Introduction
Maxillary defect or deficiency are the most common
problems in dental implantation, especially in the treatment of
patients with extensive jawbone defects caused by trauma or tumor
resection (1). Due to the rapid
development of material science, bone tissue engineering has
attracted wide attention, and extensive research has been carried
out on the subject. Bone tissue engineering strikes a balance
between the advantages and disadvantages of autogenous bone and
allogeneic bone (2). Artificial
bone substitute materials are based on bone tissue engineering
using scaffolds, growth factors and stem cells; these have good
histocompatibility and reproducibility, and are expected to be good
substitutes for bone defect repair (3).
Hydroxyapatite (HA), as an inorganic mineral, is the
main component of natural bone inorganic salts, and an
indispensable inorganic component of the human skeleton (4). Research demonstrates that 50% of the
human skeleton is comprised of homogeneous inorganic HA (4). The physicochemical properties,
chemical composition and crystal structure of HA are very similar
to human bone and are highly safe for humans. The results of our
previous study demonstrated that porous HA with a grooved structure
(HAG) scaffolds enhanced osteogenesis in vivo and in
vitro (5).
Transcriptome sequencing is a technique for
determining the expression level of the transcriptome, and is
widely used in research (6). RNA
sequencing (RNA-seq) is a high-throughput sequencing assay that can
provide detailed information regarding the working mechanisms
underlying the target tissue or the molecular pathobiology of a
disease (7-11).
Thus, RNA-seq plays an important role in studying the mechanisms
underlying osteogenesis. High-throughput transcriptome profiling
was used to further understand the molecular mechanism underlying
lithium in regulating the osteogenic fate of human mesenchymal stem
cells (hMSCs), when stimulated with lithium for 7 days (12).
According to the International Society for Cellular
Therapy (ISCT) (13), mesenchymal
stem cells (MSCs) must satisfy three requirements: i) MSCs must be
plastic-adherent when maintained in standard culture conditions;
ii) MSCs must express CD105, CD73 and CD90, and lack the expression
of CD45, CD34, CD14 and human leukocyte antigen (HLA)-DR surface
molecules and either CD14 or CD11b, and either CD79a or CD19; and
iii) MSCs must differentiate to osteoblasts, adipocytes and
chondroblasts in vitro (13). The criteria do not support the
purification of homogenous MSC populations. The isolation of MSCs
according to ISCT criteria produces heterogeneous, non-clonal
cultures of stromal cells containing stem cells with different
multipotent properties, committed progenitors and differentiated
cells. Currently, MSCs can be isolated from multiple tissues, such
as placenta (14,15).
In the present study, the RNA-seq technique was used
to study the osteogenic mechanism underlying porous HA
scaffold-based delivery of human placenta-derived (hP)MSCs in the
dorsal muscles of dogs. Samples were taken for sequencing 4, 8 and
12 weeks after heterotopic implantation. The association between
the expression and time of different genes was identified by
analyzing differentially expressed genes, and the osteogenesis
mechanism underlying the porous HAG scaffold-based delivery of
hMSCs was investigated using results of transcriptome sequencing.
The novelty of the present study was demonstrated by the analysis
of the biological properties of the porous HAG scaffold-based
delivery of hMSCs via transcriptomics, which provided novel
applications of this cell type in osteogenesis research.
Materials and methods
Preparation of cell-adhered HAG
scaffolds for transplantation
Cell culture conditions were the same as in our
previous study (5): High glucose
DMEM (Thermo Fisher Scientific, Inc.) containing 10% FBS (Thermo
Fisher Scientific, Inc.), 20 mmol/l β-glycerophosphate, 50 g/ml
vitamin C and 10 mol/l dexamethasone at 37˚C in 5% CO2.
The immortalized hPMSCs used in this study originated from the
State Key Laboratory of Biological Therapy at Sichuan University
(provided by the National Experimental Cell Resource Sharing
Platform, http://www.cellresource.cn/). The
fifth generation of hPMSCs in the exponential growth period were
counted. According to the counting results, high glucose DMEM was
added until the cell density reached 2x105/ml. According
to our previous study (5), cells
were analyzed using a flow cytometer (FACSAria III; BD Biosciences)
with related antibodies (all dilutions, 1:100; all from Abcam) for
the following surface markers: HLA-DR (cat. no. ab92511), CD11b
(cat. no. ab133357), CD14 (cat. no. ab183322), CD19 (cat. no.
ab134114), CD34 (cat. no. ab81289), CD44 (cat. no. ab189524), CD45
(cat. no. ab40763), CD73 (cat. no. ab202122), CD79 (cat. no.
ab134147), CD90 (cat. no. ab23894) and CD105 (cat. no. ab231774)
(16,17). A total of 5x105 cells
were cultured in a flow tube at 37˚C, after which cells were
re-suspended into monocytes using 100 lX PBS and antibodies (1 µl)
were added. Samples were incubated at 4˚C for 30 min in the dark
and analyzed using FlowJo 10.6.2 software (BD Biosciences).
The cells were directly dripped onto the surface of
the porous HAG scaffold until the material was just infiltrated by
the cell suspension. Following incubation for 3 h, the HAG scaffold
was turned over, cell suspension droplets were added to the other
side of the scaffold and culture medium (high glucose DMEM) was
added following a further 3 h in the incubator. Tissue engineered
bone was cultured in a constant temperature incubator at 37˚C on a
six-well plate for 7 days, and the solution was changed once every
1-2 days. hPMSCs were cultured in low-glucose DMEM supplemented
with 10% FBS and 1% penicillin-streptomycin in a 5% CO2
atmosphere at 37˚C.
Preparation and characterization of
the scaffolds
The HAG scaffolds were used in this study as
described by our previous work (5).
Experimental animals and
implantation
All animal experiments were conducted according to
the protocols approved by the Animal Care and Use Committee of
Sichuan Provincial People's Hospital [Sichuan, P.R. China; approval
no. 2019NSF(98)]. A total of 18 male beagles (age, 12 months old;
weight range, 10-12 kg) were selected as the experimental samples.
Beagles were housed at 16-28˚C (softened animal feed with milk and
water were obtained freely from 8 a.m. to 5 p.m.) and under
relative humidity of 40-80%. They were exposed to a 12 h light/dark
cycle and ammonia concentrations of <14 mg/m3, which
was administered to avoid the occurrence of disease. Beagles were
provided by the Experimental Animal Center of Sichuan Provincial
People's Hospital (Sichuan, P.R. China). All animal surgery was
approved by the Institutional Animal Care and Use Committee and
implemented according to relevant requirements. The beagles were
used for a duration of 4 months in the present study. Pentobarbital
sodium was injected intravenously into the forelimb for general
anesthesia at a concentration of 30 mg/kg. The animals were
euthanized 1 month after the experiment, using pentobarbital sodium
injected intravenously into the forelimb at a concentration of 100
mg/kg. The death of all animals was confirmed by cardiac arrest,
respiratory arrest and loss of reflex. A total of 18 beagles were
randomly allocated into one of the following groups, with 3
beagles/group: i) HAG group with stem cell implantation (HAG/hPMSC
group) and ii) HAG group without stem cell implantation. The
scaffolds were implanted separately into the dorsal muscle of each
dog as described by the heterotopic implantation discussion in our
previous study (5). The scaffold
attached by muscle from each beagle was obtained, and RNA and
protein were extracted for analysis by crushing with a grinder. A
total of three scaffolds from the dorsal muscle of each dog were
pooled together for RNA-seq. Western blotting analysis and qPCR
were repeated three times.
H&E staining and western blot
analysis
At 4, 8 and 12 weeks after surgery, the surface of
the scaffold was washed with normal saline and placed into 10%
neutral formaldehyde fixing solution at 4˚C overnight. New bone was
identified using optical microscopy of H&E-stained implanted
materials. H&E staining can detect the formation of bone
matrix, which can indicate osteogenic effects (18,19).
Images were captured using an optical microscope (CX23; Carl Zeiss
AG).
Following SDS-PAGE and membrane transfer, the
membrane was incubated overnight at 4˚C with IgG (Abcam; 1:300).
Following incubation with the secondary antibody, buffer from the
membrane was discarded and developing reagent was added. The PVDF
membrane was rocked until development of bands was apparent. The
membrane was washed three times using distilled water for 30 min.
The membrane was visualized using ChemiDocMP (Bio-Rad Laboratories,
Inc.).
Isolation and purification of RNA
For mRNA deep sequencing, total RNA was extracted
from the control and treated frozen samples using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The quality of total
RNA was checked using an Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc.). Equal amounts of RNA samples from three
control individuals were pooled to generate a mixed sample for the
control subgroup, and the same was repeated for the treated
subgroup.
cDNA synthesis and library
construction for mRNA deep sequencing
For mRNA sequencing, a cDNA library was constructed
with a NEBNext® Ultra™ RNA Library Prep kit (Illumina,
Inc.). The concentration of the library was >2 nM, indicating
that the library was effective. Samples were diluted to 1 ng/µl and
detected using a DNA Quantification kit (llumina, Inc.). Briefly,
mRNA was purified from total RNA using poly-T oligo-attached
magnetic beads. Fragmentation was carried out using divalent
cations under elevated temperature in NEBNext First Strand
Synthesis Reaction Buffer (5X) (Illumina, Inc.). Index codes were
added to attribute sequences to each sample and 150 bp paired-end
reads were generated. The obtained library was used for sequencing
with Illumina Hiseq™ 2500 (Illumina, Inc.).
Reference genome and gene model annotation files
were downloaded from genome websites (NCBI, https://www.ncbi.nlm.nih.gov/) directly. An index of
the reference genome was built using Bowtie v2.2.3 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml),
and paired-end clean reads were aligned to the reference genome
using TopHat v2.0.12 (http://ccb.jhu.edu/software/tophat/manual.shtml).
TopHat was selected as the mapping tool as it can generate a
database of splice junctions based on the gene model annotation
files. We used KOBAS software (http://kobas.cbi.pku.edu.cn/) to test the statistical
enrichment of differential expression genes in KEGG pathway.
Reverse transcription-quantitative PCR
(RT-qPCR)
To validate the RNA-seq data, differentially
expressed genes were selected for RT-qPCR analysis. All primers
were designed by the Premier5.0 software (Premier Biosoft
International; Table I). RNA was
extracted using the TRIzol kit (Thermo Fisher Scientific, Inc.).
cDNA was synthesized at 37˚C for 15 min and 85˚C for 5 sec using
the Prime Script® RT reagent kit with the gDNA Eraser
kit (all from Takara Biotechnology Co., Ltd.). RT-qPCR was carried
out with SsoFast™ EvaGreen® Supermix (Bio-Rad
Laboratories, Inc.). Two mRNAs were selected at random from
upregulated, downregulated and non-altered genes for qPCR analysis
using β-actin as a reference gene. The fold change from the qPCR
was determined using the 2-ΔΔCq method (20). The RNAs of samples were reverse
transcribed into cDNA in a 20 µl reaction containing 13 µl RNA
template, 2 µl Oligo dT primer or microRNA sequence-specific
primers (10 µM), 4 µl of 5X PrimeScript Buffer and 1 µl PrimeScript
RT Enzyme Mix Ι (Takara Bio, Inc.). For subsequent qPCR reactions
using gene-specific primers, 1 µl cDNA sample was amplified under
the following cycle conditions: 94˚C for 30 sec, followed by 39
cycles of 55˚C for 20 sec and 72˚C for 20 sec.
 | Table IPrimers for reverse
transcription-quantitative PCR. |
Table I
Primers for reverse
transcription-quantitative PCR.
| Name | Sequence
(5'-3') | Fragment, bp |
|---|
| COL-1-F |
AGGGGTCTCCATGGTGAGTT | 119 |
| COL-1-R |
GAAGGACCTCGGCTTCCAAT | |
| Runx2-F |
TTCCAGAATGCTTCCGCCAT | 110 |
| Runx2-R |
AACTGCTGTGGCTTCCATCA | |
|
β-actin-F |
CAATACAACTCTCCACAACC | 281 |
|
β-actin-R |
CAGATAGCACCTTCAGCAC | |
Statistical analysis
All the statistical data were analyzed using
Bonferroni ANOVA using SPSS 20.0 statistics software (IBM Corp.).
All data are presented as the mean ± SD. P<0.05 was considered
to indicate a statistically significant difference. All analyses
were repeated three times. Gene expression used double-screening by
q-value (adjusted P-value, ≤0.05) and normalized log2ratio
(|log2ratio |≥1; Table SI,
Table SII and Table SIII) according previous sequencing
methods (21).
Results
Osteogenesis effects of the HAG-based
delivery of hPMSCs
The beagles in the HAG/hPMSC experimental group had
HAG scaffolds implanted with hPMSCs implanted in their dorsal
muscles, while the HAG control group was implanted with the HAG
scaffolds alone. At a total of 4, 8 and 12 weeks after the
operation, two groups of scaffolds were obtained from the dorsal
muscles of the beagles and labelled as follows: HAG-4 weeks (w),
HAG-8w, HAG-12w and HAG/hPMSC-4w, HAG/hPMSC-8w and
HAG/hPMSC-12w.
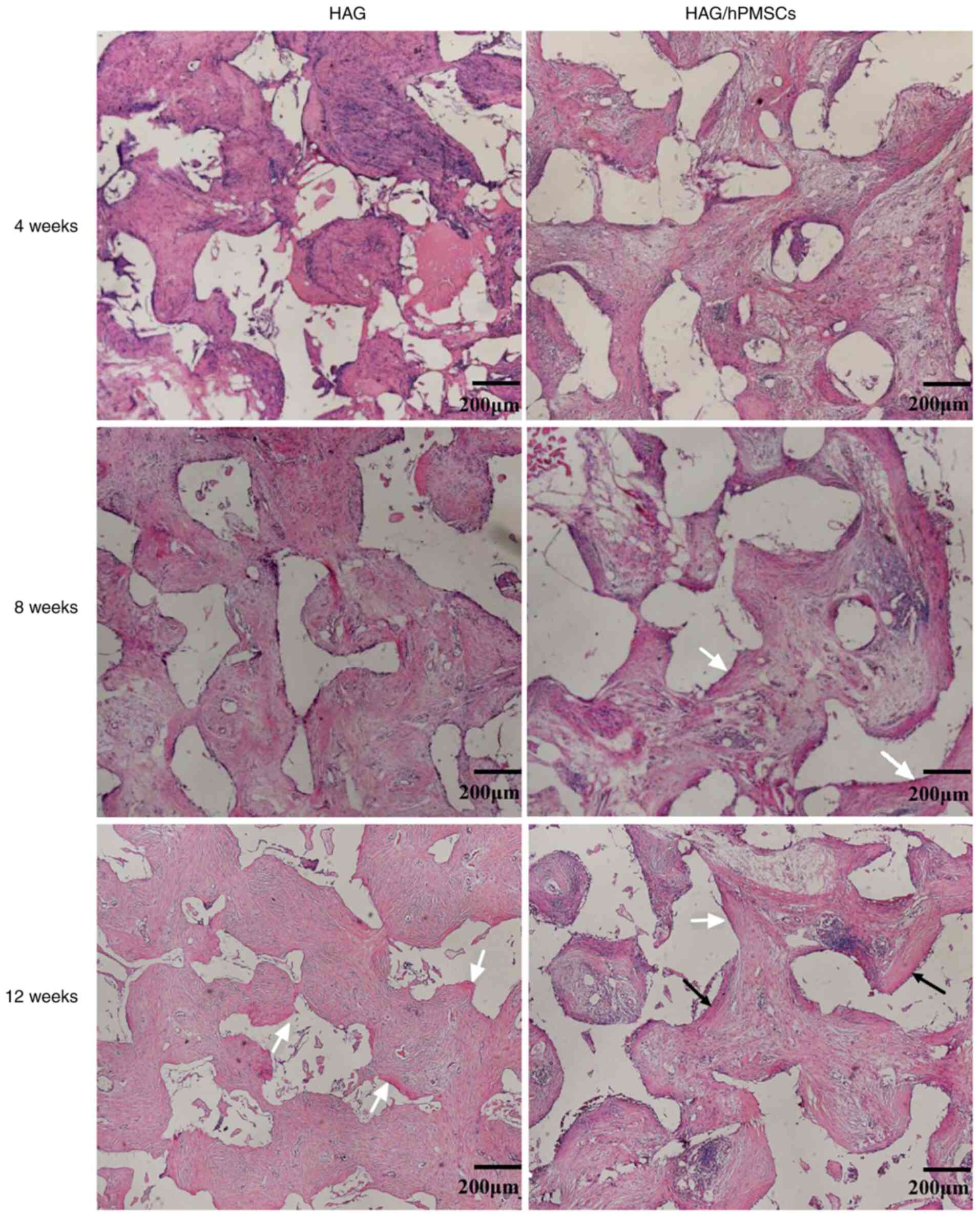
H&E staining demonstrated that a large number of
fibroblasts grew and fibrous tissue formed in both groups at 4
weeks after implantation, while osteoid formation was found on the
surface of scaffolds in the HAG/hPMSC-4w compared with the HAG-4w
group (Fig. 1). After 8 weeks, the
area of bone matrix in the HAG/hPMSC-8w group was larger than that
in the HAG group (Fig. 1). After 12
weeks, a large amount of bone matrix was formed in both groups, but
mature bone plate structure was observed in the HAG/hPMSC-12w group
(Fig. 1). The statistical results
indicated that the osteogenic ability and effects of the HAG/hPMSC
group were markedly greater than those of the HAG group (Table II).
 | Table IIStatistical anlaysis of the ratio of
total area occupied by the newborn. |
Table II
Statistical anlaysis of the ratio of
total area occupied by the newborn.
| Sample | Mean, % | Standard deviation,
% |
|---|
| HAG-4w | 3.7 | 0.3 |
| HAG/hPMSC-4w | 5.2 | 0.5 |
| HAG-8w | 6.5 | 1.2 |
| HAG/hPMSC-8w | 5.1 | 0.3 |
| HAG-12w | 11.2 | 0.3 |
| HAG/hPMSC-12w | 12.3 | 0.4 |
Illumina sequencing and mapping to
reference the genome
cDNA libraries were constructed for different RNA
samples at different times of implantation, with one treatment and
one control for Illumina sequencing. From these libraries,
49,398,934, 53,012,698, 51,462,894, 60,674,628, 53,302,412 and
48,165,136 raw reads were obtained from HAG/hPMSC-4w, HAG/hPMSC-8w,
HAG/hPMSC-12w, HAG-4w, HAG-8w and HAG-12w, respectively, which
generated 7.05, 7.59, 7.36, 8.65, 7.61 and 6.89 Gb of cleaned data.
The schematic of Illumina deep sequencing and analysis is
demonstrated in Table III.
 | Table IIIStatistics of mRNA sequencing
reads. |
Table III
Statistics of mRNA sequencing
reads.
| Sample | Raw reads | Clean reads | Clean bases, G | Error rate, % | Q20, % | Q30,% | GC content, % |
|---|
| HAG-4w | 60,674,628 | 57,642,840 | 8.65 | 0.02 | 96.65 | 91.59 | 50.29 |
| HAG/hPMSC-4w | 49,398,934 | 46,993,014 | 7.05 | 0.02 | 96.88 | 92.03 | 50.79 |
| HAG-8w | 53,302,412 | 50,746,542 | 7.61 | 0.02 | 96.96 | 92.26 | 51.79 |
| HAG/hPMSC-8w | 53,012,698 | 50,596,524 | 7.59 | 0.02 | 96.83 | 91.94 | 50.55 |
| HAG-12w | 48,165,136 | 45,911,572 | 6.89 | 0.02 | 96.55 | 91.37 | 51.01 |
| HAG/hPMSC-12w | 51,462,894 | 49,097,376 | 7.36 | 0.02 | 96.81 | 91.88 | 50.58 |
High-quality reads were mapped to the coding
sequences from the reference genome by Bowtie software, with the
default parameters for the HAG group set as 48,759,095 (84.59%),
43,082,474 (84.9%) and 38,758,173 (84.42%). For the HAG/hPMSC
group, default parameters were set as 38,443,778 (81.81%),
42,956,903 (84.9%) and 41,701,352 (84.94%; Table IV).
 | Table IVResults of reads compared with the
reference genome. |
Table IV
Results of reads compared with the
reference genome.
| Sample | Total reads | Total mapped
(%) | Multiple mapped
(%) |
|---|
| HAG-4w | 57,642,840 | 48,759,095
(84.59) | 1,043,484
(1.81) |
| HAG/hPMSC-4w | 46,993,014 | 38,443,778
(81.81) | 885,270 (1.88) |
| HAG-8w | 50,746,542 | 43,082,474
(84.9) | 866,880 (1.71) |
| HAG/hPMSC-8w | 50,596,524 | 42,956,903
(84.9) | 844,023 (1.67) |
| HAG-12w | 45,911,572 | 38,758,173
(84.42) | 701,480 (1.53) |
| HAG/hPMSC-12w | 49,097,376 | 41,701,352
(84.94) | 742,653 (1.51) |
Functions of differentially expressed
genes
The sequencing data revealed that 1,539 (HAG-4w vs.
HAG/hPMSC-4w), 1,187 (HAG-8w vs. HAG/hPMSC-8w) and 30 (HAG-12w vs.
HAG/hPMSC-12w) genes that were expressed differently between the
control and treated groups were related to osteogenesis responses.
This was due to HAG scaffold-based delivery of hPMSCs under a
double-screening using q-value (adjusted P-value, ≤0.05) and
normalized log2ratio (|log2ratio|≥1; Table SI, Table SII and Table SIII) according previous sequencing
methods (22).
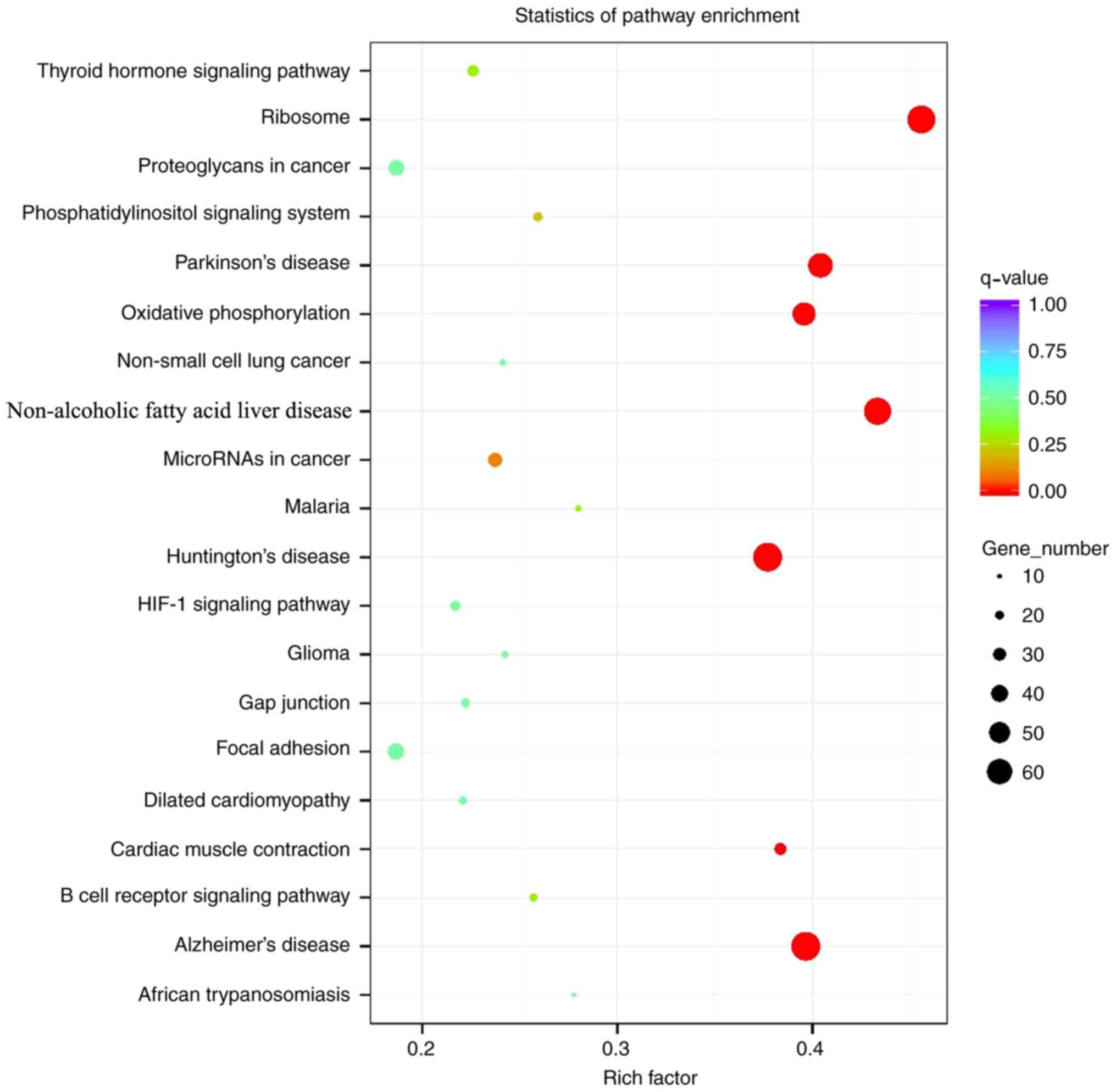
Analysis of Kyoto Encyclopedia of Genes and Genomes
(KEGG) enrichment demonstrated that the signal pathways enriched
with differentially expressed genes differed during the all periods
after implantation (Fig. 2).
Fig. 2 summarizes all of the KEGG
pathways, rather than the differences at each particular stage. At
4 weeks following implantation, the involved signaling pathways
included ‘extracellular matrix (ECM)-receptor interaction’ and
‘PI3K-AKT signaling’, compared with ‘hematopoietic cell lineage’,
‘focal adhesion’, ‘cell adhesion molecules’ (CAMs), ‘protein
digestion and absorption’, ‘platelet activation’, ‘B cell receptor
signaling pathway’ and ‘osteoclast differentiation’, which were
involved at 8 weeks following implantation. At 12 weeks following
implantation, the signaling pathways involved mainly included
‘protein digestion and absorption’, ‘ECM-receptor interaction’,
‘AMP-activated protein kinase (AMPK) signaling’ and ‘osteoclast
differentiation’.
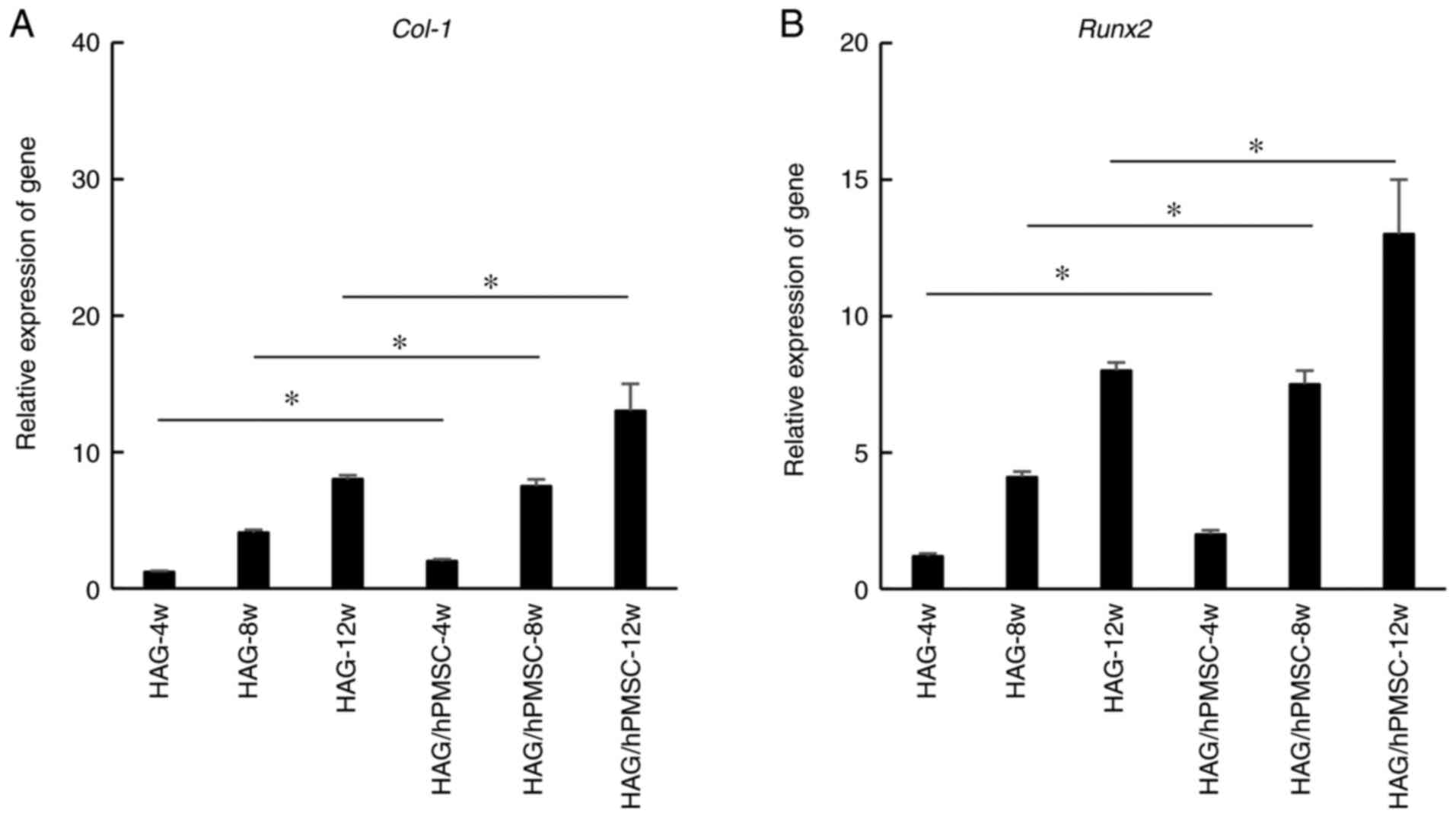
Detection and verification of
osteogenic genes
To validate the results of the Illumina sequencing,
the osteogenic genes COL-1 and RUNX2 were selected
for RT-qPCR analysis. The results revealed that COL-1
(Fig. 3A) and RUNX2
(Fig. 3B) genes were expressed in
HAG and HAG/hPMSC groups during heterotopic osteogenesis and that
the expression increased with time. The gene expression in the
HAG/hPMSC group was markedly upregulated compared with in the HAG
group at all time points (Fig. 3).
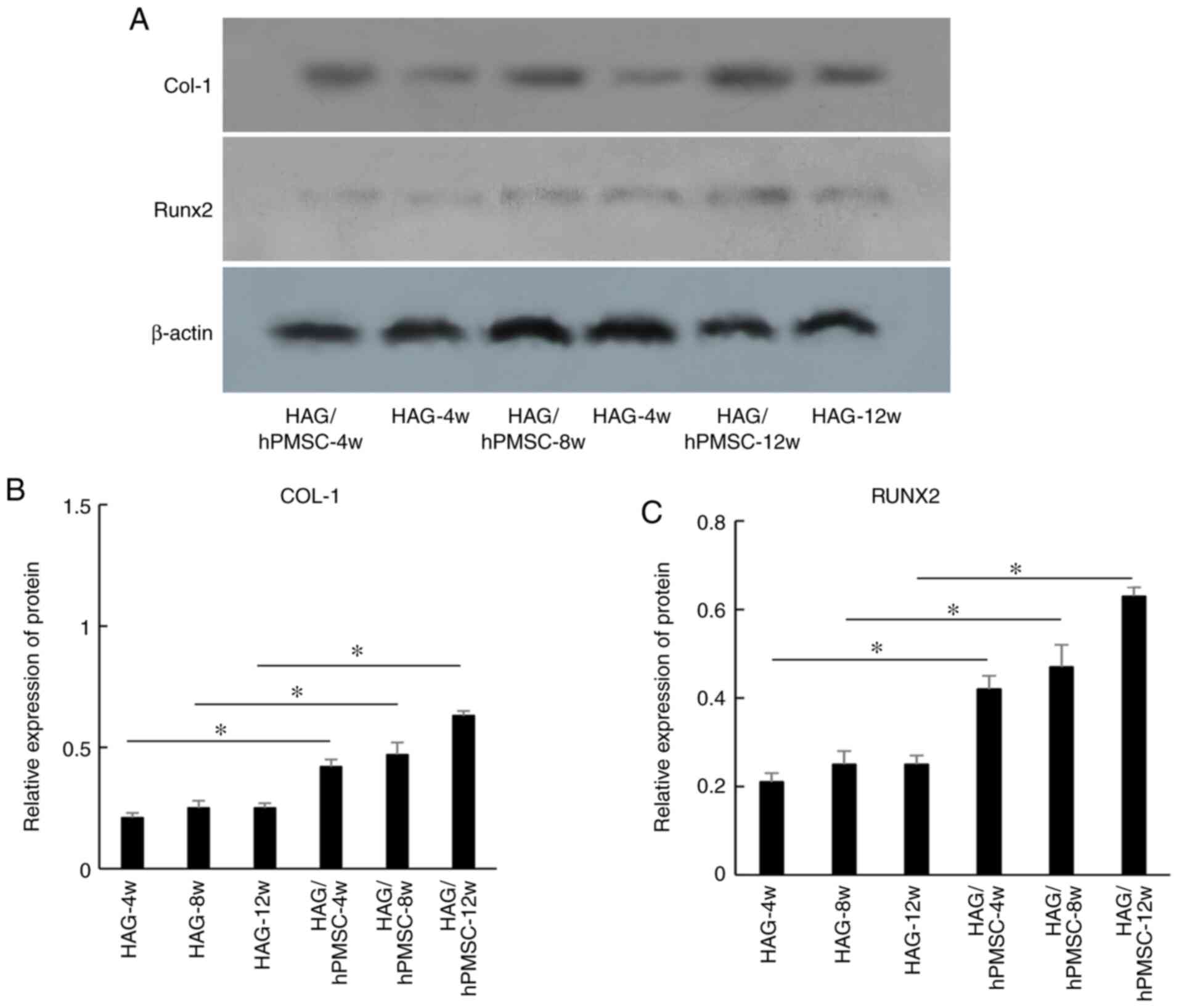
The results of western blot analysis revealed that the expression
levels of proteins COL-1 (Fig. 4A
and B) and RUNX2 (Fig. 4A and C) were similar to those of the respective
genes, but that the difference was not marked between each of the
experimental groups.
Discussion
Tissue-engineered bone consists of seed cells,
scaffolds and growth factors. Of the seed cells, bone marrow MSCs,
embryonic stem cells and adipose stem cells are the most widely
studied. Biomaterials with co-cultured stem cells exhibit good
biocompatibility for increasing osteogenesis and thus offer
potential as bone graft substitutes (23,24).
The seed cells used in the present study are MSCs derived from
human placenta, which is an ideal source of seed cells for humans
(25). To provide the growth
factors required for tissue engineering, seed cells in the present
study were co-cultured with porous scaffolds to explore whether
this method effectively improved osteogenesis. The experimental
results revealed that the scaffolds in the HAG/hPMSC group
demonstrated improved heterotopic osteogenesis in beagle dorsal
muscles. However, in contrast to other engineered cells, the stem
cells used in the present study are derived from humans, allowing
for more valuable application in future clinical use. The
human-derived stem cells used in the present study provide the
necessary growth factors for osteogenesis.
At 4 weeks following implantation, bone-like
structure was also observed on the surface of the scaffold.
Compared with canine stem cell-seeded collagen-hydroxyapatite
scaffolds, the HAG/hPMSC scaffold used in the present study
exhibited an advantage in promoting osteogenesis at 4 weeks
compared with 3 months (26).
Analysis of KEGG enrichment revealed that ECM-receptor interactions
(27) were involved in the process
of osteogenesis. Furthermore, the PI3K-AKT pathway, which inhibits
apoptosis and increases cell survival rate (28), may have improved the surface
compatibility between the scaffold and the muscle cells. At 8 and
12 weeks following implantation, the areas of bone matrix and
mature bone plate in the HAG/hPMSC group were larger than those in
the control group, suggesting that hPMSCs may continue to play an
important role in the whole process of osteogenesis. At 8 weeks
following implantation, pathways associated with cell adhesion,
hematopoietic cell lineage, focal adhesion and CAMs played
important roles, and osteoclast differentiation was activated. At
12 weeks following implantation, osteogenesis was closely
associated with protein digestion and absorption, ECM-receptor
interactions, the AMPK signaling pathway and osteoclast
differentiation. The AMPK signaling pathway plays a vital role in
osteoclast differentiation and aids in improving osteogenesis
(21,29). In the present study, differences in
the signaling pathways involved in the different stages of
osteogenesis were revealed, which are novel findings.
COL-1 is the main type of collagen secreted by
osteoblasts, accounting for ~90% of the components of the ECM
(30). The expression of COL-1 is
upregulated following the differentiation of osteoblasts (30). COL-1 is a marker gene for the
phenotypic identification and osteogenic differentiation of
osteoblasts (31). The results of
the present study revealed that the expression of COL-1 increased
with time, indicating that bone formation also increased with time.
RUNX2, a transcription factor belonging to the Runt family, plays a
key role in bone development (32).
RUNX2 regulates osteoblast differentiation and osteogenesis during
osteogenic differentiation, and the expression level of RUNX2 can
also indicate the function and differentiation of osteoblasts
(33). The results of the current
study are consistent with the qPCR results of a previous study
(5). In the present study, the
expression level of RUNX2 increased with time, suggesting that
osteoblasts continued to differentiate and drive bone formation.
However, the present study is limited by the use of only two
markers and an increase in the number of marker genes is required
for further studies.
In conclusion, the present study preliminarily
explored the effect of ectopic osteogenesis of hPMSCs combined with
porous HAG scaffolds in vivo. The results of the present
study provide the potential for novel applications of this cell
type in osteogenesis research.
Supplementary Material
Differentially expressed genes in HAG
and HAG/human placenta-derived mesenchymal stem cell scaffold
implantation after 4 weeks. HAG, hydroxyapatite with a grooved
structure.
Differentially expressed genes in HAG
and HAG/human placenta-derived mesenchymal stem cell scaffold
implantation after 8 weeks. HAG, hydroxyapatite with a grooved
structure.
Differentially expressed genes in HAG
and HAG/human placenta-derived mesenchymal stem cell scaffold
implantation after 12 weeks. HAG, hydroxyapatite with a grooved
structure.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Department of Science
and Technology of Sichuan Province (grant nos. 2016TD0008 and
2018HH0080) and Chinese National Natural Science Foundation (grant
no. 82071168).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI SRA repository (https://ncbiinsights.ncbi.nlm.nih.gov/tag/sra/) with
accession numbers SRR13236657, SRR13236658, SRR13236659,
SRR13236660, SRR13236661 and SRR13236662.
Authors' contributions
YM conceived and designed the study. XR, QW, CL, QZ,
JZ and HX performed the experiments. QW wrote the paper. XR, KT and
YM revised the manuscript and gave final approval of the version to
be published. KT reanalyzed the data. YM, QW and KT confirmed the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were conducted according to
the protocols approved by the Animal Care and Use Committee of
Sichuan Provincial People's Hospital [Sichuan, P.R. China; approval
no. 2019NSF(98)].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cheng X, Lei D, Mao T, Yang S, Chen F and
Wu W: Repair of critical bone defects with injectable platelet rich
plasma/bone marrow-derived stromal cells composite: Experimental
study in rabbits. Ulus Travma Acil Cerrahi Derg. 14:87–95.
2008.PubMed/NCBI
|
|
2
|
Marx RE, Miller RI, Ehler WJ, Hubbard G
and Malinin TI: A comparison of particulate allogeneic and
particulate autogenous bone grafts into maxillary alveolar clefts
in dogs. J Oral Maxillofac Surg. 42:3–9. 1984.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Janicki P and Schmidmaier G: What should
be the characteristics of the ideal bone graft substitute?
Combining scaffolds with growth factors and/or stem cells. Injury.
42 (Suppl 2):S77–S81. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Suresh SS, Raniga S, Shanmugam V, George M
and Zaki H: Carpal tunnel syndrome due to hydroxyapatite crystal
deposition disease. J Hand Microsurg. 5:96–99. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xiaohua R, Qiang T, Kun T, Huang GT, Li
JY, Xu T, Lv X, Wu J, Chen Z, Weng J, et al: Enhancement of
osteogenesis using a novel porous hydroxyapatite scaffold in vivo
and vitro. Ceram Int. 44:21656–21665. 2018.
|
|
6
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Carnes MU, Allingham RR, Ashley-Koch A and
Hauser MA: Transcriptome analysis of adult and fetal trabecular
meshwork, cornea, and ciliary body tissues by RNA sequencing. Exp
Eye Res. 167:91–99. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cloonan N, Forrest AR, Kolle G, Gardiner
BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G,
et al: Stem cell transcriptome profiling via massive-scale mRNA
sequencing. Nat Methods. 5:613–619. 2008.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Guttman M, Garber M, Levin JZ, Donaghey J,
Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et
al: Ab initio reconstruction of cell type-specific transcriptomes
in mouse reveals the conserved multi-exonic structure of lincRNAs.
Nat Biotechnol. 28:503–510. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Satija NK, Sharma D, Afrin F, Tripathi RP
and Gangenahalli G: High throughput transcriptome profiling of
lithium stimulated human mesenchymal stem cells reveals priming
towards osteoblastic lineage. PLoS One. 8(e55769)2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop DJ and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The international society for cellular
therapy position statement. Cytotherapy. 8:315–317. 2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Squillaro T, Peluso G and Galderisi U:
Clinical trials with mesenchymal stem cells: An Update. Cell
Transplant. 25:829–848. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Alessio N, Squillaro T, Özcan S, Di
Bernardo G, Venditti M, Melone M, Peluso G and Galderisi U: Stress
and stem cells: adult Muse cells tolerate extensive genotoxic
stimuli better than mesenchymal stromal cells. Oncotarget.
9:19328–19341. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fathi E and Farahzadi R: Zinc sulphate
mediates the stimulation of cell proliferation of rat adipose
tissue-derived mesenchymal stem cells under high intensity of EMF
exposure. Biol Trace Elem Res. 184:529–535. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fathi E, Farahzadi R and Sheikhzadeh N:
Immunophenotypic characterization, multi-lineage differentiation
and aging of zebrafish heart and liver tissue-derived mesenchymal
stem cells as a novel approach in stem cell-based therapy. Tissue
Cell. 57:15–21. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lim J, Lee J, Yun HS, Park EK and Shin HI:
Comparison of bone regeneration rate in flat and long bone defects:
Calvarial and tibial bone. Tissue Eng Regene Med. 10:336–340.
2013.
|
|
19
|
Lin YH, Chen CY, Chou LY, Chen CH, Kang L
and Wang CZ: Enhancement of bone marrow-derived mesenchymal stem
cell osteogenesis and new bone formation in rats by obtusilactone
A. Int J Mol Sci. 18(2422)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2002.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang YG, Han XG, Yang Y, Qiao H, Dai KR,
Fan QM and Tang TT: Functional differences between AMPK alpha1 and
alpha2 subunits in osteogenesis, osteoblast-associated induction of
osteoclastogenesis, and adipogenesis. Sci Rep.
6(32771)2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shu C, Zhao M, Anderson JP, Garg G, Singh
KB, Zheng W, Wang C, Yang M and Zhou E: Transcriptome analysis
reveals molecular mechanisms of sclerotial development in the rice
sheath blight pathogen Rhizoctonia solani AG1-IA. Funct Integr
Genomics. 19:743–758. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu Z, Yin X, Ye Q, He W and Zou S:
Periodontal regeneration with stem cells-seeded
collagen-hydroxyapatite scaffold. J Biomater Appl.
31(121)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang G, Yang H, Li M, Lu S, Chen X and Cai
X: The use of silk fibroin/hydroxyapatite composite co-cultured
with rabbit bone-marrow stromal cells in the healing of a segmental
bone defect. Bone Joint Surg Br. 92:320–325. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Diao Y, Ma Q, Cui F and Zhong Y: Human
umbilical cord mesenchymal stem cells: Osteogenesis in vivo as seed
cells for bone tissue engineering. J Biomed Mater Res Part A.
91A:123–131. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu Z, Yin X, Ye Q, He W, Ge M, Zhou X, Hu
J and Zou S: Periodontal regeneration with stem cells-seeded
collagen-hydroxyapatite scaffold. J Biomater Appl. 31:121–131.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y,
Zhao X, Liang C, Wang Y, Sun L, et al: Integrated profiling of
microRNAs and mRNAs: microRNAs located on Xq27.3 associate with
clear cell renal cell carcinoma. PLoS One. 5(e15224)2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hung SC, Pochampally RR, Chen SC, Hsu SC
and Prockop DJ: Angiogenic effects of human multipotent stromal
cell conditioned medium activate the PI3K-Akt pathway in hypoxic
endothelial cells to inhibit apoptosis, increase survival, and
stimulate angiogenesis. Stem Cells. 25:2363–2370. 2007.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang YG, Qu XH, Yang Y, Han XG, Wang L,
Qiao H, Fan QM, Tang TT and Dai KR: AMPK promotes osteogenesis and
inhibits adipogenesis through AMPK-Gfi1-OPN axis. Cell Signal.
28:1270–1282. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gupta V, Shah MA, Shah SK and Shah JM:
Osteoanabolics. Indian J Endocrinol Metab. 6:349–357.
2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zheng J, Mao X, Ling J, Chen C and Zhang
W: Role of magnesium transporter subtype 1 (MagT1) in the
osteogenic differentiation of rat bone marrow stem cells. Biol
Trace Elem Res. 171:131–137. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cui CB, Cooper LF, Yang X, Karsenty G and
Aukhil I: Transcriptional coactivation of bone-specific
transcription factor Cbfa1 by TAZ. Mol Cell Biol. 23:1004–1013.
2003.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kwun IS, Cho YE, Lomeda RA, Shin HI, Choi
JY, Kang YH and Beattie JH: Zinc deficiency suppresses matrix
mineralization and retards osteogenesis transiently with catch-up
possibly through Runx 2 modulation. Bone. 46:732–741.
2010.PubMed/NCBI View Article : Google Scholar
|