LGR5,
lentivirus-based LGR5 overexpression vector; sh-LGR5,
lentivirus-based short hairpin RNA vector targeting LGR5; lv-con,
lentivirus-based negative shRNA control; LGR5, leucine-rich
repeat-containing G-protein coupled receptor; Lef1, lymphoid
enhancer-binding protein 1; Tcf1, T-cell factor 1; ALP, alkaline
phosphatase; OCN, osteocalcin; Rspo-2, R-spondin-2. |
Discussion
Wnt signaling is widely known to play a pivotal role
in bone remodeling and development, particularly its major branch,
the canonical (Wnt/β-catenin) pathway (23,31).
Wnt/β-catenin signaling is initiated through the binding of Wnt to
the frizzled receptor and low-density lipoprotein receptor-related
protein (LRP)5/6 coreceptors (23).
After this, the β-catenin ‘destruction complex’ which comprises
adenomatous polyposis coli, glycogen synthase kinase 3 and the
scaffolding protein Axin, is inactivated to inhibit β-catenin
phosphorylation and proteasomal degradation. Consequently, the
amount of β-catenin that translocates into the nucleus is
increased, and target genes including Lef1 and Tcf1 are activated
(23,32). Activation of this pathway promotes
osteoblastic differentiation and bone formation (23,31)
(Fig. 6).
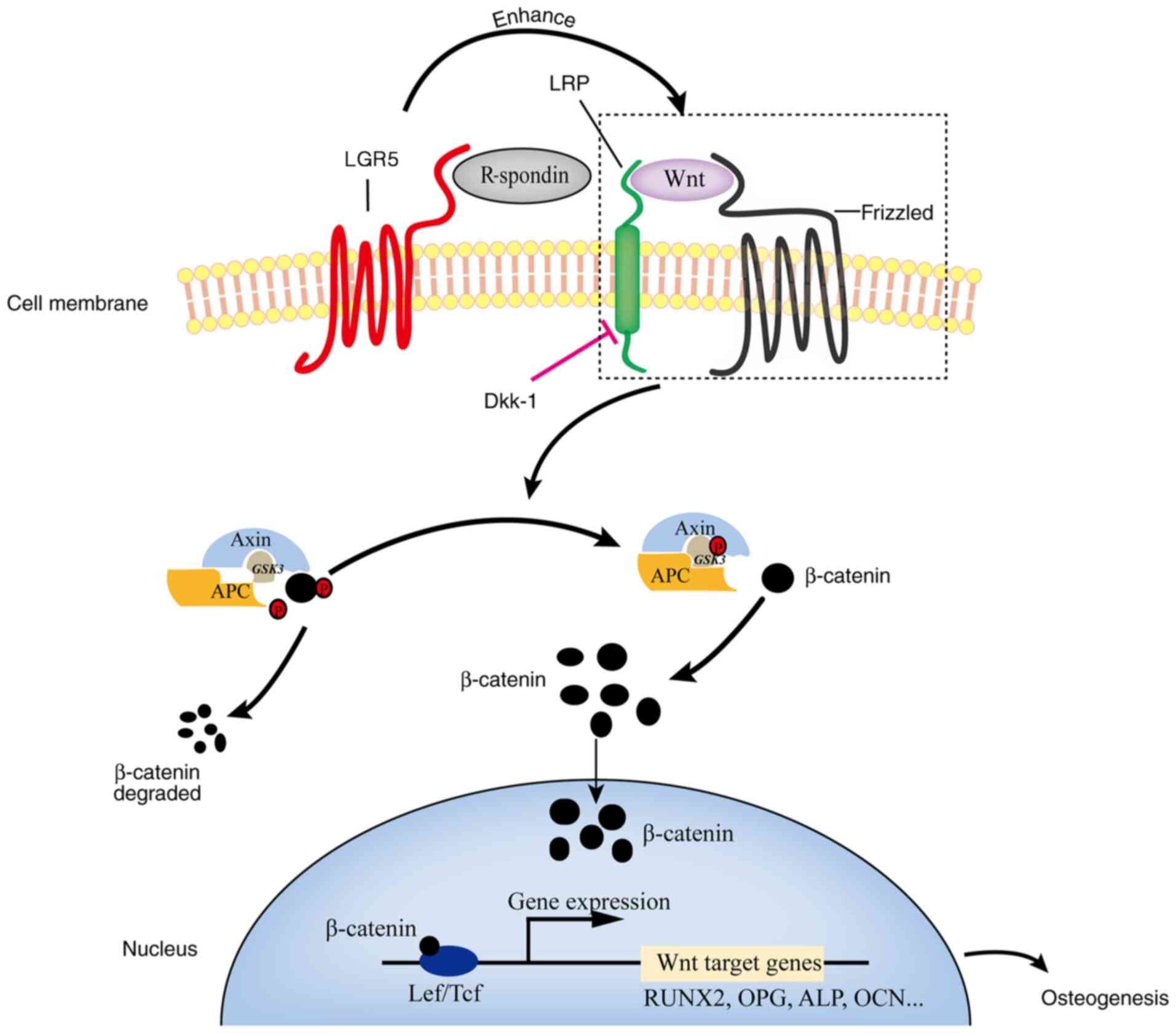 | Figure 6Schematic diagram of the molecular
mechanism by which LGR5 regulates osteogenic differentiation
through Wnt/β-catenin signaling. Once activated by its ligand
R-spondin, LGR5 recruits the frizzled/LRP Wnt receptor complex,
thereby inhibiting β-catenin degradation and increasing the nuclear
accumulation of β-catenin. Inside the cell nucleus, β-catenin binds
to Tcf/Lef transcription factors and then induces the expression of
the downstream target genes (such as RUNX2, OPG, ALP and OCN), thus
enhancing osteogenesis in osteoblasts. LGR5, leucine-rich
repeat-containing G-protein coupled receptor LRP, low density
lipoprotein receptor-related protein; Dkk-1, Dickkopfs-1; APC,
adenomatous polyposis coli; GSK3, glycogen synthase kinase 3; Lef,
lymphoid enhancer-binding factor; Tcf, T cell-factor; Runx2, runt
related transcription factor 2; OPG, osteoprotegerin; ALP, alkaline
phosphatase; OCN, osteocalcin. |
LGR5, also known as G-protein-coupled receptor 49,
is a marker of matured stem cells and is essential for the normal
embryonic development of various organs and tissues (33,34).
LGR5 drives the self-renewal of stem cells in the stomach (11), small intestine, colon (12), hair follicles (10) and mammary glands (35). Similarly, LGR5 is upregulated in the
colorectal cancer, basal cell carcinoma and glioblastoma cell
lines, and promotes the initiation and proliferation of carcinomas
(14-16).
The underlying mechanisms of LGR5 involve the promotion of cancer
stem cell proliferation and self-renewal via the potentiation of
canonical Wnt/β-catenin signaling (36).
LGR5 is a type B LGR protein, along with the closely
related receptors LGR4 and LGR6. Both LGR4 and LGR6 have been
reported to play positive roles in bone formation. LGR4 promotes
bone formation via Wnt/β-catenin signaling and inhibits bone
resorption by suppressing RANK signaling (18,37),
and LGR6 promotes osteoblastic differentiation in MC3T3-E1 cells
through Wnt/β-catenin signaling (19). Considering its homology with LGR4
and LGR6, we hypothesized LGR5 may also play a critical role in
osteoblastic differentiation. In addition, recent studies have
shown that LGR5 is upregulated in bone-associated Ewing sarcoma and
promotes tumorigenesis through Wnt/β-catenin signaling (20). Bone marrow stem cells with LGR5
overexpression have been demonstrated to have greater potential for
the promotion of fracture healing (21). In the present study, MC3T3-E1 cells
that overexpressed LGR5 exhibited enhanced differentiation
potential, as verified by the expression of osteogenic marker
genes, as well as ALP and Alizarin red staining. Since LGR5 and its
family members LGR4 and LGR6 are known as receptors of the Rspo
family, which activate Wnt/β-catenin signaling by complexing with
frizzled/LRP receptors (9,24), whether the potentiating effects of
LGR5 on osteogenesis were mediated through Wnt/β-catenin signaling
were then explored. The results demonstrated that LGR5
overexpression did not alter the transcriptional level of β-catenin
but significantly elevated the protein level of β-catenin in total
cells. Furthermore, western blotting showed that LGR5 reduced
β-catenin phosphorylation levels in the cytoplasm, and increased
the accumulation of β-catenin in the nucleus, indicating that the
degradation of β-catenin in the cytoplasm was decreased. These
results suggest that LGR5 overexpression reinforced the
Wnt/β-catenin signaling pathway by increasing the cytoplasmic
stabilization and nuclear accumulation of β-catenin. As a
consequence, the expression of osteoblastic
differentiation-associated genes was triggered and osteogenesis was
enhanced. In addition, the results indicate that the Wnt signaling
antagonist Dkk-1 blocked the interaction of Wnt ligand with
frizzled and LRP receptors, thereby abrogating the potentiating
effects of LGR5 on osteoblastic differentiation. LGR5 knockdown
antagonized the activation of Wnt/β-catenin and osteogenesis
induced by Rspo-2, while LGR5 knockdown did not affect the
osteogenesis of MC3T3-E1 cells induced by Wnt-3a, a potent
Wnt/β-catenin activator. These results together demonstrate that
LGR5 acted as the Rspo receptor. Previous studies have reported
that LGR5 recruits the LRP-frizzled receptor complex, and then
binds to Wnt ligands (9,24). Overall, the activation of canonical
Wnt signaling enhances osteogenic gene expression and promotes
osteoblastic differentiation (Fig.
6).
In summary, through LGR5 gene regulation in MC3T3-E1
cells, the present study revealed the potentiating effects of LGR5
on osteoblastic differentiation. The study demonstrated that LGR5
promotes osteoblastic differentiation through Wnt/β-catenin
signaling at the cellular level. Therefore, the regulation of
LGR5/Wnt/β-catenin signaling may offer promise as a potential
therapy for osteoporosis and other bone loss conditions. However,
the role of LGR5 in animals requires further study.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the Zhejiang Province
Technology Project (grant no. 2015C33209) and Wenzhou Technology
Project (grant no. Y20150243).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Ethics approval and consent to
participate
Not applicable.
Authors' contributions
WY performed the majority of experiments and drafted
the manuscript. WY and CRX confirm the authenticity of all the raw
data. CRX, FCC and PC assisted with the experiments. CRX and LY
analyzed the data and drafted the manuscript. LY and XYP conceived
the study, supervised the experiments and edited the manuscript.
All authors read and approved the final manuscript.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jiang M, Wang T, Yan X, Liu Z, Yan Y, Yang
K, Qi J, Zhou H, Qian N, Zhou Q, et al: A novel rhein derivative
modulates bone formation and resorption and ameliorates
estrogen-dependent bone loss. J Bone Miner Res. 34:361–374.
2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yadav VK, Balaji S, Suresh PS, Liu XS, Lu
X, Li Z, Guo XE, Mann JJ, Balapure AK, Gershon MD, et al:
Pharmacological inhibition of gut-derived serotonin synthesis is a
potential bone anabolic treatment for osteoporosis. Nat Med.
16:308–312. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Black DM and Rosen CJ: Clinical practice.
Postmenopausal osteoporosis. N Engl J Med. 374:254–262.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Canalis E: Management of endocrine
disease: Novel anabolic treatments for osteoporosis. Eur J
Endocrinol. 178:R33–R44. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Riggs BL and Hartmann LC: Selective
estrogen-receptor modulators-mechanisms of action and application
to clinical practice. N Engl J Med. 348:618–629. 2003.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Leder BZ: Parathyroid hormone and
parathyroid hormone-related protein analogs in osteoporosis
therapy. Curr Osteoporos Rep. 15:110–119. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Luo J, Zhou W, Zhou X, Li D, Weng J, Yi Z,
Cho SG, Li C, Yi T, Wu X, et al: Regulation of bone formation and
remodeling by G-protein-coupled receptor 48. Development.
136:2747–2756. 2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Xu JG, Huang C, Yang Z, Jin M, Fu P, Zhang
N, Luo J, Li D, Liu M, Zhou Y and Zhu Y: Crystal structure of
LGR4-Rspo1 complex: Insights into the divergent mechanisms of
ligand recognition by leucine-rich repeat G-protein-coupled
receptors (LGRs). J Biol Chem. 290:2455–2465. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
de Lau W, Barker N, Low TY, Koo BK, Li VS,
Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M,
et al: Lgr5 homologues associate with Wnt receptors and mediate
R-spondin signalling. Nature. 476:293–297. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jaks V, Barker N, Kasper M, van Es JH,
Snippert HJ, Clevers H and Toftgård R: Lgr5 marks cycling, yet
long-lived, hair follicle stem cells. Nat Genet. 40:1291–1299.
2008.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Barker N, Huch M, Kujala P, van de
Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H,
van den Born M, et al: Lgr5(+ve) stem cells drive self-renewal in
the stomach and build long-lived gastric units in vitro. Cell Stem
Cell. 6:25–36. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Barker N, van Es JH, Kuipers J, Kujala P,
van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H,
Peters PJ and Clevers H: Identification of stem cells in small
intestine and colon by marker gene Lgr5. Nature. 449:1003–1007.
2007.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Morita H, Mazerbourg S, Bouley DM, Luo CW,
Kawamura K, Kuwabara Y, Baribault H, Tian H and Hsueh AJ: Neonatal
lethality of LGR5 null mice is associated with ankyloglossia and
gastrointestinal distension. Mol Cell Biol. 24:9736–9743.
2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kemper K, Prasetyanti PR, De Lau W,
Rodermond H, Clevers H and Medema JP: Monoclonal antibodies against
Lgr5 identify human colorectal cancer stem cells. Stem Cells.
30:2378–2386. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tanese K, Fukuma M, Yamada T, Mori T,
Yoshikawa T, Watanabe W, Ishiko A, Amagai M, Nishikawa T and
Sakamoto M: G-protein-coupled receptor GPR49 is up-regulated in
basal cell carcinoma and promotes cell proliferation and tumor
formation. Am J Pathol. 173:835–843. 2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Nakata S, Campos B, Bageritz J, Bermejo
JL, Becker N, Engel F, Acker T, Momma S, Herold-Mende C, Lichter P,
et al: LGR5 is a marker of poor prognosis in glioblastoma and is
required for survival of brain cancer stem-like cells. Brain
Pathol. 23:60–72. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Vieira GC, Chockalingam S, Melegh Z,
Greenhough A, Malik S, Szemes M, Park JH, Kaidi A, Zhou L,
Catchpoole D, et al: LGR5 regulates pro-survival MEK/ERK and
proliferative Wnt/beta-catenin signalling in neuroblastoma.
Oncotarget. 6:40053–40067. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhu C, Zheng XF, Yang YH, Li B, Wang YR,
Jiang SD and Jiang LS: LGR4 acts as a key receptor for R-spondin 2
to promote osteogenesis through Wnt signaling pathway. Cell Signal.
28:989–1000. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu SL, Zhou YM, Tang DB, Zhou N, Zheng
WW, Tang ZH, Duan CW, Zheng L and Chen J: LGR6 promotes
osteogenesis by activating the Wnt/β-catenin signaling pathway.
Biochem Biophys Res Commun. 519:1–7. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Scannell CA, Pedersen EA, Mosher JT, Krook
MA, Nicholls LA, Wilky BA, Loeb DM and Lawlor ER: LGR5 is expressed
by ewing sarcoma and potentiates Wnt/β-catenin signaling. Front
Oncol. 3(81)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lin W, Xu L, Pan Q, Lin S, Feng L, Wang B,
Chen S, Li Y, Wang H, Li Y, et al: Lgr5-overexpressing mesenchymal
stem cells augment fracture healing through regulation of Wnt/ERK
signaling pathways and mitochondrial dynamics. FASEB J.
33:8565–8577. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Krishnan V, Bryant HU and Macdougald OA:
Regulation of bone mass by Wnt signaling. J Clin Invest.
116:1202–1209. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Carmon KS, Gong X, Lin Q, Thomas A and Liu
Q: R-spondins function as ligands of the orphan receptors LGR4 and
LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci
USA. 108:11452–11457. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Glinka A, Dolde C, Kirsch N, Huang YL,
Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM and Niehrs C:
LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and
Wnt/PCP signalling. EMBO Rep. 12:1055–1061. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gong X, Carmon KS, Lin Q, Thomas A, Yi J
and Liu Q: LGR6 is a high affinity receptor of R-spondins and
potentially functions as a tumor suppressor. PLoS One.
7(e37137)2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yoon JK and Lee JS: Cellular signaling and
biological functions of R-spondins. Cell Signal. 24:369–377.
2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yamada W, Nagao K, Horikoshi K, Fujikura
A, Ikeda E, Inagaki Y, Kakitani M, Tomizuka K, Miyazaki H, Suda T
and Takubo K: Craniofacial malformation in R-spondin2 knockout
mice. Biochem Biophys Res Commun. 381:453–458. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang H, Han X, Wei B, Fang J, Hou X, Lan
T and Wei H: RSPO2 enhances cell invasion and migration via the
WNT/β-catenin pathway in human gastric cancer. J Cell Biochem.
120:5813–5824. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Friedman MS, Oyserman SM and Hankenson KD:
Wnt11 promotes osteoblast maturation and mineralization through
R-spondin 2. J Biol Chem. 284:14117–14125. 2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Leucht P, Minear S, Ten Berge D, Nusse R
and Helms JA: Translating insights from development into
regenerative medicine: The function of Wnts in bone biology. Semin
Cell Dev Biol. 19:434–443. 2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Baron R and Kneissel M: WNT signaling in
bone homeostasis and disease: From human mutations to treatments.
Nat Med. 19:179–192. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
33
|
Barker N and Clevers H: Leucine-rich
repeat-containing G-protein-coupled receptors as markers of adult
stem cells. Gastroenterology. 138:1681–1696. 2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu D, He XC, Qian P, Barker N, Trainor
PA, Clevers H, Liu H and Li L: Leucine-rich repeat-containing
G-protein-coupled receptor 5 marks short-term hematopoietic stem
and progenitor cells during mouse embryonic development. J Biol
Chem. 289:23809–23816. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Plaks V, Brenot A, Lawson DA, Linnemann
JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD and Werb Z:
Lgr5-expressing cells are sufficient and necessary for postnatal
mammary gland organogenesis. Cell Rep. 3:70–78. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Xu L, Lin W, Wen L and Li G: Lgr5 in
cancer biology: Functional identification of Lgr5 in cancer
progression and potential opportunities for novel therapy. Stem
Cell Res Ther. 10(219)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G,
Huang J, Dai W, Li C, Zheng C, et al: LGR4 is a receptor for RANKL
and negatively regulates osteoclast differentiation and bone
resorption. Nat Med. 22:539–546. 2016.PubMed/NCBI View Article : Google Scholar
|