Introduction
Bone tissue is a typical mechanoresponsive tissue.
Lack of mechanical stimulation is the major cause of the loss of
bone mass and osteoporosis (1,2).
Exercise is able to apply mechanical stimulation to bone tissue,
and the mechanical stress produced by exercise may increase the
bone turnover rate and bone density (3), stimulate osteoblast activity, increase
bone mass and promote bone reconstruction and metabolism (4,5). Thus,
exercise is one of the primary modifiable factors associated with
improved bone health outcomes (6).
Certain types of exercise may result in improved
bone strength, even after menopause (7). An increasing number of trials have
indicated that treadmill exercise may promote bone health,
particularly by suppressing estrogen deficiency-induced
osteoporosis, for instance, treadmill training could increase bone
mineral density in specific parts of rats, such as cortical bone
(8), and treadmill exercise could
increase bone density, improve the bone trabecular microstructure
of mice, and inhibit osteoporosis and osteoclast activation in bone
tissues of ovariectomized mice (9,10). In
addition, the mechanical loading of treadmill exercise
substantially enhances the osteogenic response (11). However, these studies have not fully
revealed the response of bone tissue to running motion stimulation
at the molecular level, particularly in terms of regulation by
microRNAs (miRNAs/miRs).
miRNAs, a class of small noncoding RNAs, are able to
inhibit the expression of negative regulatory genes and
protein-coding genes by binding to target mRNAs (12). MiRNAs participate in vital
activities, including development, organ formation, tumorigenesis,
cell proliferation, differentiation and apoptosis, by modulating
gene expression (12,13). MiRNAs have multiple roles in
osteoblasts and osteoclasts in the presence of mechanical cyclical
stretch (14), fluid shear stress
(15), compressive force (16), orthodontic force (17) and microgravity (18). In addition, miRNAs regulate
osteogenesis, which may be translated into novel therapeutic
approaches for orthodontic conditions and bone fractures, as well
as for systemic diseases, such as osteoporosis (19).
Osteocytes account for >90% of the adult bone
cell population and are critical sensors of mechanical loading in
bone (20). In a previous study by
our group, differentially expressed miRNAs [40 miRNAs, 10 of which
were confirmed via reverse transcription-quantitative (RT-q)PCR]
were identified in MLO-Y4 osteocytes mechanically stimulated in
vitro (21). As osteocytes are
dominant and mechanosensitive, it was speculated that the same
miRNAs may be differentially expressed in mechanically stimulated
osteocytes in vitro and in the bone tissue of mice subjected
to exercise on a treadmill. These miRNAs are likely to be involved
in the response to mechanical strain and bone metabolism.
In the present study, to explore the effect of
treadmill exercise on the expression of miRNAs in the bone tissues
of mice, the differentially expressed miRNAs in the bone tissues of
mice trained on a treadmill were screened using a miRNA microarray
and verified by RT-qPCR, and the target genes of these
differentially expressed miRNAs were predicted. In addition, the
differentially expressed miRNAs in mechanically stimulated
osteocytes were compared with the differentially expressed miRNAs
in the bone tissues of the mice. Specific differentially expressed
miRNAs that exhibited the same expression trends were selected and
their target genes were predicted.
Materials and methods
Treadmill running exercise
A total of 22 male BALB/c mice (age, 8 weeks old;
weight, 25-30 g; purchased from Hunan Anshengmei Pharmaceutical
Research Institute Co., Ltd.) were randomly divided into two
groups. The treadmill training group (treadmill speed, 13 m/min;
slope, 9˚; training for 40 min per day at the same time each day, 6
days/week) and the control group (no treadmill training). All mice
had free access to food and water under a relative humidity of
40-70%, temperature of 22-25˚C and 12-h light/dark cycle. The
training lasted for 8 weeks. All of the experimental protocols were
performed according to the Guidelines for Animal Research of Guilin
Medical University (Guide for the Care and Use of Laboratory
Animals) and were approved by the Animal Ethics Committee of Guilin
Medical University (approval no. 2019-0013; Guilin, China).
Bone tissue sampling
After 8 weeks of treadmill running exercise, the
mice were all euthanized in transparent plastic boxes with
CO2 at a flow rate of 20% chamber air exchange/min (1.2
l/min) in December 2019. Death was confirmed by exposing the thorax
to observe the lack of heartbeat, as well as observing pupil
dilation and unresponsiveness to light. The femurs of the mice were
collected and the soft tissue of the bone surface was removed.
There were 11 mice in the treadmill training group and 11 rats in
the control group, six of them were tested for bone mechanical
properties. After testing the mechanical properties of the bone
tissues of six mice, five of them were subsequently tested for
alkaline phosphatase (ALP) activity and osteocalcin (OCN). The last
five of the mice were used for the miRNA microarray and RT-qPCR
assays. The femur bone was used for each of the assays.
Application of mechanical stimulation
to osteocytes
MLO-Y4 osteocytes (purchased from Guangzhou Jennio
Biotech Co., Ltd.) were seeded in α-MEM (Invitrogen; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Invitrogen; Thermo Fisher
Scientific, Inc.) and 1% penicillin (Invitrogen; Thermo Fisher
Scientific, Inc.) in the loading culture chamber of a four-point
bending loading device (Institute of Medical Equipment, Academy of
Military Medical Sciences, Tianjin, China). The osteocytes were
seeded at a density of 2.5x104 cells/cm2 in
mechanical loading plates and cultivated until they reached
confluence. Cyclic mechanical tensile strain (2,500 µε, 0.5 Hz, 8
h) was applied to the cells, the cells were cultured in serum-free
medium (Invitrogen; Thermo Fisher Scientific, Inc.) during the
application of mechanical load. Cells not subjected to mechanical
stimulation were used as the control group. These procedures were
performed according to a previously outlined method (21).
Mechanical properties of bone
tissue
Using a material testing machine (RGM6010; Anhui
Regal Electronic Technology Co., Ltd.), the mechanical properties
of the mouse femur bones were determined in a classical three-point
bending experiment with the following parameters: Preload, 0.5 N;
loading rate, 2 mm/min; and span, 10 mm (sufficient to break the
bone tissue). The fracture (breaking) load, maximum elastic load,
maximum bending stress and bending modulus of the mouse femurs were
determined.
ALP and OCN measurements
The bone tissues (femurs) of the mice were weighed
and fully ground with 1.5 ml PBS in a tissue grinder, and
subsequently, the grinding solution was transferred to a 2-ml
Eppendorf tube and centrifuged at 4˚C and 2x103 x g for
5 min. The supernatant was then discarded, and subsequently, 0.2 ml
RIPA lysis buffer (Invitrogen; Thermo Fisher Scientific, Inc.) was
added to the precipitate at the proportions of 1:10 (g:ml), and
then the lysate was sonicated in an ultrasonic cell grinder (200 W,
30 cycles/6 sec). After centrifugation (6x103 x g, 20
min, 4˚C), the protein content of the supernatant was determined by
a BCA protein assay kit (Beyotime Institute of Biotechnology). The
ALP activity of the supernatant was detected by using an ALP kit
(cat. no. A059-2-1; Nanjing Jiancheng Bioengineering Institute)
according to the manufacturer's protocol. In addition, the OCN
content of the bone tissue protein solution was detected using a
mouse OCN ELISA kit (cat. no. H152; Nanjing Jiancheng
Bioengineering Institute) according to the manufacturer's
protocol.
miRNA microarray and RT-qPCR
After the bone tissues of the mice were completely
shredded and ground in liquid nitrogen, the total RNA was purified
by using TRIzol® RNA extraction reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol.
Part of the total RNA was purified with the mirVana
miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) and
labeled with Cyanine 3 using the ULS™ mirVana miRNA ArrayLabeling
kit (Ambion; Thermo Fisher Scientific, Inc.). Target labeling,
hybridization, imaging and data processing were performed using a
RiboArray miDETECT mouse array (Guangzhou RiboBio Co., Ltd.) that
included all mouse miRNAs according to the manufacturer's protocol.
miRNAs were considered to be differentially expressed if expression
was >2-fold higher or lower than that in the control group
(P<0.05).
Using the total RNA as a template, the miDETECT A
Track™ miRNA qRT-PCR Start kit (Guangzhou RiboBio Co., Ltd.) was
used to perform poly(A) tailing (to tail RNA) and Uni-RT (RT based
on Uni-RT primers), followed by qPCR, according to the
manufacturer's protocols of the 7900HT Fast Real Time PCR machine
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
miRNA-specific primers miDETECT A Track™ miRNA Forward Primers and
qPCR primers for the miRNA mature chain were synthesized by
Guangzhou RiboBio Co., Ltd. Following cDNA synthesis using
Megaplex™ RNA RT mix (cat. no. 4444766; Applied Biosystems; Thermo
Fisher Scientific, Inc.), qPCR was performed using Power SYBR-Green
PCR Master Mix (cat. no. 4367659; Applied Biosystems; Thermo Fisher
Scientific, Inc.). The thermocycling conditions used were as
follows: 10 min at 95˚C, followed by 45 cycles of 15 sec at 95˚C
and 1 min at 60˚C. miRNA microarray and RT-qPCR experiments were
performed at Guangzhou RiboBio Co., Ltd.
The miRNA microarray and RT-qPCR analysis of MLO-Y4
mouse osteocytes that were stimulated with a mechanical tensile
strain of 2500 µε at 0.5 Hz were performed as described in Zeng
et al (21), this mechanical
tensile strain has been demonstrated to promote the osteoblastic
differentiation of osteoblasts in vitro.
miRNA target gene prediction
TargetScan (www.targetscan.org/), MicroRNA.org
(www.microrna.org/) and miRDB (http://www.mirdb.org/2/) were used to predict the
target genes of the miRNAs, and target genes related to osteogenic
differentiation or bone metabolism were identified.
Statistical analysis
Values are expressed as the mean ± standard
deviation from three separate experiments (n=5 or 6 mice/group).
Data were tested for normality of distribution using the
Shapiro-Wilk test and differences between groups were analyzed
using one-way ANOVA. Statistical analysis was performed using SPSS
software (version 18; SPSS, Inc.) and P<0.05 was considered to
indicate a statistically significant difference.
Results
Evaluation of mechanical
properties
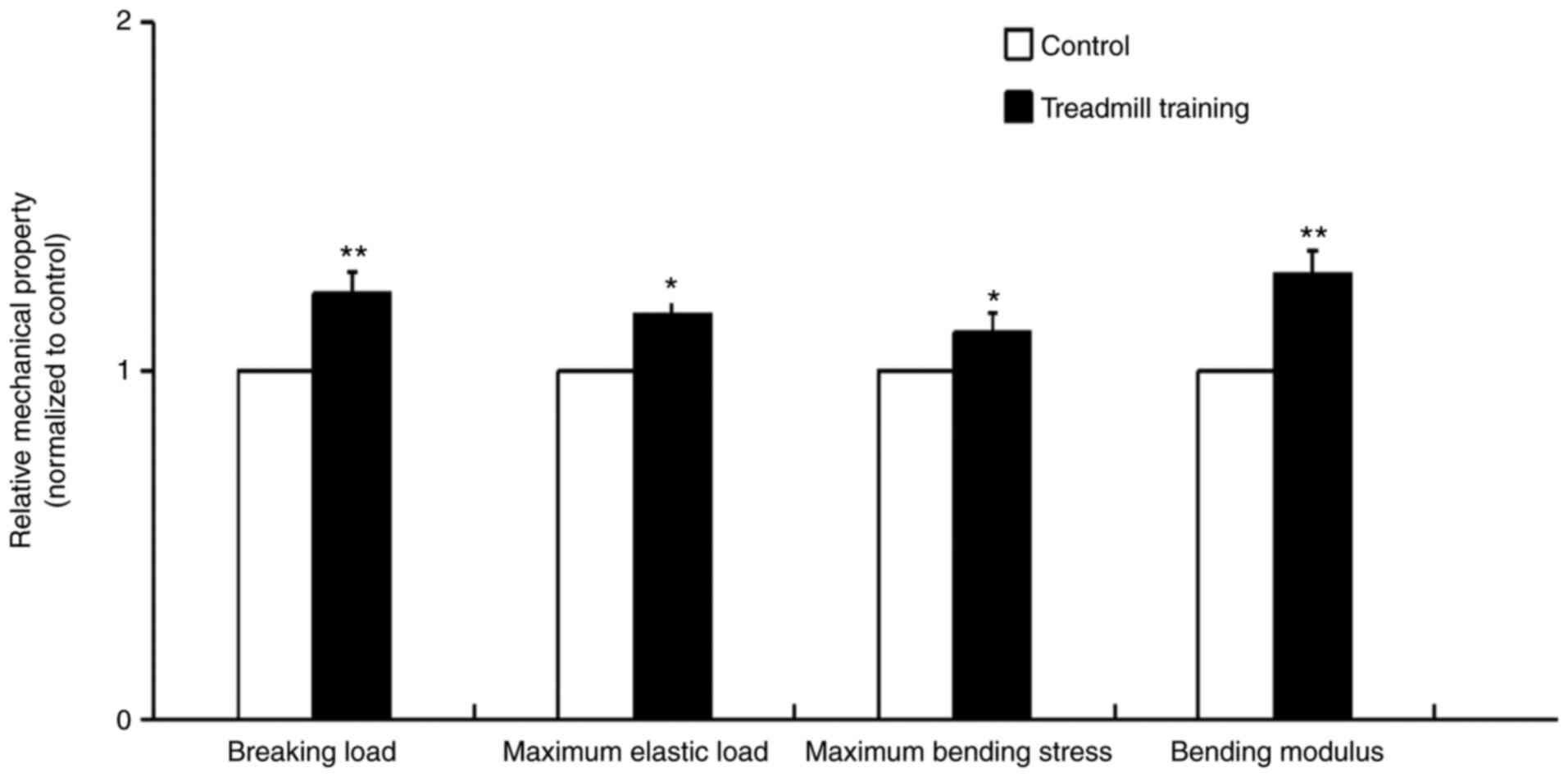
After 8 weeks of treadmill training, the mechanical
properties of the femur bone tissues of the mice were significantly
improved. These improved properties included the breaking load,
maximum elastic load, maximum bending stress and bending modulus,
and the breaking load and bending modulus were particularly
improved (Fig. 1). This result
indicated that treadmill training was able to improve the
mechanical strength and elasticity of bone tissue.
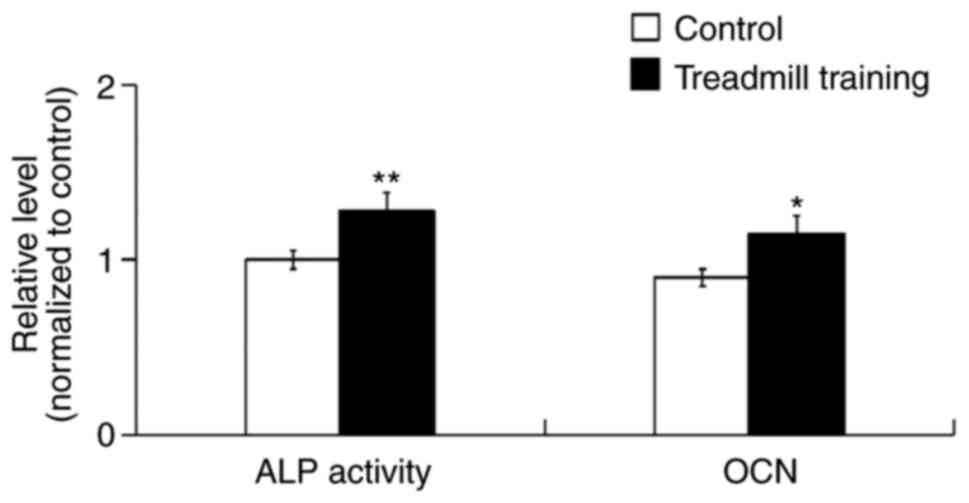
ALP activity and OCN expression
After 8 weeks of treadmill training, the ALP
activity and OCN content of the femur bone tissues were
significantly increased compared with in the control group
(Fig. 2), which indicated that
treadmill training was able to promote the osteogenic activity of
cells in tissue.
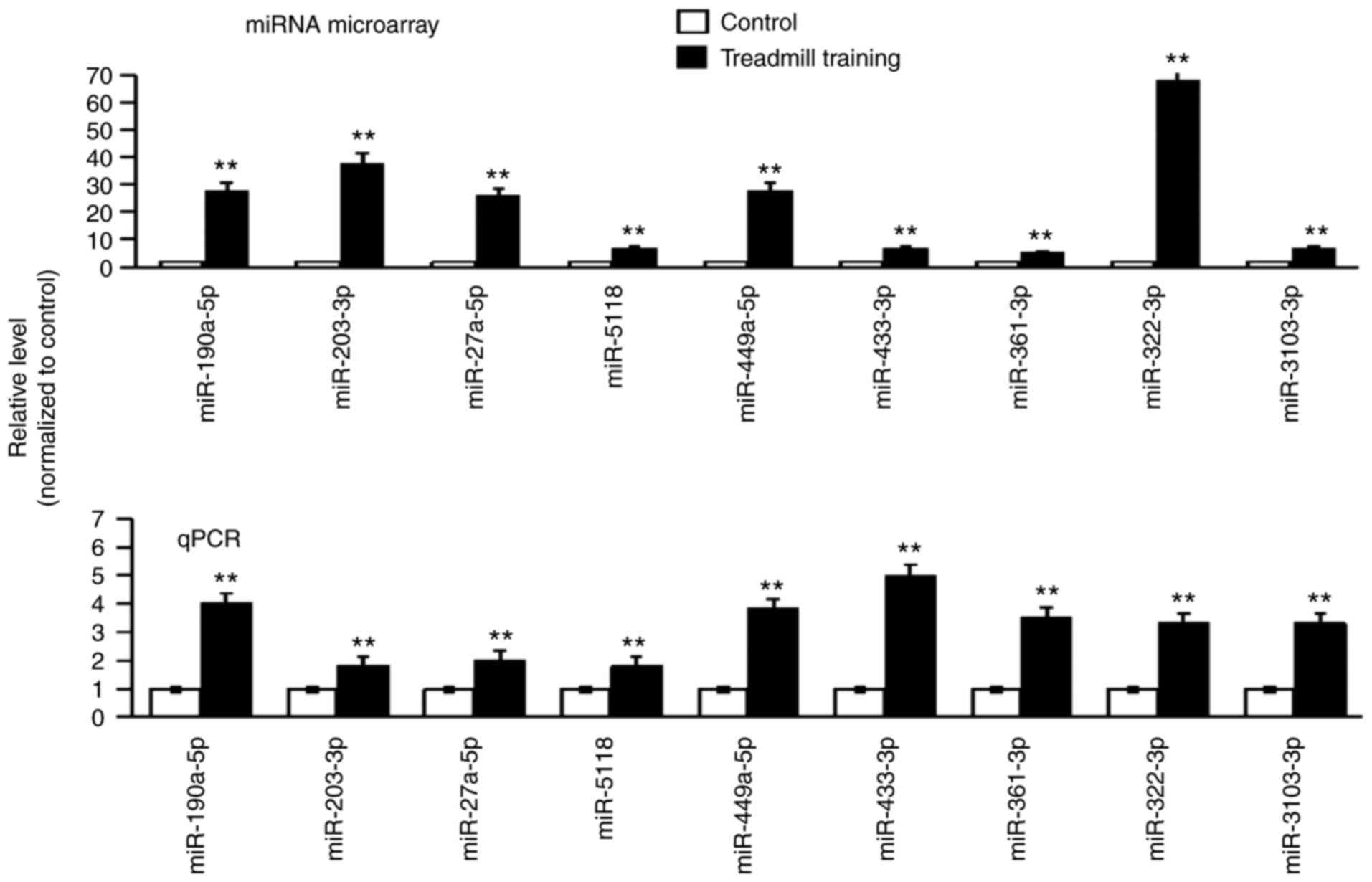
Differential expression of miRNAs
After 8 weeks of treadmill training, the expression
of all of the miRNAs in the femur bones was detected via miRNA
microarray. A total of 122 miRNAs were indicated to be
differentially expressed. From these miRNAs, 19 miRNAs with
P<0.01 were selected for further verification (data not shown).
The RT-qPCR results suggested that the expression trend of each of
the nine miRNAs was the same as that in the miRNA microarray; all
of these miRNAs were upregulated (the expression level in the
experimental group was higher compared with in the control group;
Fig. 3). The nine differentially
expressed miRNAs were as follows: miR-190a-5p, miR-203-5p,
miR-27a-5p, miR-5118, miR-449a-5p, miR-433-3p, miR-361-3p,
miR-322-3p and miR-3103-3p.
Prediction of target genes
Using bioinformatics techniques, certain osteogenic
differentiation and bone metabolism-related target genes of the 9
aforementioned differentially expressed miRNAs in bone tissue were
predicted and their details are presented in Table I.
 | Table IPredictive target genes of
differentially expressed miRNAs in bone tissue. |
Table I
Predictive target genes of
differentially expressed miRNAs in bone tissue.
| miRNA | Target gene | Gene
description | (Refs.) |
|---|
| miR-190a-5p | Smad2 | SMAD family member
2 | (39,47) |
| | Cacnb2 | Calcium channel
voltage-dependent β 2 subunit | (48,49) |
| miR-203-5p | Mmp13 | Matrix
metallopeptidase 13 | (50) |
| | Wnt7b | Wingless-related
MMTV integration site 7B | (51) |
| | Col3a1 | Collagen type III α
1 | (52,53) |
| miR-27a-5p | Ngfr | Nerve growth factor
receptor | (54) |
| | Stk38 | Serine/threonine
kinase 38 | (55) |
| | Ube2d2a |
Ubiquitin-conjugating enzyme E2D 2A | (56) |
| miR-5118 | Ngfr | Nerve growth factor
receptor | (54) |
| | Sh3kbp1 | SH3-domain kinase
binding protein 1 | (57) |
| | Ube2d2a |
Ubiquitin-conjugating enzyme E2D 2A | (56) |
| miR-449a-5p | Arhgap26 | Rho GTPase
activating protein 26 | (58) |
| | Atp2b4 | ATPase
Ca++ transporting plasma membrane 4 | (36) |
| miR-361-3p | Rptor | Regulatory
associated protein of MTOR complex 1 | (59) |
| | Map3k9 | Mitogen-activated
protein kinase kinase kinase 9 | (33,34) |
| miR-322-3p | Mettl9 | Methyltransferase
like 9 | (60) |
| | Ptpre | Protein tyrosine
phosphatase receptor type E | (37) |
| miR-3103-3p | Nfrkb | Nuclear factor
related to κB binding protein | (35) |
| | Arhgap 30 | Rho
GTPase-activating protein 30 | (58) |
| miR-433-3p | Creb1 | cAMP responsive
element binding protein 1 | (61) |
| | Map2 |
Microtubule-associated protein 2 | (62) |
Comparison of differentially expressed
miRNAs
From the results of the miRNA microarray of the bone
tissues and MLO-Y4 osteocytes, 19 miRNAs with the same expression
trends in both bones and osteocytes were selected. These miRNAs
were analyzed in bone tissues and MLO-Y4 osteocytes using RT-qPCR.
The results indicated that each of the eight differentially
expressed miRNAs in both the bone tissues and MLO-Y4 osteocytes had
the same expression trend; four miRNAs were upregulated and four
miRNAs were downregulated (Table
II). The target genes of the eight differentially expressed
miRNAs were predicted and Table
III presents the osteogenic differentiation- and bone
metabolism-related target genes, such as dickkopf homolog 2,
wingless-related MMTV integration site 2b (Wnt2b), frizzled homolog
5, transforming growth factor β receptor, cAMP responsive element
binding protein 1.
 | Table IIDifferentially expressed miRNAs in
both bone tissue and in osteocytes with the same expression level
trend, as indicated by miRNA microarray and quantitative PCR. |
Table II
Differentially expressed miRNAs in
both bone tissue and in osteocytes with the same expression level
trend, as indicated by miRNA microarray and quantitative PCR.
| A, Upregulated |
|---|
| miRNA | miRbase accession
no. |
|---|
| miR-5118 | MIMAT0020626 |
| miR-433-3p | MIMAT0001420 |
| miR-190a-5p | MIMAT0000220 |
| miR-470-5p | MIMAT0002111 |
| B,
Downregulated |
| miRNA | miRbase accession
no. |
| miR-3082-5P | MIMAT0014872 |
| miR-6348 | MIMAT0025091 |
| miR-669-5P | MIMAT0017346 |
| miR-32-3p | MIMAT0017050 |
 | Table IIIPredicted target genes of
differentially expressed miRNAs both in bone tissue and
osteocytes. |
Table III
Predicted target genes of
differentially expressed miRNAs both in bone tissue and
osteocytes.
| miRNA | Target gene | Gene
description | (Refs.) |
|---|
| miR-5118 | DKK2 | Dickkopf homolog
2 | (63) |
| | Wnt2b | Wingless related
MMTV integration site 2b | (51) |
| | Fzd5 | Frizzled homolog
5 | (64) |
| | Tgfbr1 | Transforming growth
factor β receptor | (40,41) |
| miR-433-3p | Creb1 | cAMP responsive
element binding protein 1 | (61) |
| | Map2 |
Microtubule-associated protein 2 | (62) |
| miR-190a-5p | Smad2 | SMAD family member
2 | (39,47) |
| miR-470-5p | DKK2 | Dickkopf homolog
2 | (63) |
| | ATP2b1 | Plasma membrane
calcium ATPase | (59) |
| | Runx2 | Runt related
transcription factor 2 | (65) |
| miR-3082-5p | Gria4 | Glutamate receptor
ionotropic AMPA4 | (66,67) |
| | Creb1 | cAMP responsive
element binding protein 1 | (61) |
| miR-6348 | Map3k12 | Activated protein
kinase kinase kinase 12 | (33,34) |
| | Dnm3 | Dynamin 3 | (68) |
| miR-669-5p | Creb1 | cAMP responsive
element binding protein 1 | (61) |
| | Cask |
Calcium/calmodulin-dependent serine
protein kinase | (69) |
| miR-32-3p | Tgfbr1 | Transforming growth
factor β receptor | (40,41) |
| | ATP13a3 | ATPase type
13A3 | (70,71) |
| | Dnm3 | Dynamin 3 | (68) |
Discussion
Bone is an important structure that bears mechanical
loading and has a vital role in maintaining mineral homeostasis
(22). Mechanical loading,
particularly dynamic loading, is a major determinant in the
regulation of the morphology and architecture of bone (13,23).
Suitable mechanical loading prevents bone loss or promotes bone
formation, and the absence of suitable mechanical loading results
in a decline in bone mass (24).
Exercise produces mechanical loading, which reduces bone loss,
increases bone strength and prevents osteoporosis in aging
individuals (25,26).
The effects of exercise on the structure and
metabolism of bone tissue, such as cytokines and signaling
pathways, have been studied at the cellular and molecular levels.
However, only a limited number of studies have investigated the
involvement of miRNAs induced by exercise in regulating bone
metabolism and osteoblastic differentiation, such as
mechanically-induced overexpression of miR-214 not only inhibited
the expression of these osteogenic factors, but also attenuated
mechanical strain-enhanced osteogenesis in osteoblasts (14).
Running is a normal form of exercise; it applies a
dynamic mechanical loading to the bone tissues, particularly the
femur and the tibia tissues, which is beneficial for bone health
(27). In the present study, mice
were forced to run on a treadmill. After 8 weeks of running
exercise, it was determined that running training increased the
mechanical properties of the femur, and increased the activity of
ALP and level of OCN in the bone tissues. ALP activity and OCN
levels are important markers of osteogenic differentiation
(28,29), and these markers are related to the
deposition and mineralization of bone matrix (30). The present results indicated that
treadmill training promoted the maturation and differentiation of
bone tissue and was beneficial for bone health, which was
consistent with the results of dynamic load countering
ovariectomy-induced osteoporosis in rats (31) and mechanical stretch increasing
osteogenesis-related protein expression in the bones of
ovariectomized rats (32).
Subsequently, nine of the numerous differentially
expressed miRNAs identified in the screening by miRNA microarray
were selected for verification via RT-qPCR analysis, and the target
genes of the nine miRNAs related to osteogenic differentiation were
predicted. As an example, one target gene of miR-316-3p, Map3k9, is
a MAPK signaling molecule (33).
MAPKs function to regulate the key transcriptional mediators of
osteoblast differentiation, with ERK and p38 MAPKs phosphorylating
runt-related transcription factor 2, the master regulator of
osteoblast differentiation. ERK also activates ribosomal S6 kinase
2, which in turn phosphorylates activating transcription factor 4,
a transcriptional regulator of late-stage osteoblast synthetic
functions (34). One target gene of
miR-3103-3p, nuclear factor related to kB binding protein, is
related to the NF-κB signaling pathway (35). The transcription factor NF-κB is a
member of a family of proteins involved in signaling pathways that
are essential for normal cellular functions and development
(35). Deletion of various
components of this pathway results in abnormal skeletal
development, research in the last decade has indicated that NF-κB
signaling mediates RANK ligand-induced osteoclastogenesis (35). Plasma membrane
Ca2+-transporting ATPase 4, a target gene of
miR-449a-5p, has been proven to be overexpressed during the
maturation or senescence of bone tissue cells (36). In addition, protein tyrosine
phosphatase receptor type E, a target gene of miR-322-3p, belongs
to the protein tyrosine kinase family and overexpression of this
kinase may promote osteogenic differentiation (37). These target genes and their
signaling pathways have been confirmed to be related to osteogenic
differentiation or bone metabolism.
In addition, the differentially expressed miRNAs in
the bones of the exercised mice were compared with the
differentially expressed miRNAs in mechanically strained osteocytes
in vitro. A total of eight differentially expressed miRNAs
in both bone tissues and osteocytes were identified, and each of
these miRNAs had the same expression trend in bone tissues and
osteocytes. Of these, four miRNAs (miR-5118, miR-433-3p,
miR-190a-5p and miR-470-5p) were upregulated and four miRNAs
(miR-3082-5p, miR-6438, miR-669-5p and miR-32-3p) were
downregulated, and these results were confirmed via miRNA
microarray and RT-qPCR. Furthermore, the target genes of the eight
differentially expressed miRNAs were predicted and certain target
genes were confirmed to be related to osteogenic differentiation or
bone metabolism. For instance, Wnt2b, one of the target genes of
miR-5118, is a molecule of the Wnt/β-catenin signaling pathway and
activation of Wnt/β-catenin signaling may promote osteogenic
differentiation and increase mechanically induced bone formation
(38). Smad2, a target gene of
miR-190a-5p, is a Smad signaling molecule and mechanical stress
rapidly inhibits Smad3 phosphorylation, thereby inhibiting the
activity of the key transforming growth factor-β (TGF-β) effectors
Smad2 and Smad3 in osteocytes, and maintaining bone homeostasis via
TGF-β signaling (39). In the
present study, downregulation of miR-669m-5p expression was
associated with one of its target genes, TGF-β receptor 1, and the
reduction in TGF-β signaling, through its effector SMAD3, enhanced
the mechanical properties and mineral concentration of the bone
matrix as well as the bone mass (40,41).
The present results indicated that exercise-induced
differentially expressed miRNAs in the bone most likely regulate
bone metabolism and osteoblastic differentiation, and the same
miRNAs that were differentially expressed in both mechanically
stimulated osteocytes in vitro and in the bone tissues of
mice subjected to exercise on a treadmill, also probably regulate
bone metabolism and osteoblastic differentiation.
As terminally differentiated cells, osteocytes that
are mechanically stimulated produce factors such as proteins,
peptides and signaling molecules that regulate osteogenic
differentiation or bone metabolism (42,43).
Mechanical stimuli, such as fluid shear stress, increase the number
of exosomes released by osteocytes, these exosomes contain factors
such as sclerotinins, NF-κB receptor activators and
osteoprotegerin, which regulate osteogenic differentiation
(44,45). Certain osteocyte-derived miRNAs,
such as miR-218, may influence osteoblastic differentiation
(46). Therefore, in the present
study, it was hypothesized that these differentially expressed
miRNAs in mechanically stimulated osteocytes in vitro and in
bone tissue of treadmill trained mice likely regulate osteogenic
differentiation or bone metabolism. It is hypothesized that this
regulatory mechanism may be via exosomes, thus in the future, how
these miRNAs regulate osteogenic differentiation or bone metabolism
through exosomes will be studied.
In conclusion, treadmill training resulted in the
differential expression of miRNAs in the bone tissues of mice.
Certain differentially expressed miRNAs were also observed in
osteocytes and these miRNAs probably have an important role in the
regulation of bone metabolism and osteogenic differentiation.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Nature
Science Foundation of China (grant nos. 31660261 and 32071309) and
the Natural Science Foundation of Guangxi (grant nos. 2018JIA140050
and 2018GXNSFAA281357).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The datasets generated and/or analyzed during the current
study are available in the Gene Expression Omnibus repository,
[https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179199
and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179201].
Authors' contributions
YG and JG designed the study, analyzed the data and
participated in the bioinformatics analysis. HY and ZC performed
all the RT-qPCR analyses at Guangzhou RiboBio Co., Ltd.,
(experimental equipment and reagents were provided by Guangzhou
RiboBio Co., Ltd.)., and assays for ALP activity and OCN levels,
and participated in the animal experiments. YW and JW performed the
miRNA microarray at Guangzhou RiboBio Co., Ltd., (experimental
equipment and reagents were provided by Guangzhou RiboBio Co.,
Ltd.), and performed the animal experiments. BH, FY and YQ
performed the bioinformatics analysis and detected the mechanical
properties of the mouse femur bones. YG, HY and ZC wrote and
revised the manuscript. All authors have read and approved the
final manuscript. YG, JG, HY, ZC, YW, JW, BH, FY and YQ confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The present study involved animals and was approved
by the Animal Ethics Committee of Guilin Medical University
(Guilin, China; approval no. 2019-0013). All of the authors declare
that the experiments complied with the current laws of China
(Guangxi) where they were performed.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Belavý DL, Baecker N, Armbrecht G, Beller
G, Buehlmeier J, Frings-Meuthen P, Rittweger J, Roth HJ, Heer M and
Felsenberg D: Serum sclerostin and DKK 1 in relation to exercise
against bone loss in experimental bed rest. Bone Miner Metab.
34:354–365. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhao JG, Zeng XT, Wang J and Liu L:
Association between calcium or vitamin D supplementation and
fracture incidence in community-dwelling older adults: A systematic
review and meta analysis. JAMA. 318:2466–2482. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yamazaki S, Ichimura S, Iwamoto J, Takeda
T and Toyama Y: Effect of walking exercise on bone metabolism in
postmenopausal women with osteopenia/osteoporosis. Bone Miner
Metab. 22:500–508. 2004.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wen HJ, Huang TH, Li TL, Chong PN and Ang
BS: Effects of short-term step aerobics exercise on bone metabolism
and functional fitness in postmenopausal women with low bone mass.
Osteoporos Int. 28:539–547. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Vainionpää A, Korpelainen R, Väänänen HK,
Haapalahti J, Jämsä T and Leppäluoto J: Effect of impact exercise
on bone metabolism. Osteoporos Int. 20:1725–1733. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Weaver CM, Gordon CM, Janz KF, Kalkwarf
HJ, Lappe JM, Lewis R, O'Karma M, Wallace TC and Zemel BS: The
national osteoporosis foundation's position statement on peak bone
mass development and lifestyle factors: A systematic review and
implementation recommendations. Osteoporos Int. 27:1281–1386.
2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Polidoulis I, Beyene J and Cheung AM: The
effect of exercise on pQCT parameters of bone structure and
strength in postmenopausal women-a systematic review and
meta-analysis of randomized controlled trials. Osteoporos Int.
23:39–51. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Iwamoto J, Yeh JK and Aloia JF:
Differential effect of treadmill exercise on three cancellous bone
sites in the young growing rat. Bone. 24:163–169. 1999.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ma T, Li SC, Liang XX, Luo J and Li ZX:
Effects of uphill or downhill running on bone mineral density and
the indexes of bone histomorphometry in ovariectomized mice. Sports
Sci. 31:48–55. 2011.(In Chinese).
|
|
10
|
Ma T and Li SC: Effect of uphill or
downhill running on the osteoclast differentiation of
ovariectomized mice. Chin J Sports Med. 34:468–474. 2015.(In
Chinese).
|
|
11
|
Borer KT, Zheng Q, Jafari A, Javadi S and
Kernozek T: Nutrient intake prior to exercise is necessary for
increased osteogenic marker response in diabetic postmenopausal
women. Nutrients. 11(1494)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhai Y, Tyagi SC and Tyagi N: Cross-talk
of microRNA and hydrogen sulfide: A novel therapeutic approach for
bone diseases. Biomed Pharmacother. 92:1073–1084. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lanyon LE: Control of bone architecture by
functional load bearing. Bone Miner Res. 7 (Suppl 2):S369–S375.
1992.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yuan Y, Guo J, Zhang L, Tong X, Zhang S,
Zhou X, Zhang M, Chen X, Lei L, Li H, et al: miR-214 attenuates the
osteogenic effects of mechanical loading on osteoblasts. Int J
Sports Med. 40:931–940. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang H, Sun Z, Wang Y, Hu Z, Zhou H, Zhang
L, Hong B, Zhang S and Cao X: MiR-33-5p, a novel mechano-sensitive
microRNA promotes osteoblast differentiation by targeting Hmga2.
Sci Rep. 6(23170)2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Iwawaki Y, Mizusawa N, Iwata T, Higaki N,
Goto T, Watanabe M, Tomotake Y, Ichikawa T and Yoshimoto K:
miR-494-3p induced by compressive force inhibits cell proliferation
in MC3T3-E1 cells. J Biosci Bioeng. 120:456–462. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen N, Sui BD, Hu CH, Cao J, Zheng CX,
Hou R, Yang ZK, Zhao P, Chen Q, Yang QJ, et al: MicroRNA-21
contributes to orthodontic tooth movement. J Dent Res.
95:1425–1433. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang Y, Jia L, Zheng Y and Li W: Bone
remodeling induced by mechanical forces is regulated by miRNAs.
Biosci Rep. 38(BSR20180448)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang Y, Wang K, Hu Z, Zhou H, Zhang L,
Wang H, Li G, Zhang S, Cao X and Shi F: MicroRNA-139-3p regulates
osteoblast differentiation and apoptosis by targeting ELK1 and
interacting with long noncoding RNA ODSM. Cell Death Dis.
9(1107)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Schaffler MB, Cheung WY, Majeska R and
Kennedy O: Osteocytes: Master orchestrators of bone. Calcif Tissue
Int. 94:5–24. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zeng QC, Wang Y, Gao J, Yan ZX, Li ZH, Zou
XQ, Li YN, Wang JH and Guo Y: miR-29b-3p regulated osteoblast
differentiation via regulating IGF-1 secretion of mechanically
stimulated osteocytes. BioMed Central. 24(11)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mosley JR: Osteoporosis and bone
functional adaptation: Mechanobiological regulation of bone
architecture in growing and adult bone, a review. J Rehabil Res
Dev. 37:189–199. 2000.PubMed/NCBI
|
|
23
|
Rubin CT and Lanyon LE: Regulation of bone
formation by applied dynamic loads. Bone Joint Surg Am. 66:397–402.
1984.PubMed/NCBI
|
|
24
|
Hillam RA and Skerry TM: Inhibition of
bone resorption and stimulation of formation by mechanical loading
of the modeling rat ulna in vivo. J Bone Miner Res. 5:683–689.
1995.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yoshiya S: The effects of exercise and
sports activities on bone and joint morbidities. Clin Calcium.
27:39–43. 2017.PubMed/NCBI(In Japanese).
|
|
26
|
Singh MA: Physical activity and bone
health. Aust Fam Physician. 33(125)2004.PubMed/NCBI
|
|
27
|
Santos L, Elliott-Sale KJ and Sale C:
Exercise and bone health across the lifespan. Biogerontology.
18:931–946. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jang WG, Koh JT and Kim EJ: Tunicamycin
negatively regulates BMP2-induced osteoblast differentiation
through CREBH expression in MC3T3E1 cells. BMB Rep. 44:735–740.
2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Beck GJ Jr, Zerler B and Moran E:
Phosphate is a specific signal for induction of osteopontin gene
expression. Proc Natl Acad Sci USA. 97:8352–8357. 2000.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Mahalingam CD, Datta T, Patil RV, Kreider
J, Bonfil RD, Kirkwood KL, Goldstein SA, Abou-Samra AB and Datta
NS: Mitogen-activated protein kinase phosphatase 1 regulates bone
mass, osteoblast gene expression, and responsiveness to parathyroid
hormone. J Endocrinol. 211:145–156. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li H, Li RX, Wan ZM, Xu C, Li JY, Hao QX,
Guo Y, Liu L and Zhang XZ: Counter-effect of constrained dynamic
loading on osteoporosis in ovariectomized mice. J Biomech.
46:1242–1247. 2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wu Y, Zhang P, Dai Q, Yang X, Fu R, Jiang
L and Fang B: Effect of mechanical stretch on the proliferation and
differentiation of BMSCs from ovariectomized rats. Mol Cell
Biochem. 382:273–282. 2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Cong Q, Jia H, Li P, Qiu S, Yeh J, Wang Y,
Zhang ZL, Ao J, Li B and Liu H: P38α MAPK regulates proliferation
and differentiation of osteoclast progenitors and bone remodeling
in an aging-dependent manner. Sci Rep. 7(45964)2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Greenblatt MB, Shim JH and Glimcher LH:
Mitogen-activated protein kinase pathways in osteoblasts. Annu Rev
Cell Dev Biol. 29:63–79. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Abu-Amer Y: NF-κB signaling and bone
resorption. Osteoporos Int. 24:2377–2386. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yoo JK, Choi SJ and Kim JK: Expression
profiles of subtracted mRNAs during cellular senescence in human
mesenchymal stem cells derived from bone marrow. Exp Gerontol.
5:464–471. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang H, Ye X, Xiao H, Zhu N, Wei C, Sun X,
Wang L, Wang B, Yu X, Lai X, et al: PTPN21 overexpression promotes
osteogenic and adipogenic differentiation of bone marrow-derived
mesenchymal stem cells but inhibits the immunosuppressive function.
Stem Cells Int. 2019(4686132)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Liedert A, Nemitz C, Haffner-Luntzer M,
Schick F, Jakob F and Ignatius A: Effects of estrogen receptor and
Wnt signaling activation on mechanically induced bone formation in
a mouse model of postmenopausal bone loss. Int J Mol Sci.
21(8301)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Nguyen J, Tang SY, Nguyen D and Alliston
T: Load regulates bone formation and Sclerostin expression through
a TGFβ-dependent mechanism. PLoS One. 8(e53813)2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang H, Huang W, Liu H, Zheng Y and Liao
L: Mechanical stretching of pulmonary vein stimulates matrix
metalloproteinase-9 and transforming growth factor-β1 through
stretch-activated channel/MAPK pathways in pulmonary hypertension
due to left heart disease model rats. PLoS One.
15(e0235824)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Balooch G, Balooch M, Nalla RK, Schilling
S, Filvaroff EH, Marshall GW, Marshall SJ, Ritchie RO, Derynck R
and Alliston T: TGF-beta regulates the mechanical properties and
composition of bone matrix. Proc Natl Acad Sci USA.
102:18813–18818. 2005.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Klein-Nulend J, Bakker AD, Bacabac RG,
Vatsa A and Weinbaum S: Mechanosensation and transduction in
osteocytes. Bone. 54:182–190. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yan Y, Wang L, Ge L and Pathak JL:
Osteocyte-mediated translation of mechanical stimuli to cellular
signaling and its role in bone and non-bone-related clinical
complications. Curr Osteoporos Rep. 18:67–80. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Morrell AE, Brown GN, Robinson ST, Sattler
RL, Baik AD, Zhen G, Cao X, Bonewald LF, Jin W, Kam LC and Guo XE:
Mechanically induced Ca2+ oscillations in osteocytes
release extracellular vesicles and enhance bone formation. Bone
Res. 6(6)2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Eichholz KF, Woods I, Riffault M, Johnson
GP, Corrigan M, Lowry MC, Shen N, Labour MN, Wynne K, O'Driscoll L
and Hoey DA: Human bone marrow mesenchymal stem/stromal cell
behaviour is coordinated via mechanically activated
osteocyte-derived extracellular vesicles. Stem Cells Transl Med.
9:1431–1447. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Qin Y, Peng Y, Zhao W, Pan J,
Ksiezak-Reding H, Cardozo C, Wu Y, Divieti PP, Bonewald LF, Bauman
WA and Qin W: Myostatin inhibits osteoblastic differentiation by
suppressing osteocyte-derived exosomal microRNA-218: A novel
mechanism in muscle-bone communication. J Biol Chem.
292:11021–11033. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zheng W, Chen Q, Zhang Y, Xia R, Gu X, Hao
Y, Yu Z, Sun X and Hu D: BMP9 promotes osteogenic differentiation
of SMSCs by activating the JNK/Smad2/3 signaling pathway. J Cell
Biochem. 121:2851–2863. 2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Walker LM, Publicover SJ, Preston MR, Said
Ahmed MA and El Haj AJ: Calcium-channel activation and matrix
protein upregulation in bone cells in response to mechanical
strain. J Cell Biochem. 79:648–661. 2000.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Uda Y, Azab E, Sun N, Shi C and Pajevic
PD: Osteocyte mechanobiology. Curr Osteoporos Rep. 15:318–325.
2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Mazur CM, Woo JJ, Yee CS, Fields AJ,
Acevedo C, Bailey KN, Kaya S, Fowler TW, Lotz JC, Dang A, et al:
Osteocyte dysfunction promotes osteoarthritis through
MMP13-dependent suppression of subchondral bone homeostasis. Bone
Res. 7(34)2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Lerner UH and Ohlsson C: The WNT system:
Background and its role in bone. J Intern Med. 277:630–649.
2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chen Y, Mohammed A, Oubaidin M, Evans CA,
Zhou X, Luan X, Diekwisch TG and Atsawasuwan P: Cyclic stretch and
compression forces alter microRNA-29 expression of human
periodontal ligament cells. Gene. 566:13–17. 2015.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Pozio A, Palmieri A, Girardi A, Cura F and
Carinci F: Titanium nanotubes activate genes related to bone
formation in vitro. Dent Res J (Isfahan). 9 (Suppl 2):S164–S168.
2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Yasui M, Shiraishi Y, Ozaki N, Hayashi K,
Hori K, Ichiyanagi M and Sugiura Y: Nerve growth factor and
associated nerve sprouting contribute to local mechanical
hyperalgesia in a rat model of bone injury. Eur J Pain. 16:953–965.
2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Shi J, Wang Z, Guo X, Shen J, Sun H, Bai
J, Yu B, Wang L, Zhou W, Liu Y, et al: Spirin inhibits osteoclast
formation and wear-debris-induced bone destruction by suppressing
mitogen-activated protein kinases. J Cell Physiol. 235:599–2608.
2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Vriend J and Reiter RJ: Melatonin, bone
regulation and the ubiquitin-proteasome connection: A review. Life
Sci. 145:152–160. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Matsumoto Y and Rottapel R: Bone dynamics
and inflammation: Lessons from rare diseases. Immunol Med.
43:61–64. 2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Hamamura K, Swarnkar G, Tanjung N, Cho E,
Li J, Na S and Yokota H: RhoA-mediated signaling in
mechanotransduction of osteoblasts. Connect Tissue Res. 53:398–406.
2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Rosselli-Murai LK, Almeida LO, Zagni C,
Galindo-Moreno P, Padial-Molina M, Volk SL, Murai MJ, Rios HF,
Squarize CH and Castilho RM: Periostin responds to mechanical
stress and tension by activating the MTOR signaling pathway. PLoS
One. 8(e83580)2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Lorentzon M, Eriksson AL, Nilsson S,
Mellström D and Ohlsson C: Association between physical activity
and BMD in young men is modulated by catechol-O-methyltransferase
(COMT) genotype: The GOOD study. J Bone Miner Res. 22:1165–1172.
2007.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Chen B, Lin T, Yang X, Li Y, Xie D and Cui
H: Intermittent parathyroid hormone (1-34) application regulates
cAMP-response element binding protein activity to promote the
proliferation and osteogenic differentiation of bone mesenchymal
stromal cells, via the cAMP/PKA signaling pathway. Exp Ther Med.
11:2399–2406. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Yin C, Zhang Y, Hu L, Tian Y, Chen Z, Li
D, Zhao F, Su P, Ma X, Zhang G, et al: Mechanical unloading reduces
microtubule actin crosslinking factor 1 expression to inhibit
β-catenin signaling and osteoblast proliferation. J Cell Physiol.
233:5405–5419. 2018.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Olivares-Navarrete R, Hyzy S, Wieland M,
Boyan BD and Schwartz Z: The roles of Wnt signaling modulators
Dickkopf-1 (Dkk1) and Dickkopf-2 (Dkk2) and cell maturation state
in osteogenesis on microstructured titanium surfaces. Biomaterials.
313:2015–2024. 2010.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Leijten JC, Bos SD, Landman EB, Georgi N,
Jahr H, Meulenbelt I, Post JN, van Blitterswijk CA and Karperien M:
GREM1, FRZB and DKK1 mRNA levels correlate with osteoarthritis and
are regulated by osteoarthritis-associated factors. Arthritis Res
Ther. 15(R126)2013.PubMed/NCBI View
Article : Google Scholar
|
|
65
|
Galea GL, Paradise CR, Meakin LB,
Camilleri ET, Taipaleenmaki H, Stein GS, Lanyon LE, Price JS, van
Wijnen AJ and Dudakovic A: Mechanical strain-mediated reduction in
RANKL expression is associated with RUNX2 and BRD2. Gene X.
5(100027)2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Szczesniak AM, Gilbert RW, Mukhida M and
Anderson GI: Mechanical loading modulates glutamate receptor
subunit expression in bone. Bone. 37:63–73. 2005.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Lin TH, Yang RS, Tang CH, Wu MY and Fu WM:
Regulation of the maturation of osteoblasts and osteoclastogenesis
by glutamate. Eur J Pharmacol. 589:37–44. 2008.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Eleniste PP, Huang S, Wayakanon K, Largura
HW and Bruzzaniti A: Osteoblast differentiation and migration are
regulated by dynamin GTPase activity. Int J Biochem Cell Biol.
46:9–18. 2014.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Naghii MR, Torkaman G and Mofid M: Effects
of boron and calcium supplementation on mechanical properties of
bone in rats. Biofactors. 28:195–201. 2006.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Nakano Y, Forsprecher J and Kaartinen MT:
Regulation of ATPase activity of transglutaminase 2 by MT1-MMP:
Implications for mineralization of MC3T3-E1 osteoblast cultures.
Cell Physiol. 223:260–269. 2010.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Sun D, Junger WG, Yuan C, Zang W, Bao Y,
Qin D, Wang C, Tan L, Qi B, Zhu D, et al: Shockwaves induce
osteogenic differentiation of human mesenchymal stem cells through
ATP release and activation of P2X7 receptors. Stem Cells.
31:1170–1180. 2013.PubMed/NCBI View Article : Google Scholar
|