Introduction
Acute lymphoblastic leukemia (ALL) is a
hematological malignant tumor characterized by a large number of
immature leukocytes caused by abnormal lymphoblastic proliferation
and abnormal differentiation, and it is a common type of childhood
leukemia (1,2). In recent years, despite clinical
advances in the diagnosis and treatment of ALL, patients with ALL,
especially those with refractory or relapsed ALL, still have a poor
prognosis, serious adverse events and low survival rates (3-5).
T cell ALL (T-ALL), an aggressive and heterogeneous malignancy
originating from T cell precursors (thymocytes), accounts for ~15%
of all ALL cases in children and for ~25% in adults (6,7).
Currently, the outcomes for T-ALL are still lagging behind by 5-10%
compared with B cell ALL in most studies (8,9).
Therefore, it is of great significance to further study the
pathogenesis of T-ALL and find novel effective treatment
strategies.
MicroRNAs (miRNAs/miRs) are a class of highly
conserved non-coding small RNAs of 19 to 25 nucleotide in length
that are ubiquitous in organisms and bind to the 3'-untranslated
region (UTR) of target mRNAs, inhibit mRNA translation and can
regulate gene expression (10,11).
miR-221 is one of the miRNAs discovered earlier in humans and is
located on the X chromosome p11. 3(12). In previous years, studies on miR-221
have focused on tumors and inflammation, and studies have confirmed
that miR-221 is upregulated and plays a key role in a number of
malignant tumors, such as multiple myeloma, bladder cancer and oral
squamous cell carcinoma, and inflammatory-related diseases, such as
atherosclerosis, osteoarthritis, autoimmune and degenerative
diseases (12-16).
However, to the best of our knowledge, the role of miR-221 in ALL
and its related mechanisms of action have yet not been
reported.
In the present study, the effect of miR-221 on human
T-ALL cells and its molecular mechanisms were studied.
Materials and methods
Cell culture
Human T-ALL cell line Jurkat was purchased from The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. Cells were cultured in 75-cm2 flasks with DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin (Nanjing Sunshine Biotech Co., Ltd.) and 100 µg/ml
streptomycin (Nanjing Sunshine Biotech Co., Ltd.). Cells were
incubated in a 5% CO2 incubator at 37˚C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the human T-ALL cell
line using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The concentration of RNA was detected using a NanoDrop™2000
spectrophotometer (Thermo Fisher Scientific, Inc.). The RNA samples
were stored at -80˚C for future use. Then, cDNA was synthesized
with a miScript Reverse Transcription kit (Qiagen GmbH) according
to the manufacturer's protocol. The QuantiFast SYBR Green PCR Kit
(Qiagen GmbH) was used to perform RT-qPCR using a CFX Connect
Real-Time System (Bio-Rad Laboratories, Inc.). GAPDH or U6 was used
as the internal control. Thermocycling conditions for qPCR were as
follows: Initial denaturation at 95˚C for 10 min; 37 cycles of 95˚C
for 15 sec and 55˚C for 40 sec. The 2-ΔΔCq method
(17) was applied for the
quantification of relative gene expression. Primer sequences were
obtained from GenScript and were as follows: U6 forward,
5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3';
GAPDH forward, 5'-TCAACGACCACTTTGTCAAGCTCA-3' and reverse,
5'-GCTGGTGGTCCAGGGGTCTTACT-3'; miR-221 forward,
5'-GCCGAGAGCTACATTGTCTGC-3' and reverse, 5'-CTCAACTGGTGTCGTGGA-3';
PTEN forward, 5'-ATACCAGGACCAGAGGAAACC-3' and reverse,
5'-TTGTCATTATCTGCACGCTC-3'.
Dual-luciferase reporter assay
Next, the mechanism by which miR-221 acted on human
T-ALL cell lines was investigated. The target genes of miR-221 were
searched using miRanda (microRNA.org),
and PTEN was found to be a potential target of miR-221. To confirm
the binding sites between miR-221 and the 3'-UTR of PTEN, a
dual-luciferase reporter assay was performed. The wild-type
(WT-PTEN) and mutant (MUT-PTEN) 3'-UTRs of PTEN were cloned into a
pmiR-RB-Report™ dual-luciferase reporter gene plasmid vector
(Guangzhou RiboBio Co., Ltd.). The MUT 3'-UTR of PTEN was
constructed using a QuikChange Site-Directed Mutagenesis kit
(Stratagene; Agilent Technologies, Inc.) according to the
manufacturer's instructions. Jurkat cells (5x104
cells/well) were transfected with the reporter constructs and
miR-221 mimic (5'-AGCUACAUUGUCUGCUGGGUUUC-3'; Guangzhou Ribobio
Co., Ltd.) or mimic control (5'-CGGUACGAUCGCGGCGGGAUAUC-3';
Guangzhou Ribobio Co., Ltd.) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Luminescence was
assayed 48 h later using the Dual-Luciferase Reporter Assay System
(Promega Corporation) according to the manufacturer's instructions.
Results were normalized to the Renilla luminescence from the
same vector and shown as the ratio between the various treatments
and cells transfected with control vector.
Western blotting
Jurkat cells were washed with ice-cold PBS, and then
lysed with radioimmunoprecipitation lysate buffer (Beyotime
Institute of Biotechnology), including 1% PMSF at 4˚C for 1 h. A
BCA assay (Thermo Fisher Scientific, Inc.) was used to measure the
protein concentrations. Protein samples were collected by
centrifugation for 5 min at 4˚C and 10,000 x g. Proteins (40 µg per
lane) were resolved via 10% SDS Page, electroblotted to PVDF
membranes and then blocked in 5% non-fat milk at room temperature
for 2 h. Membranes were then incubated overnight at 4˚C with
primary antibodies against: PTEN (1:1,000; cat. no. 9188; Cell
Signaling Technology, Inc.), Bcl-2 (1:1,000; cat. no. 4223; Cell
Signaling Technology, Inc.), Bax (1:1,000; cat. no. 5023; Cell
Signaling Technology, Inc.), AKT (1:1,000; cat. no. 4691; Cell
Signaling Technology, Inc.), phosphorylated (p)-AKT (1:1,000; cat.
no. 4060; Cell Signaling Technology, Inc.) and β-actin (1:1,000;
cat. no. 4970; Cell Signaling Technology, Inc.). Subsequently, the
membranes were washed with PBS with 0.1% Tween-20 (PBST) four
times. Membranes were then incubated with horseradish
peroxidase-conjugated anti-rabbit IgG secondary antibody (1:2,000;
cat. no. 7074; Cell Signaling Technology, Inc.) for 2 h at room
temperature and washed with PBST four times. Finally, ECL reagent
(EMD Millipore) was used to visualize protein bands using FluorChem
FC3 (ProteinSimple), and AlphaView 3.4.0 software (ProteinSimple)
was used for semi-quantification.
Cell transfection
Jurkat cells were seeded into 6-well plates
(1x106 cells/well) and cultured at 37˚C for 24 h. Then,
cells were transfected with 100 nM inhibitor control (the negative
control of miR-221 inhibitor; 5'-CAGUACUUUUGUGUAGUACAA-3';
Guangzhou Ribobio Co., Ltd.), 100 nM miR-221 inhibitor (miR-221
antagonist' 5'-GAAACCCAGCAGACAAUGUAGCU-3'; Guangzhou Ribobio Co.,
Ltd.), 50 nM mimic control (the negative control of miR-221 mimic;
5'-CGGUACGAUCGCGGCGGGAUAUC-3'; Guangzhou Ribobio Co., Ltd.), 50 nM
miR-221 mimic (miR-221 agonist; 5'-AGCUACAUUGUCUGCUGGGUUUC-3';
Guangzhou Ribobio Co., Ltd.), 100 nM miR-221 inhibitor + 0.2 µM
control-small interfering (si)RNA (cat. no. sc-36869; Santa Cruz
Biotechnology, Inc.) or 100 nM miR-221 inhibitor + 0.2 µM
PTEN-siRNA (cat. no. sc-29459; Santa Cruz Biotechnology, Inc.)
using Lipofectamine 3000 reagent, according to the manufacturer's
instructions. The transfection efficiency was detected 48 h later
using RT-qPCR.
3-(4,5-dimethylthiahiazol-2-y1)-2,5-diphenytetrazolium bromide
(MTT) assay
The MTT method was performed to detect cell
viability. Jurkat cells were collected after transfection for 48 h,
and then cells were seeded into plates (96-well) at a density of
2x104 cells/ml. MTT (10 µl/well) reagent (Beyotime
Institute of Biotechnology) was added to each well. Then, the wells
were incubated at 37˚C for another 4 h. DMSO (100 µl; Nanjing
KeyGen Biotech Co., Ltd.) was used to dissolve the formazan
crystals. The absorbance at a wavelength of 490 nm was measured by
Thermo Scientific™ Multiskan™ FC Microplate Photometer (Thermo
Fisher Scientific, Inc.). The experiment was repeated three
times.
Transwell migration and invasion
assays
For the invasion assay, Transwell chambers (pore
size, 8 µm; Costar; Corning Inc.) were pre-coated with 30 ml
Matrigel (R&D Systems, Inc.) at 37˚C for 30 min and loaded into
24-well flat-bottomed culture plates. The chambers were loaded into
24-well flat-bottomed plates without Matrigel for the migration
assay. Jurkat cells (2x104) were seeded into the upper
chamber with serum-free DMEM and 600 µl DMEM containing 20% FBS was
added to the lower chamber. After incubation at 37˚C with 5%
CO2 for 24 h, cells on the upper surfaces of the
Transwell chambers were scraped with cotton swabs. Then, the
membrane was fixed with 4% paraformaldehyde at room temperature for
30 min, and then stained with 0.1% crystal violet at room
temperature for 30 min. The stained cells were imaged and counted
under a light microscope (magnification, x100) in five randomly
selected fields.
Flow cytometry
The apoptosis of Jurkat cells was detected using the
Annexin V-FITC/PI kit [cat. no. 70-AP101-100; Hangzhou Multi
Sciences (Lianke) Biotech Co., Ltd.], according to the
manufacturer's instructions. Briefly, Jurkat cells were digested
using 0.2% trypsin, followed by washing with PBS three times. Then,
the cells were incubated with 5 µl Annexin V-FITC and propidium
iodide at 4˚C in the dark for 15 min. Finally, the cell apoptosis
rate (early + late apoptosis) was measured using a FACSCalibur™
flow cytometer (BD Biosciences) with Cell Quest software version
5.1 (BD Biosciences). The assay was performed in triplicate.
Statistical analysis
Experiments were repeated at least three times.
Statistical analyses were performed using GraphPad Prism 5
(GraphPad Software, Inc.). Data are presented as the mean ±
standard deviation. An unpaired Student's t-test or one-way
analysis of variance (ANOVA) followed by Tukey's test was performed
for comparisons between groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-221 upregulation promotes Jurkat
cell viability, migration and invasion, and inhibits cell
apoptosis
In order to investigate the role of miR-221 in
T-ALL, the effect of miR-221 on T-ALL cells was studied. Jurkat
cells were transfected with miR-221 mimic or mimic control for 48
h, and it was found that the miR-221 mimic significantly enhanced
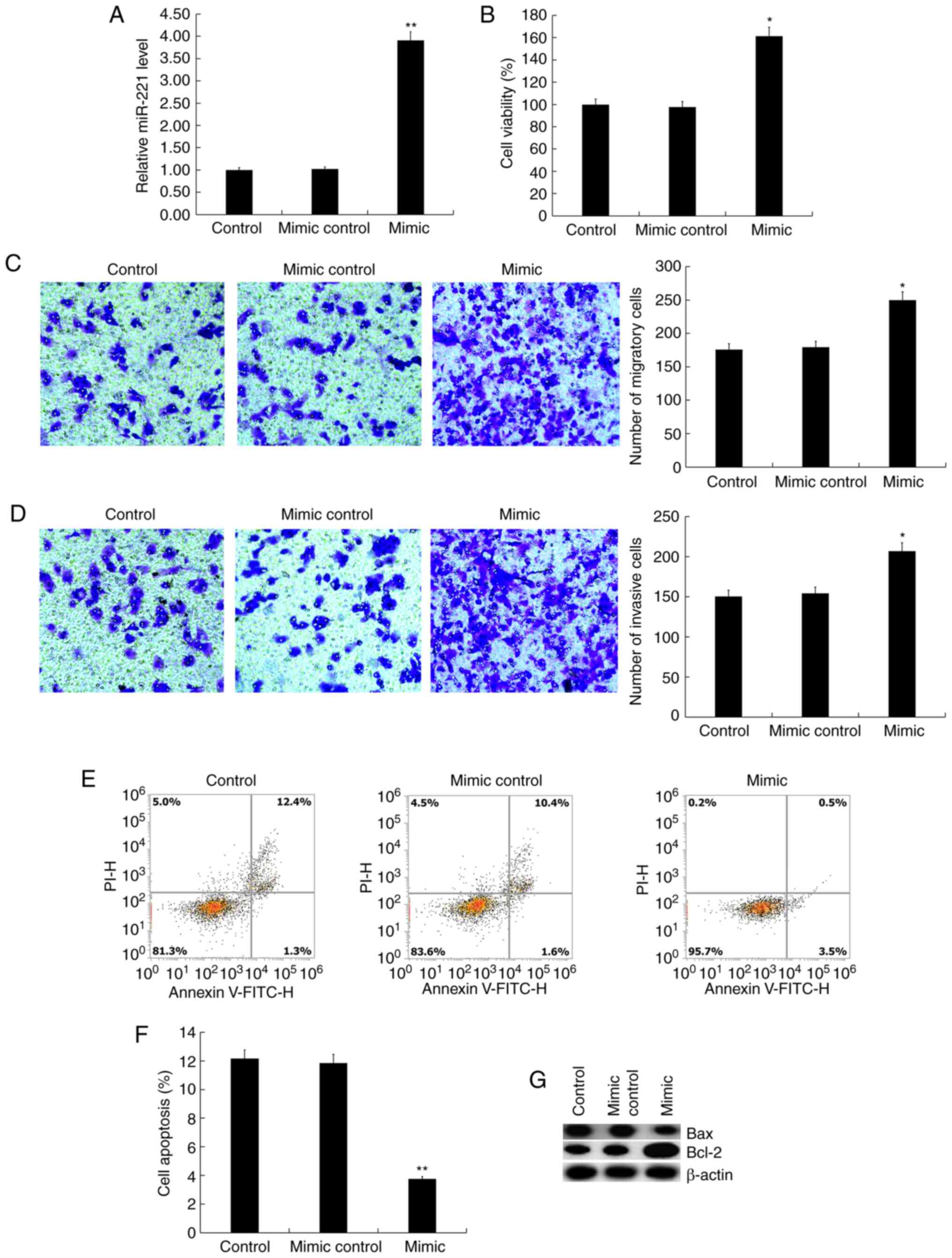
the expression of miR-221 in Jurkat cells (Fig. 1A). The MTT assay results suggested
that compared with the control group, the miR-221 mimic
significantly promoted Jurkat cell viability (Fig. 1B). The Transwell assay showed that
miR-221 mimic significantly promoted Jurkat cell migration
(Fig. 1C) and invasion (Fig. 1D). In addition, it was found that
miR-221 mimic significantly inhibited Jurkat cell apoptosis
(Fig. 1E and F), and the protein level of Bax in Jurkat
cells was notably decreased by miR-221 mimic transfection, whereas
Bcl-2 protein expression was increased (Fig. 1G).
miR-221 knockdown inhibits Jurkat cell
viability, migration and invasion, and promotes cell apoptosis
Then, the effect of miR-221 knockdown on T-ALL cells
was investigated. Jurkat cells were transfected with miR-221
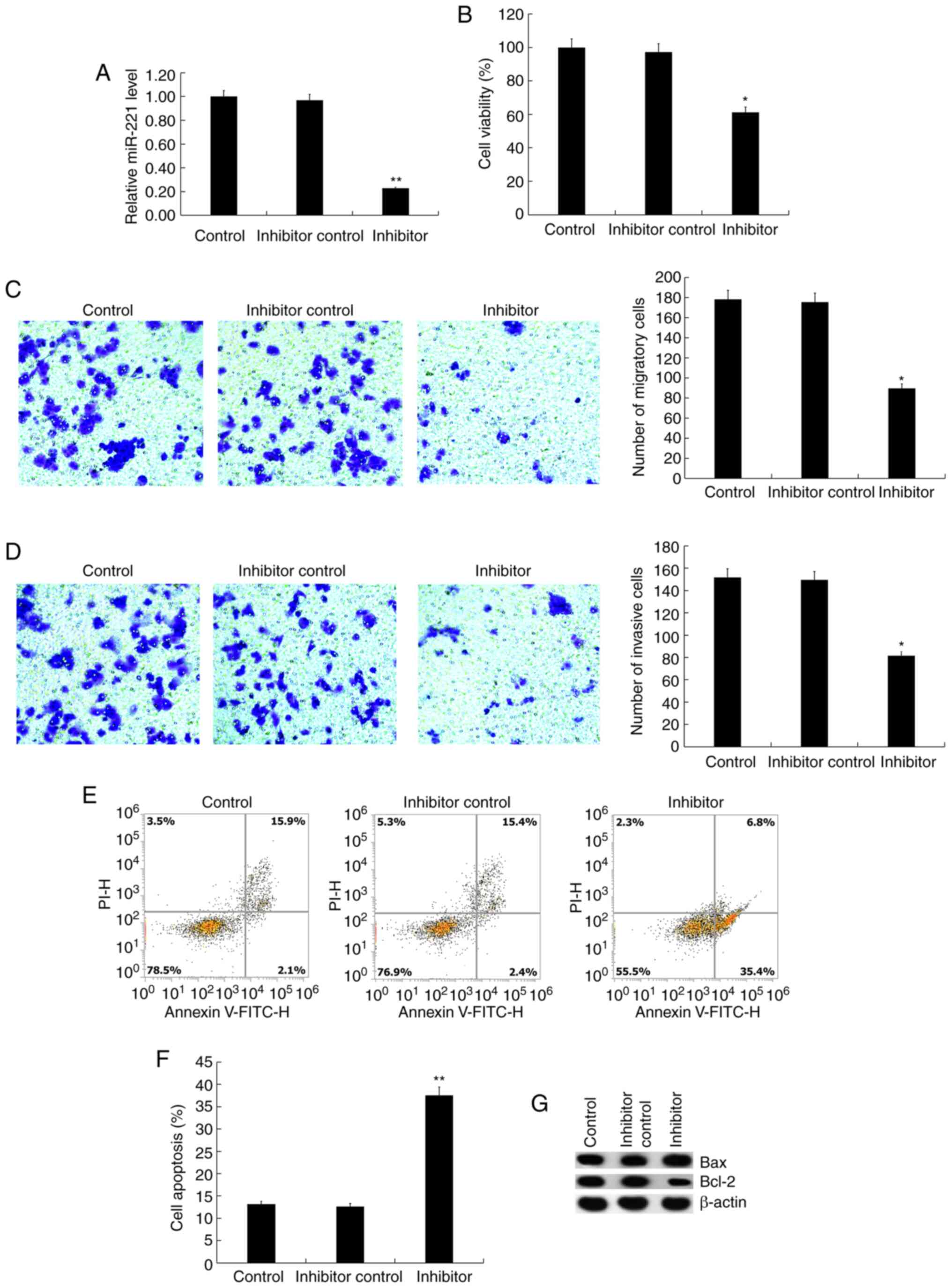
inhibitor or inhibitor control for 48 h, and it was found that the
miR-221 inhibitor significantly reduced the expression of miR-221
in Jurkat cells (Fig. 2A). Compared
with the control group, miR-221 inhibitor significantly inhibited
Jurkat cell viability (Fig. 2B),
migration (Fig. 2C) and invasion
(Fig. 2D). Besides, as expected, it
was found that the miR-221 inhibitor significantly induced Jurkat
cell apoptosis (Fig. 2E and
F), and enhanced the protein
expression level of Bax in Jurkat cells, whereas Bcl-2 protein
expression was reduced (Fig.
2G).
PTEN is a direct target of
miR-221
To explore the molecular mechanism underlying the
effect of miR-221 on human T-ALL cells, potential targets of
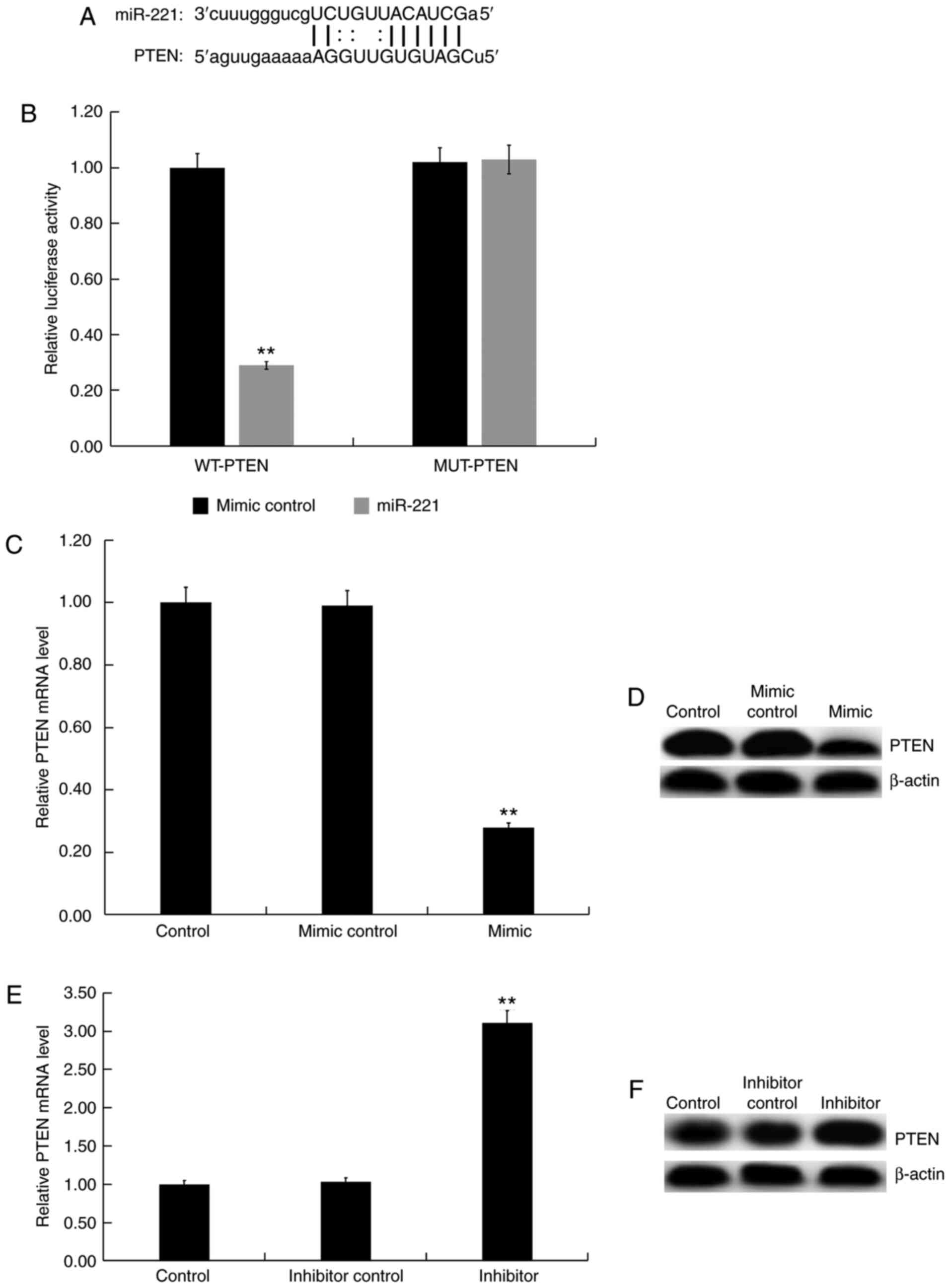
miR-221 were predicted using the bioinformatics tool microRNA.org (Fig.
3A), and a dual-luciferase reporter assay (Fig. 3B) was performed to show the binding
sites between miR-221 and PTEN. Compared with the control group,
the miR-221 mimic significantly decreased the mRNA and protein
expression of PTEN in Jurkat cells (Fig. 3C and D), whereas the miR-221 inhibitor enhanced
the mRNA and protein expression of PTEN in Jurkat cells (Fig. 3E and F). These results demonstrated that PTEN
was a direct target of miR-221.
Inhibitory effects of miR-221
inhibitor on Jurkat cells are abolished by PTEN gene silencing
As PTEN was identified as a target of miR-221, it
was hypothesized that miR-221 may play a role in the regulation of
T-ALL cells by regulating the expression of PTEN. Jurkat cells were
transfected with the inhibitor control, miR-221 inhibitor, miR-221
inhibitor + control-siRNA, or miR-221 inhibitor + PTEN-siRNA for 48
h, then cell viability, migration, invasion and cell apoptosis were
analyzed. It was first confirmed that compared with the
control-siRNA group, PTEN-siRNA significantly reduced PTEN mRNA
expression in Jurkat cells (Fig.
S1). The findings suggested that the inhibited cell viability
(Fig. 4A), migration (Fig. 4B), invasion (Fig. 4C), increased cell apoptosis
(Fig. 4D and E), increased Bax protein expression and
decreased Bcl-2 protein expression (Fig. 4F) in Jurkat cells induced by
transfection with the miR-221 inhibitor were significantly reversed
by PTEN silencing.
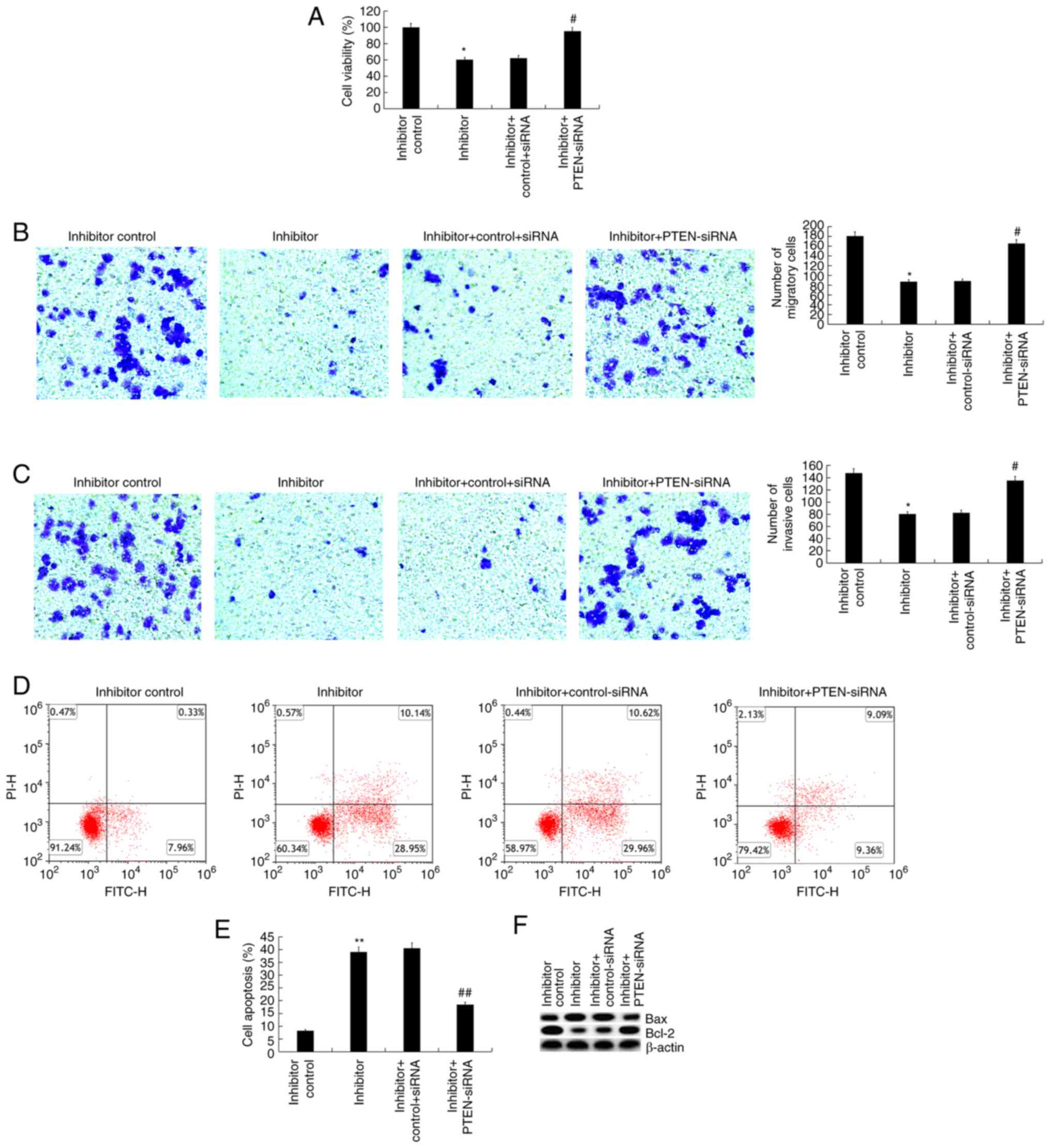 | Figure 4Effect of PTEN silencing on miR-221
inhibitor-transfected Jurkat cells. (A) 48 h after Jurkat cells
were transfected with inhibitor control, miR-221 inhibitor, miR-221
inhibitor + control-siRNA or miR-221 inhibitor + PTEN-siRNA, cell
viability was determined via
3-(4,5-dimethylthiahiazol-2-y1)-2,5-diphenytetrazolium bromide
assay. (B and C) Cell migration and invasion were determined via
Transwell assays. (D and E) Cell apoptosis was measured by flow
cytometry, and the cell apoptosis rate was calculated and
presented. (F) The protein expression levels of Bax and Bcl-2 were
detected via western blotting. Data are presented as the mean ± SD.
*P<0.05, **P<0.01 vs. inhibitor control
group; #P<0.05, ##P<0.01 vs. inhibitor
+ control-siRNA group. PTEN, phosphatase and tensin homologue
deleted on chromosome 10; miR, microRNA; siRNA, small interfering
RNA. |
Phosphatidylinositol 3-kinase
(PI3K)/AKT pathway is involved in the effects of miR-221 on Jurkat
cells
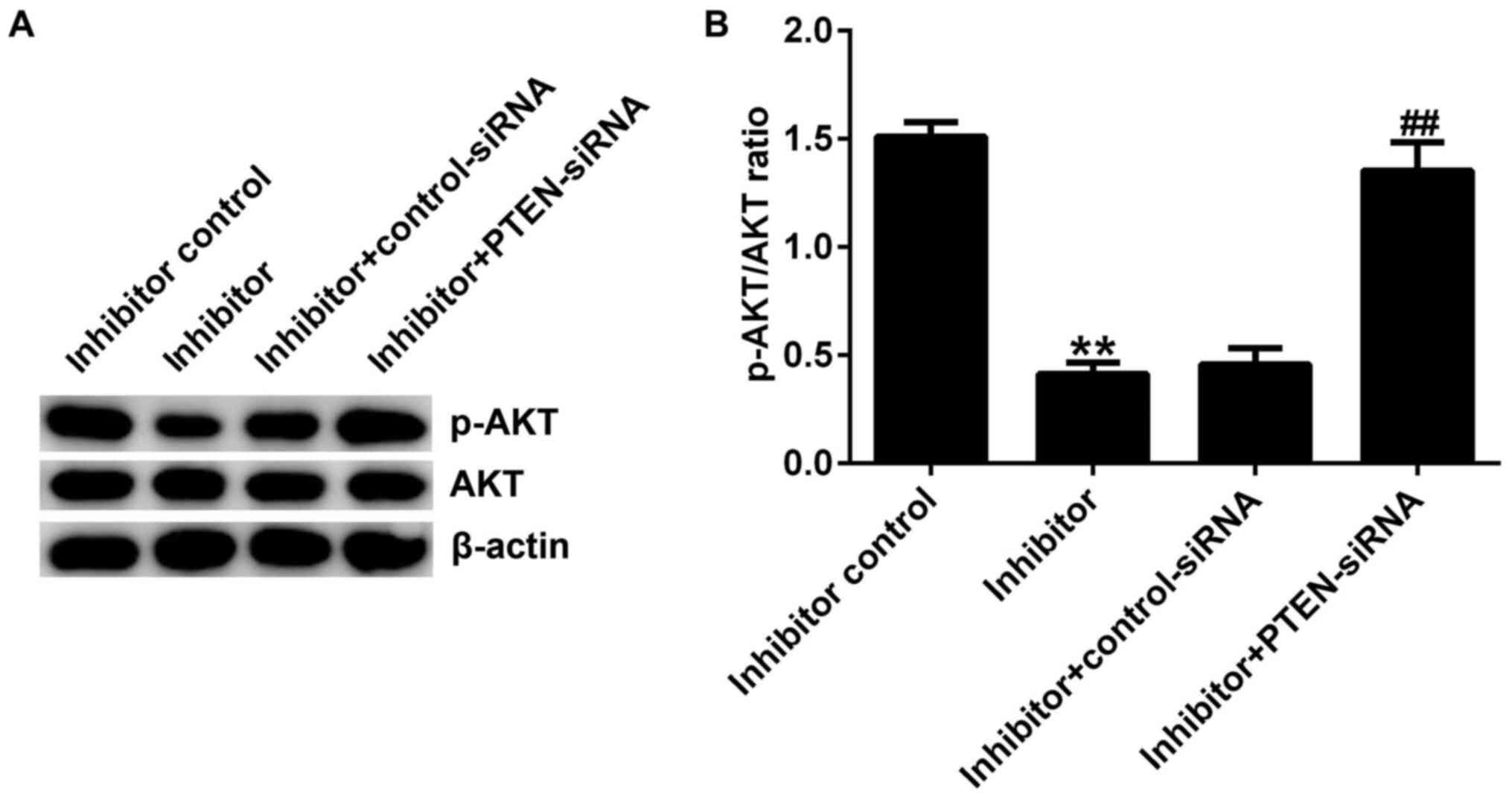
The results of western blotting showed that compared
with the inhibitor control group, miR-221 inhibitor significantly
reduced the protein expression of p-AKT in Jurkat cells.
Furthermore, the reduced p-AKT protein expression in Jurkat cells
induced by the miR-221 inhibitor was reversed by PTEN-siRNA
(Fig. 5A and B). The protein expression of AKT did not
change notably in each group.
Discussion
ALL is a common and life-threatening hematological
malignancy. Relapsed/refractory ALL has poor prognosis rates. For
relapsed ALL, the median survival ranges from 4 to 8 months, with a
5-year overall survival of <10% (18-20).
miRNAs are a type of non-coding small RNA with 19-25
nucleotides, which have roles in post-transcriptional regulation
and have been found to silence a broad range of target genes
(10,11). The abnormal expression of miRNA
plays an essential role in cancer occurrence and progression. For
instance, miR-221, an oncogenic miRNA, which belongs to the
miR-221/222 family, is involved in cancer invasion and migration in
multiple types of cancer, including colorectal cancer, renal cell
carcinoma and luminal breast cancer (21-23).
The high expression of miR-221 has been found to directly inhibit
target genes, such as PTEN (24).
PTEN, an important tumor suppressor, is mutated in the majority of
advanced tumors and plays an important role in modulating the
PI3K/AKT pathway (25).
The present study verified that miR-221 upregulation
significantly promoted human T-ALL cell viability, migration and
invasion, and inhibited cell apoptosis. Whereas, miR-221
downregulation significantly inhibited human T-ALL cell viability,
migration and invasion, and induced cell apoptosis. Then,
bioinformatics analysis using miRanda and the dual-luciferase
reporter assay revealed that PTEN was a direct target of miR-221.
Moreover, it is worth noting that the effects of miR-221 knockdown
on T-ALL cells, such as cell viability, migration, invasion and
apoptosis, were reversed by PTEN silencing.
Abnormal activation of the PI3K/AKT signaling
pathway plays an important role in tumor signaling during important
pathological processes of tumor cell growth, proliferation and
apoptosis. Studies have shown that the inhibition of PI3K protein
activation can make drug-resistant tumor cells sensitive to
chemotherapeutic drugs (26,27).
AKT plays a pivotal role in PI3K signaling, in which
phosphorylation of AKT can activate or block multiple signaling
pathways, including Bcl-2/Bax, mTOR and Caspase-9 (28,29).
Among them, the Bcl-2/Bax pathway protein plays a key role in the
process of apoptosis and is also an important downstream target of
AKT signaling (30). Apoptosis is
the process of cell death under physiological or pathological
conditions under the control of multiple genes. The Bcl-2 gene
family are important regulators of apoptosis (31). The Bcl-2 protein family can be
divided into proapoptotic proteins and anti-apoptotic proteins,
among which Bax is one of pro-apoptotic proteins and Bcl-2 is one
of the anti-apoptotic proteins (32). In the present study, flow cytometry
was performed to detect apoptosis, and the protein expression
levels of Bcl-2, Bax and p-AKT were measured via western blotting.
The results showed that transfection with a miR-221 mimic
significantly inhibited the apoptosis of Jurkat cells, enhanced
Bcl-2 and decreased Bax expression. Whereas, miR-221 inhibitor
could significantly induce apoptosis of Jurkat cells, reduce Bcl-2
and p-AKT protein expression, and increase Bax expression, and all
these changes were reversed by PTEN silencing.
In conclusion, the current study showed that miR-221
downregulation significantly reduced the viability, migration and
invasion of Jurkat cells and induced apoptosis by targeting PTEN.
miR-221 downregulation inhibited human T-ALL cell growth by
regulating the PTEN/PI3K-AKT signaling pathway. Therefore, miR-221
may be a novel potential therapeutic target for T-ALL treatment.
However, this is only a preliminary study of the role of miR-221 in
T-ALL. In order to fully elucidate the role of miR-221 in T-ALL,
further experimental research is required. For example, the
expression of miR-221 in patients with T-ALL and cell lines should
be detected. The role of miR-221 in other T-ALL cell lines should
be also investigated. Besides, the relationship between the
expression of miR-221 and the clinical features of T-ALL patients
requires further research. Moreover, the role of miR-221 in T-ALL
should be investigated in vivo. In future research, we will
further study these topics.
Supplementary Material
Proof of transfection. Jurkat cells
were transfected with control-siRNA or PTEN-siRNA for 48 h, then
PTEN mRNA expression was determined using reverse
transcription-quantitative PCR. **P<0.01 vs.
control-siRNA. siRNA, small interfering RNA; PTEN, phosphatase and
tensin homologue deleted on chromosome 10.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Zhejiang
Provincial Education Department General Research Project (grant no.
Y201737673).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ contributed to study design, data collection,
statistical analysis, data interpretation and manuscript
preparation. ZB, JS, LS and YC contributed to data collection,
statistical analysis and manuscript preparation. PZ and YW
contributed to the data collection and statistical analysis. LZ and
ZB confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hunger SP and Mullighan CG: Acute
lymphoblastic leukemia in children. N Engl J Med. 373:1541–1552.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Rose-Inman H and Kuehl D: Acute leukemia.
Hematol Oncol Clin North Am. 31:1011–1028. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tallen G, Ratei R, Mann G, Kaspers G,
Niggli F, Karachunsky A, Ebell W, Escherich G, Schrappe M,
Klingebiel T, et al: Long-term outcome in children with relapsed
acute lymphoblastic leukemia after time-point and site-of-relapse
stratification and intensified short-course multidrug chemotherapy:
Results of trial ALL-REZ BFM 90. J Clin Oncol. 28:2339–2347.
2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ronson A, Tvito A and Rowe JM: Treatment
of relapsed/refractory acute lymphoblastic leukemia in adults. Curr
Oncol Rep. 18(39)2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kuhlen M, Willasch AM, Dalle JH, Wachowiak
J, Yaniv I, Ifversen M, Sedlacek P, Guengoer T, Lang P, Bader P, et
al: Outcome of relapse after allogeneic HSCT in children with ALL
enrolled in the ALL-SCT 2003/2007 trial. Br J Haematol. 180:82–89.
2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Belver L and Ferrando A: The genetics and
mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer.
16:494–507. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hefazi M and Litzow MR: Recent advances in
the biology and treatment of T cell acute lymphoblastic leukemia.
Curr Hematol Malig Rep. 13:265–274. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Teachey DT and Pui CH: Comparative
features and outcomes between paediatric T-cell and B-cell acute
lymphoblastic leukaemia. Lancet Oncol. 20:e142–e154.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ghelli Luserna Di Rorà A, Iacobucci I,
Imbrogno E, Papayannidis C, Derenzini E, Ferrari A, Guadagnuolo V,
Robustelli V, Parisi S, Sartor C, et al: Prexasertib, a Chk1/Chk2
inhibitor, increases the effectiveness of conventional therapy in
B-/T-cell progenitor acute lymphoblastic leukemia. Oncotarget.
7:53377–53391. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bartel DP: MicroRNA: Genomics, biogenesis,
mechanism, and function. Cell. 116:281–297. 2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
12
|
Zhao JJ, Chu ZB, Hu Y, Lin J, Wang Z,
Jiang M, Chen M, Wang X, Kang Y, Zhou Y, et al: Targeting the
miR-221-222/PUMA/BAK/BAX pathway abrogates dexamethasone resistance
in multiple myeloma. Cancer Res. 75:4384–4397. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu H, Chang JK, Hou JQ, Zhao ZH and Zhang
LD: Inhibition of miR-221 influences bladder cancer cell
proliferation and apoptosis. Eur Rev Med Pharmacol Sci.
21:3193–3199. 2017.PubMed/NCBI
|
|
14
|
Zhou L, Jiang F, Chen X, Liu Z, Ouyang Y,
Zhao W and Yu D: Downregulation of miR-221/222 by a microRNA sponge
promotes apoptosis in oral squamous cell carcinoma cells through
upregulation of PTEN. Oncol Lett. 12:4419–4426. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chistiakov DA, Sobenin IA, Orekhov AN and
Bobryshev YV: Human miR-221/222 in physiological and
atherosclerotic vascular remodeling. Biomed Res Int.
2015(354517)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Marques-Rocha JL, Samblas M, Milagro FI,
Bressan J, Martínez JA and Marti A: Noncoding RNAs, cytokines, and
inflammation-related diseases. FASEB J. 29:3595–3611.
2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Göekbuget N, Dombret H, Ribera JM,
Fielding AK, Advani A, Bassan R, Chia V, Doubek M, Giebel S,
Hoelzer D, et al: International reference analysis of outcomes in
adults with B-precursor Ph-negative relapsed/refractory acute
lymphoblastic leukemia. Haematologica. 101:1524–1533.
2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gökbuget N, Stanze D, Beck J, Diedrich H,
Horst HA, Hüttmann A, Kobbe G, Kreuzer KA, Leimer L, Reichle A, et
al: Outcome of relapsed adult lymphoblastic leukemia depends on
response to salvage chemotherapy, prognostic factors, and
performance of stem cell transplantation. Blood. 120:2032–2041.
2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Oriol A, Vives S, Hernández-Rivas JM,
Tormo M, Heras I, Rivas C, Bethencourt C, Moscardó F, Bueno J,
Grande C, et al: Outcome after relapse of acute lymphoblastic
leukemia in adult patients included in four consecutive
risk-adapted trials by the PETHEMA study group. Haematologica.
95:589–596. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dentelli P, Traversa M, Rosso A, Togliatto
G, Olgasi C, Marchiò C, Provero P, Lembo A, Bon G, Annaratone L, et
al: miR-221/222 control luminal breast cancer tumor progression by
regulating different targets. Cell Cycle. 13:1811–1826.
2014.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Qin J and Luo M: MicroRNA-221 promotes
colorectal cancer cell invasion and metastasis by targeting RECK.
FEBS Lett. 588:99–104. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lu GJ, Dong YQ, Zhang QM, Di WY, Jiao LY,
Gao QZ and Zhang CG: miRNA-221 promotes proliferation, migration
and invasion by targeting TIMP2 in renal cell carcinoma. Int J Clin
Exp Pathol. 8:5224–5229. 2015.PubMed/NCBI
|
|
24
|
Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang
L, Yang A, Zhao J and Jia L: MiR-221 promotes
trastuzumab-resistance and metastasis in HER2-positive breast
cancers by targeting PTEN. BMB Rep. 47:268–273. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Salmena L, Carracedo A and Pandolfi PP:
Tenets of PTEN tumor suppression. Cell. 133:403–414.
2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Fischer B, Frei C, Moura U, Stahel R and
Felley-Bosco E: Inhibition of phosphoinositide-3 kinase pathway
down regulates ABCG2 function and sensitizes malignant pleural
mesothelioma to chemotherapy. Lung Cancer. 78:23–29.
2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rahmani M, Aust MM, Attkisson E, Williams
DC Jr, Ferreira-Gonzalez A and Grant S: Dual inhibition of Bcl-2
and Bcl-xL strikingly enhances PI3K inhibition-induced apoptosis in
human myeloid leukemia cells through a GSK3- and Bim-dependent
mechanism. Cancer Res. 73:1340–1351. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhou BH, Tan PP, Jia LS, Zhao WP, Wang JC
and Wang HW: PI3K/AKT signaling pathway involvement in
fluoride-induced apoptosis in C2C12 cells. Chemosphere.
199:297–302. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Vachhani P, Bose P, Rahmani M and Grant S:
Rational combination of dual PI3K/mTOR blockade and Bcl-2/-xL
inhibition in AML. Physiol Genomics. 46:448–456. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Radha G and Raghavan SC: BCL2: A promising
cancer therapeutic target. Biochim Biophys Acta Rev Cancer.
1868:309–314. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Brown LM, Hanna DT, Khaw SL and Ekert PG:
Dysregulation of BCL-2 family proteins by leukemia fusion genes. J
Biol Chem. 292:14325–14333. 2017.PubMed/NCBI View Article : Google Scholar
|