The central dogma is one of the most important basic
laws in modern biology that reveals the process by which genetic
information is conveyed from DNA to RNA through transcription, and
subsequently to protein through translation (1). Recent advancements in genetic
technologies have revealed that genetic information encoded in the
genome is not only an alignment of base sequences but is also
associated with other multifarious levels of regulation, such as
DNA methylation and histone modification (2). The discovery of these epigenetic
research results has driven the central dogma. Increasing evidence
suggest that RNA plays a vital role in the central dogma (3). Genetic material is regulated at
various levels, in addition to DNA and RNA levels (4). RNA modification is a type of
post-transcriptional modification that controls important pathways,
such as RNA processing and metabolism (5). With the advancements in sequencing
technology, the dynamics of RNA modification and its biological
functions have received considerable attention from scientists and
have become new research hotspots in the field of epigenetics
(6-9).
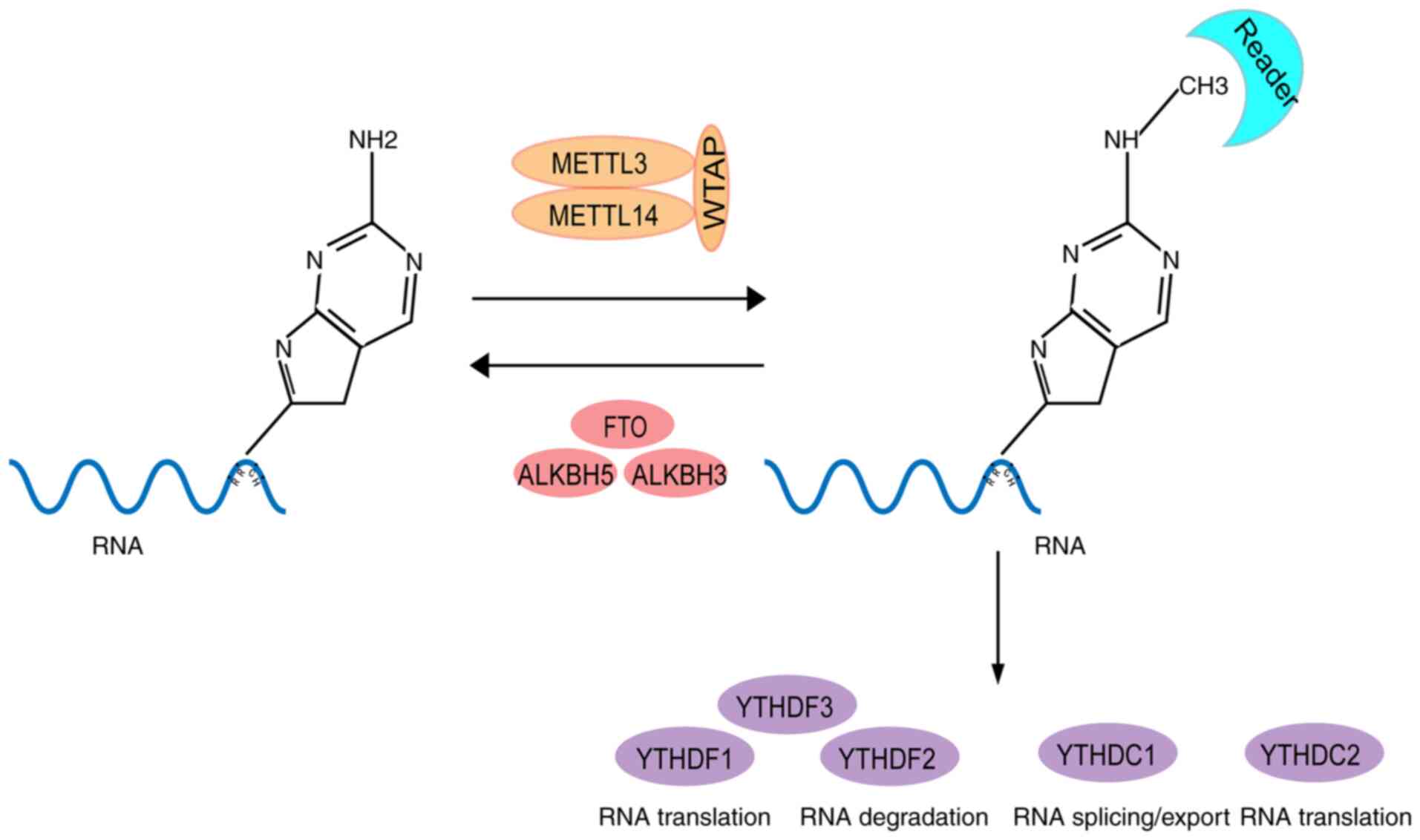
RNA methylation has been extensively identified in
vertebrates, plants, yeasts, bacteria, archaea and viruses
(10), and N6-methyladenine (m6A)
is the most common form of RNA methylation (11). Generally, m6A modifications occur at
the conserved RRACH motif (R=G or A; H=A, C or U) and are
concentrated near the stop codons of mRNAs (12) (Fig.
1). Notably, with the development of enzymatic technology, m6A
modification enzymes have been identified, among which,
methyltransferase-like (METTL)3, METTL14 and Wilms' tumor
1-associating protein (WTAP) complexes are the main m6A
methyltransferases (13-15),
whereas fat-mass and obesity-associated protein (FTO) and
α-ketoglutarate-dependent dioxygenase alkB homologue 5 (ALKBH5) are
demethylases (16,17) (Fig.
1). Currently, m6A is the most studied RNA modification
(18). m6A modification plays
various post-transcriptional regulatory roles, including
transcription regulation, selective splicing, stabilization and
translation of RNA, by binding proteins that contain the YTH domain
(19) (Fig. 1). In addition, m6a modifications
regulate cellular biological functions involved in cell
differentiation, embryonic development and disease occurrence
(20).
The discovery of m6A methyltransferases and
demethylases proved that RNA modification is dynamic and
reversible, and promoted the study of RNA modifications from
micro-regulation mechanisms to epitranscriptome levels (21). However, other modification forms,
including 5-methylcytosine (22)
and 1-methyladenine (23) are
currently at initial stages, and their modification enzymes,
dynamic regulation and biological functions require further studies
and development.
Increasing evidence suggest that modification by m6A
is mediated by methyltransferase complexes that are composed of
several proteins (35). METTL3 was
the first m6A methyltransferase that was identified (36). Methyltransferase MT-A70 was
identified as part of a large protein complex that was isolated
from enzymatic mammalian cell nuclear extracts (37), and is the leading catalytic enzyme
of the m6A system (9,38). METTL3 contains two Cys-Cys-Cys-His
(CCCH)-type zinc finger motifs at the N-terminus and one catalytic
motif [D/N/S/H]PP(Y/F/W)] in the methyltransferase domain (39,40).
It has functions in all the stages of the RNA lifecycle, including
pre-mRNA splicing (41), nuclear
export (17), translation
regulation (42), mRNA decay
(43) and microRNA (miRNA/miR)
processing (44). Generally, METTL3
forms a stable heterodimeric complex with METTL14, which is another
important methyltransferase of m6A (45), with the help of other components of
writers. This methylation was reported to occur at the N6 position
of adenosine on mRNA and was speculated to be a co-transcriptional
methylation (46). Notably,
disruption of METTL3 homologs results in severe developmental
defects in yeasts, and METTL3 homologs have lethal phenotypes in
Arabidopsis and mice (47-50).
Recent evidence suggested that METTL3 is upregulated and plays an
oncogenic role involving increased m6A expression levels in
different types of cancer, such as bladder cancer (51), lung cancer (52), colorectal cancer (53), glioma (54), breast cancer (55), leukaemia (56) and other cancers including gastric
cancer and melanoma (57,58). However, other studies have
demonstrated opposing results from the same types of cancer
(59-61).
The potential molecular mechanisms have been investigated and
include the regulation of downstream non-coding RNAs (62), modulation of miRNA processing via
DGCR8(51), regulation of apoptosis
(57) and regulation of the
PI3K/AKT pathway (62,63).
Another core writer is METTL14. Despite having ~22%
sequence identity and an almost identical topological structure
with the methyltransferase domains of METTL3, METTL14 is considered
a pseudomethyltransferase (39,45).
The function of METTL14 in the complex remains unclear. In a
surface electrostatic potential analysis, RNA binding affinity and
methyltransferase activity were revealed to moderately decrease in
the complex with double mutations in K297E and R298E, suggesting
that METTL14 may be involved in RNA interaction; however, the
binding sites of S-adenosylmethionine and the DPPW functional
domains of METTL14 are responsible for the catalysis of m6A
formation (64). METTL14 is a vital
member of the m6A methyltransferase complex (65). Notably, several studies have
demonstrated that METTL3 and METTL14 are associated with each other
(14,45,66).
Once METTL3 and METTL14 function individually, they exhibit nearly
undetectable methyltransferase activity, whereas the METTL3-METTL14
complex displays methyltransferase activity (14). However, whether METTL14 exhibits
methyltransferase activity after binding to additional factors
remains unknown. METTL14 is also involved in the regulation of
several tumour processes (67,68).
For example, the METTL14-m6A-Notch1 pathway plays a critical role
in bladder tumorigenesis and bladder tumour-initiating cells
(69). In haematopoietic stem cells
and liver tumour cells, METTL14 is downregulated and attenuates the
tumorigenesis of acute myeloid leukaemia (AML) (70).
Another component of the human m6A methyltransferase
complex is the WTAP, which plays a critical role in m6A formation
via a mechanism that differs from the mechanisms observed in METTL3
and METTL14(71). In a WTAP
knockdown study, global m6A levels in human cell lines were
reported to be markedly decreased, which indicates its significance
in producing a distinct landscape of mRNA methylation (72). WTAP predominantly acts as a
regulatory subunit that initially binds to target RNA and
subsequently recruits dimers formed by the catalytic subunits,
METTL3 and METTL14, to perform catalytic functions owing to a lack
of a catalytic domain (71).
Furthermore, METTL3 levels were revealed to be important to WTAP
protein homoeostasis, whereby METTL3 regulates WTAP expression at
various levels via different mechanisms, including mRNA translation
and stabilization (73). Taken
together, these findings suggest that the components of the
methyltransferase complex are essential in the formation of m6A. In
addition, WTAP is overexpressed in different types of tumours,
including AML (15,74,75),
and interacts with different proteins associated with cell
proliferation and RNA processing (75). Notably, disruption of WTAP results
in embryonic lethality, which is indicative of its vital biological
function in the development of vertebrates (76).
In addition to the core complex of METTL3, METTL14
and WTAP, several other proteins have been implicated in regulating
RNA m6A. For example, Virilizer and Hakai were identified as
components associated with WTAP in mammalian cells. Endogenous
protein complexes from different species in metazoans were studied
via quantitative mass spectrometry, and ZC3H13-WTAP-Virilizer-Hakai
was identified as an evolutionarily conserved complex (77). ZC3H13 plays a critical role in
anchoring WTAP, Virilizer and Hakai in the nucleus to facilitate
m6A methylation, and regulates mESC self-renewal (77). ZC3H13 also participates in the
development and progression of different types of tumours (68,78).
It has been reported that ZC3H13 expression is substantially
downregulated in clear cell renal cell carcinoma tissues (79). Conversely, ZC3H13 expression is
substantially upregulated in colon adenocarcinoma tumour tissues
compared with adjacent mucosa (80).
The reversible and dynamic regulation of m6A
modification is mediated by the functional interaction between m6A
writers and erasers; however, to identify downstream biological
functions, m6A must be recognised by several readers (46,100).
Several YTH domain family (YTHDF) members, including YTHDF1,
YTHDF2, YTHDF3, YTH domain containing (YTHDC)1 and YTHDC2, have
been identified as general ‘reader’ proteins (43).
Unlike YTHDF1, YTHDF2 and YTHDF3, which are located
in the cytoplasm, YTHDC1 is located in YT bodies adjacent to
nuclear speckles in nucleus (116). YTHDC1 cooperates with nuclear RNA
export factor 1 and the three prime repair exonuclease mRNA export
complex by interacting with SRSF3 and exports m6A-methylated mRNAs
from the nucleus (116). YTHDC1
interacts with metadherin and affects cancer development and
progression (117). YTHDC1 also
plays essential roles in the development of spermatogonia in males
and the growth and maturation of oocytes in females (118).
YTHDC2, the only RNA helicase-containing and
multi-domain m6A reader, has been demonstrated to exhibit
ATP-dependent RNA helicase activity (119,120). YTHDC2 improves the translation
efficiency of target mRNA by interacting with meiosis-specific
coiled-coil domain and 5'-3'exoribonuclease 1 after recognising m6A
(121). A previous study reported
that YTHDC2 affects the expression of drug-metabolizing P450
isoforms by mediating CYP2C8 mRNA degradation (122). YTHDC2 also plays a conserved role
in mouse germ cell fate transition (123). In addition, YTHDC2 contributes to
the metastasis of colon tumours and the proliferation of Huh7 HCC
cells (120,124).
In addition to the members of the YTHDF, other
proteins that act as m6A readers have been identified. The
RNA-binding protein, HNRNPA2B1, binds to m6A-modifying RNAs, and
its biochemical footprint is consistent with that of the m6A
consensus motif (125). HNRNPA2B1
binds to m6A in subsets of primary miRNA transcripts, interacts
with the DGCR8 microprocessor complex protein and increases primary
miRNA processing (125). In
addition, insulin-like growth factor 2 mRNA-binding proteins
(IGF2BPs), including IGF2BP1/2/3, can recognise m6A and improve the
stability and translation of mRNAs in an m6A-dependent manner;
however, this mechanism requires further investigation (126).
Osteoporosis is the most common bone disorder
worldwide, which affects >200 million people, and is
characterised by decreased bone mineral density (BMD) and increased
risk of osteoporotic fracture (127,128). The main pathological changes of
osteoporosis are characterised by low bone mass and excessive
accumulation of adipose tissue in the bone marrow milieu (129), and bone homoeostasis plays an
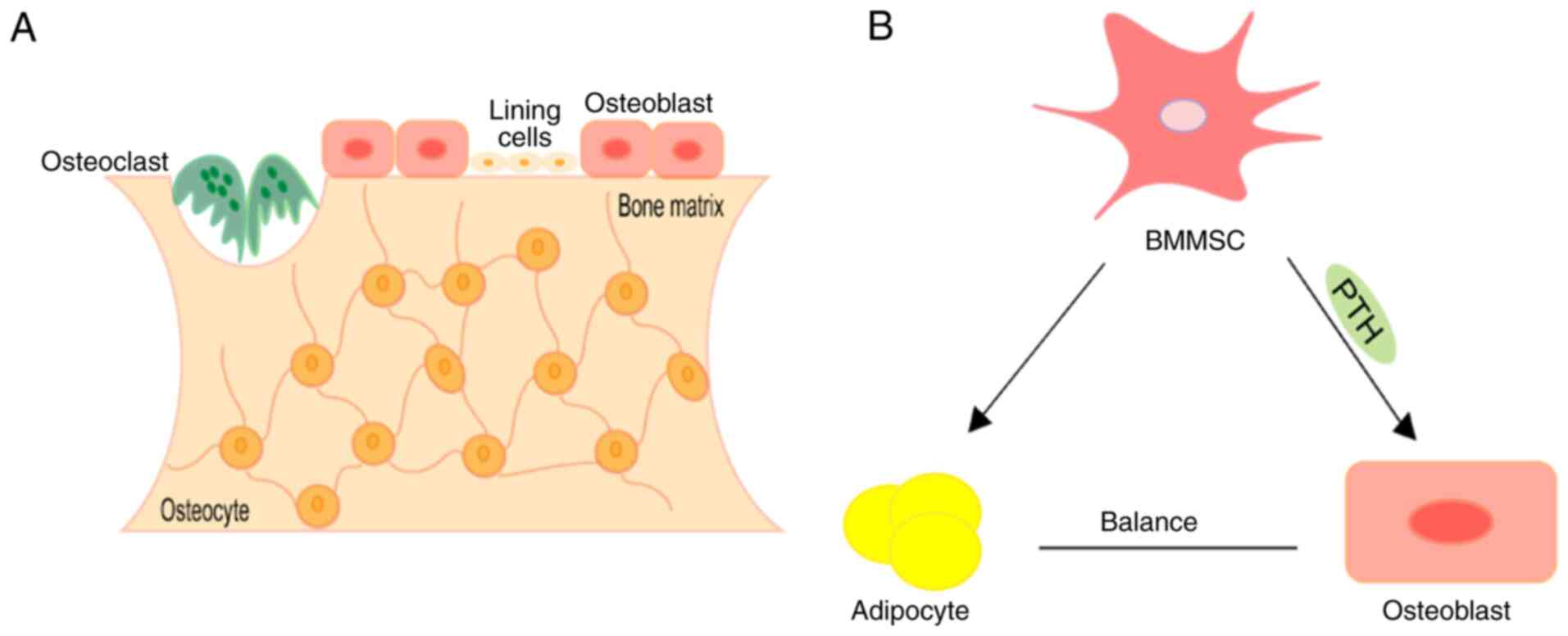
essential role in the pathogenesis of osteoporosis (11). Bone homoeostasis is mainly
maintained by osteoblasts, osteocytes and osteoclasts (130) (Fig.
2A). Osteoblasts produce bone by synthesising extracellular
matrix containing various proteins, particularly type I collagen
(131,132). The extracellular matrix is
deposited as osteoid and is subsequently mineralized through the
accumulation of calcium phosphate in the form of hydroxyapatite
(133). Conversely, osteoclasts,
through the secretion of hydrochloric acid and proteolytic enzymes,
can dissolve minerals and lysis the bone matrix (134). Osteocytes are the main cellular
component of bone tissue (135).
Osteocytes control bone homoeostasis by maintaining the balance
between the function of bone-forming osteoblasts and bone-resorbing
osteoclasts (136). The common
progenitors for osteoblasts and marrow adipocytes are bone marrow
mesenchymal stem cells (BMMSCs) (137). The osteogenic and adipogenic
differentiation of BMMSCs must maintain balance under accurate
spatio-temporal control to defend skeletal health (138) (Fig.
2B). With ageing or other pathological stimuluses, BMMSCs have
a disposition to differentiate into adipocytes, leading to the
ascendent in marrow adiposity and gradual bone loss (139,140). The variations in bone
micro-architecture result in elevated skeletal fragility and an
inclination to fracture (141).
Recently, several studies have revealed different molecular
mechanisms associated with osteoporosis, which are associated with
m6A modification (142,143).
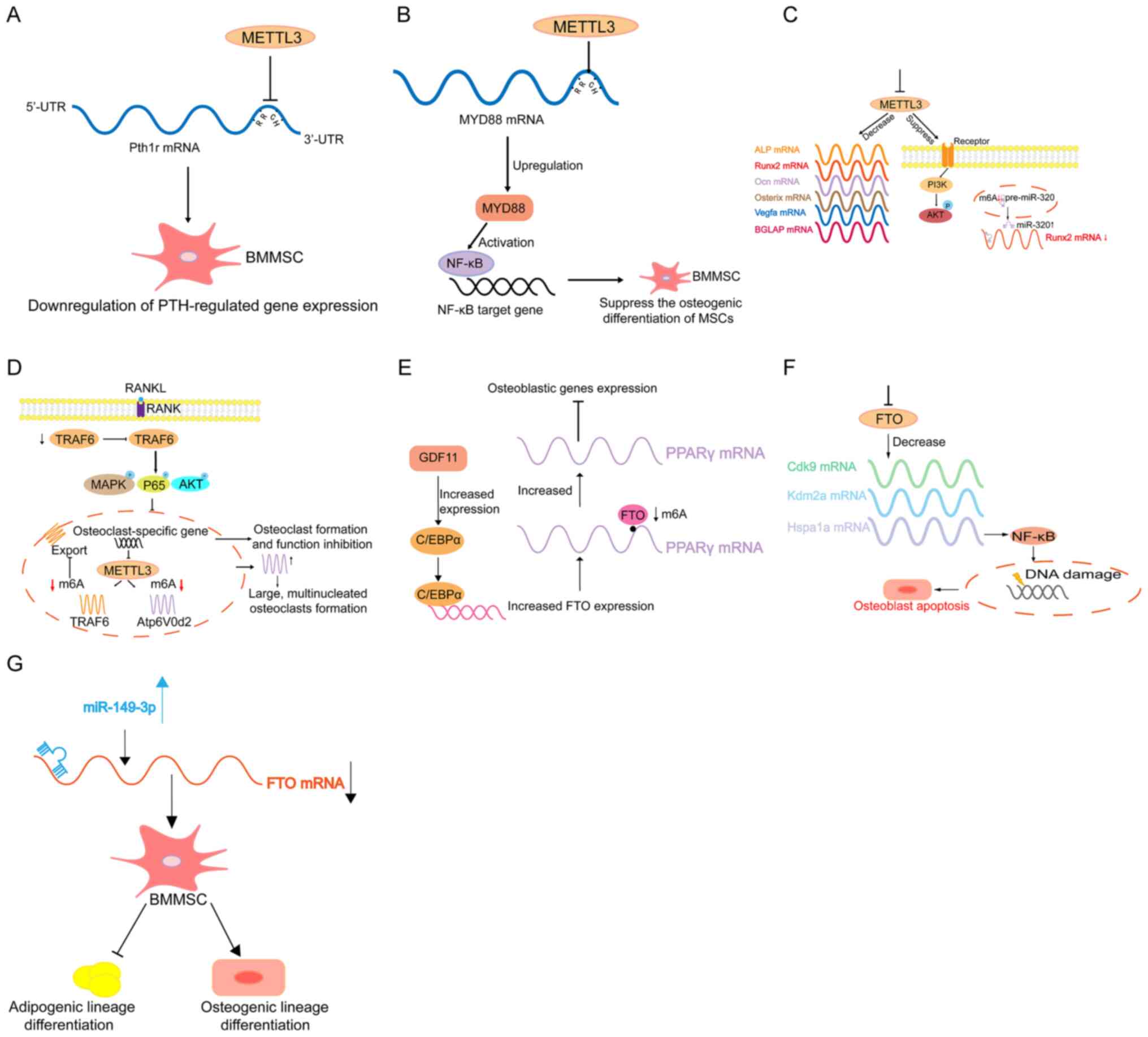
A previous study revealed that the disruption of
Mettl3 in bone marrow mesenchymal stem cells (MSCs) induces
pathological features of osteoporosis in mice (143). It was demonstrated that the
disfunction form of METTL3 resulted in destroyed bone formation,
abnormal osteogenic differentiation and improved marrow adiposity
(143). However, overexpression of
METTL3 in BMMSCs protects mice against osteoporosis caused
by oestrogen deficiency. Mechanistically, the PTH/Pth1r signalling
axis is the target downstream pathway for m6A regulation in BMMSCs
(143) (Fig. 3A). However, it has also been
reported that METTL3 upregulates MYD88 expression by enhancing m6A
modification to MYD88-RNA, consequently leading to the activation
of NF-κB, which is a repressor of osteogenesis, and inhibits
osteogenic progression (144)
(Fig. 3B). Furthermore,
METTL3 expression increases in BMMSCs undergoing osteogenic
induction, and disruption of METTL3 downregulates the expression of
bone formation-related genes, such as Runx2 and Osterix (143). In addition, following METTL3
knockdown, the alkaline phosphatase activity and mineralised nodule
formation also decline (145).
Vascular endothelial growth factor (VEGF) plays important roles in
bone formation and endothelial development (146,147). It has been reported that METTL3
knockdown decreases VEGFA expression, as well as the expression
level of its splice variants, including VEGFA-164 and VEGFA-188 in
BMMSCs (145). Another study
revealed that METTL3 disruption decreases m6A methylation levels
and hampers osteogenic differentiation of BMMSCs, thus decreasing
bone mass. In addition, METTL3 disorder can affect m6A methylation
of RUNX2 and precursor (pre-)miR-320 (Fig. 3C) (148). METTL3 disruption suppresses this
process and thus disturbs the normal osteogenic differentiation,
which results in osteoporosis (148). Recently, a study reported that
METTL3 expression increases during osteoclast differentiation,
whose deficiency results in increased size but decreased
bone-resorbing ability of osteoclasts through the mechanism
involving Atp6v0d2 mRNA degradation mediated by YTHDF2 and TRAF6
mRNA nuclear export (149). This
suggests that METTL3 may contribute to osteoporosis by regulating
osteoclast differentiation (Fig.
3D).
Arginine-316 in human FTO corresponds to
Arginine-313 in mice, which is essential for FTO catalytic activity
(150). A previous study reported
that FTO R313A/R313A mice not only decreased the body and bone
length, which was associated with a substantial reduction in BMD
and bone mineral content (BMC), but also notably abated the
alkaline phosphatase activity, indicating osteoblast function
(151). Peroxisome
proliferator-activated receptor γ (PPARγ) is a transcriptional
factor that maintains the balance between adipocyte and osteoblast
differentiation from BMMSCs, that is, accelerating the
differentiation of adipocytes and inhibiting osteoblast
differentiation (152). It has
been reported that PPARγ mRNA is targeted and demethylated by FTO,
which upregulates PPARγ mRNA expression, eventually promoting the
differentiation of osteoporotic BMMSCs to adipocytes, and decreases
bone formation in the process of osteoporosis (153) (Fig.
3E). Another study revealed that regardless of the mouse models
lacking FTO globally or selectively, both exhibited age-related
decreases in bone volume, in both the trabecular and cortical
compartments (154). The mechanism
demonstrated that FTO disruption in osteoblasts changes the Hsp70
transcripts (Hspa1a) and other genes involved in the DNA repair
pathway containing conserved m6A motifs required for demethylation
by FTO, thus affecting osteoblasts (154) (Fig.
3F). Furthermore, several FTO single nucleotide polymorphisms
(SNPs) are associated with BMD variations in Chinese populations
(155). In addition, miR-149-3p
inhibits the differentiation of the adipogenic lineage and enhances
the differentiation of the osteogenic lineage by targeting FTO
(156) (Fig. 3G). Thus, FTO may be a novel
candidate for osteoporosis (156).
Considering that inflammatory factors can hamper
osteogenesis, studies have aimed to investigate the association
between m6A, inflammation and osteoporosis. Several cytokines have
been identified that participate in the development of
osteoporosis, including RANKL, colony-stimulating factor (CSF)-1,
interleukin-34(157) and
granulocyte-macrophage-CSF (158).
A study revealed that osteoblast differentiation and Smad-dependent
signalling is inhibited after disrupting METTL3 by stabilising
Smad7 and Smurf1 mRNA transcripts, which is mediated by
YTHDF2(159). In addition, the
production of proinflammatory cytokines increases following METTL3
deficiency by activating the MAPK signalling pathway to promote the
osteoblast inflammatory response (60). Furthermore, YTHDF2 knockdown
enhances the LPS-induced expression levels of IL-6, tumor necrosis
factor (TNF)-α, IL-1β and IL-12, which contributes to bone
inflammation and osteoporosis through sophisticated mechanisms
(160,161).
Arthritis is an inflammatory disease that occurs in
several human joints and surrounding tissues. It is classified into
dozens of diseases that are triggered by multiple pathological
conditions, such as inflammation, infection, degeneration and
trauma (162). The main clinical
characteristics of arthritis include red, swollen, hot, painful,
dysfunctional and deformed joints, which all result in joint
disability and decreased quality of life (163). Osteoarthritis (121) and rheumatoid arthritis (32) are the most common types of arthritis
with different pathophysiological mechanisms and exhibit similar
clinical features. The role of epigenetics, particularly of RNA
modification in arthritis, has attracted great interest.
OA is the most common chronic joint disease
characterised by pain, stiffness and mobility difficulties
(164). OA is caused by a complex
interaction among diverse molecular factors involved in integrity,
genetic susceptibility, local inflammation, mechanical forces and
other cellular biochemical processes. The most probable cause of OA
is damage to articular cartilage via physical forces (165,166). Damage to articular cartilage may
lead to degenerative OA in which several factors, including
inflammatory response to various components of cartilage, are
involved (167-170).
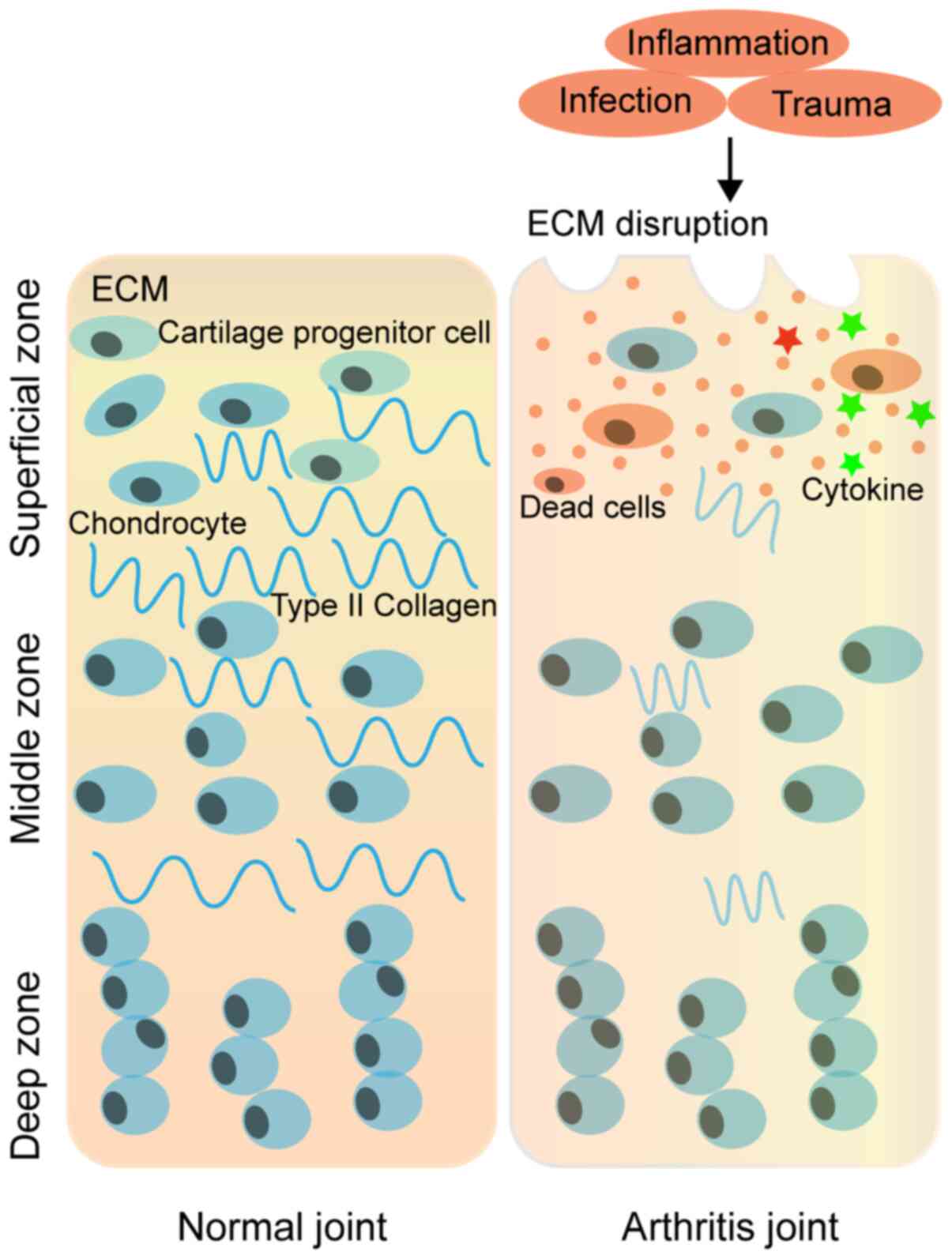
Progressive cartilage degeneration is involved in chondrocyte
reduction and in the variation of molecular components of
chondrocytes in self-synthetized extracellular matrix (ECM) in the
process of OA development (171)
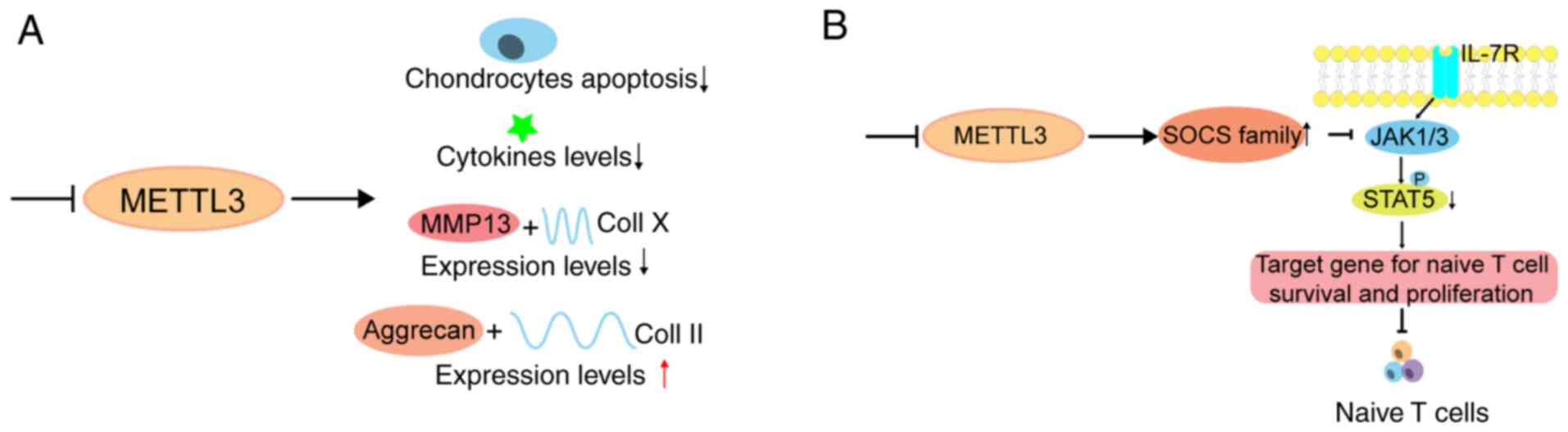
(Fig. 4). A study reported that
following treatment of ATDC5 cells with IL-1β, both the abundance
of METTL3 mRNA and the ratio of m6A methylated mRNA of total mRNA
was enhanced. Although disturbance of METTL3 lowered the proportion
of IL-1β-induced apoptosis, it inhibited IL-1β-induced increased
levels of inflammatory cytokines and activation of NF-kB signalling
in chondrocytes (172). In
addition, disruption of METTL3 improves destruction of the ECM by
decreasing matrix metalloproteinase-13 and collagen (Coll) X
expression levels, and elevating the expression levels of Aggrecan
and Coll II (172) (Fig. 5A). Simultaneously, OA is a
degenerative disease of the synovial joint. The synovial membrane
is responsible for the inflammatory response, which causes the
release of macrophage-derived pro-inflammatory cytokines, such as
RA, including IL-6(173). A study
revealed that suppressing the overexpression of IL-6 in synovial
fibroblasts is a prospective way to hamper the development and
progression of OA (173,174). Thus, it is apparent that m6A also
participates in OA by regulating several cytokines. The reasons for
the occurrence of OA are complicated, and research on the
association between m6A and OA is lacking; thus, further studies
are required.
Multiple pathological factors contribute to the
development of RA, including autoimmunity, various pathogen
infections and genetic factors (175). A study revealed that METTL3
deficiency in mouse T cells disrupts T cell homoeostasis and
differentiation (176), which
implies that m6A has a potential effect on the occurrence and
development of RA through the regulation of the immune system
(Fig. 5B). Another study revealed
that METTL3, but not other m6A methylation-related proteins,
including METTL14, FTO, ALKBH5, YTHDF1 and YTHDF2, is upregulated
in RA (177). Increased METTL3
expression is positively associated with CRP and ESR levels, which
are the two main markers of RA disease activity. It has been
reported that METTL3 participates in RA by hindering the
proliferation and inflammatory response of macrophages (177). However, whether METTL3 mediates
the progression and development of RA requires further
investigation (177). Several
studies have indicated that m6A-associated SNPs play essential
roles in gene expression and mRNA homoeostasis, which may
subsequently lead to the occurrence of disease (178,179). The RA GWAS dataset identified
several RA-associated m6A-SNPs, which may play regulatory roles in
the pathogenesis of RA, and some of them were associated with the
mRNA expression of local RA-related genes (178). Taken together, these findings
provide insight into the association between SNPs and RA. However,
further studies are required to determine the molecular mechanisms.
RA is a systemic disease with several immunological events, and the
key pathogenetic changes are the production of pro-inflammatory
cytokines from macrophage-like synoviocytes, including IL-1, IL-6
and TNF (180). YTHDF2 disruption
notably increases the expression levels of LPS-induced IL-6, TNF-α,
IL-1 β and IL-12. Thus, m6A also participates in RA pathology by
regulating several cytokines (160). Furthermore, synovium inflammation
is involved in extensive activated CD4+ T cells
(181), suggesting that disturbed
homoeostasis of CD4+ T cells plays a critical role in
the development of RA (182,183). Since T cells mediate the adaptive
immune response and contain several subgroups (184), the concrete details and molecular
mechanisms require further investigation.
Currently, the number of different types of
chemical modifications in RNA has reached ~140(7). m6A is a pivotal epitranscriptomic
modification in common orthopaedic diseases (187,193,194). Several studies have revealed the
underlying molecular mechanisms of m6A modifications in cancer
(51,63,195,196); however, research on the
association between m6A and bone-related diseases is still lacking
and should be addressed. In addition to the m6A-associated proteins
mentioned, many other proteins, including writers (METTL5, METTL16,
KIAA1429, RBM15, VIRMA and ZCCHC4) and readers (IGF2BP1, IGF2BP2,
IGF2BP3 and eIF3), have been demonstrated to participate in the
formation of m6A (19,197). However, whether these proteins are
involved in the pathogenesis and development of bone-related
diseases remains unknown. m6A methyltransferase can methylate
non-coding RNAs (198); however,
studies on the interaction between non-coding RNAs and m6A in
bone-related diseases are scarce. In addition, several bone-related
diseases are associated with the activities of m6A, such as
epigenetic m6A modification, which plays an important role in the
ossification of the ligamentum flavum (199). Recently, a review summarized the
association between m6A and musculoskeletal disorders (200). However, the roles of m6A in T cell
homoeostasis and differentiation remain unclear, both of which
closely associated with the occurrence and development of
osteoporosis, osteoarthritis, rheumatoid arthritis and
osteosarcoma, which is a subject of novel and extensive research.
Thus, further studies are required to determine the underlying
cellular and molecular mechanisms of m6A in bone-related diseases.
In conclusion, m6A may serve as a promising molecular target in
regenerative medicine, bone tissue engineering and in the treatment
of bone-related cancer.
Not applicable.
Funding: No funding was received.
Not applicable.
XZ conceived and designed the present review. YH
and XZ performed the data analysis and interpretation. YH and XZ
drafted the initial manuscript. Data authentication is not
applicable. Both authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Rits S, Olsen BR and Volloch V:
Protein-encoding RNA to RNA information transfer in mammalian
cells: RNA-dependent mRNA amplification. Identification of chimeric
RNA intermediates and putative RNA end products. Ann Integr Mol
Med. 1:23–47. 2019.PubMed/NCBI
|
|
2
|
Howie H, Rijal CM and Ressler KJ: A review
of epigenetic contributions to post-traumatic stress disorder.
Dialogues Clin Neurosci. 21:417–428. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Holt CE and Schuman EM: The central dogma
decentralized: New perspectives on RNA function and local
translation in neurons. Neuron. 80:648–657. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Maydanovych O and Beal PA: Breaking the
central dogma by RNA editing. Chem Rev. 106:3397–3411.
2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Thakur P, Estevez M, Lobue PA, Limbach PA
and Addepalli B: Improved RNA modification mapping of cellular
non-coding RNAs using C- and U-specific RNases. Analyst.
145:816–827. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and erasers. Mol Cell. 74:640–650. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu J, Dou X, Chen C, Chen C, Liu C, Xu
MM, Zhao S, Shen B, Gao Y, Han D and He C:
N6-methyladenosine of chromosome-associated regulatory
RNA regulates chromatin state and transcription. Science.
367:580–586. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13(117)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shinoda K, Suda A, Otonari K, Futaki S and
Imanishi M: Programmable RNA methylation and demethylation using
PUF RNA binding proteins. Chem Commun (Camb). 56:1365–1368.
2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen X, Chen X, Zhou Z, Mao Y, Wang Y, Ma
Z, Xu W, Qin A and Zhang S: Nirogacestat suppresses RANKL-Induced
osteoclast formation in vitro and attenuates LPS-Induced bone
resorption in vivo. Exp Cell Res. 382(111470)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
The RNA methyltransferase METTL3 promotes
oncogene translation. Cancer Discov. 6(572)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chen Y, Peng C, Chen J, Chen D, Yang B, He
B, Hu W, Zhang Y, Liu H, Dai L, et al: WTAP facilitates progression
of hepatocellular carcinoma via m6A-HuR-dependent epigenetic
silencing of ETS1. Mol Cancer. 18(127)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li J, Zhu L, Shi Y, Liu J, Lin L and Chen
X: m6A demethylase FTO promotes hepatocellular carcinoma
tumorigenesis via mediating PKM2 demethylation. Am J Transl Res.
11:6084–6092. 2019.PubMed/NCBI
|
|
17
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Parker MT, Knop K, Sherwood AV, Schurch
NJ, Mackinnon K, Gould PD, Hall AJ, Barton GJ and Simpson GG:
Nanopore direct RNA sequencing maps the complexity of Arabidopsis
mRNA processing and m6A modification. eLife.
9(e49658)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liao S, Sun H and Xu C: YTH Domain: A
family of N6-methyladenosine (m6A) readers.
Genomics Proteomics Bioinformatics. 16:99–107. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao YL, Liu YH, Wu RF, Bi Z, Yao YX, Liu
Q, Wang YZ and Wang XX: Understanding m6A function
through uncovering the diversity roles of YTH domain-containing
proteins. Mol Biotechnol. 61:355–364. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li F, Kennedy S, Hajian T, Gibson E,
Seitova A, Xu C, Arrowsmith CH and Vedadi M: A radioactivity-based
assay for screening human m6A-RNA methyltransferase, METTL3-METTL14
complex, and demethylase ALKBH5. J Biomol Screen. 21:290–297.
2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu Y and Santi DV: m5C RNA and m5C DNA
methyl transferases use different cysteine residues as catalysts.
Proc Natl Acad Sci USA. 97:8263–8265. 2000.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Roovers M, Wouters J, Bujnicki JM, Tricot
C, Stalon V, Grosjean H and Droogmans L: A primordial RNA
modification enzyme: The case of tRNA (m1A) methyltransferase.
Nucleic Scids Res. 32:465–476. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Grabowski P: Physiology of bone. Endocr
Dev. 16:32–48. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Scholtysek C, Kronke G and Schett G:
Inflammation-associated changes in bone homeostasis. Inflamm
Allergy Drug Targets. 11:188–195. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Suominen H: Muscle training for bone
strength. Aging Clin Exp Res. 18:85–93. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Fu R, Lv WC, Xu Y, Gong MY, Chen XJ, Jiang
N, Xu Y, Yao QQ, Di L, Lu T, et al: Endothelial ZEB1 promotes
angiogenesis-dependent bone formation and reverses osteoporosis.
Nat Commun. 11(460)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Landete-Castillejos T, Kierdorf H, Gomez
S, Luna S, García AJ, Cappelli J, Pérez-Serrano M, Pérez-Barbería
J, Gallego L and Kierdorf U: Antlers-Evolution, development,
structure, composition, and biomechanics of an outstanding type of
bone. Bone. 128(115046)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hassan MQ, Tye CE, Stein GS and Lian JB:
Non-coding RNAs: Epigenetic regulators of bone development and
homeostasis. Bone. 81:746–756. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Bocheva G and Boyadjieva N: Epigenetic
regulation of fetal bone development and placental transfer of
nutrients: Progress for osteoporosis. Interdiscip Toxicol.
4:167–172. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Adamik J, Roodman GD and Galson DL:
Epigenetic-based mechanisms of osteoblast suppression in multiple
myeloma bone disease. JBMR Plus. 3(e10183)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Marini F, Cianferotti L and Brandi ML:
Epigenetic mechanisms in bone biology and osteoporosis: Can they
drive therapeutic choices? Int J Mol Sci. 17(1329)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ghayor C and Weber FE: Epigenetic
regulation of bone remodeling and its impacts in osteoporosis. Int
J Mol Sci. 17(1446)2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Montecino M, Stein G, Stein J, Zaidi K and
Aguilar R: Multiple levels of epigenetic control for bone biology
and pathology. Bone. 81:733–738. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kobayashi M, Ohsugi M, Sasako T, Awazawa
M, Umehara T, Iwane A, Kobayashi N, Okazaki Y, Kubota N, Suzuki R,
et al: The RNA methyltransferase complex of WTAP, METTL3, and
METTL14 regulates mitotic clonal expansion in adipogenesis. Mol
Cell Biol. 38:e00116–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhou L, Yang C, Zhang N, Zhang X, Zhao T
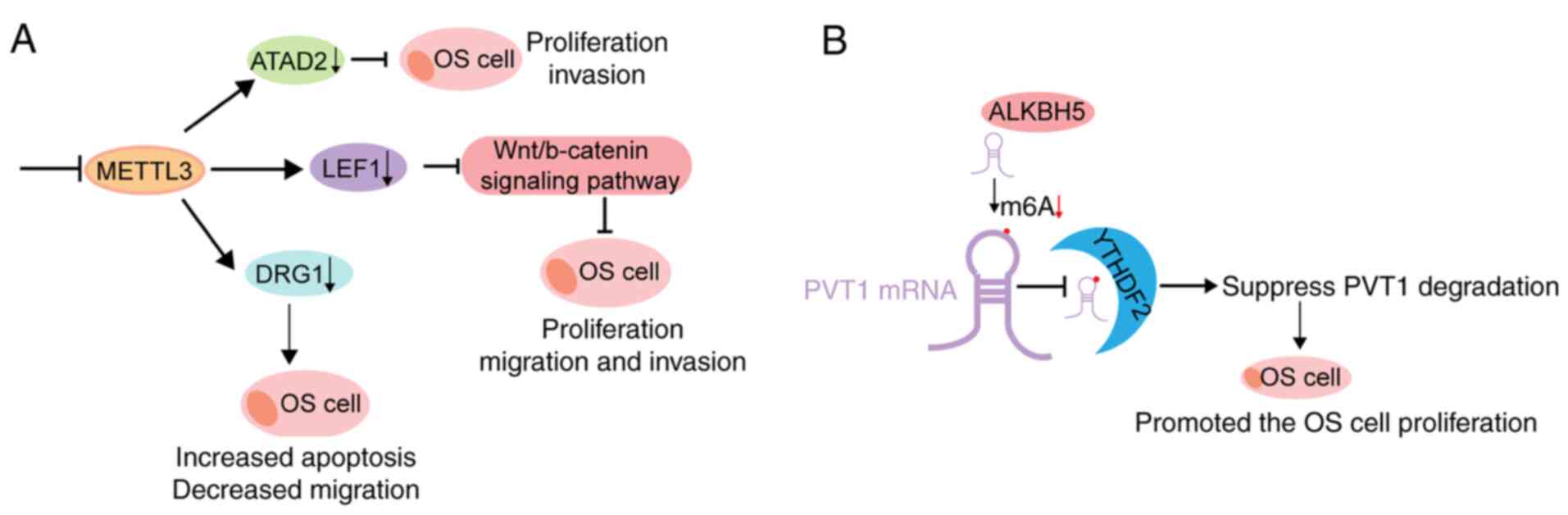
and Yu J: Silencing METTL3 inhibits the proliferation and invasion
of osteosarcoma by regulating ATAD2. Biomed Pharmacother.
125(109964)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bujnicki JM, Feder M, Radlinska M and
Blumenthal RM: Structure prediction and phylogenetic analysis of a
functionally diverse family of proteins homologous to the MT-A70
subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol.
55:431–444. 2002.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Scholler E, Weichmann F, Treiber T, Ringle
S, Treiber N, Flatley A, Feederle R, Bruckmann A and Meister G:
Interactions, localization, and phosphorylation of the
m6A generating METTL3-METTL14-WTAP complex. RNA.
24:499–512. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lence T, Paolantoni C, Worpenberg L and
Roignant JY: Mechanistic insights into m6A RNA enzymes.
Biochim Biophys Acta Gene Regul Mech. 1862:222–229. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Haussmann IU, Bodi Z, Sanchez-Moran E,
Mongan NP, Archer N, Fray RG and Soller M: m6A
potentiates Sxl alternative pre-mRNA splicing for robust drosophila
sex determination. Nature. 540:301–304. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Alarcon CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Structural basis of
N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature.
534:575–578. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Meyer KD and Jaffrey SR: Rethinking
m6A readers, writers, and erasers. Annu Rev Cell Dev
Biol. 33:319–342. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Geula S, Moshitch-Moshkovitz S,
Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V,
Peer E, Mor N, Manor YS, et al: Stem cells. m6A mRNA methylation
facilitates resolution of naive pluripotency toward
differentiation. Science. 347:1002–1006. 2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hongay CF and Orr-Weaver TL: Drosophila
Inducer of MEiosis 4 (IME4) is required for Notch signaling during
oogenesis. Proc Natl Acad Sci USA. 108:14855–14860. 2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Zhong S, Li H, Bodi Z, Button J, Vespa L,
Herzog M and Fray RG: MTA is an Arabidopsis messenger RNA adenosine
methylase and interacts with a homolog of a sex-specific splicing
factor. Plant Cell. 20:1278–1288. 2008.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Clancy MJ, Shambaugh ME, Timpte CS and
Bokar JA: Induction of sporulation in Saccharomyces cerevisiae
leads to the formation of N6-methyladenosine in mRNA: A potential
mechanism for the activity of the IME4 gene. Nucleic Acids Res.
30:4509–4518. 2002.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu
HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al: METTL3 promote tumor
proliferation of bladder cancer by accelerating pri-miR221/222
maturation in m6A-dependent manner. Mol Cancer.
18(110)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Choe J, Lin S, Zhang W, Liu Q, Wang L,
Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al: mRNA
circularization by METTL3-eIF3h enhances translation and promotes
oncogenesis. Nature. 561:556–560. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X,
Ren F, Cui G and Sun R: Expression patterns and prognostic value of
m6A-related genes in colorectal cancer. Am J Transl Res.
11:3972–3991. 2019.PubMed/NCBI
|
|
54
|
Visvanathan A, Patil V, Arora A, Hegde AS,
Arivazhagan A, Santosh V and Somasundaram K: Essential role of
METTL3-mediated m6A modification in glioma stem-like
cells maintenance and radioresistance. Oncogene. 37:522–533.
2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wang H, Xu B and Shi J: N6-methyladenosine
METTL3 promotes the breast cancer progression via targeting Bcl-2.
Gene. 722(144076)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme
METTL3 controls myeloid differentiation of normal hematopoietic and
leukemia cells. Nat Med. 23:1369–1376. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Lin S, Liu J, Jiang W, Wang P, Sun C, Wang
X, Chen Y and Wang H: METTL3 promotes the proliferation and
mobility of gastric cancer cells. Open Med (Wars). 14:25–31.
2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dahal U, Le K and Gupta M: RNA m6A
methyltransferase METTL3 regulates invasiveness of melanoma cells
by matrix metallopeptidase 2. Melanoma Res. 29:382–389.
2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zheng W, Dong X, Zhao Y, Wang S, Jiang H,
Zhang M, Zheng X and Gu M: Multiple functions and mechanisms
underlying the role of METTL3 in Human Cancers. Front Oncol.
9(1403)2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wu L, Wu D, Ning J, Liu W and Zhang D:
Changes of N6-methyladenosine modulators promote breast cancer
progression. BMC Cancer. 19(326)2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Li X, Tang J, Huang W, Wang F, Li P, Qin
C, Qin Z, Zou Q, Wei J, Hua L, et al: The M6A methyltransferase
METTL3: Acting as a tumor suppressor in renal cell carcinoma.
Oncotarget. 8:96103–96116. 2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wei W, Huo B and Shi X: miR-600 inhibits
lung cancer via downregulating the expression of METTL3. Cancer
Manag Res. 11:1177–1187. 2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhang C, Zhang M, Ge S, Huang W, Lin X,
Gao J, Gong J and Shen L: Reduced m6A modification predicts
malignant phenotypes and augmented Wnt/PI3K-Akt signaling in
gastric cancer. Cancer Med. 8:4766–4781. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Iyer LM, Zhang D and Aravind L: Adenine
methylation in eukaryotes: Apprehending the complex evolutionary
history and functional potential of an epigenetic modification.
Bioessays. 38:27–40. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Liu X, Qin J, Gao T, Li C, Chen X, Zeng K,
Xu M, He B, Pan B, Xu X, et al: Analysis of METTL3 and METTL14 in
hepatocellular carcinoma. Aging (Albany NY). 12:21638–21659.
2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Buker SM, Gurard-Levin ZA, Wheeler BD,
Scholle MD, Case AW, Hirsch JL, Ribich S, Copeland RA and
Boriack-Sjodin PA: A mass spectrometric assay of METTL3/METTL14
methyltransferase activity. SLAS Discov. 25:361–371.
2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B,
Li C, Sun L, Qin J, Xu T, et al: METTL14-mediated
N6-methyladenosine modification of SOX4 mRNA inhibits tumor
metastasis in colorectal cancer. Mol Cancer. 19(106)2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Gong PJ, Shao YC, Yang Y, Song WJ, He X,
Zeng YF, Huang SR, Wei L and Zhang JW: Analysis of
N6-methyladenosine methyltransferase reveals METTL14 and ZC3H13 as
tumor suppressor genes in breast cancer. Front Oncol.
10(578963)2020.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y,
Wang Y, Yang J and Tian F: Mettl14 inhibits bladder TIC
self-renewal and bladder tumorigenesis through
N6-methyladenosine of Notch1. Mol Cancer.
18(168)2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Weng H, Huang H, Wu H, Qin X, Zhao BS,
Dong L, Shi H, Skibbe J, Shen C, Hu C, et al: METTL14 inhibits
hematopoietic stem/progenitor differentiation and promotes
leukemogenesis via mRNA m6A modification. Cell Stem
Cell. 22:191–205 e9. 2018.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5' sites. Cell
Rep. 8:284–296. 2014.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Sorci M, Ianniello Z, Cruciani S, Larivera
S, Ginistrelli LC, Capuano E, Marchioni M, Fazi F and Fatica A:
METTL3 regulates WTAP protein homeostasis. Cell Death Dis.
9(796)2018.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Li H, Su Q, Li B, Lan L, Wang C, Li W,
Wang G, Chen W, He Y and Zhang C: High expression of WTAP leads to
poor prognosis of gastric cancer by influencing tumour-associated T
lymphocyte infiltration. J Cell Mol Med. 24:4452–4465.
2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Bansal H, Yihua Q, Iyer SP, Ganapathy S,
Proia DA, Penalva LO, Uren PJ, Suresh U, Carew JS, Karnad AB, et
al: WTAP is a novel oncogenic protein in acute myeloid leukemia.
Leukemia. 28:1171–1174. 2014.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Horiuchi K, Umetani M, Minami T, Okayama
H, Takada S, Yamamoto M, Aburatani H, Reid PC, Housman DE, Hamakubo
T and Kodama T: Wilms' tumor 1-associating protein regulates G2/M
transition through stabilization of cyclin A2 mRNA. Proc Natl Acad
Sci USA. 103:17278–17283. 2006.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Wen J, Lv R, Ma H, Shen H, He C, Wang J,
Jiao F, Liu H, Yang P, Tan L, et al: Zc3h13 regulates nuclear RNA
m6A methylation and mouse embryonic stem cell
self-renewal. Mol Cell. 69:1028–1038 e6. 2018.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Zhu D, Zhou J, Zhao J, Jiang G, Zhang X,
Zhang Y and Dong M: ZC3H13 suppresses colorectal cancer
proliferation and invasion via inactivating Ras-ERK signaling. J
Cell Physiol. 234:8899–8907. 2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Chen J, Yu K, Zhong G and Shen W:
Identification of a m6A RNA methylation regulators-based
signature for predicting the prognosis of clear cell renal
carcinoma. Cancer Cell Int. 20(157)2020.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Liu T, Li C, Jin L, Li C and Wang L: The
prognostic value of m6A RNA methylation regulators in colon
adenocarcinoma. Med Sci Monit. 25:9435–9445. 2019.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR
and Qian SB: Dynamic m(6)A mRNA methylation directs translational
control of heat shock response. Nature. 526:591–594.
2015.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Dina C, Meyre D, Gallina S, Durand E,
Körner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et
al: Variation in FTO contributes to childhood obesity and severe
adult obesity. Nat Genet. 39:724–726. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
84
|
Frayling TM, Timpson NJ, Weedon MN,
Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H,
Rayner NW, et al: A common variant in the FTO gene is associated
with body mass index and predisposes to childhood and adult
obesity. Science. 316:889–894. 2007.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun
L, Wang Y, Li X, Xiong XF, Wei B, et al: RNA N6-methyladenosine
demethylase FTO promotes breast tumor progression through
inhibiting BNIP3. Mol Cancer. 18(46)2019.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C,
Huang H, Nachtergaele S, Dong L, Hu C, et al: FTO plays an
oncogenic role in acute myeloid leukemia as a
N6-Methyladenosine RNA demethylase. Cancer Cell.
31:127–141. 2017.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Yang S, Wei J, Cui YH, Park G, Shah P,
Deng Y, Aplin AE, Lu Z, Hwang S, He C and He YY: m6A
mRNA demethylase FTO regulates melanoma tumorigenicity and response
to anti-PD-1 blockade. Nat Commun. 10(2782)2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Li J, Han Y, Zhang H, Qian Z, Jia W, Gao
Y, Zheng H and Li B: The m6A demethylase FTO promotes the growth of
lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem
Biophys Res Commun. 512:479–485. 2019.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Baltz AG, Munschauer M, Schwanhausser B,
Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew
K, Milek M, et al: The mRNA-bound proteome and its global occupancy
profile on protein-coding transcripts. Mol Cell. 46:674–690.
2012.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Fedeles BI, Singh V, Delaney JC, Li D and
Essigmann JM: The AlkB family of Fe(II)/α-Ketoglutarate-dependent
dioxygenases: Repairing nucleic acid alkylation damage and beyond.
J Biol Chem. 290:20734–20742. 2015.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Pilzys T, Marcinkowski M, Kukwa W, Garbicz
D, Dylewska M, Ferenc K, Mieczkowski A, Kukwa A, Migacz E, Wołosz
D, et al: ALKBH overexpression in head and neck cancer: Potential
target for novel anticancer therapy. Sci Rep.
9(13249)2019.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang
Y, Feng Y, Pan Q and Wan R: RNA demethylase ALKBH5 prevents
pancreatic cancer progression by posttranscriptional activation of
PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer.
19(91)2020.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Shen C, Sheng Y, Zhu AC, Robinson S, Jiang
X, Dong L, Chen H, Su R, Yin Z, Li W, et al: RNA demethylase ALKBH5
selectively promotes tumorigenesis and cancer stem cell
self-renewal in acute myeloid leukemia. Cell Stem Cell.
27:64–80.e9. 2020.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A demethylase
ALKBH5 maintains tumorigenicity of glioblastoma Stem-like cells by
sustaining FOXM1 expression and cell proliferation program. Cancer
Cell. 31:591–606.e6. 2017.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Zhu Z, Qian Q, Zhao X, Ma L and Chen P:
N6-methyladenosine ALKBH5 promotes non-small cell lung
cancer progress by regulating TIMP3 stability. Gene.
731(144348)2020.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Zhang J, Guo S, Piao HY, Wang Y, Wu Y,
Meng XY, Yang D, Zheng ZC and Zhao Y: ALKBH5 promotes invasion and
metastasis of gastric cancer by decreasing methylation of the
lncRNA NEAT1. J Physiol Biochem. 75:379–389. 2019.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Ueda Y, Ooshio I, Fusamae Y, Kitae K,
Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K and Tsujikawa
K: AlkB homolog 3-mediated tRNA demethylation promotes protein
synthesis in cancer cells. Sci Rep. 7(42271)2017.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Mohan M, Akula D, Dhillon A, Goyal A and
Anindya R: Human RAD51 paralogue RAD51C fosters repair of alkylated
DNA by interacting with the ALKBH3 demethylase. Nucleic Acids Res.
47:11729–11745. 2019.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J,
Lu Z, Zheng Z, Dai Q and Wang H: Transfer RNA demethylase ALKBH3
promotes cancer progression via induction of tRNA-derived small
RNAs. Nucleic Acids Res. 47:2533–2545. 2019.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306.
2014.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Yue Y, Liu J and He C: RNA
N6-methyladenosine methylation in post-transcriptional gene
expression regulation. Genes Dev. 29:1343–1355. 2015.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Zhu T, Roundtree IA, Wang P, Wang X, Wang
L, Sun C, Tian Y, Li J, He C and Xu Y: Crystal structure of the YTH
domain of YTHDF2 reveals mechanism for recognition of
N6-methyladenosine. Cell Res. 24:1493–1496. 2014.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Lee Y, Choe J, Park OH and Kim YK:
Molecular mechanisms driving mRNA degradation by m6A
modification. Trends Genet. 36:177–188. 2020.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z,
Perez SP, Suganthan R, He C, Bjørås M and Klungland A:
Ythdf2-mediated m6A mRNA clearance modulates neural
development in mice. Genome Biol. 19(69)2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X,
Liu B, Xiao F, Wang W, Huang G, et al: Loss of YTHDF2-mediated
m6A-dependent mRNA clearance facilitates hematopoietic
stem cell regeneration. Cell Res. 28:1035–1038. 2018.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Huang T, Liu Z, Zheng Y, Feng T, Gao Q and
Zeng W: YTHDF2 promotes spermagonial adhesion through modulating
MMPs decay via m6A/mRNA pathway. Cell Death Dis.
11(37)2020.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5'UTR
m(6)A promotes cap-independent translation. Cell. 163:999–1010.
2015.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Hu L, Wang J, Huang H, Yu Y, Ding J, Yu Y,
Li K, Wei D, Ye Q, Wang F, et al: YTHDF1 regulates pulmonary
hypertension through translational control of MAGED1. Am J Respir
Crit Care Med. 203:1158–1172. 2021.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu
Z, Li J, Hao P, Zhang Y, Zhang F, et al: m6A facilitates
hippocampus-dependent learning and memory through YTHDF1. Nature.
563:249–253. 2018.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Nishizawa Y, Konno M, Asai A, Koseki J,
Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai
D, et al: Oncogene c-Myc promotes epitranscriptome m6A
reader YTHDF1 expression in colorectal cancer. Oncotarget.
9:7476–7486. 2018.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Zhao X, Chen Y, Mao Q, Jiang X, Jiang W,
Chen J, Xu W, Zhong L and Sun X: Overexpression of YTHDF1 is
associated with poor prognosis in patients with hepatocellular
carcinoma. Cancer Biomark. 21:859–868. 2018.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Zhang Y, Wang X, Zhang X, Wang J, Ma Y,
Zhang L and Cao X: RNA-binding protein YTHDF3 suppresses
interferon-dependent antiviral responses by promoting FOXO3
translation. Proc Natl Acad Sci USA. 116:976–981. 2019.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou
A, Liu J, Che L and Li J: Long noncoding RNA GAS5 inhibits
progression of colorectal cancer by interacting with and triggering
YAP phosphorylation and degradation and is negatively regulated by
the m6A reader YTHDF3. Mol Cancer.
18(143)2019.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Jurczyszak D, Zhang W, Terry SN, Kehrer T,
Bermúdez González MC, McGregor E, Mulder LCF, Eckwahl MJ, Pan T and
Simon V: HIV protease cleaves the antiviral m6A reader protein
YTHDF3 in the viral particle. PLoS Pathog.
16(e1008305)2020.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519.
2016.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Luxton HJ, Simpson BS, Mills IG, Brindle
NR, Ahmed Z, Stavrinides V, Heavey S, Stamm S and Whitaker HC: The
oncogene metadherin interacts with the known splicing proteins
YTHDC1, Sam68 and T-STAR and plays a novel role in alternative mRNA
splicing. Cancers (Basel). 11(1233)2019.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu
Y, Gregory BD, Schultz RM and Wang PJ: Nuclear m6A reader YTHDC1
regulates alternative polyadenylation and splicing during mouse
oocyte development. PLoS Genet. 14(e1007412)2018.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B
and Qian SB: m6A in mRNA coding regions promotes
translation via the RNA helicase-containing YTHDC2. Nat Commun.
10(5332)2019.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Tanabe A, Tanikawa K, Tsunetomi M, Takai
K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, et al:
RNA helicase YTHDC2 promotes cancer metastasis via the enhancement
of the efficiency by which HIF-1α mRNA is translated. Cancer Lett.
376:34–42. 2016.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Kretschmer J, Rao H, Hackert P, Sloan KE,
Hobartner C and Bohnsack MT: The m6A reader protein
YTHDC2 interacts with the small ribosomal subunit and the 5'-3'
exoribonuclease XRN1. RNA. 24:1339–1350. 2018.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Nakano M, Ondo K, Takemoto S, Fukami T and
Nakajima M: Methylation of adenosine at the N6 position
post-transcriptionally regulates hepatic P450s expression. Biochem
Pharmacol. 171(113697)2020.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Bailey AS, Batista PJ, Gold RS, Chen YG,
de Rooij DG, Chang HY and Fuller MT: The conserved RNA helicase
YTHDC2 regulates the transition from proliferation to
differentiation in the germline. eLife. 6(e26116)2017.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Tanabe A, Konno J, Tanikawa K and Sahara
H: Transcriptional machinery of TNF-α-inducible YTH domain
containing 2 (YTHDC2) gene. Gene. 535:24–32. 2014.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Alarcon CR, Goodarzi H, Lee H, Liu X,
Tavazoie S and Tavazoie SF: HNRNPA2B1 is a mediator of
m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308.
2015.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295.
2018.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Srivastava M and Deal C: Osteoporosis in
elderly: Prevention and treatment. Clin Geriatr Med. 18:529–555.
2002.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Langdahl BL: Overview of treatment
approaches to osteoporosis. Br J Pharmacol. 178:1891–1906.
2021.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Rosen CJ and Bouxsein ML: Mechanisms of
disease: Is osteoporosis the obesity of bone? Nat Clin Pract
Rheumatol. 2:35–43. 2006.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Raisz LG: Pathogenesis of osteoporosis:
Concepts, conflicts, and prospects. J Clin Invest. 115:3318–3325.
2005.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Palmieri D, Valli M, Viglio S, Ferrari N,
Ledda B, Volta C and Manduca P: Osteoblasts extracellular matrix
induces vessel like structures through glycosylated collagen I. Exp
Cell Res. 316:789–799. 2010.PubMed/NCBI View Article : Google Scholar
|
|
132
|
DeNichilo MO, Shoubridge AJ, Panagopoulos
V, Liapis V, Zysk A, Zinonos I, Hay S, Atkins GJ, Findlay DM and
Evdokiou A: Peroxidase enzymes regulate collagen biosynthesis and
matrix mineralization by cultured human osteoblasts. Calcif Tissue
Int. 98:294–305. 2016.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Long F: Building strong bones: Molecular
regulation of the osteoblast lineage. Nat Rev Mol Cell Biol.
13:27–38. 2011.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Boyle WJ, Simonet WS and Lacey DL:
Osteoclast differentiation and activation. Nature. 423:337–342.
2003.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Villaseñor A, Aedo-Martín D, Obeso D,
Erjavec I, Rodríguez-Coira J, Buendía I, Ardura JA, Barbas C and
Gortazar AR: Metabolomics reveals citric acid secretion in
mechanically-stimulated osteocytes is inhibited by high glucose.
Sci Rep. 9(2295)2019.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Dallas SL, Prideaux M and Bonewald LF: The
osteocyte: An endocrine cell... and more. Endocr Rev. 34:658–690.
2013.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Chen Q, Shou P, Zheng C, Jiang M, Cao G,
Yang Q, Cao J, Xie N, Velletri T, Zhang X, et al: Fate decision of
mesenchymal stem cells: Adipocytes or osteoblasts? Cell Death
Differ. 23:1128–1139. 2016.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Kawai M, Devlin MJ and Rosen CJ: Fat
targets for skeletal health. Nat Rev Rheumatol. 5:365–372.
2009.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Rosen CJ, Ackert-Bicknell C, Rodriguez JP
and Pino AM: Marrow fat and the bone microenvironment:
Developmental, functional, and pathological implications. Crit Rev
Eukaryot Gene Expr. 19:109–124. 2009.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Scheller EL and Rosen CJ: What's the
matter with MAT? Marrow adipose tissue, metabolism, and skeletal
health. Ann N Y Acad Sci. 1311:14–30. 2014.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Garcia-Gomez MC and Vilahur G:
Osteoporosis and vascular calcification: A shared scenario. Clin
Investig Arterioscler. 32:33–42. 2020.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Chen X, Hua W, Huang X, Chen Y, Zhang J
and Li G: Regulatory role of RNA N6-methyladenosine
modification in bone biology and osteoporosis. Front Endocrinol
(Lausanne). 10(911)2019.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang
Y, Li J, Sheng R, Deng P, Wang Y, et al: Mettl3-mediated
m6A RNA methylation regulates the fate of bone marrow
mesenchymal stem cells and osteoporosis. Nat Commun.
9(4772)2018.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Yu J, Shen L, Liu Y, Ming H, Zhu X, Chu M
and Lin J: The m6A methyltransferase METTL3 cooperates with
demethylase ALKBH5 to regulate osteogenic differentiation through
NF-κB signaling. Mol Cell Biochem. 463:203–210. 2020.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Tian C, Huang Y, Li Q, Feng Z and Xu Q:
Mettl3 regulates osteogenic differentiation and alternative
splicing of vegfa in bone marrow mesenchymal stem cells. Int J Mol
Sci. 20(551)2019.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Busilacchi A, Gigante A, Mattioli-Belmonte
M, Manzotti S and Muzzarelli RA: Chitosan stabilizes platelet
growth factors and modulates stem cell differentiation toward
tissue regeneration. Carbohydr Polym. 98:665–676. 2013.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Hu K and Olsen BR: Osteoblast-derived VEGF
regulates osteoblast differentiation and bone formation during bone
repair. J Clin Invest. 126:509–526. 2016.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Yan G, Yuan Y, He M, Gong R, Lei H, Zhou
H, Wang W, Du W, Ma T, Liu S, et al: m6A methylation of
precursor-miR-320/RUNX2 controls osteogenic potential of bone
marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids.
19:421–436. 2020.PubMed/NCBI View Article : Google Scholar
|
|
149
|
Li D, Cai L, Meng R, Feng Z and Xu Q:
METTL3 modulates osteoclast differentiation and function by
controlling RNA stability and nuclear export. Int J Mol Sci.
21(1660)2020.PubMed/NCBI View Article : Google Scholar
|
|
150
|
Gerken T, Girard CA, Tung YC, Webby CJ,
Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill
LA, et al: The obesity-associated FTO gene encodes a
2-oxoglutarate-dependent nucleic acid demethylase. Science.
318:1469–1472. 2007.PubMed/NCBI View Article : Google Scholar
|
|
151
|
Eyre DR: Bone biomarkers as tools in
osteoporosis management. Spine (Phila Pa 1976). 22 (24
Suppl):17S–24S. 1997.PubMed/NCBI View Article : Google Scholar
|
|
152
|
Takada I, Kouzmenko AP and Kato S: Wnt and
PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat Rev
Rheumatol. 5:442–447. 2009.PubMed/NCBI View Article : Google Scholar
|
|
153
|
Shen GS, Zhou HB, Zhang H, Chen B, Liu ZP,
Yuan Y, Zhou XZ and Xu YJ: The GDF11-FTO-PPARγ axis controls the
shift of osteoporotic MSC fate to adipocyte and inhibits bone
formation during osteoporosis. Biochim Biophys Acta Mol Basis Dis.
1864:3644–3654. 2018.PubMed/NCBI View Article : Google Scholar
|
|
154
|
Zhang Q, Riddle RC, Yang Q, Rosen CR,
Guttridge DC, Dirckx N, Faugere MC, Farber CR and Clemens TL: The
RNA demethylase FTO is required for maintenance of bone mass and
functions to protect osteoblasts from genotoxic damage. Proc Natl
Acad Sci USA. 116:17980–17989. 2019.PubMed/NCBI View Article : Google Scholar
|
|
155
|
Guo Y, Liu H, Yang TL, Li SM, Li SK, Tian
Q, Liu YJ and Deng HW: The fat mass and obesity associated gene,
FTO, is also associated with osteoporosis phenotypes. PLoS One.
6(e27312)2011.PubMed/NCBI View Article : Google Scholar
|
|
156
|
Li Y, Yang F, Gao M, Gong R, Jin M, Liu T,
Sun Y, Fu Y, Huang Q, Zhang W, et al: miR-149-3p regulates the
switch between adipogenic and osteogenic differentiation of BMSCs
by targeting FTO. Mol Ther Nucleic Acids. 17:590–600.
2019.PubMed/NCBI View Article : Google Scholar
|
|
157
|
Mannerstrom B, Kornilov R, Abu-Shahba AG,
Chowdhury IM, Sinha S, Seppänen-Kaijansinkko R and Kaur S:
Epigenetic alterations in mesenchymal stem cells by
osteosarcoma-derived extracellular vesicles. Epigenetics.
14:352–364. 2019.PubMed/NCBI View Article : Google Scholar
|
|
158
|
Lorenzo J: Cytokines and bone:
Osteoimmunology. Handb Exp Pharmacol. 262:177–230. 2020.PubMed/NCBI View Article : Google Scholar
|
|
159
|
Zhang Y, Gu X, Li D, Cai L and Xu Q:
METTL3 regulates osteoblast differentiation and inflammatory
response via Smad signaling and MAPK Signaling. Int J Mol Sci.
21(199)2019.PubMed/NCBI View Article : Google Scholar
|
|
160
|
Yu R, Li Q, Feng Z, Cai L and Xu Q: m6A
reader YTHDF2 regulates LPS-induced inflammatory response. Int J
Mol Sci. 20(1323)2019.PubMed/NCBI View Article : Google Scholar
|
|
161
|
Wang T and He C: TNF-α and IL-6: The link
between immune and bone system. Curr Drug Targets. 20:213–227.
2020.PubMed/NCBI View Article : Google Scholar
|
|
162
|
Mathew AJ and Ravindran V: Infections and
arthritis. Best Pract Res Clin Rheumatol. 28:935–959.
2014.PubMed/NCBI View Article : Google Scholar
|
|
163
|
Harth M and Nielson WR: Pain and affective
distress in arthritis: Relationship to immunity and inflammation.
Expert Rev Clin Immunol. 15:541–552. 2019.PubMed/NCBI View Article : Google Scholar
|
|
164
|
Parkinson L, Waters DL and Franck L:
Systematic review of the impact of osteoarthritis on health
outcomes for comorbid disease in older people. Osteoarthritis
Cartilage. 25:1751–1770. 2017.PubMed/NCBI View Article : Google Scholar
|
|
165
|
Glyn-Jones S, Palmer AJ, Agricola R, Price
AJ, Vincent TL, Weinans H and Carr AJ: Osteoarthritis. Lancet.
386:376–387. 2015.PubMed/NCBI View Article : Google Scholar
|
|
166
|
Abramoff B and Caldera FE: Osteoarthritis:
Pathology, diagnosis, and treatment options. Med Clin North Am.
104:293–311. 2020.PubMed/NCBI View Article : Google Scholar
|
|
167
|
Liossis SN and Tsokos GC: Cellular
immunity in osteoarthritis: Novel concepts for an old disease. Clin
Diagn Lab Immunol. 5:427–429. 1998.PubMed/NCBI View Article : Google Scholar
|
|
168
|
Sakata M, Tsuruha JI, Masuko-Hongo K,
Nakamura H, Matsui T, Sudo A, Nishioka K and Kato T: Autoantibodies
to osteopontin in patients with osteoarthritis and rheumatoid
arthritis. J Rheumatol. 28:1492–1495. 2001.PubMed/NCBI
|
|
169
|
Walker J, Gordon T, Lester S, Downie-Doyle
S, McEvoy D, Pile K, Waterman S and Rischmueller M: Increased
severity of lower urinary tract symptoms and daytime somnolence in
primary Sjogren's syndrome. J Rheumatol. 30:2406–2412.
2003.PubMed/NCBI
|
|
170
|
Kato T, Xiang Y, Nakamura H and Nishioka
K: Neoantigens in osteoarthritic cartilage. Curr Opin Rheumatol.
16:604–608. 2004.PubMed/NCBI View Article : Google Scholar
|
|
171
|
Zhao W, Wang T, Luo Q, Chen Y, Leung VY,
Wen C, Shah MF, Pan H, Chiu K, Cao X and Lu WW: Cartilage
degeneration and excessive subchondral bone formation in
spontaneous osteoarthritis involves altered TGF-β signaling. J
Orthop Res. 34:763–770. 2016.PubMed/NCBI View Article : Google Scholar
|
|
172
|
Liu Q, Li M, Jiang L, Jiang R and Fu B:
METTL3 promotes experimental osteoarthritis development by
regulating inflammatory response and apoptosis in chondrocyte.
Biochem Biophys Res Commun. 516:22–27. 2019.PubMed/NCBI View Article : Google Scholar
|
|
173
|
Goldring SR and Goldring MB: The role of
cytokines in cartilage matrix degeneration in osteoarthritis. Clin
Orthop Relat Res. (427 Suppl):S27–S36. 2004.PubMed/NCBI View Article : Google Scholar
|
|
174
|
Yang F, Zhou S, Wang C, Huang Y, Li H,
Wang Y, Zhu Z, Tang J and Yan M: Epigenetic modifications of
interleukin-6 in synovial fibroblasts from osteoarthritis patients.
Sci Rep. 7(43592)2017.PubMed/NCBI View Article : Google Scholar
|
|
175
|
Guo Q, Wang Y, Xu D, Nossent J, Pavlos NJ
and Xu J: Rheumatoid arthritis: Pathological mechanisms and modern
pharmacologic therapies. Bone Res. 6(15)2018.PubMed/NCBI View Article : Google Scholar
|
|
176
|
Li HB, Tong J, Zhu S, Batista PJ, Duffy
EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, et al:
m6A mRNA methylation controls T cell homeostasis by
targeting the IL-7/STAT5/SOCS pathways. Nature. 548:338–342.
2017.PubMed/NCBI View Article : Google Scholar
|
|
177
|
Wang J, Yan S, Lu H, Wang S and Xu D:
METTL3 attenuates LPS-induced inflammatory response in macrophages
via NF-κB signaling pathway. Mediators Inflamm.
2019(3120391)2019.PubMed/NCBI View Article : Google Scholar
|
|
178
|
Mo XB, Zhang YH and Lei SF: Genome-wide
identification of N6-methyladenosine (m6A)
SNPs associated with rheumatoid arthritis. Front Genet.
9(299)2018.PubMed/NCBI View Article : Google Scholar
|
|
179
|
Zheng Y, Nie P, Peng D, He Z, Liu M, Xie
Y, Miao Y, Zuo Z and Ren J: m6AVar: A database of functional
variants involved in m6A modification. Nucleic Acids Res.
46:D139–D145. 2018.PubMed/NCBI View Article : Google Scholar
|
|
180
|
Smolen JS, Aletaha D, Barton A, Burmester
GR, Emery P, Firestein GS, Kavanaugh A, McInnes IB, Solomon DH,
Strand V and Yamamoto K: Rheumatoid arthritis. Nat Rev Dis Primers.
4(18001)2018.PubMed/NCBI View Article : Google Scholar
|
|
181
|
Kondo Y, Yokosawa M, Kaneko S, Furuyama K,
Segawa S, Tsuboi H, Matsumoto I and Sumida T: Review:
Transcriptional regulation of CD4+ T cell differentiation in
experimentally induced arthritis and rheumatoid arthritis.
Arthritis Rheumatol. 70:653–661. 2018.PubMed/NCBI View Article : Google Scholar
|
|
182
|
Noack M and Miossec P: Th17 and regulatory
T cell balance in autoimmune and inflammatory diseases. Autoimmun
Rev. 13:668–677. 2014.PubMed/NCBI View Article : Google Scholar
|
|
183
|
Hunt L, Hensor EM, Nam J, Burska AN,
Parmar R, Emery P and Ponchel F: T cell subsets: An immunological
biomarker to predict progression to clinical arthritis in
ACPA-positive individuals. Ann Rheum Dis. 75:1884–1889.
2016.PubMed/NCBI View Article : Google Scholar
|
|
184
|
Kumar BV, Connors TJ and Farber DL: Human
T cell development, localization, and function throughout life.
Immunity. 48:202–213. 2018.PubMed/NCBI View Article : Google Scholar
|
|
185
|
Abada A and Elazar Z: Getting ready for
building: Signaling and autophagosome biogenesis. Embo Rep.
15:839–852. 2014.PubMed/NCBI View Article : Google Scholar
|
|
186
|
Wang DW, Wu LW, Cao Y, Yang L, Liu W, E
XQ, Ji G and Bi ZG: A novel mechanism of mTORC1-mediated
serine/glycine metabolism in osteosarcoma development. Cell Signal.
29:107–114. 2017.PubMed/NCBI View Article : Google Scholar
|
|
187
|
Miao W, Chen J, Jia L, Ma J and Song D:
The m6A methyltransferase METTL3 promotes osteosarcoma progression
by regulating the m6A level of LEF1. Biochem Biophys Res Commun.
516:719–725. 2019.PubMed/NCBI View Article : Google Scholar
|
|
188
|
Nguyen DX, Chiang AC, Zhang XH, Kim JY,
Kris MG, Ladanyi M, Gerald WL and Massagué J: WNT/TCF signaling
through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis.
Cell. 138:51–62. 2009.PubMed/NCBI View Article : Google Scholar
|
|
189
|
Jia P, Wei G, Zhou C, Gao Q, Wu Y, Sun X
and Li X: Upregulation of miR-212 inhibits migration and
tumorigenicity and inactivates Wnt/β-catenin signaling in human
hepatocellular carcinoma. Technol Cancer Res Treat.
17(1533034618765221)2018.PubMed/NCBI View Article : Google Scholar
|
|
190
|
Wu L, Zhao JC, Kim J, Jin HJ, Wang CY and
Yu J: ERG is a critical regulator of Wnt/LEF1 signaling in prostate
cancer. Cancer Res. 73:6068–6079. 2013.PubMed/NCBI View Article : Google Scholar
|
|
191
|
Ling Z, Chen L and Zhao J: m6A-dependent
up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth,
migration, and colony formation in osteosarcoma. Biosci Rep.
40(BSR20200282)2020.PubMed/NCBI View Article : Google Scholar
|
|
192
|
Coker H, Wei G and Brockdorff N: m6A
modification of non-coding RNA and the control of mammalian gene
expression. Biochim Biophys Acta Gene Regul Mech. 1862:310–318.
2019.PubMed/NCBI View Article : Google Scholar
|
|
193
|
Li J, Rao B, Yang J, Liu L, Huang M, Liu
X, Cui G, Li C, Han Q, Yang H, et al: Dysregulated m6A-related
regulators are associated with tumor metastasis and poor prognosis
in osteosarcoma. Front Oncol. 10(769)2020.PubMed/NCBI View Article : Google Scholar
|
|
194
|
Fan D, Xia Y, Lu C, Ye Q, Xi X, Wang Q,
Wang Z, Wang C and Xiao C: Regulatory role of the RNA
N6-methyladenosine modification in immunoregulatory
cells and immune-related bone homeostasis associated with
rheumatoid arthritis. Front Cell Dev Biol. 8(627893)2020.PubMed/NCBI View Article : Google Scholar
|
|
195
|
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y,
Cheng C, Li L, Pi J, Si Y, et al: The m6A reader YTHDF1 promotes
ovarian cancer progression via augmenting EIF3C translation.
Nucleic Acids Res. 48:3816–3831. 2020.PubMed/NCBI View Article : Google Scholar
|
|
196
|
Ma Z and Ji J: N6-methyladenosine (m6A)
RNA modification in cancer stem cells. Stem Cells: Sep 27, 2020
(Epub Ahead of Print).
|
|
197
|
Patil DP, Pickering BF and Jaffrey SR:
Reading m6A in the transcriptome: m6A-binding
proteins. Trends Cell Biol. 28:113–127. 2018.PubMed/NCBI View Article : Google Scholar
|
|
198
|
Dai DJ, Wang HY, Zhu LY, Jin HC and Wang
X: N6-methyladenosine links RNA metabolism to cancer
progression. Cell Death Dis. 9(124)2018.PubMed/NCBI View Article : Google Scholar
|
|
199
|
Wang HF, Kuang MJ, Han SJ, Wang AB, Qiu J,
Wang F, Tan BY and Wang DC: BMP2 modified by the m6A
demethylation enzyme ALKBH5 in the ossification of the ligamentum
flavum through the AKT signaling pathway. Calcified Tissue Int.
106:486–493. 2020.PubMed/NCBI View Article : Google Scholar
|
|
200
|
Zhang W, He L, Liu Z, Ren X, Qi L, Wan L,
Wang W, Tu C and Li Z: Multifaceted functions and novel insight
into the regulatory role of RNA N6-methyladenosine
modification in musculoskeletal disorders. Front Cell Dev Biol.
8(870)2020.PubMed/NCBI View Article : Google Scholar
|