Introduction
Varicocele, an abnormal varix of the pampiniform
plexus vein, is the most common cause of male infertility. The
global incidence of varicocele is 15-20% in the general population
and ~40% in patients who are infertile (1-4);
this may have a negative impact on human evolution. For
spermatogenesis to occur, the testis must descend into the scrotum
during the embryonic period to provide proper the conditions of a
temperature ~2˚C lower than the central body temperature (5). The upright human posture requires
spermatic veins to work against gravity to return deoxygenated
blood back to the heart (6). If the
valves inside these veins fail, gravity can make the blood pool
inside the testicle, eventually leading to enlargement of the veins
and formation of a varicocele (7).
In ~90% of cases, the disease occurs on the left side, due to the
longer left testicular vein, hemodynamics and higher incidence of
abnormal venous valves (8,9). Effective therapies for varicocele have
yet to be determined (8,10-12).
The experimental rat varicocele model, first established in
1981(13) by partly ligating the
left renal vein, is widely used to investigate the pathophysiology,
diagnosis and treatment of the disease. Researchers have shown that
the following mechanisms contribute to the pathogenesis of
varicocele: i) Neuroendocrine system dysfunction; ii) hypoxia; iii)
accumulation of metabolites and toxicants; iv) oxidative stress; v)
disruption of the blood-testis barrier (BTB); and vi) cell damage
resulting from increased testicular temperature (14,15).
In China, Morinda officinalis F.C.How grows
in the Guangdong, Guangxi and Fujiang provinces. The roots of M.
officinalis are used in a Chinese herbal medicine known as
Bajitian; it has a long history of use for the improvement of male
sexual function and the treatment of male reproductive system
defects in China (16). M.
officinalis polysaccharide (MOP) is one of the main active
components of M. officinalis. Our previous research found
that MOP can repair varicocele-induced damage to the male rat
reproductive system by promoting spermatogenesis, reconstructing
the BTB, increasing the expression of tight junction (TJ) proteins
and restoring hormonal balance, with 300 mg/kg being the most
effective dosage (17). However,
the molecular mechanisms underlying the pathophysiology of
varicocele and the physiological functions and therapeutic effects
of MOP in varicocele are yet to be explored in detail.
Advances in high-throughput RNA sequencing (RNA-Seq)
technology and bioinformatics analysis methods have enabled
researchers to gain insight into the nature of diseases at the RNA
level by studying the dynamics of mRNA, microRNA and long
non-coding RNA (lncRNA) (18)
expression levels. According to previous research, lncRNAs, which
are >200 nucleotides long and found in multiple organisms
(19), play notable roles at almost
every step of gene expression and take part in various disease
processes (20); for example, in
Parkinson's disease (21), leukemia
(22), diabetes (23), cardiovascular disease (24), colon cancer (25,26),
lung cancer (27) and prostate
cancer (28). Due to their diverse
bioactivities, lncRNAs are regarded as potential targets for the
diagnosis and treatment of varicocele.
In the present study, rats with surgically induced
varicocele were treated with either saline or 300 mg/kg MOP by
gavage. Identification of differentially expressed (DE) mRNAs and
lncRNAs in rat left testicular tissue was conducted by RNA-Seq, and
the results were verified by reverse transcription-quantitative PCR
(RT-qPCR). Bioinformatics resources, such as Gene Ontology (GO),
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and
co-expression network analysis, were utilized to explore the
mechanisms underlying varicocele pathophysiology and the
therapeutic effect of MOP, in addition to the interactions between
DE mRNAs and lncRNAs in varicocele. The aim of the current study
was to identify novel targets and methods for varicocele diagnosis
and therapy, and establish a theoretical basis for the use of MOP
in clinical practice.
Materials and methods
Extraction of MOP
The plant M. officinalis used in the present
study was grown (via artificial cultivation) in the Nanjing county
of Fujian province in China. MOP was extracted from dried root
according to a previously described protocol (17,29,30).
In brief, after being ground, the dried root was boiled in ethyl
alcohol to remove oligosaccharides and lipids, extracted using
deionized water and precipitated with 60% alcohol; thereafter,
Sevag reagent (Sinopharm Chemical Reagent Co., Ltd) was added to
remove the protein.
Experimental design
Male, 6- to 7-week-old Sprague-Dawley rats, weighing
200±10 g, were purchased from and kept in The Laboratory Animal
Center of Fujian Medical University (Fuzhou, China). All rats were
housed in normal atmosphere (N2, 78%; O2,
21%; CO2, 0.03%) and specific pathogen-free controlled
environmental conditions, with a temperature of ~23˚C, a 12-h
day/night cycle, a humidity of 40-70% and free access to standard
rat food and water. All procedures conformed to the Guidelines for
the Care and Use of Laboratory Animals established by Fujian
Medical University. The study was approved by The Animal Approval
Committee of Fujian Medical University (approval no.
SYXK-2012-0001).
A total of 24 rats were randomly divided into three
equal groups: Control group, varicocele non-treatment group (VC)
and varicocele treatment group (VC + MOP). Induction of varicocele
was attempted by partial ligation of the left renal vein under
anesthesia using 30 mg/kg sodium pentobarbital by intraperitoneal
injection (31). Control rats
underwent the same operation without the partial ligation, and were
fed a conventional diet for 12 weeks. Subsequently, 8 weeks
post-surgery, rats in the VC and VC+MOP groups were given a daily
oral gavage of 2 ml normal saline or 300 mg/kg MOP, respectively,
for 4 weeks; rats in the Control group were given nothing except
conventional diet. The rats (weight, 500±25 g) were sacrificed by
cervical dislocation after being anesthetized using 30 mg/kg sodium
pentobarbital by intraperitoneal injection to collect left
testicular tissue, which was immediately placed in liquid nitrogen.
Varicocele rats in which vascular dilation did not occur or left
kidney atrophy occurred were excluded from the follow-up
experiment.
mRNA and lncRNA sequencing
analysis
RNA-Seq analysis was performed by Kangchen BioTech
Co., Ltd. The process used was divided into three parts: i)
Extraction of total RNA from rat left testicular tissue; ii)
construction of a cDNA library; and iii) RNA sequencing.
In brief, total RNAs were extracted using
TRIzol® Reagent (Takara Bio Europe SAS) according to the
manufacturer's instructions, followed by purification by Ribo-Zero™
Magnetic Gold Kit (Human/Mouse/Rat) (cat. no. MRZG12324; Epicentre,
Illumina, Inc.). The quantity, integrity and concentration of the
extracted RNAs were verified by 1% agarose gel electrophoresis and
NanoDrop ND-1000 spectrophotometry (Thermo Fisher Scientific,
Inc.). For library preparation, the KAPA Stranded RNA-Seq Library
Prep kit (cat. no. KK8401; Illumina, Inc.) was used (paired-ended
sequencing; 500 bp), with 1-2 µg of sample RNA. The established
libraries were examined using an Agilent 2100 Bioanalyzer G2938C
(Agilent Technologies, Inc.) and quantified by RT-qPCR. The samples
were denatured to single-stranded DNA with 0.1 M NaOH (final
density, 8 pM; concentrations measured by RT-qPCR) and
amplified using TruSeqSR Cluster Kit v3-cBot-HS (cat. no.
GD-401-3001; Illumina, Inc.). The libraries were pooled and
sequenced on the Illumina HiSeq 4000 system (Illumina, Inc.) using
150 cycles of paired-end sequencing.
The quality of the raw sequencing data was assessed
using FastQC software v0.11.7 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/),
before the adapter sequences and poor quality bases were trimmed
from the reads using the Cutadapt software v1.17 (http://dx.doi.org/10.14806/ej.17.1.200)
(32). The Hisat2 software v2.0.4
(http://ccb.jhu.edu/software/hisat2)
and StringTie software v1.2.2 (http://ccb.jhu.edu/software/stringtie) (33) were used to calculate fragments per
kilobase of transcript per million mapped reads (FPKM) values,
which represented the final expression levels of the genes.
Differential gene expression level analyses were performed using
the Ballgown package in R v2.10.0 (https://www.r-project.org) (32). An average FPKM value >0.5 was
chosen as the cut-off for gene expression in the samples.
RT-qPCR
To confirm the results of RNA-Seq, specific DE
patterns were validated by RT-qPCR, with three biological
replicates for each group. TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) was used to extract the total RNAs
from rat testicular tissue, according to the manufacturer's
instructions. cDNA was synthesized using 3 µg total RNAs,
SuperScript III reverse transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.), 5X reverse transcription buffer solution
(Invitrogen; Thermo Fisher Scientific, Inc.), 2.5 mM dNTPs (HyTest,
Ltd), primers (Invitrogen; Thermo Fisher Scientific, Inc.), and the
GeneAmp PCR 9700 System (50˚C for 60 min, 70˚C for 15 min and
stored at 4˚C for infinite time; Applied Biosystems; Thermo Fisher
Scientific, Inc.). The RT-qPCR analysis was performed using the
SYBR®-Green Real-time PCR Master mix (Arraystar, Inc.)
and ViiA™ 7 Real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.), with three technical replicates set for each
sample. The thermocycling conditions were as follows: 95˚C for 10
min; 95˚C for 10 sec, then 60˚C for 1 min (40 cycles); 95˚C for 10
sec; 60˚C for 1 min; and 95˚C for 15 sec. The primers were designed
and purchased from Kangchen BioTech Co., Ltd. The primer sequences
are shown in Table I. The relative
expression of each gene was quantified by comparing its Cq value
(2-ΔΔCq method) with that of the housekeeping gene,
β-actin (34).
 | Table IQuantitative PCR primer
sequences. |
Table I
Quantitative PCR primer
sequences.
| Gene | Primer sequence,
5'-3' |
|---|
| YIPF7 | |
|
Forward |
ATAATGATTCTAATGCTTACGGA |
|
Reverse |
ACAAGAACATCTCTGGTGGAAC |
| WNT9B | |
|
Forward |
GTGTGTGGTGACAACCTGAAGTA |
|
Reverse |
TGACACGCCATGACACTTGC |
|
TMEM255B | |
|
Forward |
GCTTGTGCCCTCCGTCTATGA |
|
Reverse |
AGGGCTGTAGTGGCAGAGGGT |
| P2RX4 | |
|
Forward |
ATATTCCGTCTTGGCACAATC |
|
Reverse |
CTCTATCCAGGTTGCAGTCCC |
| GPR162 | |
|
Forward |
F:TTGCCGTGGAAACCTTGGTG |
|
Reverse |
R:CCTAAGCCCATTTCTCCTGCC |
| CLSTN2 | |
|
Forward |
TCAAAGAACCAGCCTACAAAG |
|
Reverse |
AAGCAGTTACCAGGATCTCATAC |
| AKR1B8 | |
|
Forward |
CAGTTGAGCGACCAGGAGATG |
|
Reverse |
CTGCGTCATAGGGAAACTCTT |
| NILR1 | |
|
Forward |
CCAGGAGGAAAGCGTTTATGC |
|
Reverse |
GGGTTTTACTTGGGCGTATGTCT |
|
AABR07007833.1 | |
|
Forward |
GTGTTCACTACCTCATTCCGTC |
|
Reverse |
TCCTGCTCCTCTTGGTTCTTA |
|
AABR07014649.1 | |
|
Forward |
TGATGAGAAGACTATGAAGAATGC |
|
Reverse |
AAGTTTGGTTTGAATGCTGC |
|
AABR07050146.1 | |
|
Forward |
CTGAGGCAAAGGGACTCTGTA |
|
Reverse |
CTTTGTTGGAGGGACAGCTC |
|
AABR07058711.1 | |
|
Forward |
TGCTCCATAAGTATCGAAGGC |
|
Reverse |
CAGATATTAGCCACAAACCTCA |
|
AABR07069067.1 | |
|
Forward |
GCAGCTTCTTGACATCACATT |
|
Reverse | A
TAGGCATTCCTTAGAGCATTT |
|
AC125688.1 | |
|
Forward |
CTATTCATAGCACCTCTGTGCTG |
|
Reverse |
CGTGGAGTTCTCTGATCTTTGTA |
|
AABR07004428.1 | |
|
Forward |
CACCCACGGAGATCCCACT |
|
Reverse |
GAGCCTTCCCTGTAGCTGGTT |
| β-ACTIN | |
|
Forward |
CGAGTACAACCTTCTTGCAGC |
|
Reverse |
ACCCATACCCACCATCACAC |
Statistical analyses
RNA-Seq and RT-qPCR were performed in three
independent biological replicates for each group. Only genes with
an absolute fold-change >1.2 and P-value ≤0.05 were considered
to be differentially expressed between the groups. The data were
analyzed in SPSS v21.0 (IBM Corp.), and the results are presented
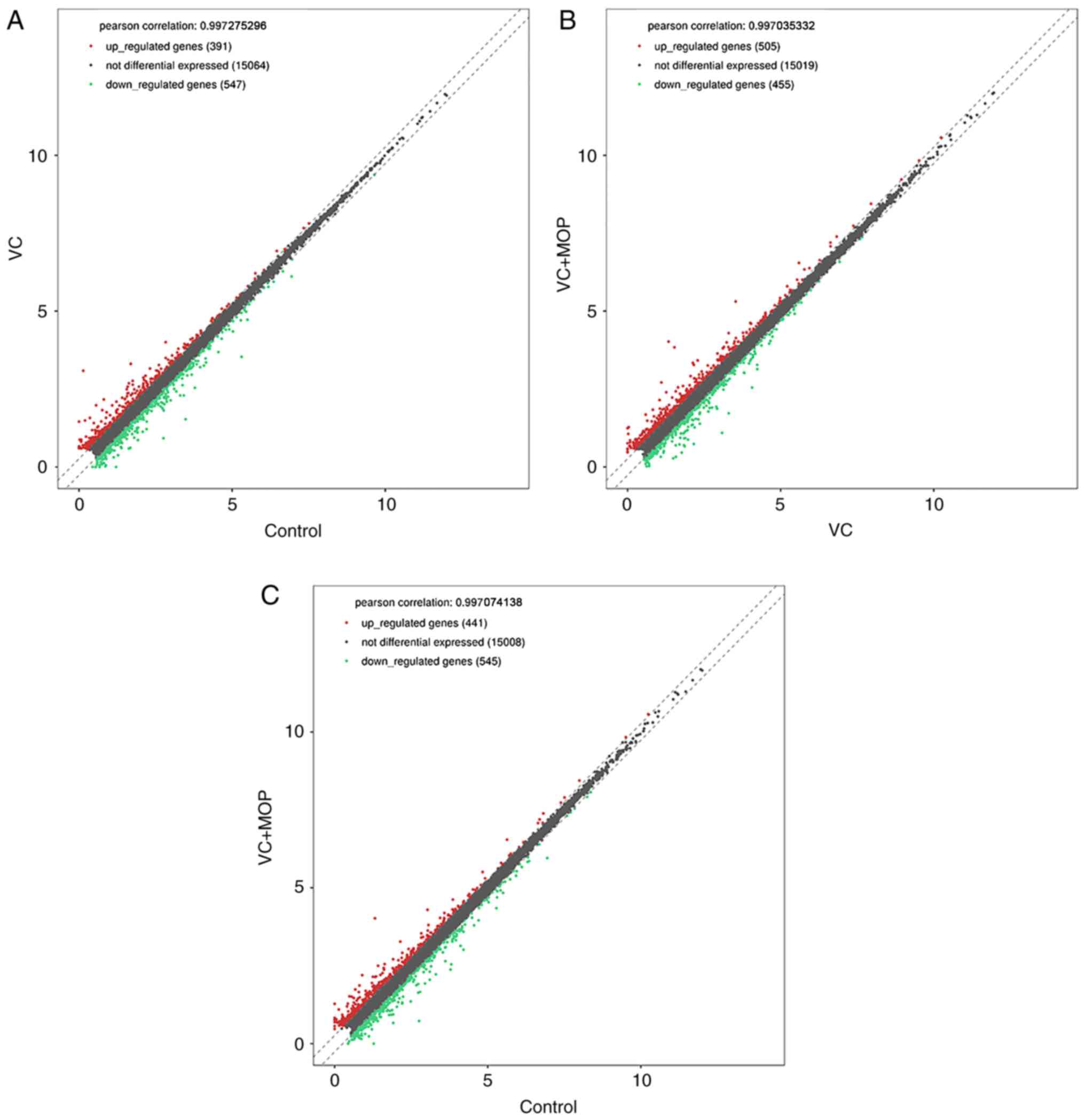
as the mean ± standard deviation. Pearson's correlation was used in
scatter plot of DE genes. One-way ANOVA was used to compare
gene expression levels between two groups, and P≤0.05 was
considered to indicate a statistically significant difference.
Bioinformatic analyses
GO classification and KEGG pathway enrichment
analyses were performed using the website of Gene Ontology Resource
(http://www.geneontology.org) and KEGG
pathway database (https://www.kegg.jp/kegg/pathway.html) respectively.
The co-expression of DE mRNA (coding genes) and lncRNA (non-coding
genes) networks were visualized using Cytoscape v2.8.3 (http://cytoscape.org). The present study data were
uploaded to the Gene Expression Omnibus (GEO) website (https://www.ncbi.nlm.nih.gov/geo/; accession no.
GSE139447; secure token: sfaxocskhpoxnix).
Results
Identification of DE mRNAs and
lncRNAs
The present study data were uploaded to the Gene
Expression Omnibus website. The top 10 upregulated and
downregulated DE genes (including mRNAs and lncRNAs) are listed in
Tables II and III, respectively. Hierarchical cluster
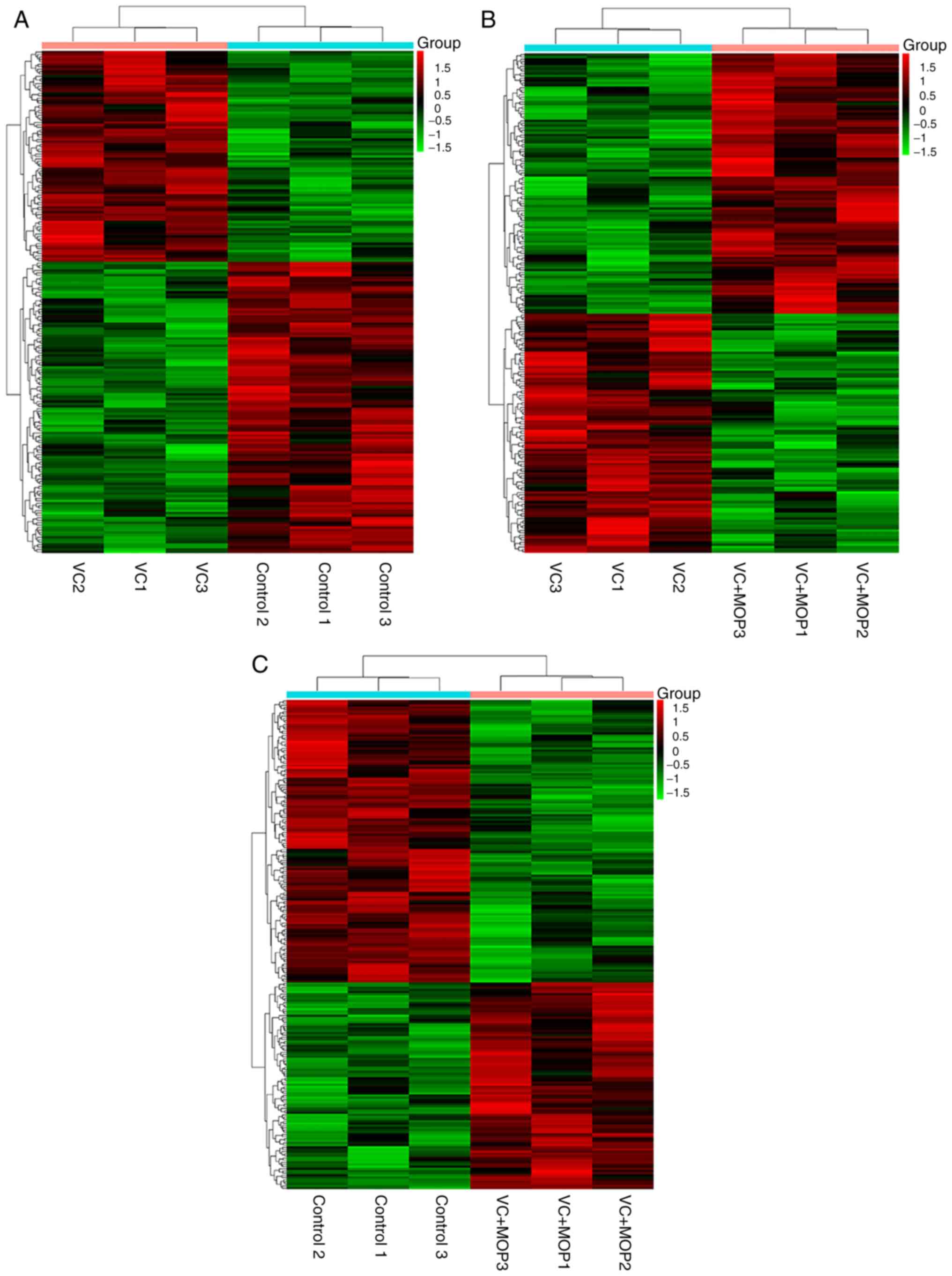
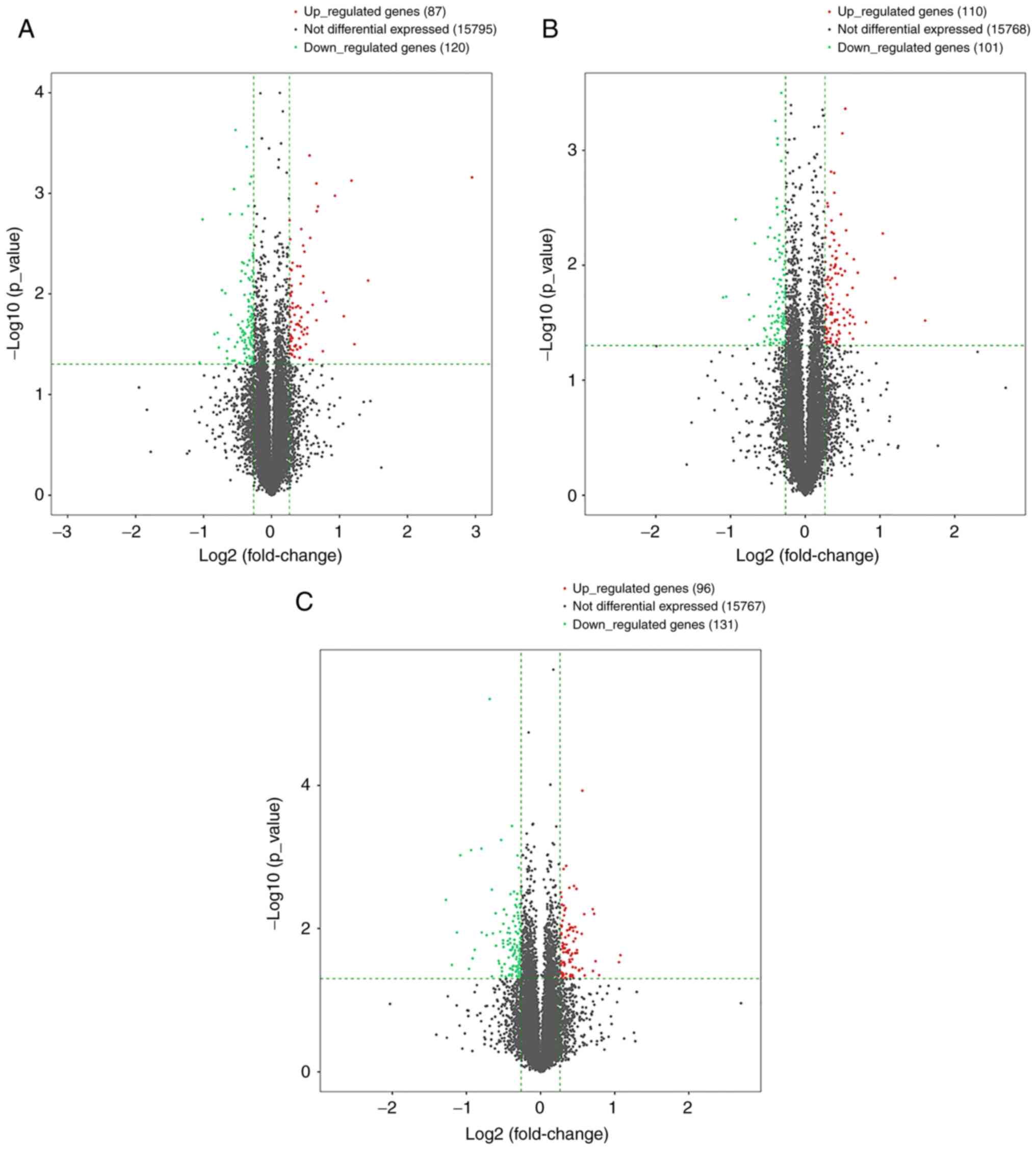
(Fig. 1), scatter plot (Fig. 2) and volcano plot (Fig. 3) analyses of the DE genes are
presented. Overall, there were 144 DE mRNAs and 63 DE lncRNAs in
the VC group vs. the Control group, 63 DE mRNAs and 148 DE lncRNAs
in the VC + MOP group vs. the VC group and 173 DE mRNAs and 54 DE
lncRNAs in the VC + MOP group vs. the Control group. The most
significantly upregulated DE genes in VC group vs. the Control
group, VC + MOP group vs. the VC group, and VC + MOP group vs. the
Control group were TMEM255B, actin g 2 smooth muscle and
calsyntenin 2, respectively. The most significantly downregulated
DE genes in VC group vs. the Control group, VC + MOP group vs. the
VC group, and VC + MOP group vs. the Control group were
AABR07050146.1, YIPF7 and ATP-dependent helicase ATRX
chromatin remodeler, respectively.
 | Table IITop 10 upregulated differentially
expressed genes between groups by RNA-sequencing analysis. |
Table II
Top 10 upregulated differentially
expressed genes between groups by RNA-sequencing analysis.
| A, VC vs.
Control |
|---|
| Tract ID | Gene name | Gene type | Fold-change | P-value |
|---|
|
ENSRNOG00000026336 |
TMEM255B | Protein-coding | 7.72 |
6.9x10-4 |
|
ENSRNOG00000006494 | TUBG2 | Protein coding | 2.68 |
7.4x10-3 |
|
ENSRNOG00000054954 | NILR1 | Protein coding | 2.33 |
3.2x10-2 |
|
ENSRNOG00000026497 | PIGC | Protein coding | 2.26 |
7.5x10-4 |
|
ENSRNOG00000051890 |
LOC102549170 | lincRNA | 2.09 |
1.7x10-2 |
|
ENSRNOG00000016143 | GPR162 | Protein coding | 1.91 |
1.1x10-3 |
|
ENSRNOG00000053415 |
AABR07014649.1 | lincRNA | 1.74 |
1.2x10-2 |
|
ENSRNOG00000058993 |
AABR07062183.1 | Protein coding | 1.70 |
9.7x10-3 |
|
ENSRNOG00000002207 | GUF1 | Protein coding | 1.69 |
3.7x10-2 |
|
ENSRNOG00000052173 | RANBP3L | Protein coding | 1.61 |
1.4x10-3 |
| B, VC + MOP vs.
VC |
| Tract ID | Gene name | Gene type | Fold-change | P-value |
|
ENSRNOG00000029401 | ACTG2 | Protein coding | 3.04 |
3.0x10-2 |
|
ENSRNOG00000017669 | CHD1L | Protein coding | 2.30 |
1.3x10-2 |
|
ENSRNOG00000061865 |
AABR07019334.1 | lincRNA | 2.05 |
5.3x10-3 |
|
ENSRNOG00000060166 |
AABR07069067.1 | lincRNA | 1.76 |
3.1x10-2 |
|
ENSRNOG00000059729 |
AC116236.2 | lincRNA | 1.63 |
1.2x10-2 |
|
ENSRNOG00000007335 | CCL11 | Protein coding | 1.58 |
3.2x10-2 |
|
ENSRNOG00000057837 |
AABR07017236.1 | Protein coding | 1.57 |
8.9x10-3 |
|
ENSRNOG00000057135 |
AABR07070714.2 | lincRNA | 1.56 |
4.4x10-2 |
|
ENSRNOG00000042665 |
AABR07007130.2 | Protein coding | 1.56 |
2.8x10-2 |
|
ENSRNOG00000052005 |
AABR07028945.1 | lincRNA | 1.55 |
1.6x10-2 |
| C, VC + MOP vs.
Control |
| Tract ID | Gene name | Gene type | Fold-change | P-value |
|
ENSRNOG00000043085 | CLSTN2 | Protein coding | 2.11 |
2.4x10-2 |
|
ENSRNOG00000023861 | SNAP91 | Protein coding | 2.08 |
2.9x10-2 |
|
ENSRNOG00000059602 |
AABR07072984.1 | lincRNA | 1.73 |
4.5x10-2 |
|
ENSRNOG00000059504 |
AABR07015078.2 | Protein coding | 1.67 |
2.9x10-2 |
|
ENSRNOG00000052211 |
AABR07058167.1 | lincRNA | 1.65 |
6.2x10-3 |
|
ENSRNOG00000052668 | TCF24 | Protein coding | 1.63 |
3.9x10-2 |
|
ENSRNOG00000057070 |
AABR07014350.1 | lincRNA | 1.63 |
5.4x10-3 |
|
ENSRNOG00000034129 |
AABR07061382.1 | Protein coding | 1.51 |
4.5x10-2 |
|
ENSRNOG00000023035 | SMIM8 | Protein coding | 1.50 |
6.3x10-3 |
|
ENSRNOG00000011969 | DOCK9 | Protein coding | 1.48 |
1.2x10-4 |
 | Table IIITotal of 10 downregulated
differentially expressed genes between groups by RNA-sequencing
analysis. |
Table III
Total of 10 downregulated
differentially expressed genes between groups by RNA-sequencing
analysis.
| A, VC vs.
Control |
|---|
| Tract ID | Gene name | Gene type | Fold-change | P-value |
|---|
|
ENSRNOG00000054630 |
AABR07050146.1 | lincRNA | 0.48 |
4.8x10-2 |
|
ENSRNOG00000004517 | IGF1 | Protein coding | 0.49 |
1.8x10-3 |
|
ENSRNOG00000061865 |
AABR07019334.1 | lincRNA | 0.55 |
2.5x10-2 |
|
ENSRNOG00000001300 | P2RX4 | Protein coding | 0.58 |
2.4x10-2 |
|
ENSRNOG00000053160 |
AABR07071659.2 | lincRNA | 0.58 |
3.4x10-2 |
|
ENSRNOG00000009734 | AKR1B10 | Protein coding | 0.60 |
9.2x10-3 |
|
ENSRNOG00000060166 |
AABR07069067.1 | lincRNA | 0.62 |
9.9x10-3 |
|
ENSRNOG00000020945 | MS4A1 | Protein coding | 0.63 |
3.7x10-2 |
|
ENSRNOG00000060865 |
AABR07013167.1 | lincRNA | 0.64 |
4.4x10-2 |
|
ENSRNOG00000056171 |
AC107505.1 | lincRNA | 0.64 |
5.0x10-2 |
| B, VC + MOP vs.
VC |
| Tract ID | Gene name | Gene type | Fold-change | P-value |
|
ENSRNOG00000002224 | YIPF7 | Protein coding | 0.47 |
1.9x10-2 |
|
ENSRNOG00000002207 | GUF1 | Protein coding | 0.48 |
1.9x10-2 |
|
ENSRNOG00000026976 | VOM2R57 | Protein coding | 0.52 |
4.0x10-3 |
|
ENSRNOG00000058242 | EPS8L3 | Protein coding | 0.59 |
1.8x10-2 |
|
ENSRNOG00000048209 | EPS8L3 | Protein coding | 0.60 |
3.0x10-2 |
|
ENSRNOG00000059026 |
AABR07055885.1 | lincRNA | 0.62 |
2.8x10-2 |
|
ENSRNOG00000027142 | OOG1 | Protein coding | 0.63 |
6.5x10-3 |
|
ENSRNOG00000003409 |
NEWGENE_1565644 | Protein coding | 0.64 |
4.9x10-2 |
|
ENSRNOG00000021010 | ARL2 | Protein coding | 0.68 |
3.5x10-2 |
|
ENSRNOG00000054516 |
AABR07056680.1 | lincRNA | 0.68 |
4.6x10-2 |
| C, VC + MOP vs.
Control |
| Tract ID | Gene name | Gene type | Fold-change | P-value |
|
ENSRNOG00000046897 | ATRX | Protein coding | 0.41 |
4.0x10-3 |
|
ENSRNOG00000008074 | CYP11A1 | Protein coding | 0.44 |
3.2x10-2 |
|
ENSRNOG00000053160 |
AABR07071659.2 | lincRNA | 0.46 |
1.1x10-2 |
|
ENSRNOG00000004517 | IGF1 | Protein coding | 0.47 |
9.5x10-4 |
|
ENSRNOG00000002079 | MAPK10 | Protein coding | 0.51 |
3.7x10-2 |
|
ENSRNOG00000060545 |
AC125688.1 | lincRNA | 0.52 |
8.0x10-4 |
|
ENSRNOG00000036641 |
LOC689065 | protein coding | 0.53 |
2.6x10-2 |
|
ENSRNOG00000061906 |
AABR07024261.1 | protein coding | 0.54 |
2.0x10-2 |
|
ENSRNOG00000057691 |
AC112557.1 | lincRNA | 0.58 |
7.7x10-4 |
|
ENSRNOG00000009734 | AKR1B10 | protein coding | 0.58 |
1.1x10-2 |
GO and KEGG analyses of DE genes
To elucidate the functions of the DE genes, GO term
enrichment and KEGG pathway analyses were performed. The GO
functional annotations were classified into three categories:
Molecular function, cellular components and biological processes.
The top 10 enriched GO terms of DE genes, both upregulated and
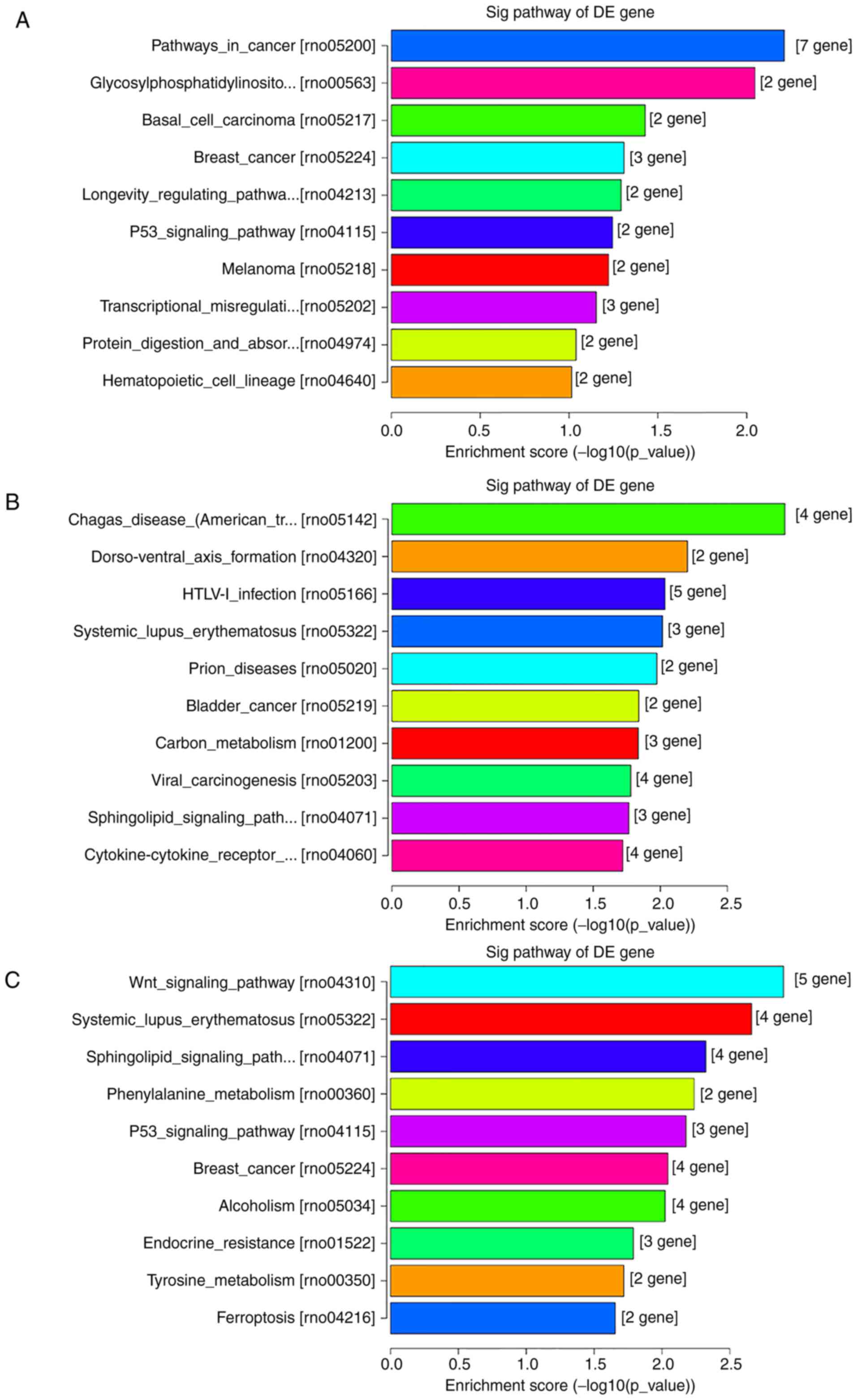
downregulated (Figs. 4 and 5) and the top 10 enriched KEGG pathways of
DE genes, both upregulated and downregulated, (Fig. 6) are shown.
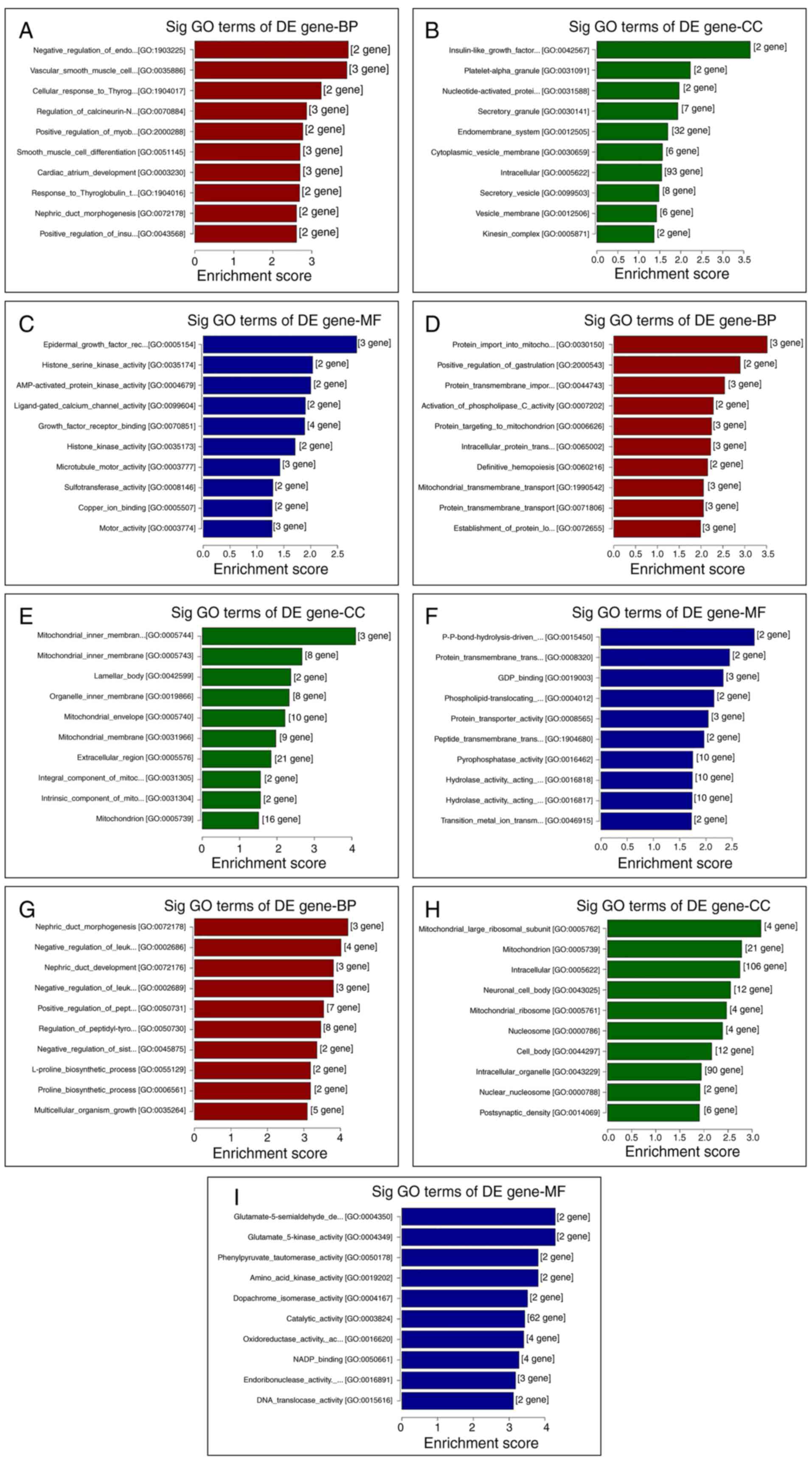 | Figure 4GO analysis of the top 10 DE genes
between different groups, as ranked by their enrichment score. Top
10 (A) BP, (B) CC and (C) MF terms between the VC and control
groups. Top 10 (D) BP, (E) CC and (F) MF terms between the VC + MOP
and VC groups. Top 10 (G) BP, (H) CC and (I) MF terms between the
VC + MOP and control groups. DE genes with fold-change >1.2 and
P≤0.05 were used. BP, biological process; CC, cellular component;
MF, molecular function; GO, Gene Ontology; DE, differentially
expressed; Sig, significant; VC, varicocele; MOP, Morinda
officinalis polysaccharide. |
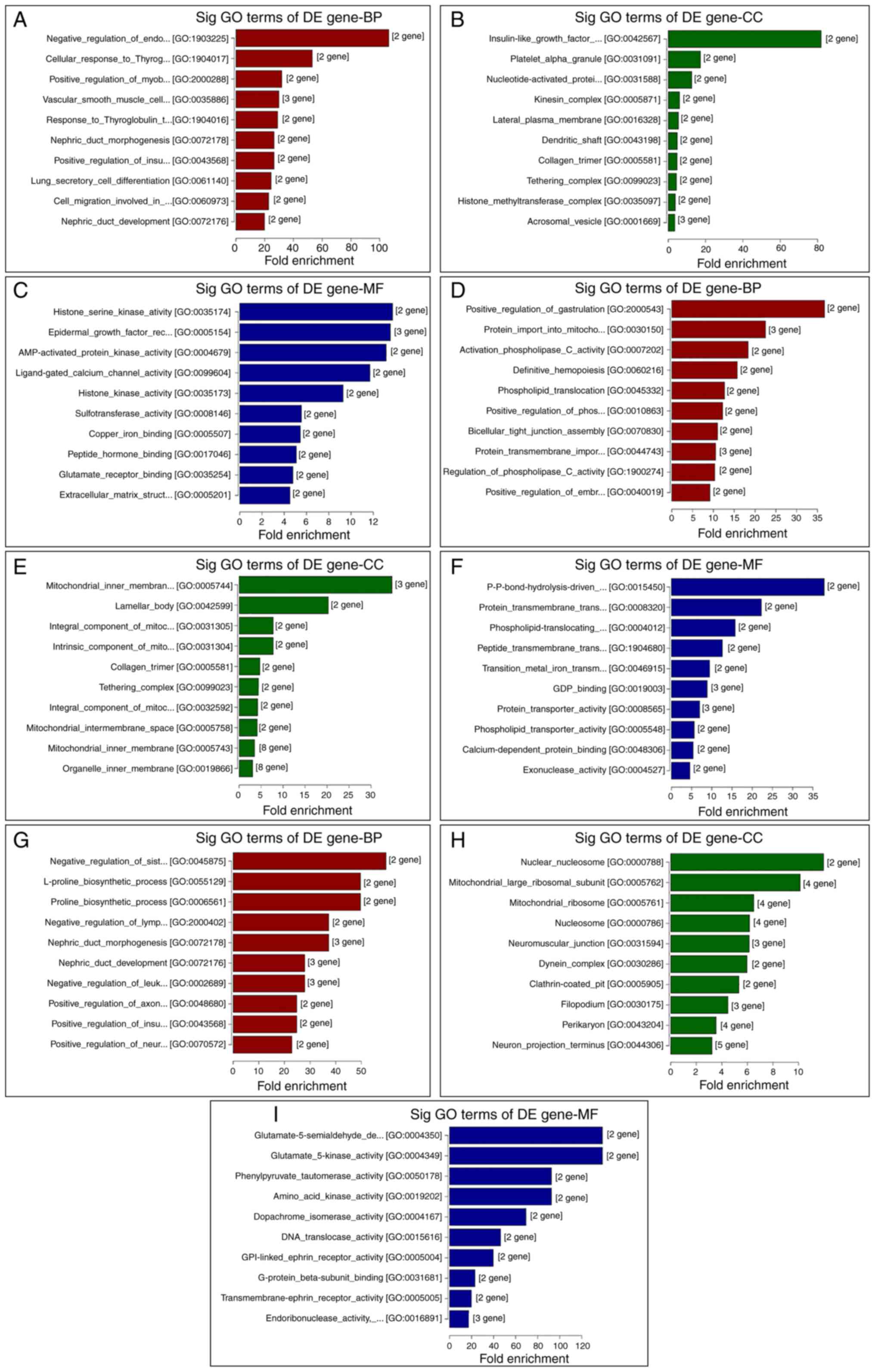 | Figure 5GO analysis of the top 10 DE genes
between different groups, as ranked by fold-enrichment. Top 10 (A)
BP, (B) CC and (C) MF between the VC and control groups. Top 10 (D)
BP, (E) CC and (F) MF terms between the VC + MOP and VC groups. Top
10 (G) BP, (H) CC and (I) MF terms, respectively, between the VC +
MOP and control groups. DE genes with fold-change >1.2 and
P≤0.05 were used. BP, biological process; CC, cellular component;
MF, molecular function; GO, Gene Ontology; DE, differentially
expressed; Sig, significant; VC, varicocele; MOP, Morinda
officinalis polysaccharide. |
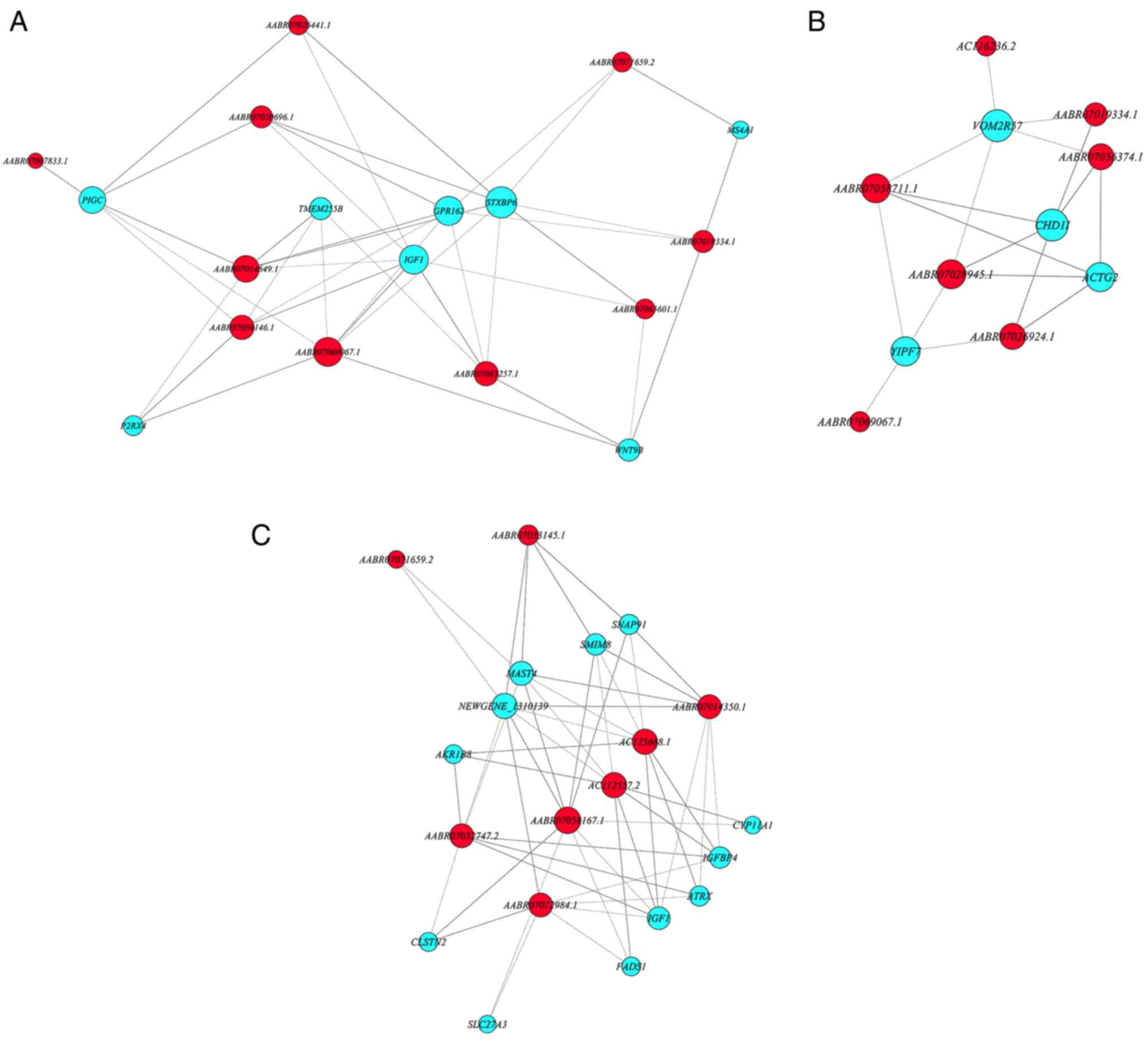
Co-expression network of the chosen DE
mRNAs and lncRNAs
Correlation coefficients of selected DE mRNAs
(protein-coding genes) and lncRNAs (non-protein-coding genes) were
calculated to build a co-expression network (Pearson correlation
coefficient ≥0.8; P≤0.05; false discovery rate, ≤1; Fig. 7). These DE mRNAs and lncRNAs
presented in Fig. 7 were selected
according to their function based on the GO term enrichment result,
but not the total DE genes. The enriched DE genes were selected
based on the GO terms the present study were interested in, such as
the ‘response to steroid hormone’, ‘oxidation-reduction process’,
‘positive regulation of MAPK cascade’ and ‘DNA repair’ terms.
Validation of DE mRNAs and lncRNAs
through RT-qPCR
The DE mRNAs and lncRNAs identified by RNA-Seq were
validated using RT-qPCR. The results are presented in Table IV and Fig. 8. There were six DE mRNAs and four DE
lncRNAs in the VC group vs. the Control group, one DE mRNA and four
DE lncRNAs in the VC + MOP group vs. the VC group and four DE mRNAs
and two DE lncRNAs in the VC + MOP group vs. the Control group.
Varicocele and MOP treatment appeared to have opposing effects on
the expression levels of YIPF7, AABR07007833.1,
AABR07014649.1 and AABR07069067.1. To be specific,
the varicocele increased the expression levels of YIPF7,
AABR07007833.1 and AABR07014649.1, and decreased the
expression level of AABR07069067.1, while MOP treatment
reversed these changes.
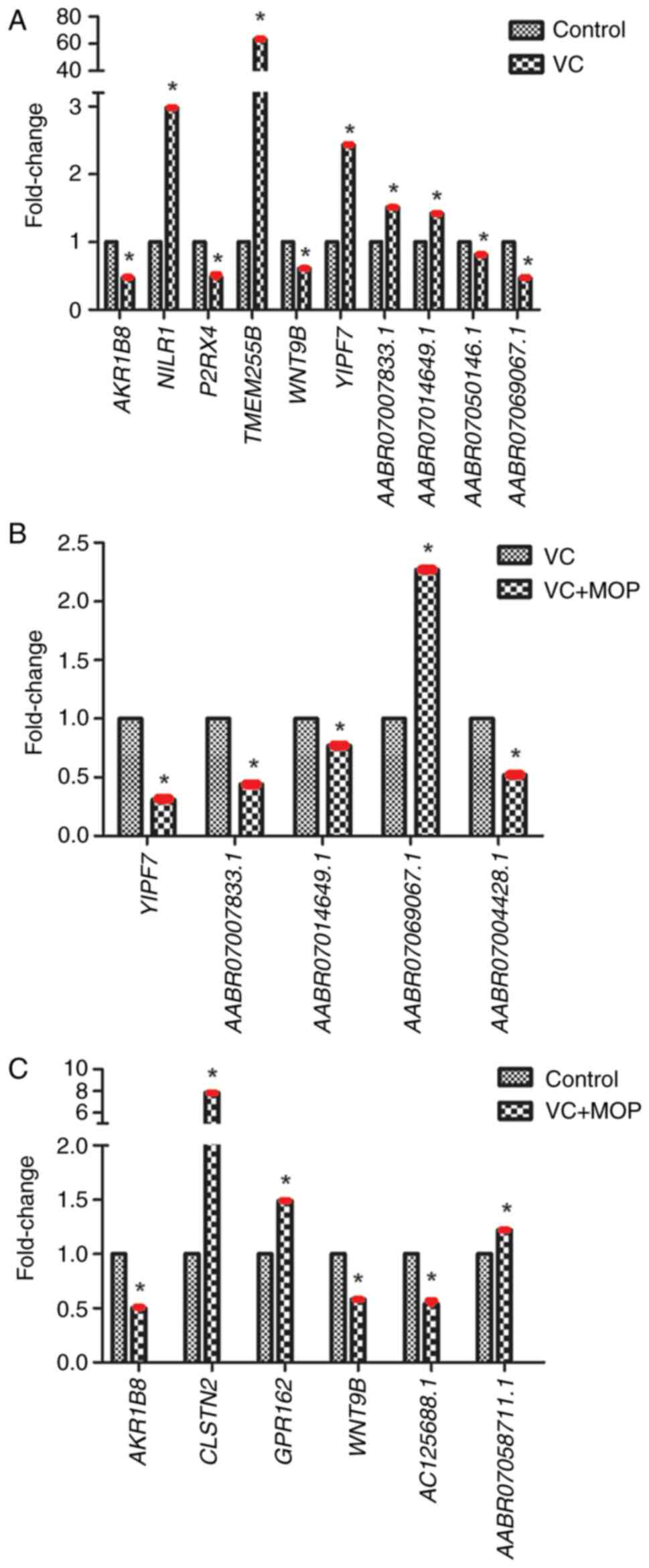 | Figure 8Expression levels of DE genes
assessed by reverse transcription-quantitative PCR. (A) DE genes
between the VC and control groups. (B) DE genes between the VC +
MOP and VC groups. (C) DE genes between the VC + MOP and control
groups. *P≤0.05 vs. respective control groups. All data
are presented as mean ± SD, bars indicate SD and are shown in red,
n=3. VC, varicocele; MOP, Morinda officinalis
polysaccharide; DE, differentially expressed; AKR1B8, Aldo-keto
reductase family 1, member B8; CLSTN2, calsyntenin 2; GPR162, G
protein-coupled receptor 162; NILR1, leukocyte immunoglobulin like
receptor B2; P2RX4, purinergic receptor P2X 4; TMEM255B,
transmembrane protein 225B; WNT9B, Wnt family member 9B; YIPF7,
Yip1 domain family member 7. |
 | Table IVExpression of differentially
expressed genes by reverse transcription-quantitative PCR. |
Table IV
Expression of differentially
expressed genes by reverse transcription-quantitative PCR.
| A, VC vs.
Control |
|---|
| Gene name | Gene type | Mean fold-change ±
SD | P-value |
|---|
| AKR1B8 | Protein coding |
0.47±1.1x10-2 | 0.024 |
| NILR1 | Protein coding |
2.98±6.2x10-4 | 0.044 |
| P2RX4 | Protein coding |
0.48±4.1x10-2 | 0.041 |
| TMEM255B | Protein coding |
63.34±1.4x10-2 | 0.044 |
| WNT9B | Protein coding |
0.61±1.0x10-3 | 0.026 |
| YIPF7 | Protein coding |
2.43±6.3x10-3 | 0.031 |
| AABR07007833.1 | lincRNA |
1.51±2.6x10-5 | 0.033 |
| AABR07014649.1 | lincRNA |
1.42±1.5x10-3 | 0.025 |
| AABR07050146.1 | lincRNA |
0.81±5.1x10-5 | 0.035 |
| AABR07069067.1 | lincRNA |
0.47±6.5x10-4 | 0.014 |
| B, VC + MOP vs.
VC |
| Gene name | Gene type | Mean fold-change ±
SD | P-value |
| YIPF7 | Protein coding |
0.31±7.3x10-3 | 0.025 |
|
AABR07007833.1 | lincRNA |
0.44±3.9x10-5 | 0.0092 |
|
AABR07014649.1 | lincRNA |
0.77±1.0x10-3 | 0.0011 |
|
AABR07069067.1 | lincRNA |
2.27±7.3x10-4 | 0.0047 |
|
AABR07004428.1 | lincRNA |
0.52±1.4x10-3 | 0.049 |
| C, VC + MOP vs.
Control |
| Gene name | Gene type | Mean fold-change ±
SD | P-value |
| AKR1B8 | Protein coding |
0.50±9.4x10-3 | 0.0072 |
| CLSTN2 | Protein coding |
7.82±1.5x10-3 | 0.023 |
| GPR162 | Protein coding |
1.49±2.0x10-4 | 0.038 |
| WNT9B | Protein coding |
0.58±1.1x10-3 | 0.030 |
|
AC125688.1 | lincRNA |
0.54±3.0x10-2 | 0.0022 |
|
AABR07058711.1 | lincRNA |
1.22±2.2x10-4 | 0.040 |
Discussion
In our previous study, varicocele induced
reproductive dysfunction in male rats via mechanisms such as
destruction of the seminiferous epithelium and TJ structure,
downregulation of TJ proteins (occludin, claudin-11 and zona
occludens protein 1), deregulation of hormone levels and an
increase in cytokine (TGF-β3 and TNF-α) levels. MOP repaired
varicocele-induced damage and promoted spermatogenesis (the most
effective dose was found to be 300 mg/kg) (17). However, the molecular mechanisms
underlying the pathophysiology of varicocele and the therapeutic
effect of MOP are still unknown. In the present study, mRNA and
lncRNA sequencing analyses, combined with validation of DE genes
through RT-qPCR, were performed to bridge these data gaps.
According to GO results, the pathophysiology of
varicocele may be associated with ‘epidermal growth factor receptor
binding’, ‘ligand-gated calcium channel activity’, ‘growth factor
receptor binding’, ‘histone kinase activity’ and ‘microtubule motor
activity’. Notably, numerous DE genes between the VC and control
groups were enriched in cancer-related pathways, such as the ‘p53
signaling pathway’, which is downregulated in breast cancer and
melanoma; this warrants further research and validation.
According to the KEGG pathway analysis,
‘cytokine-cytokine receptor interaction’, ‘Wnt signaling pathway’
and ‘p53 signaling pathway’ were all implicated in the
varicocele-therapeutic effect of MOP. In our previous study, the
levels of TGF-β3 and TNF-α were upregulated in experimental rat
left testicular tissue and downregulated following MOP treatment,
potentially through the cytokine-cytokine receptor interaction
pathway (17). The Wnt signaling
pathway plays a notable role in the normal physiology and pathology
of the male reproductive system. The Wnt/β-catenin pathway is
involved in the annexin 5-mediated stimulation of testosterone
synthesis (35), and its
dysregulation is linked to the development of granulosa cell tumors
in the testis (36). Additionally,
the activation of Wnt/β-catenin signaling in Sertoli cells results
in germ cell loss and seminiferous tubule degeneration (37). Moreover, the Wnt/β-catenin signaling
pathway facilitates the proliferation of spermatogonial stem cells
in the testis (38). Thus, proper
regulation of Wnt/β-catenin signaling is necessary for adult
spermatogenesis, and its disruption may result in infertility
(39). The results of the present
study provide compelling evidence for the involvement of
Wnt/β-catenin signaling in varicocele progression and the
therapeutic effect of MOP in varicocele; this evidence is worthy of
further research.
The RT-qPCR analysis revealed that one coding gene,
YIPF7, and three non-coding genes, AABR07007833.1,
AABR07014649.1 and AABR07069067.1, were DE between
the VC and Control groups, and between the VC + MOP and VC groups.
Varicocele and MOP treatment appeared to have opposing effects on
the expression of these genes, implying that these genes may play a
role in varicocele pathophysiology and the repair effect of MOP.
Specifically, varicocele increased the expression levels of
YIPF7, AABR07007833.1 and AABR07014649.1, and
decreased the expression level of AABR07069067.1, while MOP
treatment reversed these effects. The YIP1 family of proteins are
involved in protein transport between the endoplasmic reticulum and
Golgi apparatus (40), as well as
the regulation of membrane dynamics (41). Decreasing the expression of
YIPF7 enhances the intestinal inflammatory response and
upregulates the TNF mRNA level (42). The change in YIPF7 expression
may be associated with the testicular inflammatory response and the
fluctuation of the TNF-α expression level induced by varicocele and
MOP. This finding is in accordance with the results of our previous
study; thus, we hypothesize that YIPF7 is involved in the
process of varicocele development by increasing the TNF-α level in
the left testicular tissue, which MOP treatment is able to decrease
(17). Due to the lack of extensive
research on lncRNA, the functions of AABR07007833.1,
AABR07014649.1 and AABR07069067.1 remain elusive. In
the present study, according to the co-expression network analysis,
these non-coding genes were positively associated with
phosphatidylinositol glycan anchor biosynthesis class C
insulin-like growth factor 1 (IGF1), P2RX4,
TMEM255B and WNT9B, as well as others.
Aldo-keto reductase family 1, member B8
(AKR1B8), leukocyte immunoglobulin-like receptor B2
(NILR1), P2RX4, TMEM255B and WNT9B were
also DE between the VC and control groups. TMEM255B, also
known as FAM70B, encodes the transmembrane protein 255b,
which has been proposed as a prognostic marker for muscle-invasive
bladder cancer (43). The
expression level of TMEM255B in the VC group was ~63-fold
higher compared with that of the control group. Such a large
difference in expression levels suggests that TMEM255B could
be used as a novel diagnostic maker for varicocele; however,
further clinical research is required to confirm this hypothesis.
WNT9B takes part in the Wnt/β-catenin signaling pathway,
described as aforementioned. P2RX4, also known as
P2X4 or P2X4R, encodes the ATP-gated P2RX4 ion
channel. According to a previous study, extracellular ATP is a
danger molecule for peritubular cells and aggravates inflammatory
responses in the testis (44).
P2RX4 is involved in pain processing (45-47),
and its downregulation has been shown to moderate allergen-induced
airway inflammation (48). Although
the function of P2RX4 in the male reproductive system is unclear,
according to its diverse biological activities under physiological
and pathological conditions (44-49),
it is hypothesized that changes in P2RX4 expression levels are
involved in varicocele pathogenesis via the stimulation of
inflammatory responses and disruption of transmembrane ion channel
function.
In the present study, RNA sequencing and RT-qPCR
were performed on rat testicular tissue to reveal the molecular
mechanisms underlying varicocele pathophysiology and the
restorative effect of MOP on varicocele-induced damage in male
reproductive systems. The results of GO and KEGG pathway enrichment
analyses showed that ‘ligand-gated calcium channel activity’,
‘cytokine-cytokine receptor interaction’ and the ‘Wnt signaling
pathway’ may all be implicated to underlie this effect. RT-qPCR
confirmed that YIPF7 is upregulated in varicocele and
downregulated by MOP. Based on its diverse biological activities
and intimate connection to TNF-α, YIPF7 is considered a key
mediator of varicocele pathogenesis and therapeutic effects of MOP.
Furthermore, differential expression of AKR1B8,
NILR1, P2RX4, TMEM255B and WNT9B was
detected between the VC group and the control group.
TMEM255B may be a potential novel diagnostic marker for
varicocele; the role of WNT9B and P2RX4 in varicocele
is possibly mediated by the activation of Wnt signaling and the
regulation of transmembrane ion channels and the inflammatory
response, respectively.
In summary, the present study provides a foundation
for understanding the molecular basis of varicocele pathophysiology
and the varicocele-therapeutic effect of MOP, offers insights into
novel strategies for varicocele diagnosis and treatment, and
suggests directions for further study.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The Training Program
Foundation for the Talents of Health and Family Planning Commission
in Fujian Province (grant no. 2018-1-74), The Start-up Foundation
of Fujian Medical University (grant no. 2017XQ1005), The Research
Foundation for High-Level Talents of Fujian Medical University
(grant. no. XRCZX2017033), The Natural Science Foundation of Fujian
Province (grant no. 2017J01819) and The Co-construction Science
Foundation of National Health and Family Planning Commission-Joint
Program for Tackling Key Problems of Health and Education in Fujian
Province (grant no. WKJ2016-2-35).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding authors upon reasonable
request. The datasets generated and analyzed during the current
study are available in the Gene Expression Omnibus repository,
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi
(accession no. GSE139447; secure token: sfaxocskhpoxnix).
Authors' contributions
LZ performed the experiments and the statistical
analysis, and drafted the manuscript. WW and XZ conceived the study
and participated in its design and coordination. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by The Animal Approval
Committee of Fujian Medical University (approval no.
SYXK-2012-0001).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Leslie SW, Sajjad H and Siref LE:
Varicocele. In: StatPearls. StatPearls Publishing, Treasure Island,
FL, 2021.
|
|
2
|
Wang NN, Dallas K, Li S, Baker L and
Eisenberg ML: The association between varicocoeles and vascular
disease: An analysis of U.S. Claims data. Andrology. 6:99–103.
2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Reesink DJ, Huisman PM, Wiltink J, Kruger
AE and Lock TMT: Sneeze and pop: A ruptured varicocele; analysis of
literature, guided by a well-documented case-report. BMC Urol.
19(14)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sigalos JT and Pastuszak AW: Chronic
orchialgia: Epidemiology, diagnosis and evaluation. Transl Androl
Urol. 6 (Suppl 1):S37–S43. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Afshar A, Aliaghaei A, Nazarian H,
Abbaszadeh HA, Naserzadeh P, Fathabadi FF, Abdi S, Raee P,
Aghajanpour F, Norouzian M and Abdollahifar MA: Curcumin-loaded
iron particle improvement of spermatogenesis in azoospermic mouse
induced by long-term scrotal hyperthermia. Reprod Sci. 28:371–380.
2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jarow JP: Effects of varicocele on male
fertility. Hum Reprod Update. 7:59–64. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mehta A and Goldstein M: Microsurgical
varicocelectomy: A review. Asian J Androl. 15:56–60.
2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ahlberg NE, Bartley O and Chidekel N:
Right and left gonadal veins. An anatomical and statistical study.
Acta Radiol Diagn (Stockh). 4:593–601. 1966.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bach PV, Najari BB and Goldstein M:
Varicocele-a case for early intervention. F1000Res.
5(1792)2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chiba K, Ramasamy R, Lamb DJ and Lipshultz
LI: The varicocele: Diagnostic dilemmas, therapeutic challenges and
future perspectives. Asian J Androl. 18:276–281. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fretz PC and Sandlow JI: Varicocele:
Current concepts in pathophysiology, diagnosis, and treatment. Urol
Clin North Am. 29:921–937. 2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Luo DY, Yang G, Liu JJ, Yang YR and Dong
Q: Effects of varicocele on testosterone, apoptosis and expression
of StAR mRNA in rat Leydig cells. Asian J Androl. 13:287–291.
2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Saypol DC, Howards SS, Turner TT and
Miller ED Jr: Influence of surgically induced varicocele on
testicular blood flow, temperature, and histology in adult rats and
dogs. J Clin Invest. 68:39–45. 1981.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Pastuszak AW and Wang R: Varicocele and
testicular function. Asian J Androl. 17:659–667. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ozbek E, Yurekli M, Soylu A, Davarci M and
Balbay MD: The role of adrenomedullin in varicocele and impotence.
BJU Int. 86:694–698. 2000.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kim IT, Park HJ, Nam JH, Park YM, Won JH,
Choi J, Choe BK and Lee KT: In-vitro and in-vivo anti-inflammatory
and antinociceptive effects of the methanol extract of the roots of
Morinda officinalis. J Pharm Pharmacol. 57:607–615. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang L, Zhao X, Wang F, Lin Q and Wang W:
Effects of morinda officinalis polysaccharide on experimental
varicocele rats. Evid Based Complement Alternat Med.
2016(5365291)2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Marinov GK: On the design and prospects of
direct RNA sequencing. Brief Funct Genomics. 16:326–335.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kazimierczyk M, Kasprowicz MK, Kasprzyk ME
and Wrzesinski J: Human long noncoding RNA interactome: Detection,
characterization and function. Int J Mol Sci.
21(1027)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li L, Zhuang Y, Zhao X and Li X: Long
non-coding RNA in neuronal development and neurological disorders.
Front Genet. 9(744)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hu Y, Lin J, Fang H, Fang J, Li C, Chen W,
Liu S, Ondrejka S, Gong Z, Reu F, et al: Targeting the
MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in
multiple myeloma. Leukemia. 32:2250–2262. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mirza AH, Kaur S and Pociot F: Long
non-coding RNAs as novel players in β cell function and type 1
diabetes. Hum Genomics. 11(17)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang Y, Hou YM, Gao F, Xiao JW, Li CC and
Tang Y: lncRNA GAS5 regulates myocardial infarction by targeting
the miR-525-5p/CALM2 axis. J Cell Biochem. 120:18678–18688.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Morlando M and Fatica A: Alteration of
epigenetic regulation by long noncoding RNAs in cancer. Int J Mol
Sci. 19(570)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang Y, Junjie P, Sanjun C and Ma Y: Long
non-coding RNAs in colorectal cancer: Progression and future
directions. J Cancer. 8:3212–3225. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Loewen G, Jayawickramarajah J, Zhuo Y and
Shan B: Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol.
7(90)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang X, Ruan Y, Wang X, Zhao W, Jiang Q,
Jiang C, Zhao Y, Xu Y, Sun F, Zhu Y, et al: Long intragenic
non-coding RNA lincRNA-p21 suppresses development of human prostate
cancer. Cell Prolif. 50(e12318)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Dong H, Zhang Q, Li Y, Li L, Lan W, He J,
Li H, Xiong Y and Qin W: Extraction, characterization and
antioxidant activities of polysaccharides of Chuanminshen
violaceum. Int J Biol Macromol. 86:224–232. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xu SY, Liu JP, Huang X, Du LP, Shi FL,
Dong R, Huang XT, Zheng K, Liu Y and Cheong KL:
Ultrasonic-microwave assisted extraction, characterization and
biological activity of pectin from jackfruit peel. Lwt. 90:577–582.
2018.
|
|
31
|
Turner TT: The study of varicocele through
the use of animal models. Hum Reprod Update. 7:78–84.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang T, Zheng T, Wang C, Zhang W, Jia D,
Wang R and Qiao B: Effects of Wnt/β-catenin signaling pathway and
star D7 on testosterone synthesis. Acta Endocrinol (Buchar).
14:155–162. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Boyer A, Paquet M, Lague MN, Hermo L and
Boerboom D: Dysregulation of WNT/CTNNB1 and PI3K/AKT signaling in
testicular stromal cells causes granulosa cell tumor of the testis.
Carcinogenesis. 30:869–878. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Boyer A, Hermo L, Paquet M, Robaire B and
Boerboom D: Seminiferous tubule degeneration and infertility in
mice with sustained activation of WNT/CTNNB1 signaling in sertoli
cells. Biol Reprod. 79:475–485. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Takase HM and Nusse R: Paracrine
Wnt/β-catenin signaling mediates proliferation of undifferentiated
spermatogonia in the adult mouse testis. Proc Natl Acad Sci USA.
113:E1489–E1497. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kerr GE, Young JC, Horvay K, Abud HE and
Loveland KL: Regulated Wnt/beta-catenin signaling sustains adult
spermatogenesis in mice. Biol Reprod. 90(3)2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Tanimoto K, Suzuki K, Jokitalo E, Sakai N,
Sakaguchi T, Tamura D, Fujii G, Aoki K, Takada S, Ishida R, et al:
Characterization of YIPF3 and YIPF4, cis-golgi localizing yip
domain family proteins. Cell Struct Funct. 36:171–185.
2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kranjc T, Dempsey E, Cagney G, Nakamura N,
Shields DC and Simpson JC: Functional characterisation of the YIPF
protein family in mammalian cells. Histochem Cell Biol.
147:439–451. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kim SW, Jung YS, Ahn JB, Shin ES, Jang HW,
Lee HJ, Kim T, Kim DY, Bang D, Kim WH and Cheon JH: Identification
of genetic susceptibility loci for intestinal Behcet's disease. Sci
Rep. 7(39850)2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kang HW, Yoon HY, Ha YS, Kim WT, Kim YJ,
Yun SJ, Lee SC and Kim WJ: FAM70B as a novel prognostic marker for
cancer progression and cancer-specific death in muscle-invasive
bladder cancer. Korean J Urol. 53:598–606. 2012.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Walenta L, Fleck D, Fröhlich T, von
Eysmondt H, Arnold GJ, Spehr J, Schwarzer JU, Köhn FM, Spehr M and
Mayerhofer A: ATP-mediated events in peritubular cells contribute
to sterile testicular inflammation. Sci Rep. 8(1431)2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Jurga AM, Piotrowska A, Makuch W,
Przewlocka B and Mika J: Blockade of P2X4 receptors inhibits
neuropathic pain-related behavior by preventing MMP-9 activation
and, consequently, pronociceptive interleukin release in a rat
model. Front Pharmacol. 8(48)2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Tsuda M, Masuda T, Tozaki-Saitoh H and
Inoue K: P2X4 receptors and neuropathic pain. Front Cell Neurosci.
7(191)2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Lalisse S, Hua J, Lenoir M, Linck N,
Rassendren F and Ulmann L: Sensory neuronal P2RX4 receptors
controls BDNF signaling in inflammatory pain. Sci Rep.
8(964)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zech A, Wiesler B, Ayata CK, Schlaich T,
Dürk T, Hoßfeld M, Ehrat N, Cicko S and Idzko M: P2rx4 deficiency
in mice alleviates allergen-induced airway inflammation.
Oncotarget. 7:80288–80297. 2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Sadovnick AD, Gu BJ, Traboulsee AL,
Bernales CQ, Encarnacion M, Yee IM, Criscuoli MG, Huang X, Ou A,
Milligan CJ, et al: Purinergic receptors P2RX4 and P2RX7 in
familial multiple sclerosis. Hum Mutat. 38:736–744. 2017.PubMed/NCBI View Article : Google Scholar
|