Introduction
Sudden coronary death (SCD) is the most common type
of sudden death in adults, particularly in middle-aged and elderly
individuals (1). SCD primarily
occurs when coronary atherosclerosis results in acute myocardial
ischemia and there is a sudden interruption to coronary blood flow
(2-4).
SCD frequently occurs after an inducement, such as drinking
alcohol, fatigue, smoking or exercise, and may be fatal within a
small number of hours, whereby certain individuals pass away during
their sleep (5). Previous studies
have reported that ~80% of SCDs are associated with the existence
of coronary artery disease (CAD), which is the most common
underlying cause in adults (6,7).
Several epidemiological studies have demonstrated that a family
history of SCD is an independent risk factor of SCD and patients
with different gene mutations may have similar symptoms (8,9).
Genetic studies in the past three decades have highlighted the
genetic changes of patients with inherited cardiac disease that
cause SCD (10,11). Genome-wide association studies
(GWAS) are a systemic tool used to evaluate the whole genome to
determine the association between gene variants and diseases
(12,13). Sudden cardiac arrest caused by CAD
has been identified in GWAS (14,15).
However, whether these single nucleotide polymorphisms (SNPs)
obtained by GWAS that are associated with sudden cardiac arrest or
coronary heart disease (CHD) are directly associated with SCD has
remained elusive.
Studies on SCD have not been performed in terms of
risk gene variations in the Chinese Han population and the
underlying genetics that contribute to the SCD single nucleotide
variations are of significant interest. When using comparable
population groups, it is important to determine the genetic
background of an alive individual with CHD and that of individuals
who experienced SCD with CHD, which may help understand the
predictive value and association between the genetic changes and
incidence of SCD to improve SCD prevention measures. Forensic
practitioners frequently encounter cases of sudden death for
unknown reasons during autopsy. Thus, genetic predictive studies
will help determine the cause of death, particularly for those
patients who died of CHD.
The majority of biological tissue specimens degrade
due to external exposure prior to examination and improper
treatment, such as implementation of paraffin-embedded tissues and
formalin-fixed human tissues (16,17).
Degraded biological specimens are not used for GWAS, whole-genome
sequencing (WGS) or whole-exome sequencing (WES), which impedes the
application of these techniques in forensic examinations of cases
of SCD (18-21).
It has been reported that SCD-related SNPs may contribute to SCD,
which is easily detected in degraded samples.
The present study developed an assay with
mini-sequencing techniques based on the GWAS results of clinical
samples reported by Aouizerat et al (14) to investigate the genetic background
of SCD in the Chinese Han population. This assay applies to most
degraded tissues and is yet to be confirmed for use in other
populations to identify risk factors, as well as provide accurate
diagnoses and prevention advice for first-degree relatives.
Materials and methods
Sample collection
A total of 198 samples were collected from the
Chinese Han population. The clinical characteristics of all
participants are presented in Table
SI. Samples were classified into three groups: The CHD (n=70),
SCD (n=53) and control (n=75) groups. The CHD group consisted of
blood samples from 70 patients with CHD. The SCD group consisted of
53 cases in which SCD was confirmed following autopsy after death.
The control group consisted of 24 cases that died from other causes
(such as trauma, poisoning, suffocation, etc.) and 51 volunteers
who had no family history of SCD and CHD. The clinical
characteristics were not available for the anonymous healthy
controls. Sample data are presented in Table I. The sex ratio (male/female) was
3.67 (55/15), 4.89 (44/9) and 3.69 (59/16) in the CHD, SCD and
control groups, respectively, with respective median ages (range)
in each group of 52 (40-69), 52.5 (37-71) and 53 (34-69) years for
the males, and 58 (45-70), 61 (47-68) and 55.5 (48-68) years for
the females. Comparisons between the three groups demonstrated no
significant differences in sex, age and other complicated diseases.
Genomic DNA was extracted from venous blood using the E.Z.N.A™ SE
Blood DNA kit (Omega Bio-Tek, Inc.). Tissues were treated using the
Mag-Blind® Tissue DNA kit M6223 (Omega Bio-Tek, Inc.)
according to the manufacturer's instructions. DNA samples were
quantified at an optical density of 260 nm using a BioSpectrometer
(22331; Eppendorf AG). The extracted DNA samples were subsequently
diluted with highly purified water to the final concentration of 10
ng/µl and stored at -20˚C until subsequent analysis.
 | Table ISample data of the present study. |
Table I
Sample data of the present study.
| Group | Sex | N (%) | Sex ratio
(M/F) | Age, years | Patients
complicated with other diseases, % |
|---|
| CHD | M | 55 (78.57) | 3.67:1 | 52 (40-69) | 20 |
| | F | 15 (21.43) | | 58 (45-70) | 20 |
| SCD | M | 44 (83.02) | 4.89:1 | 52.5 (37-71) | 25 |
| | F | 9 (16.98) | | 61 (47-68) | 22 |
| Control | M | 59 (78.67) | 3.69:1 | 53 (34-69) | None recorded |
| | F | 16 (21.33) | | 55.5 (48-68) | None recorded |
Establishment of the SNaPshot
assay
The SNaPshot kit (ABI PRISM® SNaPshot™
Multiplex kit; Thermo Fisher Scientific, Inc.) was used to
establish a mini-sequencing method for SNP screening in a
combination of the multiplex PCR, single base extension (SBE) and
capillary electrophoresis (CE) techniques.
Construction of a multiplex PCR system
Candidate SNPs
The sudden cardiac arrest-related gene variations
were selected from GWAS results. To determine whether the
polymorphisms of these SNPs are prone to induce SCD in the Chinese
Han population, 21 SNPs with P<1x10-7 were selected
from GWAS, based solely on previous studies (22-25).
The results revealed that the 21 SNPs were independent and did not
exhibit any linkage disequilibrium cluster. Data on the SNPs are
presented in Table II.
 | Table IIInformation of 21 SNPs in the present
study. |
Table II
Information of 21 SNPs in the present
study.
| SNP | Allele | Context | Gene | Location
(GRCh38.p12) | P-value from
GWAS | Population by
GWAS |
|---|
| rs2389202 | A/T | Intergenic | MIR1973,
TRMT112P1 | Chr4:
116333133 |
4.000x10-7 | European |
| rs11624056 | A/T | Intergenic | FLRT2, GALC | Chr14:
87039904 |
3.000x10-8 | European |
| rs2982694 | G/T | Intron | ESR1 | Chr6:
151964552 |
7.000x10-10 | European |
| rs4665058 | A/C | Intron | BAZ2B | Chr2:
159333698 |
2.000x10-10 | European |
| rs17718586 | G/T | Intergenic | CALM2P1,
CASC17 | Chr17:
70648048 |
2.000x10-8 | European |
| rs12429889 | T/C | Intergenic | KLF12, RNY1P5 | Chr13:
74168185 |
5.000x10-20 | European |
| rs16866933 | G/A | Intron | ZNF385B | Chr2:
179701951 |
6.000x10-14 | European |
| rs7307780 | C/T | Intergenic | RPL10P13,
PHLDA1 | Chr12:
75826838 |
5.000x10-15 | European |
| rs17291650 | A/G | Cds-synon | ATF1 | Chr12:
50819650 |
3.000x10-7 | European |
| rs9581094 | T/C | Intron | PARP4 | Chr13:
24508492 |
7.000x10-7 | European |
| rs10183640 | G/A | Intergenic | ACVR1, UPP2 | Chr2:
157923692 |
5.000x10-7 | European |
| rs12189362 | C/T | Intron | GRIA1 | Chr5:
153677988 |
3.000x10-10 | European |
| rs11187837 | A/G | Intron | PLCE1 | Chr10:
94276223 |
4.000x10-7 | European |
| rs4621553 | G/A | Intergenic | YTHDC2, KCNN2 | Chr5:
113694467 |
4.000x10-8 | European |
| rs1559040 | C/T | Intron | ACYP2 | Chr2: 54120613 |
4.000x10-8 | European |
| rs10829156 | A/G | Intron | ARL5B | Chr10:
18661626 |
4.000x10-7 | European |
| rs2281680 | G/A | Intron/ncRNA | AP1G2 | Chr14:
23563861 |
6.000x10-8 | European |
| rs597503 | G/A/C | Intergenic | SCML2P1, LAMA1 | Chr18: 6939948 |
2.000x10-8 | European |
| rs16942421 | G/A/T | Intron | KCTD1 | Chr18:
26576461 |
8.000x10-10 | European |
| rs12155623 | A/C/T | Intergenic | EFCAB1, SNAI2 | Chr8: 48899642 |
3.000x10-7 | European |
| rs2251393 | G/A/C | UTR-3 | 10-Mar | Chr17:
62701571 |
4.000x10-7 | European |
Multiplex PCR
The multiplex PCR primers were designed using the
online software Primer 3.0 (http://primer3.ut.ee). The specificity of the primers
was confirmed using National Center for Biotechnology Information
Primer Blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).
Auto Dimer v1 software was used to assess the primer-dimer and
hairpin structures (26,27). In forensic medicine, most samples
experience a period of ex vivo degradation prior to the DNA
test and even during the fixation procedure with formaldehyde
(28,29). Thus, PCR with short fragment
amplification (132-280 bp) was performed, which was amenable to
type-degraded DNA samples in forensic casework. The primer
sequences are listed in Table
III. PCR amplification was performed using the GeneAmp PCR
System 9700 (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The PCR mixture had a total volume of 10 µl, containing 5 µl 2X
Multiplex PCR mix (M5 HiPer Multiplex PCR MasterMix; Mei5
Bioservices Co., Ltd.), 1 µl PCR primer mix (the final
concentration of each primer was 0.2 µM) and 5 ng DNA template. The
following thermocycling conditions were used: Initial denaturation
at 95˚C for 10 min, followed by 30 cycles of 94˚C for 20 sec, 58˚C
for 20 sec and 72˚C for 30 sec, and a final extension at 72˚C for 5
min (30).
 | Table IIIPCR and SBE primers used in the
study. |
Table III
PCR and SBE primers used in the
study.
| SNP | PCR primer sequence
(5'-3') | Product length,
bp | SBE primer sequence
(5'-3') | SBE primer
concentration, µM |
|---|
| rs2389202 | F:
TGAACTTCATTGCCATAGTCTCC | 185 |
TTGGAAAAGATAAAGTCACA | 0.20 |
| | R:
GAAGAGACACTGGCCCTCT | | | |
| rs11624056 | F:
TGTACACTGCTCGGTGATGG | 170 |
(GACT)1AAATCTCAGAAATCACCACT | 0.01 |
| | R:
ACGTCTCTCAGGCTTCTCCA | | | |
| rs2982694 | F:
CCAAGTATTTTGCTGTTGTTGCT | 280 |
(GACT)2TTACTGCATTTGTTTATCAG | 1.65 |
| | R:
CTGGGTGACAGAGTGAGACT | | | |
| rs4665058 | F:
CGCGACATGTAACCAGAAATCA | 145 |
(CT)5TCTTAAAAACAAAATAGCTT | 1.25 |
| | R:
CCGACCATTTTAGACTTTCCCAG | | | |
| rs17718586 | F:
CCATGTCTTCAGCTACACACAG | 198 |
(GACT)3CTACCTGTATCAAAGTAAAT | 0.02 |
| | R:
GCAGCATATACAACACCTAGCATAG | | | |
| rs12429889 | F:
AGCGTGCATCTTTCATTTCCT | 178 |
(GACT)4GCTTTGAAACGGTGGCTGTT | 0.50 |
| | R:
GGCAAAGAATGGCTCACAGATAC | | | |
| rs16866933 | F:
CGTGGAAAGGAATGGGCAC | 143 |
(CT)9TCCATCCTAAGCCTCCCAGA | 0.03 |
| | R:
GCAATCTGGTCTCTTTGGGC | | | |
| rs7307780 | F:
CCCAGAGTGTTTGCTGTTCC | 176 |
(GACT)5ATTAGTCTGTTCTCTCATTG | 0.35 |
| | R:
GACATGCCTTTCACCTTCCAC | | | |
| rs17291650 | F:
AGTGACCACGGAAAATTACTGAAG | 166 |
(CT)11TTTAGAGAAGCTGCTCGAGA | 0.06 |
| | R:
TTTTCCAGGACTGCAACTCG | | | |
| rs9581094 | F:
GTGTTTCCTGGAAAAGTGACTCAT | 205 |
(GACT)6ATCTTGTTTTGCATTTTTCT | 0.40 |
| | R:
GCCTAAGTGACAAAAGCGAGA | | | |
| rs10183640 | F:
CGATCAGTTTGGCTGGAGAGA | 177 |
(CT)13AGGAATTTGAACTTTATCTT | 0.10 |
| | R:
AAGCCTGGACAACATAGCGA | | | |
| rs12189362 | F:
CTCTTGGGGCTCCTGTAGAT | 200 |
(GACT)7TGCTAGAGAAGCTGTATTTC | 0.50 |
| | R:
TCTTGCTGTGCTGGTTTGTC | | | |
| rs11187837 | F:
AGTTGCCCTTGAGTCAGCC | 190 |
(CT)15CACTCTGGGAAATGCAGGCT | 0.55 |
| | R:
CACAAGTGGCCAGGTTTCA | | | |
| rs4621553 | F:
GTTCAGATGCCTTTAGTTGCTGA | 174 |
(CT)17TAGTTATACATTACTCAAGG | 0.10 |
| | R:
TGCTCATCTTGCCCAGATTTC | | | |
| rs1559040 | F:
CGCATTGTGACTATCTGTTGGTA | 234 |
(GACT)9TTGCCAGCCAGAAATCTCCA | 0.04 |
| | R:
CAGACCAGTAGCACAGCCT | | | |
| rs10829156 | F:
GCCAGTCTTCAGAGTTTAGCATA | 244 |
(CT)19TCTCGTTTATTGATGTTTGA | 0.25 |
| | R:
ACACGTCCCTTCTATTCGGT | | | |
| rs2281680 | F:
GAGGGCAGGACTCCAGAAAG | 150 |
(CT)21CATGGAAACCTCTTTCTCCT | 0.02 |
| | R:
TGAGGCATGGACCAGGATG | | | |
| rs597503 | F:
GGAGATGAATGGTAGTGGTTGC | 164 |
(CT)23TGAATTTCATGGAAATGTAC | 0.15 |
| | R:
TGGTGCCAAAAGTCCTTGTT | | | |
| rs16942421 | F:
CCCTTGCTGAGATTTGGGTG | 203 |
(CT)27AAAATACATTTGAATGTACT | 0.30 |
| | R:
CGTTCGAAATGGCTGCTAGG | | | |
| rs12155623 | F:
GTAGGGCTGAAGAACATGCAAT | 132 |
(CT)29GGCTTTGGGTGGAAAAGAAC | 0.04 |
| | R:
GCTTCAGCACCCCACAAAAC | | | |
| rs2251393 | F:
GCTGCCCATAGATGCTCAAG | 134 |
(CT)31ACCCCAAAAGAGAGTGGCAC | 0.05 |
| | R:
AGCCCTTCTTTCTACGTCCC | | | |
Construction of the mini-sequencing
system SBE primers
SBE primers were designed to pair the bases adjacent
to the expected position of the SNP. To isolate each SBE product in
the following CE, -(CT)n or -(AGCT)n (‘n’
indicates the number of repetitions) tails were added at the 5'-end
of each SBE primer, according to the length to be detected. In this
experiment, the length of expected SBE products ranged between
20-82 bp. SBE primer sequences are listed in Table III.
Purification and SBE reaction
Recombinant shrimp alkaline phosphatase (rSAP; New
England BioLabs, Inc.) was adopted to remove excessive
deoxyribonucleoside triphosphate (dNTP) from the PCR product and
exonuclease Ⅰ (Exo I; New England BioLabs, Inc.) for excessive PCR
primers. The purification reaction was performed in a total volume
of 5 µl, containing 3.5 µl PCR product, 0.5 U rSAP and 4 U Exo I.
The reaction mixture was incubated at 37˚C for 1 h, followed by
95˚C for 15 min. The purified PCR products were subjected to an SBE
reaction in a total volume of 5 µl, containing 2 µl purified PCR
products, 2.5 µl SNaPshot reaction mix (SNaPshot™ Multiplex kit;
Applied Biosystems; Thermo Fisher Scientific, Inc.) and 0.5 µl SBE
primer mixtures (the concentration of the primers was displayed in
Table III). The SBE reaction
conditions were as follows: 96˚C for 5 sec, 50˚C for 10 sec and
60˚C for 15 sec for 35 cycles. rSAP was adopted to remove excessive
dideoxyNTP (ddNTP) prior to CE.
Separation and visualization by
CE
To visualize the SBE products, 1.5 µl SNaPshot
reaction products were mixed with 10 µl Hi-Di formamide (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and 0.1 µl of GeneScan™
120 LIZ™ size standard (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The mixture was denatured at 95˚C for 5 min,
followed by incubation at 0˚C for 3 min. A 3130 genetics analyzer
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with a 36-cm
capillary filled with POP-4 gel (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used to detect the SNP genotype. Mutated or
normal SNPs were analyzed using GeneMapper ID v3.2 software
(Applied Biosystems; Thermo Fisher Scientific, Inc.), based on
different fluorescent signals.
Sensitivity of the SNaPshot assay
To further evaluate the detection sensitivity of the
mini-sequencing system, multiplex PCR and SBE procedures were
performed using 10, 1, 0.1 and 0.01 ng template DNA, respectively.
The results are presented in Fig.
S1. All experiments were performed in triplicate. Full SNP
profiles were detected with 0.1 ng human genomic DNA.
Accuracy of the mini-sequencing
system
Samples were randomly sampled for single PCR and
sent to Thermo Fisher Scientific, Inc. for Sanger sequencing to
verify the accuracy of the experiment. The results were consistent
with those of the mini-sequencing assay established in this
experiment (Fig. S2).
Statistical analysis
Microsoft Excel 2010 (Microsoft Corp.) and SPSS 23.0
software (IBM Corp.) were used to record and analyze the data. The
prediction data of CHD were obtained by comparing the control and
CHD groups, while the prediction data of SCD were obtained by
comparing the control and SCD groups and the prediction data of
sudden death from CHD were obtained by comparing the CHD and SCD
groups. Odds ratio (OR) values were obtained using the
χ2 test. P<0.05 was considered to indicate a
statistically significant difference. To evaluate the contribution
of SNPs to CHD, SCD or sudden death from CHD, two types of
prediction models were established: i) The statistically
significant SNPs obtained via the χ2 test was added into
the prediction model to assess the risk of disease. The predicted
probabilities were compared with observed disease status, whereas
the area under the receiver operating characteristic curve (AUC)
was derived as an overall measurement of prediction accuracy,
including sensitivity and specificity measured at different
probability thresholds, and the true area was set at 0.5 (an AUC
value of 0.5 signified complete lack of prediction and 1.0 perfect
prediction). Multifactor dimensionality reduction (MDR) was used to
assess the potential effect of whether the model improved
prediction accuracy and SNP-SNP interactions. ii) All polymorphic
SNPs as variables or covariates evaluated by binary logistic
regression and obtained association SNPs were incorporated into the
prediction model. The AUC and MDR were used to evaluate the ability
of the multi-SNPs to identify and predict diseases.
Results
Establishment of the mini-sequencing
assay
The present study established a mini-sequencing
system for screening SCD-related SNPs following extensive
adjustment of the multiplex PCR and primers. PCR with short
fragment amplification (132-280 bp) was performed, which is easily
detected even in degraded samples in forensic casework. All
degraded samples were fully typed in the present study. Fig. 1 presents the results of genotyping
of 21 SNPs from a random individual sample in the control
group.
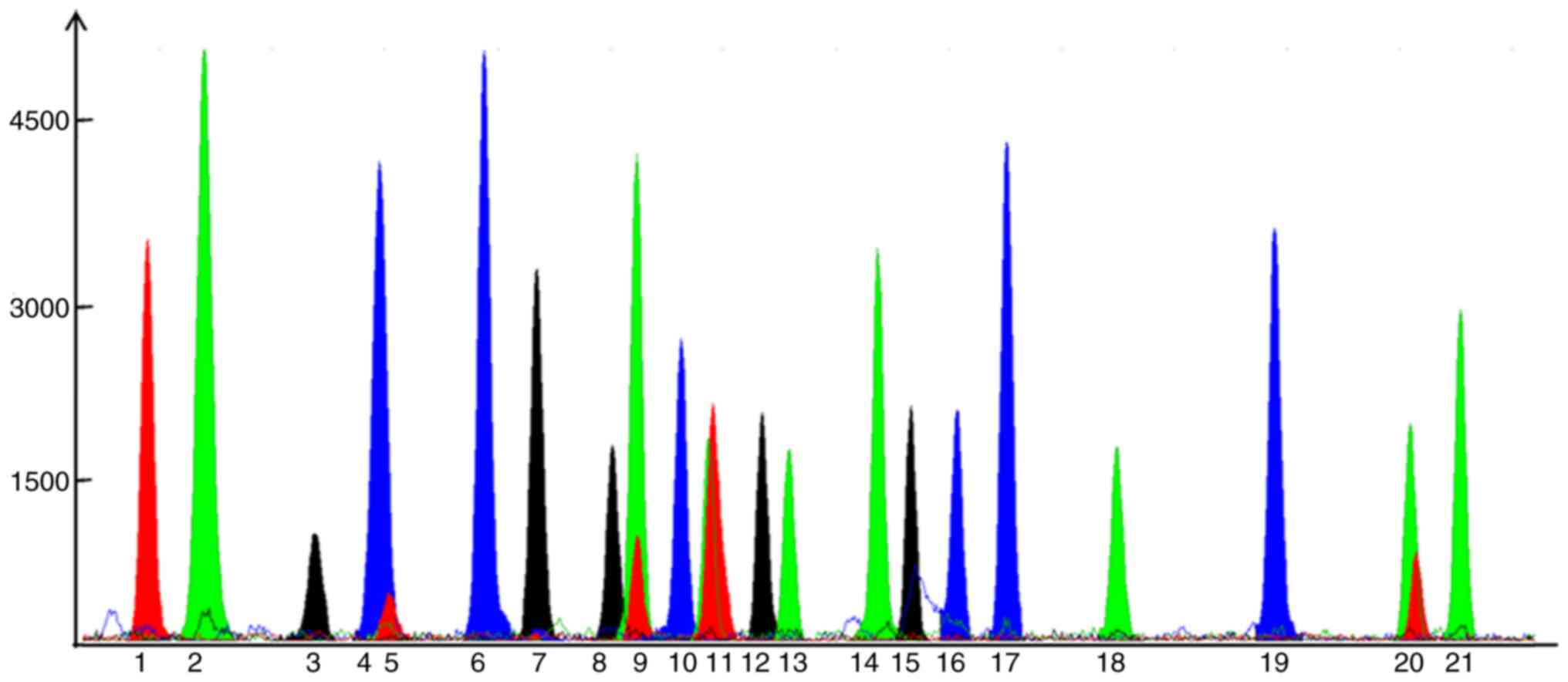 | Figure 1Results of 21 single nucleotide
polymorphisms from a random individual from the control group. Blue
represents guanine, green represents adenine, black represents
cytosine and red represents thymine. Signals: 1, rs2389202; 2,
rs11624056; 3, rs4665058; 4, rs17718586; 5, rs2982694; 6,
rs16866933; 7, rs12429889; 8, rs7307780; 9, rs17291650; 10,
rs10183640; 11, rs9581094; 12, rs12189362; 13, rs11187837; 14,
rs4621553; 15, rs1559040; 16, rs10829156; 17, rs2281680; 18,
rs597503; 19, rs16942421; 20, rs12155623; and 21, rs2251393. |
Genotype data in the Chinese Han
population
The present study focused on 21 candidate SNPs from
difference loci previously reported by Aouizerat et al
(14) via GWAS. A total of six
SNPs (rs11624056, rs4665058, rs17718586, rs17291650, rs12189362 and
rs1559040) that were reported as disease-prone in the GWAS did not
exhibit any polymorphism in the population assessed, suggesting a
different genetic background between Chinese and European
populations. A total of 15 SNPs were polymorphic in the Chinese Han
population, including rs2389202, rs12429889, rs16866933,
rs10183640, rs11187837, rs597503, rs12155623, rs2982694, rs7307780,
rs9581094, rs4621553, rs10829156, rs2281680, rs2251393 and
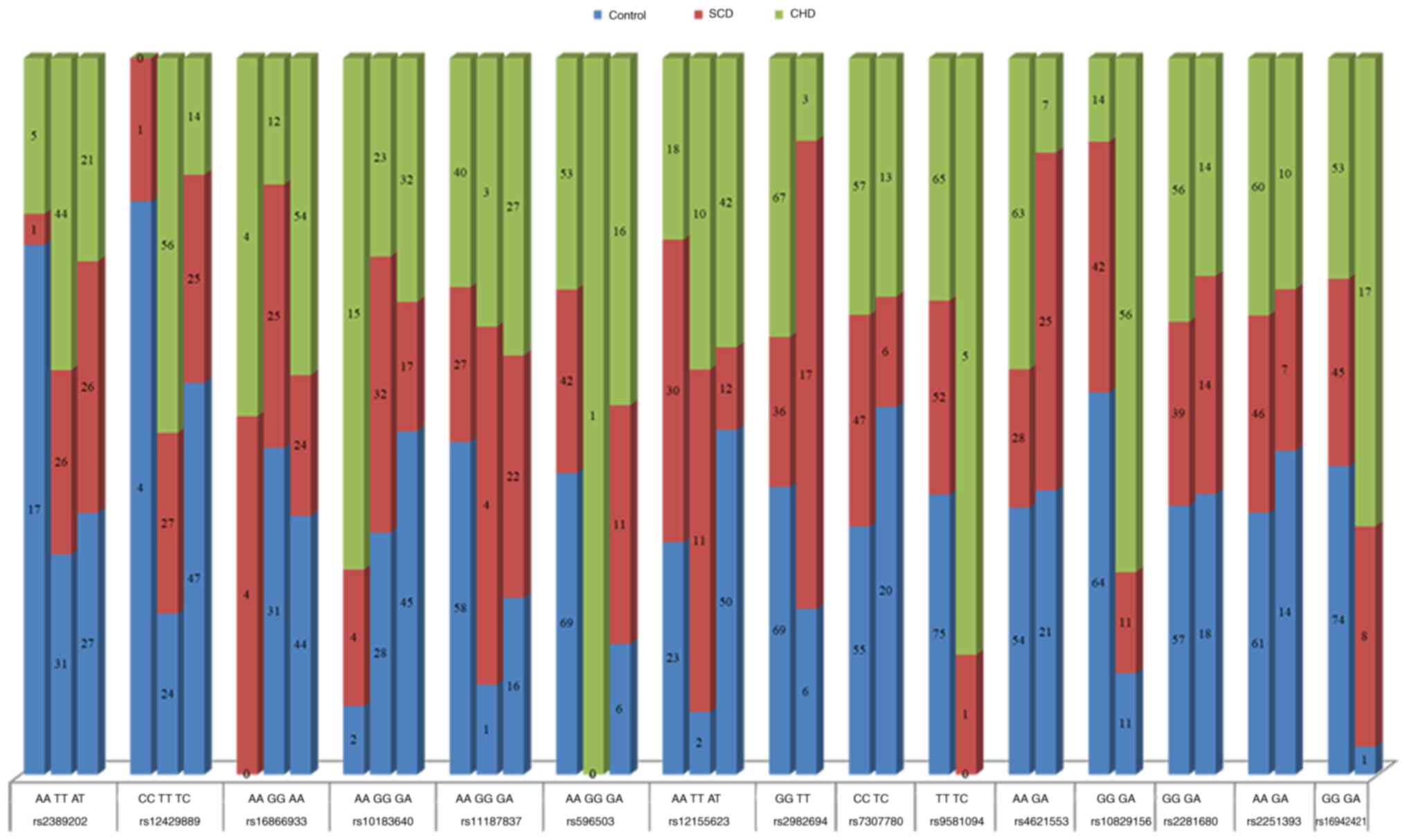
rs16942421. Genotype data for these 15 SNPs are presented in
Fig. 2 and Table SII. A total of 10 SNPs (rs2389202,
rs12429889, rs16866933, rs10183640, rs11187837, rs597503,
rs9581094, rs4621553, rs10829156 and rs16942421) were associated
with CHD and four SNPs (rs2389202, rs11187837, rs2982694 and
rs16942421) were associated with SCD. Furthermore, six SNPs
(rs12429889, rs16866933, rs10183640, rs2982694, rs4621553 and
rs10829156) were identified as risk factors for sudden death in
patients with CHD. The OR values of the 15 SNPs in the three groups
are presented in Table IV. These
genotype data contribute to the accumulating evidence on the
influence of genetic variation on the risk of CHD and SCD.
 | Table IVResults of 15 SNPs compared with the
χ2 test among three groups of samples. |
Table IV
Results of 15 SNPs compared with the
χ2 test among three groups of samples.
| A, Comparison
between control group and CHD group |
|---|
| SNP | Allele | P-value | Risk allele | Odds ratio | 95% CI |
|---|
| rs2389202 | A>T | 0.001 | A | 1.837 | 1.274-2.648 |
| rs12429889 | T>C | <0.0001 | C | 3.667 | 2.138-6.290 |
| rs16866933 | G>A | 0.008 | G | 1.268 | 1.059-1.519 |
| rs10183640 | G>A | 0.042 | G | 1.209 | 1.004-1.454 |
| rs11187837 | A>G | 0.010 | A | 1.151 | 1.032-1.284 |
| rs597503 | G>A | 0.006 | A | 1.102 | 1.026-1.183 |
| rs12155623 | A>T | 0.150 | A | 1.149 | 0.950-1.390 |
| rs2982694 | G>T | 0.190 | T | 1.867 | 0.720-4.839 |
| rs7307780 | C>T | 0.278 | T | 1.436 | 0.743-2.776 |
| rs9581094 | T>C | 0.020 | T | 1.037 | 1.004-1.071 |
| rs4621553 | G>A | 0.010 | G | 2.800 | 1.229-6.382 |
| rs10829156 | G>A | <0.0001 | G | 1.544 | 1.339-1.781 |
| rs2281680 | G>A | 0.587 | G | 0.978 | 0.902-1.060 |
| rs2251393 | G>A | 0.499 | G | 1.307 | 0.600-2.845 |
| rs16942421 | G>A | <0.0001 | G | 1.131 | 1.062-1.204 |
| B, Comparison
between control group and SCD group |
| SNP | Allele | P-value | Risk allele | Odds ratio | 95% CI |
| rs2389202 | A>T | 0.018 | A | 1.540 | 1.061-2.233 |
| rs12429889 | T>C | 0.059 | C | 1.440 | 0.977-2.121 |
| rs16866933 | G>A | 0.883 | G | 1.012 | 0.861-1.190 |
| rs10183640 | G>A | 0.114 | G | 0.881 | 0.756-1.028 |
| rs11187837 | A>G | 0.001 | A | 1.227 | 1.074-1.403 |
| rs597503 | G>A | 0.044 | A | 1.071 | 0.996-1.152 |
| rs12155623 | A>T | 0.515 | A | 0.942 | 0.789-1.125 |
| rs2982694 | G>T | <0.0001 | T | 0.249 | 0.136-0.459 |
| rs7307780 | C>T | 0.045 | T | 2.356 | 0.979-5.666 |
| rs9581094 | T>C | 0.233 | T | 1.010 | 0.991-1.028 |
| rs4621553 | G>A | 0.049 | G | 0.594 | 0.351-1.003 |
| rs10829156 | G>A | 0.392 | G | 1.034 | 0.956-1.119 |
| rs2281680 | G>A | 0.774 | G | 1.014 | 0.922-1.115 |
| rs2251393 | G>A | 0.433 | G | 1.413 | 0.591-3.382 |
| rs16942421 | G>A | 0.003 | G | 1.074 | 1.016-1.136 |
| C, Comparison
between CHD group and SCD group |
| SNP | Allele | P-value | Risk allele | Odds ratio | 95% CI |
| rs2389202 | A>T | 0.437 | A | 1.193 | 0.765-1.860 |
| rs12429889 | T>C | 0.001 | C | 2.547 | 1.406-4.614 |
| rs16866933 | G>A | 0.024 | G | 1.253 | 1.032-1.521 |
| rs10183640 | G>A | 0.001 | G | 1.372 | 1.144-1.645 |
| rs11187837 | A>G | 0.400 | A | 0.938 | 0.807-1.091 |
| rs597503 | G>A | 0.550 | A | 1.028 | 0.939-1.126 |
| rs12155623 | A>T | 0.052 | A | 1.219 | 1.001-1.485 |
| rs2982694 | G>T | <0.0001 | T | 7.484 | 3.262-17.171 |
| rs7307780 | C>T | 0.292 | T | 0.610 | 0.240-1.551 |
| rs9581094 | T>C | 0.186 | T | 1.027 | 0.990-1.066 |
| rs4621553 | G>A | <0.0001 | G | 4.717 | 2.121-10.490 |
| rs10829156 | G>A | <0.0001 | G | 1.494 | 1.286-1.735 |
| rs2281680 | G>A | 0.433 | G | 0.964 | 0.879-1.058 |
| rs2251393 | G>A | 0.869 | G | 0.925 | 0.364-2.349 |
| rs16942421 | G>A | 0.237 | G | 1.052 | 0.969-1.142 |
Prediction model analysis via the
χ2 test and binary logistic regression
The statistically significant SNPs were sequentially
added into the prediction model to evaluate the accuracy by the AUC
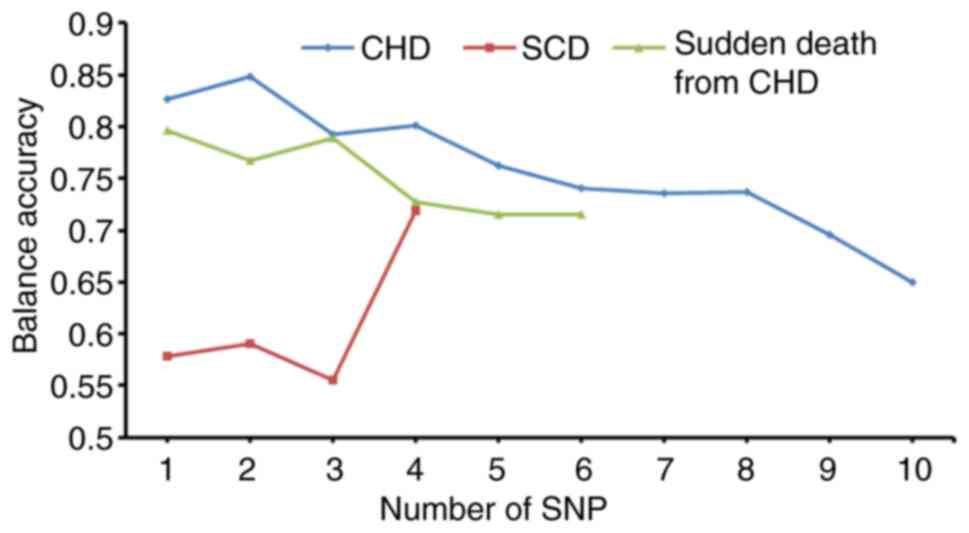
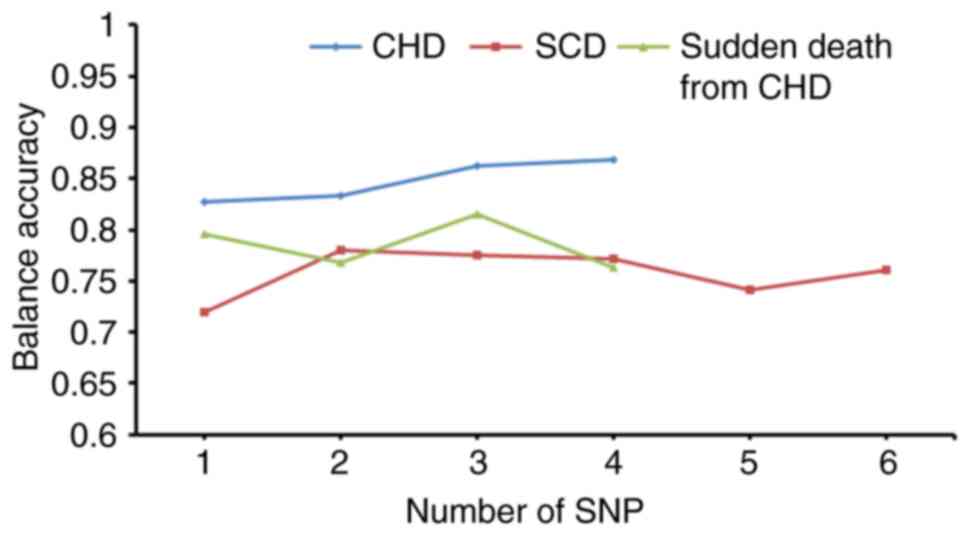
and MDR. The results demonstrated that, as the number of predictive
model SNPs increased, the testing accuracy of models exhibited a
downward trend by MDR (Fig. 3);
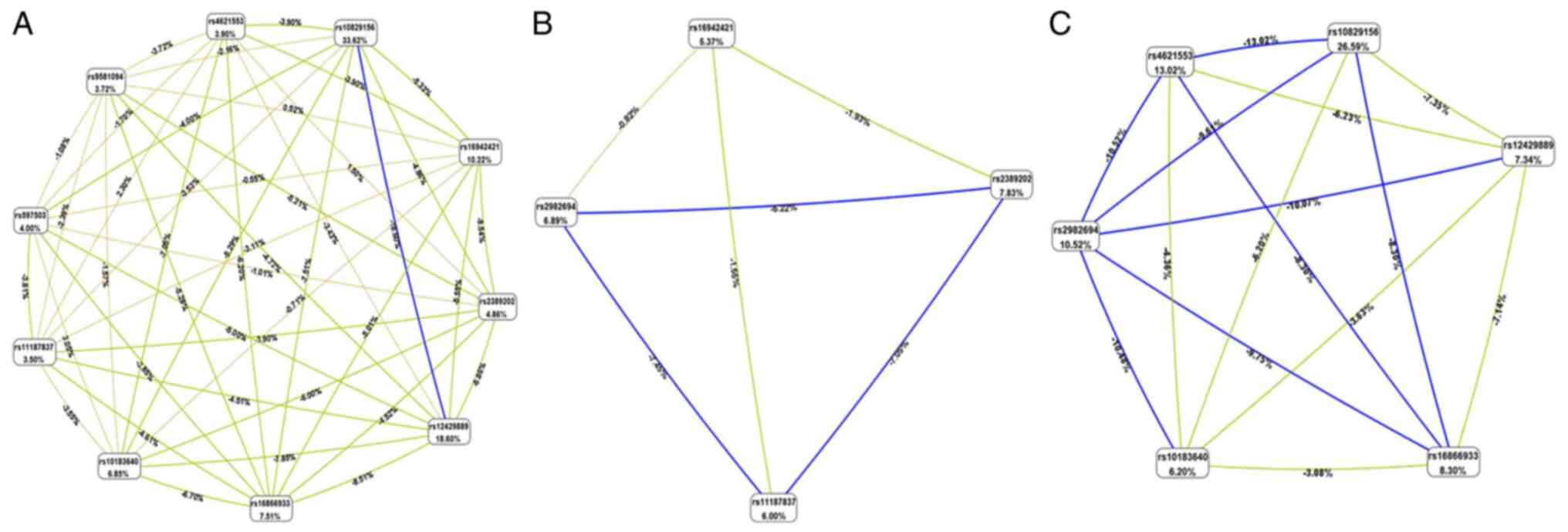
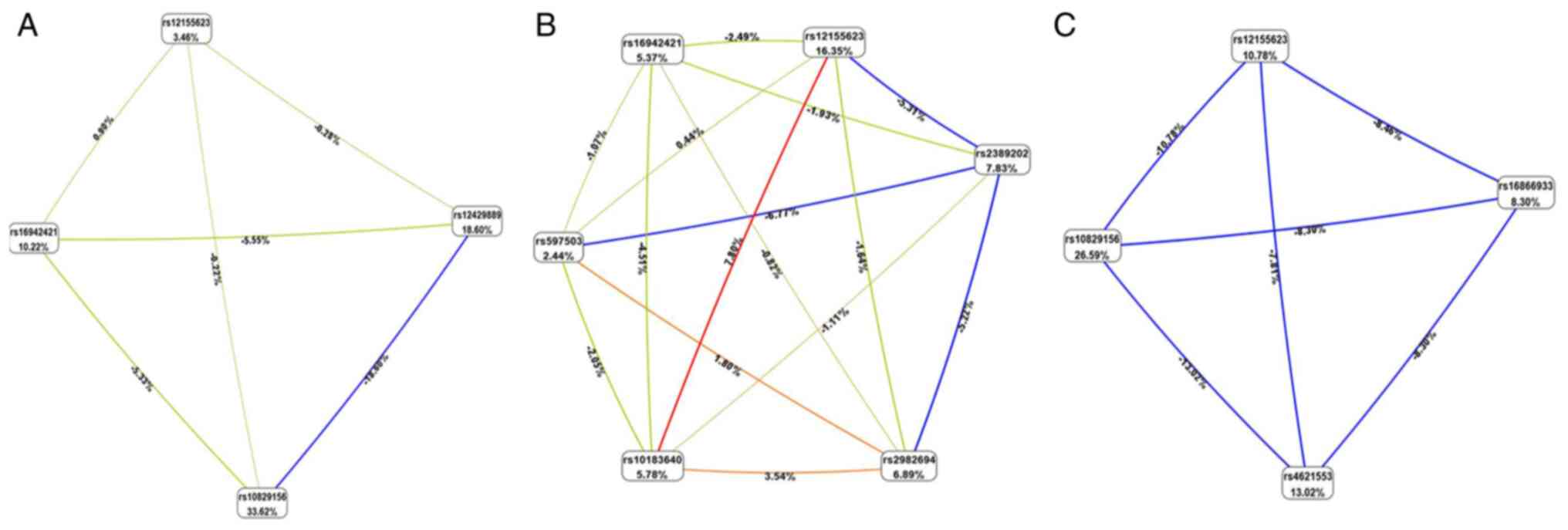
however, the AUC exhibited an upward trend (Fig. 4). MDR data are presented in
Fig. 5 and Table SIII. According to entropy-based
analysis, this interaction is redundant (redundant information from
both factors). Rs10829156 and rs12429889 treated independently
explains 33.62 and 18.60% of entropy (it removes 33.62 and 18.60%
of ‘uncertainty’ in CHD determination), respectively. The entropy
of interaction between these two SNPs was -18.60%, suggesting that
this part of the variation in the determination of CHD is common
for both SNPs (Fig. 5A).
Similarly, rs2982694 and rs11187837 treated independently explains
6.89 and 6.00% of entropy (it removes 6.89 and 6.00% of
‘uncertainty’ in the SCD determination), respectively. The entropy
of interaction between these two SNPs was -7.45%, suggesting that
this part of the variation in the determination of SCD is common
for both SNPs (Fig. 5B). The AUC
values of the prediction models for CHD, SCD and sudden death from
CHD are presented in Table SIV.
Taken together, these results suggested that feasibly incorporated
statistically significant SNPs in the prediction models may be used
to predict the occurrence of CHD or SCD.
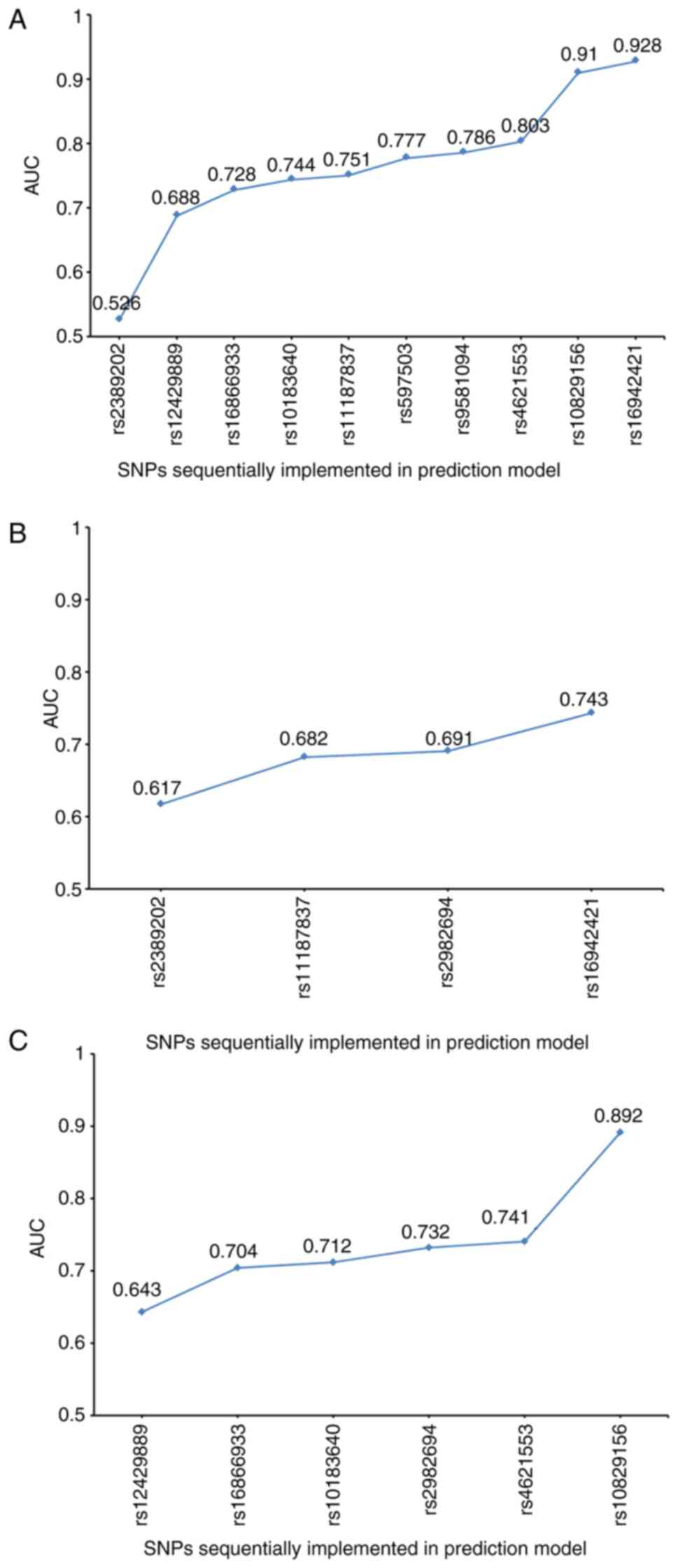
To identify a better prediction model, the role of
the 15 polymorphic SNPs in CHD or SCD was assessed via binary
logistic regression. The predicted probabilities were compared with
the observed disease status and the AUC was derived as an overall
measurement of prediction accuracy. These SNPs were added to the
prediction model and interactions were analyzed by MDR. The results
demonstrated that as the number of SNPs increased, the testing
accuracy of models exhibited a steady or slight upward trend
(Fig. 6), which achieved an
improved prediction effect. MDR data are presented in Fig. 7 and Table V. The AUC values of these
prediction models determined via binary logistic regression were
>90% (Fig. 8). The SNPs of each
prediction model are listed in Table
VI. Taken together, these results suggested that the high
efficiency of these models may provide prediction results for
certain individuals.
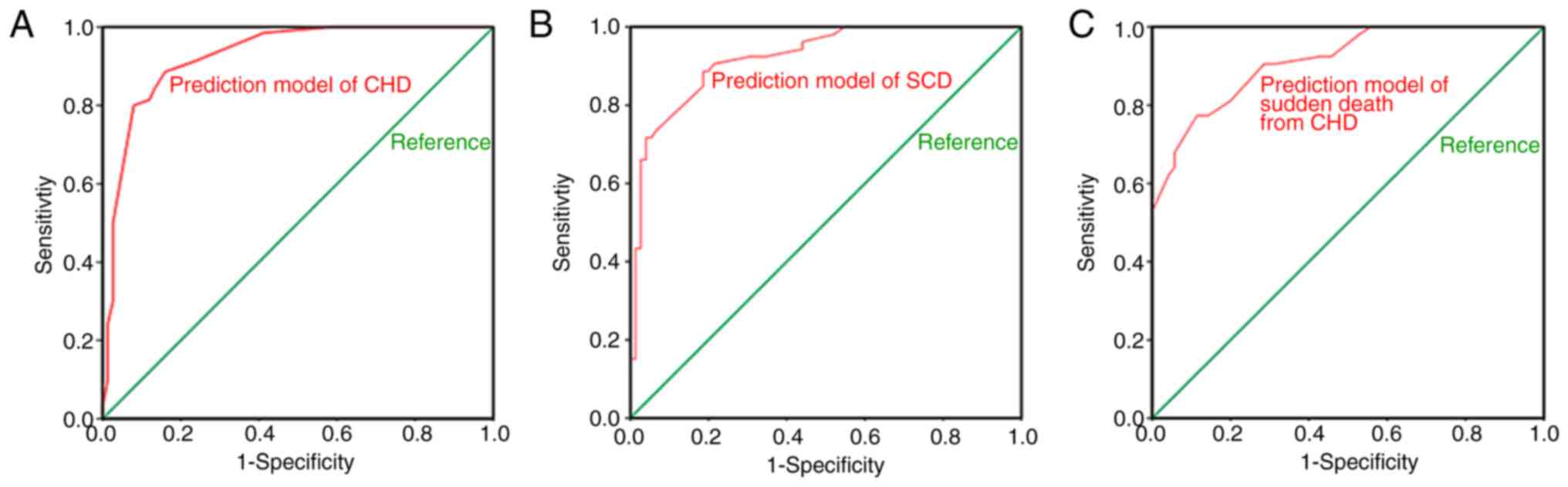 | Figure 8AUC values of the prediction models
that added SNPs via binary logistic regression. (A) Prediction
model of CHD. (B) Prediction model of SCD. (C) Prediction model of
sudden death from CHD. The SNPs of each prediction model and their
AUC values were as follows: Four SNPs (rs12429889, rs10829156,
rs16942421 and rs12155623) that predict CHD, 0.928; six SNPs
(rs2389202, rs2982694, rs10183640, rs597503, rs16942421 and
rs12155623) that predict SCD, 0.922; and four SNPs (rs16866933,
rs4621553, rs10829156 and rs12155623) that predict sudden death
from CHD, 0.912. SNP, single nucleotide polymorphism; CHD, coronary
heart disease; SCD, sudden coronary death; MDR, multifactor
dimensionality reduction; AUC, area under the receiver operating
characteristic curve. |
 | Table VMultifactor dimensionality reduction
results of the prediction models which added SNPs obtained by
binary logistic regression. |
Table V
Multifactor dimensionality reduction
results of the prediction models which added SNPs obtained by
binary logistic regression.
| A, Prediction model
of CHD |
|---|
| Step | SNPs | Balance accuracy
training | Balance accuracy
testing | CV consistency | χ2
(P-value) |
|---|
| 1 | rs10829156 | 0.8267 | 0.8267 | 10/10 | 62.1767
(<0.0001) |
| 2 | rs10829156,
rs16942421 | 0.8482 | 0.8338 | 8/10 | 70.2788
(<0.0001) |
| 3 | rs10829156,
rs16942421, rs12155623 | 0.8629 | 0.8629 | 10/10 | 76.3077
(<0.0001) |
| 4 | rs12429889,
rs10829156, rs16942421, rs12155623 | 0.8757 | 0.869 | 10/10 | 81.8735
(<0.0001) |
| B, Prediction model
of SCD |
| Step | SNPs | Balance accuracy
training | Balance accuracy
testing | CV consistency | χ2
(P-value) |
| 1 | rs12155623 | 0.7201 | 0.7201 | 10/10 | 24.0998
(<0.0001) |
| 2 | rs10183640,
rs12155623 | 0.8066 | 0.7796 | 10/10 | 50.2438
(<0.0001) |
| 3 | rs10183640,
rs597503, rs12155623 | 0.8392 | 0.7757 | 8/10 | 58.5882
(<0.0001) |
| 4 | rs2982694,
rs10183640, rs597503, rs12155623 | 0.8634 | 0.7718 | 7/10 | 64.916
(<0.0001) |
| 5 | rs2389202,
rs10183640, rs597503, rs16942421, rs12155623 | 0.8851 | 0.7413 | 6/10 | 73.5285
(<0.0001) |
| 6 | rs2389202,
rs2982694, rs10183640, rs597503, rs16942421, rs12155623 | 0.899 | 0.7601 | 10/10 | 83.8712
(<0.0001) |
| C, Prediction model
of sudden death from CHD |
| Step | SNPs | Balance accuracy
training | Balance accuracy
testing | CV consistency | χ2
(P-value) |
| 1 | rs10829156 | 0.7962 | 0.7962 | 10/10 | 42.6898
(<0.0001) |
| 2 | rs16866933,
rs10829156 | 0.7975 | 0.7677 | 8/10 | 42.6898
(<0.0001) |
| 3 | rs16866933,
rs4621553, rs10829156 | 0.8245 | 0.8151 | 9/10 | 50.9111
(<0.0001) |
| 4 | rs16866933,
rs4621553, rs10829156, rs12155623 | 0.8364 | 0.7628 | 10/10 | 53.857
(<0.0001) |
 | Table VISNPs included in each prediction
model. |
Table VI
SNPs included in each prediction
model.
| | Prediction model
obtained by χ2 test | Prediction model
obtained by binary logistic regression |
|---|
| Predicted
disease | SNP | AUC | MDR | SNP | AUC | MDR |
|---|
| CHD | rs2389202,
rs12429889, rs16866933, rs10183640, rs11187837, rs597503,
rs9581094, rs4621553, rs10829156, rs16942421 | 0.928 | 0.65 | rs12429889,
rs10829156, rs16942421, rs12155623 | 0.928 | 0.869 |
| SCD | rs2389202,
rs11187837, rs2982694, rs16942421 | 0.743 | 0.7191 | rs2389202,
rs2982694, rs10183640, rs597503, rs16942421, rs12155623 | 0.922 | 0.7601 |
| Sudden death from
CHD | rs12429889,
rs16866933, rs10183640, rs2982694, rs4621553, rs10829156 | 0.892 | 0.7156 | rs16866933,
rs4621553, rs10829156, rs12155623 | 0.912 | 0.7628 |
Discussion
Despite extensive research on the etiology,
pathogenesis and risk factors of SCD in recent years, strategies
for its prevention and treatment remain insufficient. In addition,
the interpretation of GWAS remains difficult. A key limitation of
GWAS is that it only depicts associations, not causation.
Furthermore, their clinical utility remains to be evaluated in
prospective studies together with established risk factors in the
present study. In the present study, a mini-sequencing detection
system was established containing 21 putative SNPs that have been
reported to be associated with sudden cardiac arrest via GWAS. The
association of these SNPs with the risk of CHD or SCD in the
Chinese Han population was assessed. The results confirmed that 15
SNPs were polymorphic in the Chinese Han population. Different
prediction models were constructed for these 15 SNPs to evaluate
the risk of CHD, SCD and sudden death from CHD. These models may
provide a prediction for a significant proportion of individuals
and may be useful to provide a novel detection and evaluation
method for the prevention and treatment of CHD and SCD in the
future.
The genetic architecture of human complex traits
substantially differs. Given that the majority of GWAS were
performed in European-descent individuals, SNPs from these GWAS may
not be transferrable to individuals from other populations,
highlighting the importance of diversification of GWAS into
non-European populations. No polymorphism was detected in six SNPs
in the Chinese Han population, including rs11624056, rs4665058,
rs17718586, rs17291650, rs12189362 and rs1559040. In addition, 10
SNPs were associated with CHD, four SNPs were associated with SCD
and six SNPs were associated with sudden death from CHD. This
confirmed that gene variations exert different effects in different
populations. Furthermore, whether these results represent an
association of genetic variation with the phenotype CHD or SCD
requires further investigation.
In the present study, the prediction model obtained
via the χ2 test was required to be combined with
additional SNPs to achieve the same prediction probability as the
prediction model obtained via binary logistic regression.
Furthermore, the prediction model obtained via the χ2
test was less capable of predicting SCD compared with the
prediction model obtained via binary logistic regression. Thus, the
prediction model obtained via binary logistic regression is more
effective to assess and predict the risk of CHD, SCD or sudden
death from CHD.
Previous studies have demonstrated that differences
in gene expression result in different physical and pathological
traits (31). The rs11187837 locus
is situated in the intron region of the phospholipase C ε 1 (PLCE1)
gene (GRCh38.p12). A previous study reported that differentially
expressed genes of PLCE1 may have a critical role in atrial myocyte
hypertrophy (32). The rs10183640
variation is located at the junction of activin A receptor type 1
(ACVR1) and the uridine phosphorylase 2 gene (GRCh38.p12). Previous
studies have demonstrated that the ACVR1 gene was closely
associated with cardiac fibroblasts, aortic valve development and
endocardia cushion formation (33-35).
Differentially expressed genes of ACVR1 may be predictors for
decreased left ventricular ejection fraction and help diagnose
congenital heart defects (36-38).
The rs4621553 locus is located at the junction of the YTH domain
containing 2 and potassium calcium-activated channel subfamily N
member 2 (KCNN2) genes (GRCh38.p12). Previous studies reported that
KCNN2 gene variation may have an important role in the development
of coronary artery aneurysms and may act as adjunctive markers for
risk stratification in patients susceptible to SCD (39,40).
The rs12429889 locus is located at the junction of the Kruppel-like
factor 12 (KLF12) and RNY1 pseudogene 5 genes (GRCh38.p12). GWAS
reported that KLF12 genes are associated with an increased risk of
ventricular arrhythmia, syncope and SCD (41,42).
The rs597503 locus is located at the junction of the SCML2
pseudogene 1 and laminin subunit α 1 genes (GRCh38.p12). A study on
1,414 Hispanics demonstrated that LAMA1 gene variants are strongly
associated with cardio-metabolic traits (43). In addition, Aouizerat et al
(14) confirmed that rs2389202,
rs16942421, rs16866933, rs9581094 and rs10829156 are risk factors
of CAD. Of note, rs2982694 was reported to be associated with SCD
rather than CAD in the present study. Several genetic association
studies of estrogen receptor 1 (ESR1) gene variants concerning CAD
have been published (44,45). It has been suggested that ESR1 is a
potential candidate gene during acute coronary events, such as
acute thrombotic cardiovascular diseases and atherosclerosis to
plaque rupture (46-48).
In addition, the ESR1 gene regulates the expression of multiple
genes following activation by estrogen in cardiovascular disease
(49-51).
Previous studies have demonstrated that ESR1 polymorphism was
associated with atherosclerosis in coronary arteries, the presence
of coronary thrombosis and myocardial infarction, and was
associated with an elevated risk of CHD in males (46,52,53).
Whether rs2982694 is associated with CAD or other cardiac diseases
requires further investigation.
CHD and SCD are associated with genetic
complexities. The results of the present study indicated that the
four SNPs, rs12429889, rs10829156, rs16942421 and rs12155623, are
able to predict CHD, the six SNPs, rs2389202, rs2982694,
rs10183640, rs597503, rs16942421 and rs12155623, are able to
predict SCD and the four SNPs, rs16866933, rs4621553, rs10829156
and rs12155623, are able to predict sudden death from CHD. In the
present study, the AUC values of these prediction models were
>90% (P<0.0001; Table VI).
Taken together, the results of the present study have achieved
considerable progress in assessing and predicting CHD, SCD and
sudden death from CHD. However, improving the prediction accuracy
for CHD or SCD will need to rely on the identification of more
associated DNA variants and additional sample sizes, which will be
a continuous and accumulative effort, but is certainly achievable
in the future.
These SNPs have so far not been used in clinical and
forensic prediction and diagnosis of CHD, SCD or sudden death from
CHD. The 15 polymorphic SNPs were confirmed in the Chinese Han
population (n=198), which may help further understand the
pathogenesis of SCD. The purpose of establishing this detection
system and prediction models was to achieve a forensic assistant
identification and genetic diagnosis of SCD, and to provide a basis
for gene therapy. The screening of susceptible genes of CHD or SCD
in relatives may provide guidance for their lifestyle and
medication, as well as a basis for precision medicine.
A key limitation of the present study is that the
CHD and SCD samples were collected from different individuals. In
addition, the prediction model is only based on 198 samples from
the North Chinese Han population and certain patients with CHD and
those who passed away from SCD cannot be traced. Furthermore, it is
difficult to predict whether patients with CHD may experience
sudden death in the future.
The present study focused on SNPs associated with
SCD and the SCD samples collected had an uneven sex ratio (4:1).
Thus, CHD and normal control samples of patients whose clinical
characteristics [with an uneven sex ratio (4:1)] were consistent
with those of the SCD group were collected to control for the
variables. No statistically significant differences were observed
in terms of age or sex distribution and the presence of any
underlying diseases among the three groups, suggesting that these
variables were evenly distributed among the groups. The purpose of
this process is to minimize the interference of other factors with
the prediction model to improve the accuracy of prediction.
In the present study, the SNaPshot assay was
performed using multiplex PCR, SBE and CE techniques. The
mini-sequencing technology uses a dideoxynucleotide termination
method to ligate a fluorescent ddNTP to the 3'-end of an SBE primer
to obtain a fluorescent deoxynucleotide sequence. This technique is
widely adopted in DNA laboratories due to its high accuracy and
sensitivity. There are several technical methods for screening
SNPs, such as Sanger sequencing of exons, WGS and WES based on
next-generation sequencing (NGS) techniques. Sanger sequencing and
mini-sequencing methods are considered more accurate compared with
NGS. NGS is expensive and time-consuming and is not applicable to
degraded DNA. In clinical and forensic practice, delay-examination
and environmental exposure of formaldehyde-fixed samples frequently
lead to degradation of tissues, which results in decreased DNA
quality for WGS or WES detection (54). In the present experiment, tissues
from the SCD group and subjects with death for other causes were
degraded due to formaldehyde fixation (fixation period varies from
1 month to 3 years). Not every DNA laboratory is equipped with an
NGS instrument to detect this prevailing disease. Using the method
of the present study is more feasible to detect degraded DNA
samples and more compatible with regular DNA laboratories to meet
the increasing requirement of detecting this global disease.
In the present study, 15 polymorphic SNPs associated
with CHD or SCD were identified and their predictive value was
determined from 198 samples from the Chinese Han population.
Prediction accuracies were expressed as AUC values >0.9.
Although these prediction models have not been introduced in
clinical practice, the preliminary genetic model may assist
decision-making on CHD, SCD or sudden death from CHD for
preventative actions and forensic investigations of the cause of
death. Furthermore, the results of the present study suggest that
with more genome-wide associated SNPs identified in the future and
included in the prediction model together with the SNPs presented
here, CHD, SCD or sudden death from CHD will become predictable
from DNA, with high accuracy to allow routine practical
applications, such as in medicine and forensics.
In conclusion, the present study established a
mini-sequencing detection system containing 21 putative SNPs that
have been reported to be associated with sudden cardiac arrest. The
results of the χ2 test demonstrated significant
associations for 10, 4 and 6 SNPs, in CHD, SCD and sudden death
from CHD, respectively. Furthermore, prediction models were
established to assess and predict the risk of CHD, SCD or sudden
death from CHD. The combination of SNP-associated loci in each
group enables the development of a test model with a good
predictive performance. Taken together, these results confirm the
influence of genetic variation on the risk of SCD in patients with
CHD.
Supplementary Material
Results of the system sensitivity.
Capillary electrophoresis typing with (A) 10, (B) 1, (C) 0.1 and
(D) 0.01 ng of input DNA template.
Results of Sanger sequencing using
random samples. These results were consistent with those of the
SNaPshot method.
Clinical characteristics of the 198
samples in this study.
Typing results of 15 SNPs and their
percentages in each group.
Multifactor dimensionality reduction
results of the prediction models that added SNPs identified by the
χ2 test with 10, 4 and 6 SNPs, respectively.
Results of area under the curve of the
prediction models that added SNPs identified by the χ2
test with 10, 4 and 6 SNPs, respectively.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Basic Public
Welfare Planning Project of Zhejiang Province, China (grant no.
LGD19C040001), the Sci-Tech Planning Project of Jiaxing, China
(grant no. 2020AY30004), the Natural Science Foundation of China
(grant no. 30900593), the Shanxi Scholarship Council of China
(grant no. 2016-055) and the Key Research and Development Projects
of Shanxi Province (grant no. 201803D31069).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GZ and DC conceived and designed the study. GZ and
NZ administratively supported the present study. DC and XC
performed the majority of the experiments and drafted the
manuscript. XL, JW, JDL, JS, JL, BH and DC provided, selected,
assembled, analyzed and interpreted the data. GZ and DC confirm the
authenticity of all the raw data. All authors contributed toward
data analysis, drafting and critically revising the manuscript, and
agree to be accountable for all aspects of the work. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shanxi Medical University (Jinzhong, China; approval
no. ZX201601) and written informed consent was provided by all
participants or their family members prior to the start of the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zegard A, Okafor O, de Bono J, Kalla M,
Lencioni M, Marshall H, Hudsmith L, Qiu T, Steeds R, Stegemann B
and Leyva F: Myocardial fibrosis as a predictor of sudden death in
patients with coronary artery disease. J Am Coll Cardiol. 77:29–41.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pannone L, Falasconi G, Cianfanelli L,
Baldetti L, Moroni F, Spoladore R and Vergara P: Sudden cardiac
death in patients with heart disease and preserved systolic
function: Current options for risk stratification. J Clin Med.
10(1823)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Michaud K, Magnin V, Faouzi M, Fracasso T,
Aguiar D, Dedouit F and Grabherr S: Postmortem coronary artery
calcium score in cases of myocardial infarction. Int J Legal Med:
Apr 13, 2021. doi: 10.1007/s00414-021-02586-z.
|
|
4
|
Lynge TH, Risgaard B, Banner J, Nielsen
JL, Jespersen T, Stampe NK, Albert CM, Winkel BG and Tfelt-Hansen
J: Nationwide burden of sudden cardiac death: A study of 54,028
deaths in Denmark. Heart Rhythm: May 7, 2021. doi:
10.1016/j.hrthm.2021.05.005.
|
|
5
|
Silverman MG, Yeri A, Moorthy MV, Camacho
Garcia F, Chatterjee NA, Glinge CSA, Tfelt-Hansen J, Salvador AM,
Pico AR, Shah R, et al: Circulating miRNAs and risk of sudden death
in patients with coronary heart disease. JACC Clin Electrophysiol.
6:70–79. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Goldberger JJ, Basu A, Boineau R, Buxton
AE, Cain ME, Canty JM Jr, Chen PS, Chugh SS, Costantini O, Exner
DV, et al: Risk stratification for sudden cardiac death: A plan for
the future. Circulation. 129:516–526. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Khera AV, Mason-Suares H, Brockman D, Wang
M, VanDenburgh MJ, Senol-Cosar O, Patterson C, Newton-Cheh C,
Zekavat SM, Pester J, et al: Rare genetic variants associated with
sudden cardiac death in adults. J Am Coll Cardiol. 74:2623–2634.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Stattin EL, Westin IM, Cederquist K,
Jonasson J, Jonsson BA, Mörner S, Norberg A, Krantz P and Wisten A:
Genetic screening in sudden cardiac death in the young can save
future lives. Int J Legal Med. 130:59–66. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tsuda T, Fitzgerald KK and Temple J:
Sudden cardiac death in children and young adults without
structural heart disease: A comprehensive review. Rev Cardiovasc
Med. 21:205–216. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lacaze P, Sebra R, Riaz M, Ingles J,
Tiller J, Thompson BA, James PA, Fatkin D, Semsarian C, Reid CM, et
al: Genetic variants associated with inherited cardiovascular
disorders among 13,131 asymptomatic older adults of European
descent. NPJ Genom Med. 6(51)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Johannsen EB, Baughn LB, Sharma N, Zjacic
N, Pirooznia M and Elhaik E: The genetics of sudden infant death
syndrome-towards a gene reference resource. Genes (Basel).
12(216)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Valdisser PAMR, Müller BSF, de Almeida
Filho JE, Morais Júnior OP, Guimarães CM, Borba TCO, de Souza IP,
Zucchi MI, Neves LG, Coelho ASG, et al: Genome-wide association
studies detect multiple QTLs for productivity in mesoamerican
diversity panel of common bean under drought stress. Front Plant
Sci. 11(574674)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yayla C, Yayla KG and Demirtaş K:
Genome-wide association studies in patients with coronary artery
disease. Angiology: June 3, 2021. doi:
10.1177/00033197211022076.
|
|
14
|
Aouizerat BE, Vittinghoff E, Musone SL,
Pawlikowska L, Kwok PY, Olgin JE and Tseng ZH: GWAS for discovery
and replication of genetic loci associated with sudden cardiac
arrest in patients with coronary artery disease. BMC Cardiovasc
Disord. 11(29)2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ashar FN, Mitchell RN, Albert CM,
Newton-Cheh C, Brody JA, Müller-Nurasyid M, Moes A, Meitinger T,
Mak A, Huikuri H, et al: A comprehensive evaluation of the genetic
architecture of sudden cardiac arrest. Eur Heart J. 39:3961–3969.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Andersen JD, Jacobsen SB, Trudso LC,
Kampmann ML, Banner J and Morling N: Whole genome and transcriptome
sequencing of post-mortem cardiac tissues from sudden cardiac death
victims identifies a gene regulatory variant in NEXN. Int J Legal
Med. 133:1699–1709. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bastos da Silva I, Batista TP, Martines
RB, Kanamura CT, Ferreira IM, Vidal JE and Pereira-Chioccola VL:
Genotyping of Toxoplasma gondii: DNA extraction from formalin-fixed
paraffin-embedded autopsy tissues from AIDS patients who died by
severe disseminated toxoplasmosis. Exp Parasitol. 165:16–21.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tam V, Patel N, Turcotte M, Bosse Y, Pare
G and Meyre D: Benefits and limitations of genome-wide association
studies. Nat Rev Genet. 20:467–484. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
De R, Bush WS and Moore JH: Bioinformatics
challenges in genome-wide association studies (GWAS). Method Mol
Biol. 1168:63–81. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hehir-Kwa JY, Pfundt R and Veltman JA:
Exome sequencing and whole genome sequencing for the detection of
copy number variation. Expert Rev Mol Diagn. 15:1023–1032.
2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Meienberg J, Bruggmann R, Oexle K and
Matyas G: Clinical sequencing: Is WGS the better WES? Hum Genet.
135:359–362. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Emdin CA, Haas M, Ajmera V, Simon TG,
Homburger J, Neben C, Jiang L, Wei WQ, Feng Q, Zhou A, et al:
Association of genetic variation with cirrhosis: A multi-trait
genome-wide association and gene-environment interaction study.
Gastroenterology. 160:1620–1633.e13. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Levin MG, Klarin D, Walker VM, Gill D,
Lynch J, Hellwege JN, Keaton JM, Lee KM, Assimes TL, Natarajan P,
et al: Association between genetic variation in blood pressure and
increased lifetime risk of peripheral artery disease. Arterioscler
Thromb Vasc Biol. 41:2027–2034. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Nasu T, Satoh M, Hachiya T, Sutoh Y,
Ohmomo H, Hitomi S, Taguchi S, Kikuchi H, Kobayashi T, Takahashi Y,
et al: A genome-wide association study for highly sensitive cardiac
troponin T levels identified a novel genetic variation near a
RBAK-ZNF890P locus in the Japanese general population. Int J
Cardiol. 329:186–191. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zinelabidine LH, Torres-Pérez R, Grimplet
J, Baroja E, Ibáñez S, Carbonell-Bejerano P, Martínez-Zapater JM,
Ibáñez J and Tello J: Genetic variation and association analyses
identify genes linked to fruit set-related traits in grapevine.
Plant Sci. 306(110875)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu J, Li W, Wang J, Chen D, Liu Z, Shi J,
Cheng F, Li Z, Ren J, Zhang G and Yun K: A new set of DIP-SNP
markers for detection of unbalanced and degraded DNA mixtures.
Electrophoresis. 40:1795–1804. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hu B, Wu T, Zhao Y, Xu G, Shen R and Chen
G: Physiological signatures of dual embryonic origins in mouse
skull vault. Cell Physiol Biochem. 43:2525–2534. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Dear JD, Sykes JE and Bannasch DL: Quality
of DNA extracted from formalin-fixed, paraffin-embedded canine
tissues. J Vet Diagn Invest. 32:556–559. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Amemiya K, Hirotsu Y, Oyama T and Omata M:
Simple and rapid method to obtain high-quality tumor DNA from
clinical-pathological specimens using touch imprint cytology. J Vis
Expe. 133(56943)2018.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Shi M, Bai R, Yu X, Lv J and Hu B:
Haplotype diversity of 22 Y-chromosomal STRs in a southeast China
population sample (Chaoshan area). Forensic Sci Int Genet.
3:e45–e47. 2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hindricks G, Potpara T, Dagres N, Arbelo
E, Bax JJ, Blomström-Lundqvist C, Boriani G, Castella M, Dan GA,
Dilaveris PE, et al: 2020 ESC Guidelines for the diagnosis and
management of atrial fibrillation developed in collaboration with
the European Association for Cardio-Thoracic Surgery (EACTS): The
Task Force for the diagnosis and management of atrial fibrillation
of the European Society of Cardiology (ESC) Developed with the
special contribution of the European Heart Rhythm Association
(EHRA) of the ESC. Eur Heart J. 42:373–498. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chang TH, Chen MC, Chang JP, Huang HD, Ho
WC, Lin YS, Pan KL, Huang YK, Liu WH and Wu CC: Exploring
regulatory mechanisms of atrial myocyte hypertrophy of mitral
regurgitation through gene expression profiling analysis: Role of
NFAT in cardiac hypertrophy. PLoS One. 11(e0166791)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hu J, Wang X, Wei SM, Tang YH, Zhou Q and
Huang CX: Activin A stimulates the proliferation and
differentiation of cardiac fibroblasts via the ERK1/2 and p38-MAPK
pathways. Eur J Pharmacol. 789:319–327. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Thomas PS, Sridurongrit S, Ruiz-Lozano P
and Kaartinen V: Deficient signaling via Alk2 (Acvr1) leads to
bicuspid aortic valve development. PLoS One.
7(e35539)2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Thomas PS, Rajderkar S, Lane J, Mishina Y
and Kaartinen V: AcvR1-mediated BMP signaling in second heart field
is required for arterial pole development: Implications for
myocardial differentiation and regional identity. Dev Biol.
390:191–207. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Gorący I, Safranow K, Dawid G,
Skonieczna-Żydecka K, Kaczmarczyk M, Gorący J, Loniewska B and
Ciechanowicz A: Common genetic variants of the BMP4, BMPR1A,
BMPR1B, and ACVR1 genes, left ventricular mass, and other
parameters of the heart in newborns. Genet Test Mol Biomarkers.
16:1309–1316. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Smith KA, Joziasse IC, Chocron S, van
Dinther M, Guryev V, Verhoeven MC, Rehmann H, van der Smagt JJ,
Doevendans PA, Cuppen E, et al: Dominant-negative ALK2 allele
associates with congenital heart defects. Circulation.
119:3062–3069. 2009.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Joziasse IC, Smith KA, Chocron S, van
Dinther M, Guryev V, van de Smagt JJ, Cuppen E, Ten Dijke P, Mulder
BJ, Maslen CL, et al: ALK2 mutation in a patient with Down's
syndrome and a congenital heart defect. Eur J Hum Genet.
19:389–393. 2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kim JJ, Park YM, Yoon D, Lee KY, Seob Song
M, Doo Lee H, Kim KJ, Park IS, Nam HK, Weon Yun S, et al:
Identification of KCNN2 as a susceptibility locus for coronary
artery aneurysms in Kawasaki disease using genome-wide association
analysis. J Hum Genet. 58:521–525. 2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yu CC, Chia-Ti T, Chen PL, Wu CK, Chiu FC,
Chiang FT, Chen PS, Chen CL, Lin LY, Juang JM, et al: KCNN2
polymorphisms and cardiac tachyarrhythmias. Medicine (Baltimore).
95(e4312)2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Lahrouchi N, Tadros R, Crotti L, Mizusawa
Y, Postema PG, Beekman L, Walsh R, Hasegawa K, Barc J, Ernsting M,
et al: Transethnic genome-wide association study provides insights
in the genetic architecture and heritability of long QT syndrome.
Circulation. 142:324–338. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Naik A: Long QT syndrome revisited. J
Assoc Physicians India. 55 (Suppl):S58–S61. 2007.PubMed/NCBI
|
|
43
|
Hellwege JN, Palmer ND, Dimitrov L, Keaton
JM, Tabb KL, Sajuthi S, Taylor KD, Ng MC, Speliotes EK, Hawkins GA,
et al: Genome-wide linkage and association analysis of
cardiometabolic phenotypes in Hispanic Americans. J Hum Genet.
62:175–184. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Maruyama H, Toji H, Harrington CR, Sasaki
K, Izumi Y, Ohnuma T, Arai H, Yasuda M, Tanaka C, Emson PC, et al:
Lack of an association of estrogen receptor alpha gene
polymorphisms and transcriptional activity with Alzheimer disease.
Arch Neurol. 57:236–240. 2000.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Matsubara Y, Murata M, Kawano K, Zama T,
Aoki N, Yoshino H, Watanabe G, Ishikawa K and Ikeda Y: Genotype
distribution of estrogen receptor polymorphisms in men and
postmenopausal women from healthy and coronary populations and its
relation to serum lipid levels. Arterioscler Thromb Vasc Biol.
17:3006–3012. 1997.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lehtimäki T, Kunnas TA, Mattila KM, Perola
M, Penttilä A, Koivula T and Karhunen PJ: Coronary artery wall
atherosclerosis in relation to the estrogen receptor 1 gene
polymorphism: An autopsy study. J Mol Med (Berl). 80:176–180.
2002.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Shearman AM, Cupples LA, Demissie S, Peter
I, Schmid CH, Karas RH, Mendelsohn ME, Housman DE and Levy D:
Association between estrogen receptor alpha gene variation and
cardiovascular disease. JAMA. 290:2263–2270. 2003.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Herrington DM, Howard TD, Brosnihan KB,
McDonnell DP, Li X, Hawkins GA, Reboussin DM, Xu J, Zheng SL,
Meyers DA and Bleecker ER: Common estrogen receptor polymorphism
augments effects of hormone replacement therapy on E-selectin but
not C-reactive protein. Circulation. 105:1879–1882. 2002.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Henttonen AT, Kortelainen ML, Kunnas TA
and Nikkari ST: Estrogen receptor-1 genotype is related to coronary
intima thickness in young to middle-aged women. Scand J Clin Lab
Invest. 67:380–386. 2007.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yilmaz A, Menevse S, Erkan AF, Ergun MA,
Ilhan MN, Cengel A and Yalcin R: The relationship of the ESR1 gene
polymorphisms with the presence of coronary artery disease
determined by coronary angiography. Genet Test. 11:367–371.
2007.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Boroumand M, Ghaedi M, Mohammadtaghvaei N,
Pourgholi L, Anvari MS, Davoodi G, Amirzadegan A, Saadat S,
Sheikhfathollahi M and Goodarzynejad H: Association of estrogen
receptor alpha gene polymorphism with the presence of coronary
artery disease documented by coronary angiography. Clin Biochem.
42:835–839. 2009.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Kunnas T, Silander K, Karvanen J,
Valkeapaa M, Salomaa V and Nikkari S: ESR1 genetic variants,
haplotypes and the risk of coronary heart disease and ischemic
stroke in the Finnish population: A prospective follow-up study.
Atherosclerosis. 211:200–202. 2010.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Roszkowska-Gancarz M, Kurylowicz A,
Polosak J, Ambroziak M and Puzianowska-Kuznicka M: The -351A/G
polymorphism of ESR1 is associated with risk of myocardial
infarction but not with extreme longevity. Clin Chim Acta.
411:1883–1887. 2010.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Nouws S, Bogaerts B, Verhaegen B, Denayer
S, Van Braekel J, Winand R, Fu Q, Crombé F, Piérard D, Marchal K,
et al: Impact of DNA extraction on whole genome sequencing analysis
for characterization and relatedness of Shiga toxin-producing
Escherichia coli isolates. Sci Rep. 10(14649)2020.PubMed/NCBI View Article : Google Scholar
|