1. Introduction
Multiple Sclerosis (MS) is a chronic inflammatory
demyelinating disease of the central nervous system (CNS), commonly
affecting young adults. MS usually manifests as a
relapsing-remitting (RR) process, predominantly characterized by
inflammatory demyelination that secondarily evolves to a
progressive stage with neurodegeneration, gliosis and accumulating
disability. Although the etiology of MS is largely unknown, it is
considered primarily an autoimmune disease, in which activated
myelin-specific T-cells, migrate from the periphery to the CNS, by
crossing the blood brain barrier (BBB) inducing the formation of
new inflammatory demyelinating lesions (1,2).
Clinically isolated syndrome (CIS) describes a clinical episode of
at least 24 h, suggestive of an inflammatory demyelinating disorder
of the CNS. CIS usually occurs in young adults and represents a
monophasic episode, isolated in time, and usually isolated in
space, affecting optic nerves, the brainstem, or the spinal cord,
while there are no signs of fever or infection. Although patients
usually recover from their presenting episode, CIS is often the
first manifestation of MS. The course of MS after CIS is variable:
after 15-20 years, one third of patients have a benign course with
minimal or no disability, while half will have developed secondary
progressive MS with increasing disability (3,4). MS
and CIS clinical studies, need to be accompanied by sensitive and
reliable imaging methods, in order to investigate the specific
pathological alterations of the white matter (WM). Imaging methods
for MS include optical computed tomography and coherent anti-Stokes
Raman scattering microscopy (5).
Magnetic Resonance Imaging (MRI) is the modality of choice for
diagnosis and monitoring MS pathology, by exploiting both
conventional and quantitative protocols (6).
Conventional MR techniques for MS and CIS imaging,
include T1-w images since in this protocol lesions are usually
depicted by lower signal intensity compared to the surrounding
tissues, while T1 severe hypointense lesions (black holes)
represent the most severe stage of MS lesions with irreversible
axonal loss (5,7). Lesions in earlier stages of the
disease can be captured using Gd enhanced T1-w MRI sequences, based
on the fact that contrast agent (CA) uptake indicates premature
stage of inflammation and blood brain barrier (BBB) disruption.
Furthermore, T2-w MRI sequences are also sensitive in detecting
lesions, since these are demonstrated as bright areas in a dark
background. T2 hyperintense lesions represent tissues that are
characterized by edema, inflammation, demyelination, axonal loss
and gliosis. Sensitivity is further enhanced by using the
fluid-attenuated inversion recovery (FLAIR) protocol, which can be
used for highlighting subcortical and periventricular lesions by
suppressing T2 signal from cerebrospinal fluid (CSF) (8). The aforementioned techniques
constitute the gold standard for MS and CIS diagnosis and
monitoring (9).
However, conventional MRI do not provide sufficient
sensitivity that could enable early diagnosis or appropriate
specificity to predict disease severity. Quantitative MRI
techniques provide enhanced insights in disease severity and tissue
damage. These techniques include: i) MR spectroscopy which provides
a non-invasive method to examine the biochemical changes in MS
(10); ii) magnetization transfer
imaging which offers improved sensitivity and specificity for MS
studies (11,12); iii) diffusion weighted imaging
(DWI) and diffusion tensor imaging (DTI) which are quantitative MRI
techniques, providing information on size, integrity, geometry, and
orientation of tissue fibres by capturing the motion of tissue
water (13); iv) dynamic contrast
enhanced MRI which enables quantification of BBB disruption, a
therapeutic target in MS (14);
and v) dynamic susceptibility contrast MRI that provides
quantitative maps of cerebral blood flow, cerebral blood volume and
temporal parameters such as mean transit time by intravenous
administration of CA (14).
Conventional clinical protocols for MS diagnosis and
monitoring, utilize imaging data and measurements, such as signal
intensities and volumetric results from user defined regions of
interest, in order to compare healthy and MS subjects or to examine
longitudinal changes during therapy, and evaluate the clinical
outcome. However, these results are prone to errors due to
inter-observer variability, while the lack of ability to compare
studies from different modalities is a major limitation. Moreover,
procedures that involve human interplay, besides that are time
consuming and employ human experts, these may be characterized by
high inter observer variability that may hinder the quality of the
final results. Therefore, during the last years artificial
intelligence (AI) techniques have opened new horizons in computer
aided diagnosis (CAD) systems, by automatically generalizing rules
and patterns that exist in labeled imaging data, while by utilizing
this information they are able to generate predictions and
classifications on independent datasets that were not used in the
model training process (15).
Furthermore, segmentation techniques in MS imaging
are gaining ground during the last years, considering the necessity
of accurate algorithms for automatic delineation of anatomical
structures (16). Performing these
tasks manually is time-consuming and prone to errors, thus there is
a lot of interest to accomplish this task using automated computer
algorithms, towards increased accuracy and precision, while at the
same time minimize human involvement.
Finally, computer algorithms able to assess image
quality of the acquired data, as well as methods that are able to
compute optimal protocol parameters prior to image acquisition, are
tools that can save time and provide noise free and qualitative
data with increased diagnostic information (17). To this purpose, AI techniques
constitute a valuable methodology for assessing MR image quality as
well as determining the necessity to repeat the acquisition, while
there are attempts to provide algorithms aiming to image protocol
optimization.
The focus of this review paper, is to study the
current literature regarding MR imaging deep learning (DL)
applications focused on MS and CIS imaging. The reviewed
publications were examined from different standpoints including the
different DL architectures, the patient cohorts and the end-point
of the reported studies in the field.
2. Selection criteria
Based on a PubMed search using keywords: ‘deep
learning’ AND ‘multiple sclerosis’, 74 articles were initially
identified. Subsequently, screening for MR imaging relevance and
removing duplicates led to 32 original research articles that
applied DL techniques in MS and CIS for diagnosis tasks,
segmentation tasks and clinical protocol optimization. Afterwards,
a similar search in google scholar, using the same keywords, and
keeping only original research journal papers, led to 13 additional
articles making a total of 45 original papers.
After an initial review, selected articles were
separated in three broad categories i.e., i) segmentation of MS and
CIS lesions; ii) classification of different pathologies and
anatomical structures; and iii) post processing techniques and
image enhancement methods with application in MS and CIS imaging.
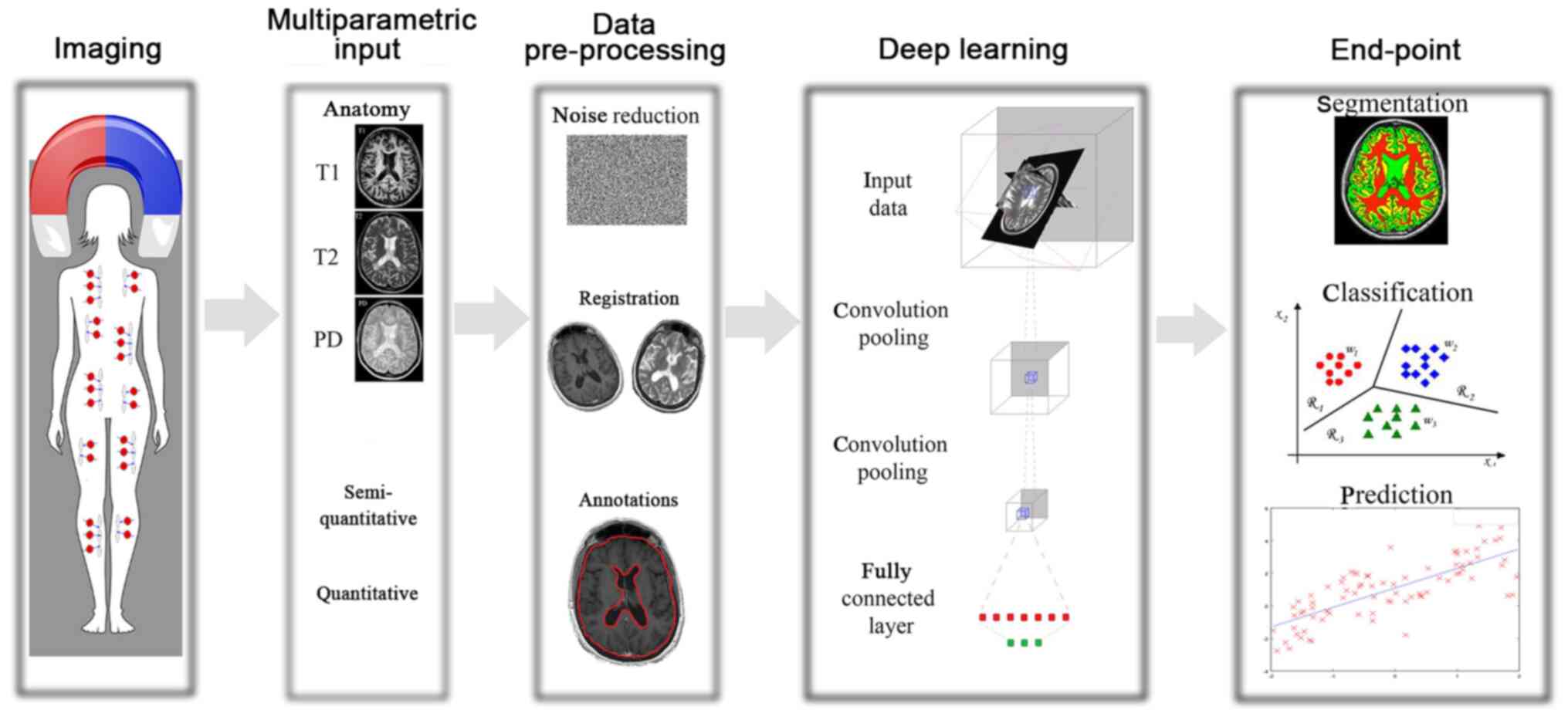
In Fig. 1 presents the general
workflow of the DL techniques along with the final goal, as was
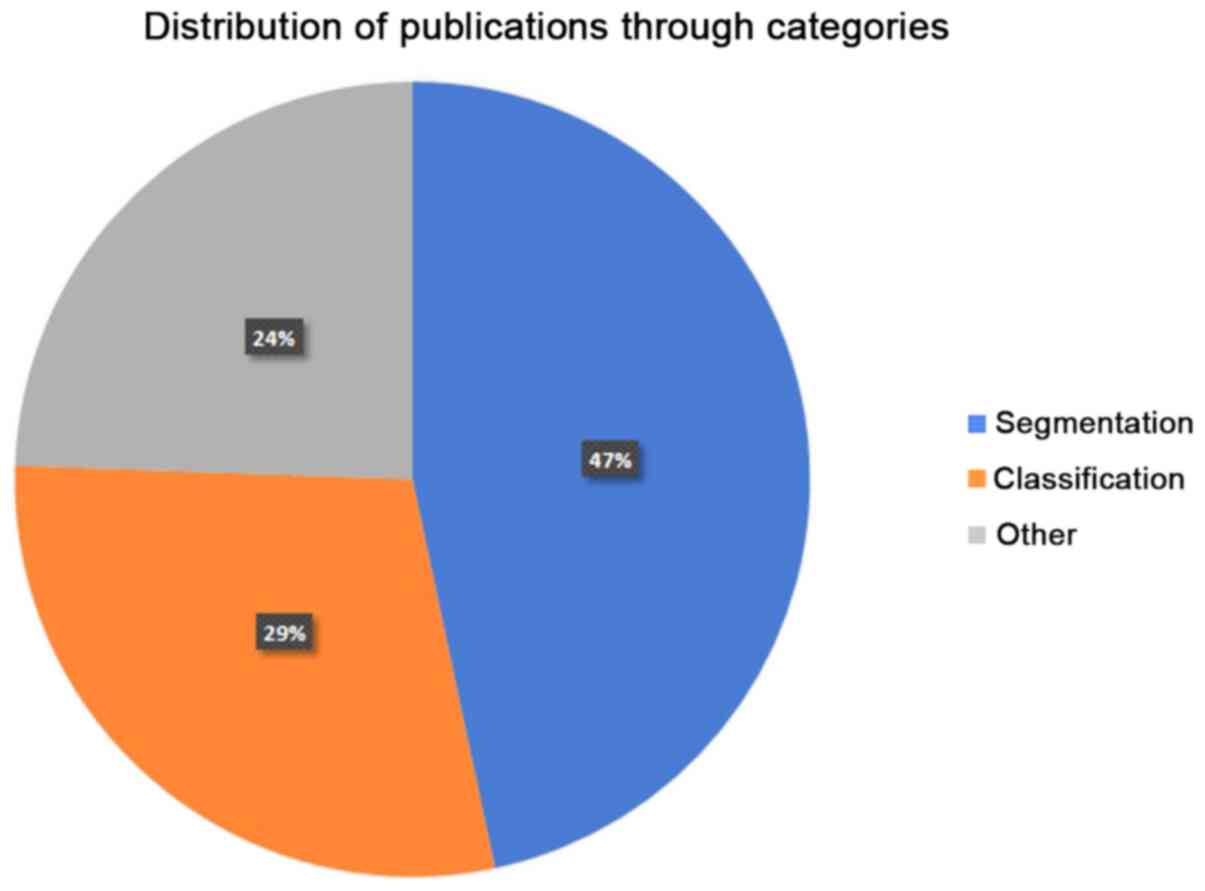
determined in the present review manuscript. In Fig. 2, the distribution of the reviewed
papers in each of the three abovementioned categories is presented.
Obviously, segmentation and classification techniques are the main
areas of interest, accounting for the 76% of the overall
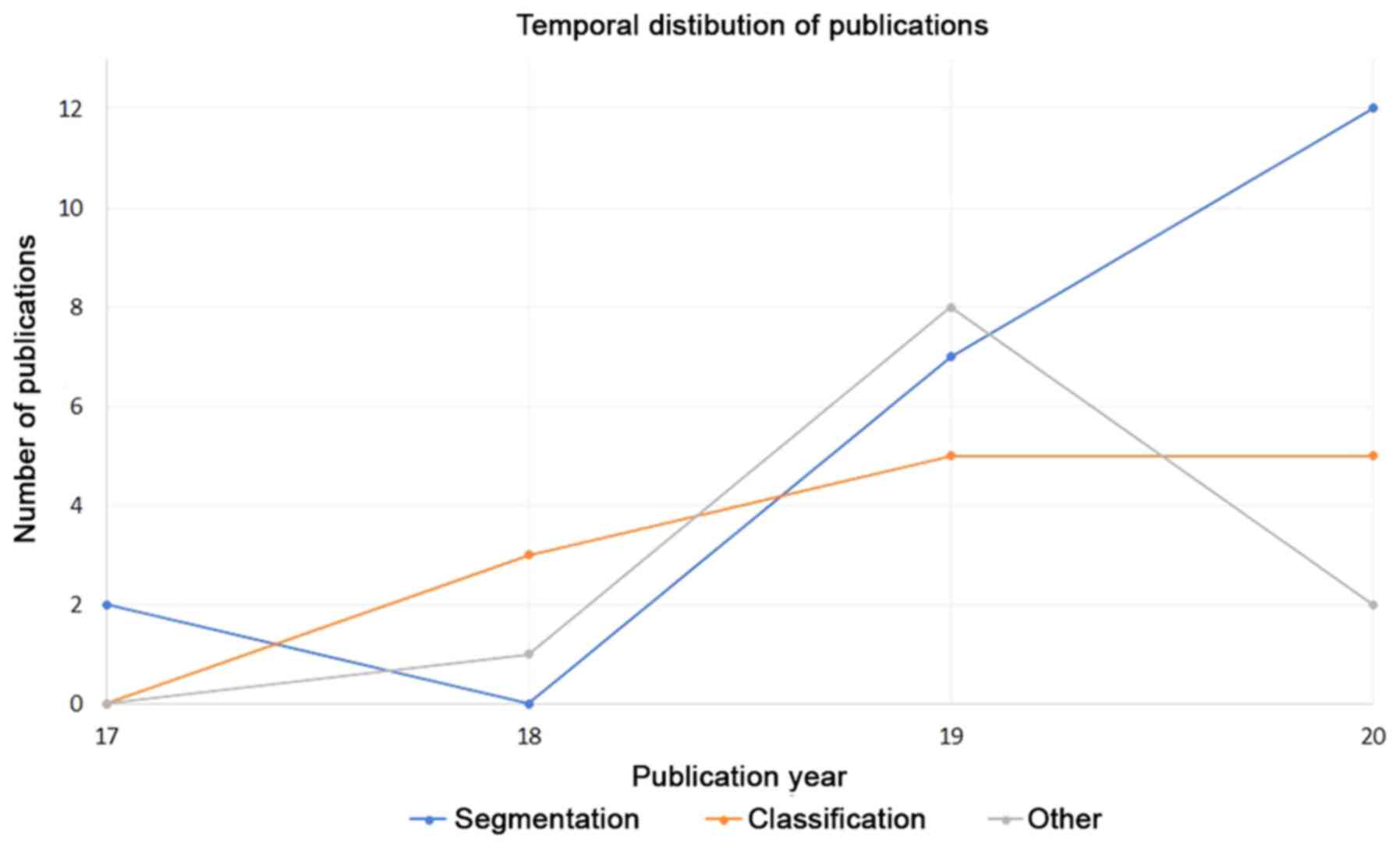
publications. Moreover, in Fig. 3
the number of publications through years, for the three
abovementioned application fields, is presented.
3. Detection: Segmentation methods
MS is a progressive disease of CNS affecting the
myelin sheath, a fatty material that envelops neuron axons, and as
a result alters the morphology and structure of the brain.
Furthermore, brain atrophy is taking place in cortical and
subcortical regions, whereas the gray matter (GM) atrophy and its
association with disability and cognitive impairment is under
investigation (18,19). Nevertheless, measurement of GM
volume is a useful method for assessing the overall GM damage and
can be estimated through conventional MRI techniques (6). Another established measurement for
prediction of neurological disorders is the lesion load (20), since it has been proved that both
lesion volume and lesion count are strong predictors of disease
course and progressive timepoints. Additionally, detection of MS
lesion is of paramount importance since the diagnosis of MS is
based on the spatial and temporal distribution of focal
demyelination lesions (21). Since
manual detection of MS lesions is time consuming and prone to
errors as well as inter observer variability, algorithms aiming to
automatically delineate these tissues may constitute a great tool
for clinical practice. Considering all the aforementioned,
automated computer algorithms aiming to segment anatomical
structures as well as to detect MS lesions, are valuable tools for
clinicians offering objective and repeatable results. Accurate
segmentation of WM lesions is hindered due to overlapping
intensities of these lesions with GM, while finite resolution of MR
images, complicated shapes that vary among different lesions as
well as the partial volume effect, hamper the accurate delineation
of MS tissues. Additionally, gray matter lesions are often not
visible in T2-w images due to size, magnetic relaxation
characteristics and partial volume effects with CSF. Finally, these
lesions may not be visible even in FLAIR sequences, despite the
increased sensitivity of this protocol compared to a conventional
T2-w one.
Many previous studies have proposed DL methods
aiming to provide a robust and reliable framework for detection and
segmentation of MS lesions, while the most significant of are
summarized below. Valverde et al (22) presented an automated WM lesions
segmentation method, by utilizing a cascade of two 3D patch-wise
convolution neural networks (CNN), the first was sensitive for
revealing possible candidate lesions voxels while the second one
was responsible for reducing the number of misclassified voxels
resulted from the first network. The accuracy of the proposed
approach was evaluated in the medical image computing and computer
assisted intervention society (MICCAI) 2008 database (http://www.ia.unc.edu/MSseg), as well as on two
private MS clinical datasets, while results were also compared with
other lesion segmentation tools and methods. It was reported that
the proposed approach is the best ranked on the MICCAI2008
challenge, and in the clinical datasets it exhibited an increased
accuracy in the WM lesions segmentations.
Birendaum et al (23) developed an automated DL method for
MS lesion segmentation, utilizing a CNN that was mainly based on
the Single View CNN (V-Net) and the Longitudinal Network (L-Net).
For this work, the 2015 Longitudinal MS Lesion Segmentation
Challenge dataset (24) was used
while the network resulted in Dice Similarity Coefficient 0.627.
Finally, authors reported that the aforementioned method achieved
performance level comparable to a trained human rater.
Gros et al (25), proposed a fully-automated framework
for segmenting spinal cord and intramedullary MS lesions using
conventional MRI data. The aim of this study was to provide a
robust and automated framework, by overcoming the technical
limitations of large variabilities related to acquisition
parameters and image artifacts, and furthermore to eliminate the
inter-rater variability as well as to optimize the large-throughput
analysis pipeline. The cohort of this study consisted of 1,042
subjects; 459 healthy controls, 471 MS patients and 112 with other
spinal pathologies. Authors used a sequence of two CNNs, the first
intended for the detection of spinal cord centerline using 2D
convolutions, while the second segmented the spinal cord and/or
lesions using 3D convolutions. These networks were trained
independently using the Dice loss (26), considering that it is not sensitive
to high class imbalances, while comparing the proposed technique
with the PropSeg method, an unsupervised technique for spinal cord
segmentation (27), resulted in
median Dice 95% when using manual vs 88% when using PropSeg.
Moreover, regarding MS lesion segmentation, the proposed technique
provided Dice of 60%, while sensitivity and precision were 83% and
77% respectively.
Aslani et al (28) presented an automated approach for
MS lesion segmentation, based on a Deep end-to-end 2D CNN, a
technique that incorporates a multi-branch down-sampling path which
enables the network to encode information stemming from multiple
modalities in a separate way. The proposed model was trained and
tested by using orthogonal plane orientations for every 3D
modality, in order to exploit information from all directions.
Evaluation of the proposed method was on two datasets, one private
(37 MS patients) as well as the International Symposium on
Biomedical Imaging (ISBI) 2015 longitudinal MS lesion segmentation
challenge dataset (14 MS patients) (24). Results on the ISBI challenge ranked
the proposed methodology among the top (DSC 0.6114), while on the
private dataset the proposed method achieved the best results (DSC
0.6655).
Sander et al (29), employed a fully automated
segmentation method, based on multi-dimensional gated recurrent
units (MD-GRU). MD-GRU, is a DL approach that employs a
convolutional adaptation of a recurrent neural architecture. This
network was trained on 50 MS patients and 17 healthy controls,
while a refinement procedure of the algorithm parameters took place
in a cohort of 20 independent MS patients. For accuracy testing, 20
MS or CIS patients from the same cohort were used, while accuracy
was further assessed in a dataset including 80 independent MS or
CIS patients from the same cohort. Reproducibility assessment was
followed by considering T1-w images from 33 healthy controls.
Finally, robustness of the segmentation approach was assessed using
images from 50 Alzheimer patients. According to the results, the
proposed segmentation approach provided accurate, highly
reproducible and a robust fully-automated segmentation framework
for the brainstem and its substructure in both healthy controls as
well as in MS and Alzheimer disease patients. In more detail, study
results were compared with FreeSurfer (30) software results, while authors
reported that the mean % change/SD between test-retest brainstem
volumes were 0.45%/0.005 for MD-GRU and 0.95%/0.009 for FreeSurfer
and 0.86%/0.007 for manual segmentations. MD-GRU segmentations were
compared to segmentations performed from experts, resulted in mean
Dice score/SD of 0.97/0.005 for brainstem, 0.95/0.0131 for
mesencephalon, 0.98/0.006 for pons and 0.95/0.015 for medulla
oblongata.
Hashemi et al (31), developed a 3D fully connected
convolution neural network (FCNN) with DenseNet blocks
(FC-DenseNet), using an asymmetric similarity loss function Tversky
index, aiming to mitigate the data imbalance problem and to achieve
a better trade-off among precision and recall. This method used 15
patients from the MS lesion segmentation challenge (MSSEG) of the
2016 Medical Image Computing and Computer Assisted Intervention
conference (32), as well as 5
patients from the longitudinal MS lesion segmentation challenge of
the IEEE International Symposium on Biomedical Imaging (ISBI)
conference (33). For training
purposes, a 5-fold cross validation strategy was utilized, in which
five instances of the training were performed on 4/5th of each
dataset and validated on the remaining 1/5th. For testing the
implemented architecture, due to limited number of subjects 5-fold
cross validation was used. Study results report that MS lesion
segmentation presented improved performance evaluation metrics by
using asymmetry similarity loss function rather than using the Dice
similarity in the loss layer. Furthermore, authors reported that
the proposed framework achieved improved precision-recall trade-off
and average DSC scores of 69.9% and 65.74% for MSSEG 2016 and ISBI
longitudinal MS lesion segmentation challenge correspondingly.
Gabr et al (34) investigated the performance of FCNN
in segmenting brain tissues using a larger cohort of MS patients.
The MRI protocols and the patient cohort used in this study,
included a dual echo fast spin echo (FSE) sequence, a T1-w sequence
and FLAIR sequence from a cohort of 1,000 RRMS patients emanating
from the CombiRx clinical trial, while training, validation and
testing of the FCNN were based on the aforementioned data.
Moreover, automated segmentation results were validated by two
experts, while for cross-validation of the results, the
leave-one-centre-out approach was used. Results from this study,
report high average Dice similarity coefficient for all segmented
tissues (0.95 for WM, 0.96 for GM, 0.82 for CSF and 0.82 for T2
lesions), and high correlations among the DL segmented tissues and
the annotations from the expert (R2>0.92). Finally, cross
validation reported consistency of the results among the different
centers.
Weeda et al (35) compared four different lesion
segmentation methods to investigate the suitability of the DL CNN
method nicMSlesions (36), in an
independent dataset aiming to determine whether this method is
suitable for larger, multi-center studies. For this purpose, manual
segmentation was performed in 14 MS subjects, while five different
automated lesion segmentation methods were compared: i)
unsupervised-untrained LesionTOADS (37); ii) supervised-untrained methods
with threshold adjustment LST-LPA (38) and nicMSlesions with default
settings; iii) supervised-untrained LST-LPA with threshold
adjustment; iv) supervised-trained with leave-one-out
cross-validation nicMSlesions and BIANCA (39); and v) supervised-trained on a
single subject nicMSlesions. Results from this study report that
the best volumetric and spatial agreement with manual annotations
was obtained with the supervised and trained methods nicMSlesions
and BIANCA, while the worst results was provided by the
unsupervised, untrained method lesionTOADS, concluding that input
from a single subject to fine tune a DL CNN is sufficient for
lesion segmentation.
McKinley et al (40) presented a segmentation framework
based on a cascade of two CNNs, providing multimodal MR images
segmentation into lesion, normal appearing white matter (NAWM) and
normal appearing gray matter (NAGM) structures. For this purpose,
MRI exams from 122 RRMS patients were used (90 patients for
training and validation of the classifier, 32 patients for
testing), while a second dataset including 10 MRI datasets was used
for direct comparison with other centers. Results in the current
study reported that both Dice coefficient as well as specificity
and sensitivity are improved compared to previous approaches, while
a good agreement with individual human raters was achieved.
Moreover, authors concluded that the proposed method performed well
on data emanating from different centers, scanners and
field-strengths that were not represented in the training dataset
indicating good generalizability, while it was also reported that
the classifier achieved to identify lesions missed by human
raters.
Narayana et al (41) studied the dependence of DL
segmentation accuracy on the training size, aiming to define the
required training set for an adequate accuracy in brain MRI
segmentation in MS patients. For this purpose, a 2D FCNN was
trained using 16 different training sizes, while segmentation
accuracy was determined according to the training size, and network
performance was evaluated by the dice similarity coefficient and
lesion true-positive and false positive-rates. The presented
methodology was evaluated in MRI data from a cohort of 1,008 MS
patients, while the automatic segmentation results were assessed by
two neuroimaging experts. Study results report that lesion
segmentation showed stronger dependency to the sample size
comparing with the GM, WM and CSF, concluding that excellent
results were provided by a training set of 10 image volumes for GM,
WM and CSF. Finally training size of at least 50 images was
necessary for adequate lesion segmentation with DSC <0.68 while
for 150 sample size the DSC was 0.82.
Nair et al (42) presented a 3D MS lesion detection
and segmentation CNN. The network was trained using a large scale,
multi-site, multi-scanner clinical MS dataset consisted of 1,064
RRMS patients scanned annually over a period of 24 months, while
imaging protocol included T2w, T1-w, FLAIR and proton density (PD)
images. The network was trained using a weighted binary
cross-entropy loss, while the performance of the network was
evaluated separately in i) a voxel-level analysis; and ii)
lesion-level analysis. Study results report that uncertainty
filtering vastly improves lesion detection accuracy for small
lesions.
In the same dataset, McKinley et al (43) trained two state-of-the-art CNNs
architectures, a 3D Unet consisting a reference implementation and
a more recently proposed architecture, the DeepSCAN, in order to
segment multimodal MR images into lesion classes and NAWM and NAGM
structures. Results from this study reported that both examined
methods outperformed previous approaches in the literature related
to the MSSEG dataset, while a good agreement among the automated
segmentations and results from human ratters is reported. Moreover,
DeepSCAN network found to achieve the best performance in both
lesion and anatomical labelling, while there is reported that this
classifier recognized lesions that were missed by human
ratters.
Narayana et al (44) investigated the effects that
different combinations of multi-contrast MR images may infer when
serving as input to a CNN. To this end, U-net, a fully CNN was used
in order to automatically segment GM, WM, CSF and lesions in 1,000
MS patients. Image datasets included T1-w images, FLAIR and dual
echo turbo spin echo or fast spin echo images. For the assessment
of the segmentation performance, the DSC was evaluated, while for
lesions the true positive rate (TPR) and false positive rate (FPR)
were also reported. Results from this study report that when
segmentation input was a combination of all four image modality
data, it was achieved the highest DSC for all tissue volumes, while
high DSC results were also obtained when FLAIR protocol was
included in the segmentation input. Finally, it was reported that
lesion segmentation was poor when considering very small
lesions.
Salem et al (45), proposed a FCNN in order to detect
early T2-w lesions in MR images. Patient cohort included 60 early
MS and CIS patients, with a baseline and a follow-up study for each
patient, while new T2-w lesions were found in the follow-up exam.
Authors concluded that the proposed methodology achieved increased
accuracy regarding the detected newly formed T2-w lesions, and
therefore it has the potential to be used in the clinical practice
for monitoring disease progression.
Brown et al (46) implemented a FCNN for automatic
segmentation of orbital fat, tested on pediatric-inset MS patients,
while they introduced a preprocessing step of image calibration
aiming to remove technical intensity artefacts. This method was
evaluated on 1,018 scans form 256 participants. Study results
report that automatic segmentations agreed with manual segmentation
from an expert, reporting estimated mean Jacard index 0.74, while
the proposed image calibration contributed significantly in the
performance of segmentation. Finally, the proposed methodology
provided a robust serial calibration framework that allowed
comparison of follow-up studies of the patient, while it is
considered to be a fast technique that may be applied to large as
well as to small datasets.
Ackaouy et al (47) proposed an unsupervised domain
adaptation framework based on optimal transport (3D-Unet),
Seg-JDOT, adapting a deep model to samples from a source domain to
a target domain that share similar representations. This study used
the MICCAI 2016 MS lesion segmentation challenge dataset (48) that contains 53 MRI images from MS
patients, splitted to 15 train and 38 test images. Results from
this study with single and multi-source training, indicated that
model adaptation to a target site, can yield better model
performance compared to the standard training.
Coronado et al (49) utilized a 3D CNN model based on
multispectral MRI data of 1,006 RRMS patients, aiming to
automatically segment gadolinium-enhanced T1 lesions. The dice
similarity coefficient, true-positive rate, false-positive rate
indexes over all the enhancing lesions were 0.77/0.90/0.23 when
using FLAIR, T2 and pre- and post-contrast T1-w images,
0.72/0.86/0.31 when using only pre and post contrast T1-w images
and comparable performance when using only post-contrast
images.
La Rosa et al (50) proposed a framework for automatic
segmentation of cortical and white matter lesions based on T1 and
T2-weighted MRI data, in a cohort of 90 MS patients including 728
gray and 3,856 white matter lesions. For this purpose, an FCNN
architecture based on the 3D-U-Net was used. Results of the present
study reported that the proposed framework was able to achieve a
detection rate of 76% for both cortical and white matter lesions
with a false positive rate of 29% in comparison to manual
segmentation. Finally, it is reported that the proposed methodology
achieved to adequately generalize the exams acquired in two
hospitals with different scanners.
Gessert et al (51) examined a CNN model for lesion
segmentation from two time points, using two path architectures.
They stratified a cohort of 89 MS patients, with two MRI exams
each, baseline and follow-up respectively, while for validation and
testing they used a 3-fold cross-validation. Results indicate that
the proposed model outperformed classic methodologies, while it was
reported a lesion-wise false-positive rate of 26.4% at a
true-positive rate of 74.2%.
Essa et al (52) proposed an automatic segmentation
framework, based on region-based CNN (R-CNN) model applied on T2-w
and FLAIR MR images, evaluated on the MICCAI 2008 MS challenge
(http://www.ia.unc.edu/MSseg). The
proposed model shows competitive results compared with the
state-of-the-art MS segmentation methods, with average total score
83.25 and average sensitivity 61.8% on the testing set.
Finally, Barquero et al (53) developed a CNN architecture (RimNet)
for automated rim lesion detection in MS. MR imaging data were
acquired from three different scanners, in a cohort of 124
patients. The multimodal RimNet architecture achieved to better
classify lesions [area under curve (AUC)=0.943] compared to
unimodal approaches, while sensitivity and specificity (70.6 and
94.9%, respectively) were comparable to the experts scores
4. Classification: Diagnosis
During the last years, DL techniques have been
successfully applied for the diagnosis and classification of MS
disease (54). These techniques
can identify patterns of the imaging data without using an explicit
feature extraction method, but instead using automatic feature
extraction. Advantages stemming from the use of DL techniques in MS
diagnosis include the increment of diagnostic accuracy, early stage
diagnosis, increase the reliability of the diagnosis, decrease
disease related cost, and improvement in patient quality of life.
In this section, the most significant research articles in the MS
classification using DL techniques are reviewed.
Yoo et al (55) presented an automatic framework for
detection of MS pathology at an early stage. In this approach, 3D
image patches extracted from myelin maps and the corresponding T1-w
MR images was used to train a latent joint myelin feature
representation using an unsupervised four-layer deep belief network
(DBN) framework. This framework used 55 RRMS patients and 44
healthy controls, while by using 11-fold cross validation, it
achieved 87.9% average classification accuracy, and authors
reported that suggested method was able to identify MS image
features from normal appearing brain tissues.
Wang et al (56) implemented a 14-layer CNN combined
with batch normalization, dropout and stochastic pooling techniques
aiming towards early diagnosis and treatment of MS disease. Imaging
data for the current study, included 38 MS patients from eHealth
laboratory and 26 healthy controls (HC). Results reported that the
proposed 14-layer CNN network had sensitivity of 98.77%,
specificity 98.76% and accuracy 98.77%, while after comparison of
the aforementioned method with maximum pooling, average pooling,
five traditional AI methods and a DL method, it was shown that the
proposed method outperforms all the above-mentioned techniques.
Zhang et al (57) developed a DL algorithm based on CNN
that combined parametric rectified linear unit (PReLU) and dropout
techniques, in order to accurately separate MS from HC. This study
stratified 676 MS brain slices and 681 HC slices, while data
augmentation technique was used in order to increase the size of
the training set. The 10-layer deep convolutional neural network
used in this study was consisted of 7 convolution layers and 3
fully-connected layers. The proposed method achieved 98.22%
sensitivity, 98.24% specificity and 98.23% accuracy, while the
dropout method increased accuracy by 0.88%, PReLU increased
accuracy by 1.92% compared to ordinary ReLU and by 1.48% compared
to leaky ReLU.
Talo et al (58) implemented a deep transfer learning
framework to automatically classify normal and abnormal brain MR
images via using a ResNet34 CNN DL model featuring data
augmentation, optimal learning rate finder and fine-tuning to adapt
a pre-trained model. This study stratified a cohort of 42 subjects
(2 HC and 40 patients with cerebrovascular, neoplastic,
degenerative, and inflammatory disease types), resulting in 613
images, 27 normal and 513 abnormal, for training and validation.
Results from the current study report that they achieved 100%
classification accuracy.
Lu et al (59) presented a transfer learning
technique for automatic detection of pathological brain, by
utilizing a pre-trained model featuring AlexNet architecture. In
this study brain MR images from 38 HC and 177 pathological were
used, including pathologies such as Alzheimer, Glioma, Huntington,
AIDS dementia, MS, and Pick. Results reported that the proposed
method achieved accuracy of 100% which, outperformed
state-of-the-art approaches.
McKinley et al (60), investigated the ability of DL
algorithm to discriminate radiologically progressive from
radiologically stable patients, in 3 different private datasets. To
this end, DeepSCAN MS classifier, a fully-convolutional neural
network was utilized. The methodology that was used in this study,
achieved to temporally track lesion load changes by leveraging
measures of uncertainty in the location of lesion boundaries.
Results from the present study, disclosed that the proposed
framework was able to separate progressive from stable time-points
(AUC=0.999) and changes in lesion volume (AUC=0.71). Moreover,
method validation on two external datasets confirmed the
performance of this method by achieving accuracies 75 and 85% in
separating stable and progressive time-points.
Marzullo et al (61) introduced a graph convolutional
neural network (GCNN) to classify MS patients on four clinical
profiles, (CIS, RR, secondary-progressive SP, primary-progressive
PP). Methodology in the present study used structural connectivity
information by DWI and evaluated the classification performance
using unweighted and weighted connectivity matrices. Moreover, the
role of graph-based features aiming to better characterization and
classification of the MS pathology were investigated. For the
purposes of this study, 90 MS patients (12 CIS, 30 RRMS, 28 SPMS
and 20 PPMS) and 24 healthy controls were included in the analysis.
Results demonstrated that the aforementioned NN framework offers
very good performance in clinical profiles classification, while
graph weights representation of brain connections offer significant
information for clinical profiles discrimination.
Eitel et al (62) presented a DL framework based on a
3D CNN and a layer-wise relevance propagation (LRP) in order to
accurately diagnose MS disease. In this study, LRP was used as an
additional tool aiming to reveal further relevant image features
that will be afterwards forwarded to the trained CNN. The CNN model
was trained using MRI data from the Alzheimer Disease Neuroimaging
Initiative (921 patients) and afterwards the CNN was used to
discriminate among MS (76 patients) and controls (71 healthy
controls). Authors reported that the presented methodology showed
accuracy 87% and AUC 96%, concluding that LRP in conjunction with
proposed CNN is a framework capable of sufficiently classify MS
patients and healthy controls.
Narayana et al (63) used a CNN in order to evaluate the
performance in predicting enhanced MR lesions without using
contrast agents. To this end, a CNN was used for classifying
lesions from MR images as enhanced or unenhanced. In this study,
1,008 MS patients were involved, while at least one enhancing
lesion was observed for 519 participants. MR demyelinating lesions
were classified in two classes, enhanced and unenhanced, while
performance was assessed by using fivefold cross-validation.
Results report sensitivity 78%, specificity 73%, AUC 0.82 for
slice-wise prediction, while for participant-wise these were 72%,
70% and 0.75, respectively.
Maggi et al (64) proposed a DL based prototype for
automated assessment of the central vein sign (CVS) in WM MS
lesions. This study included 80 subjects from three different
sites, 42 MS, 33 MS mimics with diseases that are described by WM
abnormalities similar to MS, and 5 patients with uncertain
diagnosis. To this end, a 3D CNN (‘CVSnet’) was designed, trained
on 47 examinations and tested on the remaining 33. Authors reported
that the proposed framework achieved human expert performance,
achieving lesion-wise median balanced accuracy of 81% and
subject-wise balanced accuracy of 89% on the validation and 91% on
the test set, while evaluation on data from different
hospitals-scanners is promising for larger multi-center trials
using the CVS marker in the MS diagnostic criteria.
Wang et al (65), utilized a CNN framework in order to
discriminate neuromyelitis optical spectrum disorder (NMOSD) (41
patients) from MS (47 patients), a challenging classification
problem considering that NMOSD is a rare disease thus there is a
limitation in the availability of exams, as well as the lesions in
the above mentioned diseases are scattered and overlapping, adding
an extra embodiment in the classification procedure. Study results
reported that the novel proposed CNN model achieved to better
discriminate NMOSD from MS compared to conventional CNN models.
More specifically, the proposed method exhibited accuracy 0.75,
sensitivity 0.707, and specificity 0.759, while traditional 3D CNNs
that tested on the same problem did not achieve to distinguish the
different classes.
Roca et al (66) used a DL network in order to predict
the expanded disability status scale (EDSS) in a cohort of 1,446 MS
patients, partitioned in 971 subjects for training, and 475
subjects for testing. The proposed framework consisted of a CNN
model and a classical ML predictor. They finally reported that the
proposed method was able to predict two-year EDSS score, based on
FLAIR MRI imaging data, while the more informative variables were
age, volume of lateral ventricles and the lesion load in main white
matter tracts.
Lopatina et al (67) utilized a CNN model aiming to
identify MS patients by analyzing susceptibility weighted images,
considering that in this protocol vein patterns are identifiable
and it is also able to indicate extensive demyelination and iron
accumulation. The stratified cohort was composed by 66 MS patients
and 66 healthy controls, while results indicate that veins located
in the anterior medial and lower peripheral regions, are most
relevant for the classification decision.
5. Post processing techniques and image
enhancement methods
Reducing rescans and recalls is of great importance
for the optimization of health care management, since low image
quality MR acquisitions often need to be repeated, thus leading to
increased hospital costs and extended MR examination times.
Furthermore, visual inspection of the related examinations aiming
to assess image quality, is impractical in large multi-center
studies as well as prone to errors emanating from intra-observer
variability. In recent years, computational tools that assess the
image quality of the examinations have been developed based on
automated assessment of image quality metrics. To this purpose,
computer algorithms have been implemented during the last years,
aiming to automatically assess image quality as well as acquisition
parameters. In this section, recent studies focusing on solving the
aforementioned problems by using DL techniques in MR imaging of
neurodegenerative diseases are reviewed.
Sreekumari et al (68) developed an automated method for
assessing the need of rescan, in motion corrupted brain scans.
Authors developed a CNN with 7 convolutional layers, 4 max pooling
layers, and 3 batch normalization layers that computed the
probability for a MR series to be clinically useful, while by
combining this probability with a scan dependent and radiologist
defined threshold, they determined whether a series need to be
rescanned. Moreover, the classification performance was compared
with that of 4 technologists and 5 radiologists in 49 series,
stemming from MS and stroke patients, characterized by low and
moderate motion artefacts. Results indicated that radiologists -
technologists produced mean ratio of rescans/recalls of
(4.7_5.1)/(9.5_6.8) for MS and (8.6_7.7)/(1.6_1.9) for stroke,
while DL produced (7.3_2.2)/(3.2_2.5) for MS, and
(3.6_1.5)/(2.8_1.6) for stroke, concluding that this technology
independent method can reliably decrease rescan and recall
rates.
Sujit et al (69) developed a DCNN aiming to
automatically evaluate the quality of multicenter structural brain
MRI images, using 1,064 images from autism patients from ABIDE
database (60% training, 20% validation and 20% test) while they
tested on a cohort of 110 MS patients from the CombiRx dataset
(70). The results demonstrated
the high accuracy of the proposed method to evaluate image quality
of structural brain MRI in multi-center studies (ABIDE dataset
achieved AUC 0.90, sensitivity 0.77, specificity 0.85, accuracy
0.84, PPV 0.42, and NPV 0.96 while for the CombiRx there were AUC
0.71, sensitivity 0.41, specificity 0.84, accuracy 0.73, PPV 0.48,
and NPV 0.80).
In many applications, the compromise between spatial
resolution, SNR, as well as temporal resolution in specific
protocols, is limiting the clinical and research applicability of
the MR modality. An ordinary approach to overcome this problem is
to acquire images with adequate in-plane resolution and low
through-plane resolution and afterwards interpolate data, by using
super-resolution techniques, in order to obtain isotropic voxels.
To this purpose, Zhao et al (71) developed a DL method, called SMORE,
that accomplished both anti-aliasing and super-resolution using no
external atlases. Authors demonstrated the performance of the
proposed algorithm in four applications: i) improve visualization
of the brain WM lesions in MS patients; ii) improve the
visualization of scarring in cardiac left ventricular remodeling
after myocardial infarction; iii) performance on multi-view images
of the tongue; and iv) improve performance in brain ventricular
system parcellation. Regarding the MRI visualization of MS lesions,
that is speculated in the present review article, authors examined
whether the SMORE computationally enhanced FLAIR images can provide
additional diagnostic information compared to a conventional
interpolation. For this purpose, FLAIR data was reconstructed from
initial resolution of 0.828 x 0.828 mm x 4.4 mm onto a 0.828 x
0.828 mm x 0.828 mm digital grid by a conventional b-spline
interpolation, the JogSSR approach that is a super resolution
method which improves resolution in the through-plane direction
(72), as well as the proposed
SMORE algorithm. Authors reported that both JogSSR and SMORE
resulted in sharper edges compared to the conventional b-splines
interpolation, while SMORE results appeared to be more realistic,
however results in this part of the study were assessed only by
visual inspection while authors do not provide any quantitative
measurement.
Another aspect that has recently gained interest, is
the generation of synthetic MR images by using computational
techniques. This is an interesting and emerging area of research
especially considering that some protocols may be time-consuming
and some sequences may be missed due to limited scanning time or
patients' interruptions in case of anxiety and confusion. Also, the
ability to synthesize information related to different imaging
modalities directly from MRI has gained the attention of many
researchers in the field of DL in medical imaging.
To this purpose, Wei et al (73) used a 3D FCNN to predict FLAIR pulse
sequence from other MRI protocols. For this study, 20 MS patients
and 4 healthy controls were involved, including T1-w, T2-w, PD,
FLAIR, T1 SE and double inversion echo sequences. The performance
of the proposed method was compared qualitatively and
quantitatively with four state-of-the-art approaches: modality
propagation (74), random forests
including 60 trees (75), U-Net
(76) and voxel wise multilayer
perceptron consisted of two hidden layers and 100 hidden neurons
for every layer. Results from this study indicated that the
proposed FLAIR synthesis method provides competitive performance to
previous techniques. In more detail, the proposed technique was
statistically significantly better than the other methods
(P<0.05) in average, by providing MSE (SD) 918.07 (41.70) and
SSIM(SD) 0.860 (0.031). Finally, authors compared the
aforementioned methods in a MS lesion detection task, by evaluating
the MS lesion to NAWM as well as to the surrounding NAWM tissue
contrast. Also, in this task the proposed method outperformed the
competitive techniques on both ratios, revealing that it provides
better contrast for MS lesions.
Salem et al (77) proposed a synthesis method for MS
lesions on MR images aiming to improve the performance of
supervised machine learning algorithms, thus avoiding the lack of
ground truth. To this end, they used a two-input two-output FCNN
for MS lesion synthesis, in which the lesion information was
encoded as discrete binary intensities level masks. For this
purpose, authors assessed the proposed methodology in two cohorts
of patients, by evaluating the similarities among real and
synthetic images, as well as by measuring the performance of lesion
detection by segmenting both the original and the synthetic images.
The two cohorts used included 15 HC and 65 patients with CIS or
RRMS, and the ISBI2015 dataset (24) comprising 5 training and 14 testing
subjects with 4 to 5 follow-up studies for every subject, while
they also used data augmentation techniques. The effect of data
augmentation was demonstrated in both analyzed datasets, while
results showed the effectiveness of using synthetic MS lesions.
Finally, regarding the ISBI2015 challenge, the model was trained
using a single image and the synthetic data augmentation and
performed similarly to other CNN methods that were fully
trained.
Wei et al (78) proposed a method to predict positron
emission tomography (PET)-derived myelin content map from
multimodal MR imaging data, by introducing a new approach called
Sketcher-refiner GANs with a specially designed adversarial loss
function, in which the Sketcher network generated global anatomical
and physiological information, while the Refiner model refined and
generated the tissue myelin content. For this purpose, a dataset
including 18 MS patients and 10 HC was used, while both MR and PET
images were available. It was reported that regarding image quality
and myelin content, the proposed approach outperformed the
state-of-the-art methods, as well as that prediction results were
comparable to PET derived gold standard both in global and
voxel-wise levels.
Finck et al (79) implemented a diamond shape topology
generative adversarial NN (DiamondGAN) in order to produce
synthetic double inversion recovery (synthDIR) images while the
diagnostic performance of the proposed images was compared to
conventional MR images. This study recruited 100 MS patients, while
the DiamondGAN was trained in a subset of 50 MS patients and an
additional 50 images of generated synthetic data. Results indicate
that synthetically generated DIR images resulted in improved
detection of juxtacortical lesions, leading to improvement on
lesion detection.
Finally, the last category is related to
computational methods that are focusing on post-processing MR
images for image reconstruction, and computational analysis. These
methods are of great interest considering that they can bypass
unreliable computational methods and ill-conditioned mathematical
problems, as well as combine large-scale information from
multi-site studies by providing quantitatively and qualitatively
consistent images to automated algorithms.
Yoon et al (80) presented a DL network (QSMnet) with
a modified U-net structure able to generate high quality
susceptibility source map from single orientation data. A total of
12 healthy controls were used for training, using COSMOS QSM maps,
while for evaluation data from a microbleed, a MS and a patient
with intraparenchymal hemorrhage were used. Results demonstrated
that QSMnet provided superior image quality results compared to TKD
and MEDI, while the image quality is comparable to COSMOS.
Bollman et al (81) trained a FCNN, DeepQSM, in order to
invert the magnetic dipole kernel convolution, and to provide a QSM
framework that determines the composition of myelin sheaths of
nerve fibers in the brain, as well as to assess quantitative
information on iron homeostasis and its dysregulation. This network
was trained on synthetic examples, while for testing procedure
there were performed four experiments with synthetic data, a single
orientation background field corrected tissue phase image and an
STI susceptibility map, and finally clinical data from a patient
with MS. Study results showed that the proposed methodology enabled
identification of deep brain structures, not visible in MRI data.
Furthermore, study results revealed that the presented methodology
provides information for magnetic tissue properties, and finally
showed increased sensitivity in identifying WM lesions regarding
the MS patient.
Dewey et al (82) proposed a contrast harmonization
method, DeepHarmony, by using U-Net DL technique in order to
provide images with consistent contrast. To this end, 12 subjects
(10 MS and 2 HC) were scanned twice within 30 days, and
longitudinal data were retrospectively collected from 45 RRMS
patients, while both cohorts were based on two different scanning
protocols. Results suggested that DeepHarmony harmonized images
showed significant improvement, as well as reduced dependency of
atrophy calculations when using DeepHarmony method.
Liu et al (83) implemented a fast myelin water
fraction (MWF) maps data analysis method, that was based on Myelin
Water Imaging data acquired by a 32-echo 3D gradient and spin echo
sequence. For this purpose, an NN model was used on data stemming
from 4 healthy controls, while for testing there were utilized 1 MS
brain, 1 healthy spinal cord, and 2 healthy brains acquired from a
different scanner. Results proposed that time for calculation of
MWF maps was dramatically reduced while their quality was similar
to ground truth levels.
6. Discussion
This review paper aims to shed light on DL
applications related to MR imaging in MS and CIS. For this purpose,
an extensive search led to 45 original research papers focused on
this subject, that were further grouped in three categories i.e.
segmentation, classification and a broader category related to
image optimization and post processing techniques in MS and CIS.
Findings of the current review indicate that there is a growing
interest during the last years towards applying DL techniques for
segmentation and classification related to MS imaging studies.
A performance analysis was conducted by grouping the
studies into three main categories based on imaging tasks such as
segmentation in Table I,
classification in Table II and
various image processing tasks (synthesis, quality assessment,
image enhancement, etc) in Table
III. A few important issues in comparing these studies namely
are: i) the lack of a clear data stratification protocol (57-59);
ii) applying ‘good practices’ such as use of separate internal and
external validation sets (28);
iii) incorporating an adequate number of patients for DL model
convergence (35,83); iv) evaluating with appropriate
metrics (42,59) like AUC in addition to ACC in binary
classification tasks; and v) incorporating different metrics for
the same task such as DSC (23)
versus VD (22) in segmentation.
Thus, reporting the aforementioned performance measurements was a
difficult task which included interpreting figures, highly variable
experimental settings and, most importantly, identifying metrics on
unseen testing sets (if available) versus internal validation sets
which were also used in optimizing the models. Nevertheless,
Narayana et al (44)
proposed a FLAIR-based lesion segmentation with a testing DSC of
0.91 and Sander et al (29)
utilized a multi-dimensional gated recurrent unit model achieving a
performance up-to DSC 0.97. In MS classification, McKinley et
al (60) achieved sensitivity
up-to 0.72 on two different external validation sets, similar to
Narayana et al (63) (SN
0.72) on patient-basis versus SN 0.78 on a slice-basis in the same
study.
 | Table ISummary and performance metrics of
the reviewed publications relevant to detection-segmentation
tasks. |
Table I
Summary and performance metrics of
the reviewed publications relevant to detection-segmentation
tasks.
| Author (Ref.) | Year | Performance | Architecture | Dataset, #patients
(training/test) |
|---|
| Valverde et
al (22) | 2017 | Competition score
87.12 Volume difference down to 40.8 TPR up to 68.7 | A cascade of two 3D
patch-wise CNNs | 45 patients with MS
from MICCAI 2008 dataset, and two private MS clinical datasets |
| Birendaum et
al (23) | 2017 | Dice similarity
coefficient 0.627 | CNNs based on the
single view CNN (V-Net) and the longitudinal network (L-Net) | 2015 Longitudinal
Multiple Sclerosis Segmentation Challenge |
| Gros et al
(25) | 2019 | DSC up-to
0.604 | A sequence of two
CNNs | 1,042 subjects; 459
HC, 471 MS, 112 with other spinal pathologies |
| Aslani et al
(28) | 2019 | DSC up-to 0.611
(ISBI) DSC up-to 0.6655 (private) | Deep end-to-end 2D
CNN | 37 MS private
dataset, and 14 patients with MS from ISBI 2015 longitudinal MS
lesion segmentation challenge dataset |
| Sander et al
(29) | 2019 | Mean % change/SD
test- retest 0.45%/0.005 (brainstem) Dice score/SD 0.97/0.005 for
brainstem, 0.95/0.0131 for mesencephalon, 0.98/0.006 for pons,
0.95/0.015 for medulla oblongata | MD-GRU | 50 patients with MS
and 17 HC, 20 independent patients with MS, 50 patients with
Alzheimer's |
| Hashemi et
al (31) | 2019 | DSC 0.703 | 3D FCNN
(FC-DenseNet) | 15 patients from
the MSSEG of the 2016 Medical Image Computing and Computer Assisted
Intervention conference, five patient data from the longitudinal MS
lesion segmentation challenge of ISBI conference |
| Gabr et al
(34) | 2019 | T2 lesion 0.82 | FCNN | 1,000 patients with
RRMS from the CombiRx clinical trial |
| Weeda et al
(35) | 2019 | SN up-to 0.698 | DL CNN method
nicMSlesions | 14 patients with
MS |
| McKinley et
al (40) | 2019 | DSC 0.58 | A cascade of two
convolutional neural networks CNNs | 122 MRI exams from
patients with RRMS (90 patients for training and validation of the
classifier, 32 patients for testing), while a second dataset
including 10 MRI datasets was used for direct comparison with other
centers |
| Narayana et
al (41) | 2020 | DSC up-to 0.86 | FCNN | 1,008 patients with
clinically definite MS |
| Nair et al
(42) | 2020 | TPR 0.8, FDR
0.2 | 3D CNN WITH
dropout | 1,064 patients with
RRMS |
| McKinley et
al (43) | 2020 | DSC 0.661 | Two
state-of-the-art CNNs architectures, a 3D Unet consisting of a
reference implementation and a more recently proposed architecture,
the DeepSCAN | 2016 MSSEG training
dataset, retrained on a larger dataset comprising of 122 patients
with MS (50 training, 40 validation and 32 patients for
testing) |
| Narayana et
al (44) | 2020 | DSC up-to 0.91
(FLAIR) DSC up-to 0.90 (FLAIR, PD, T1, T2) DSC up-to 0.60 (PD) | U-net, a fully
CNN | 1,000 patients with
MS |
| Salem et al
(45) | 2020 | DSC for detection
0.83, DSC for segmentation 0.55 | FCNN | 60 early patients
with MS and CIS |
| Brown et al
(46) | 2020 | Not
applicable. | FCNN similar to
U-net | 1,018 scans form
256 participants in a study of pediatric-onset MS |
| Ackaouy et
al (47) | 2020 | No table with
absolute measurements, too many combinations of multi-centric
experiments, only boxplot results | 3D-Unet | 53 patients with MS
from MICCAI 2016 MS lesion segmentation challenge |
| Coronado et
al (49) | 2020 | DSC 0.77 | 3D CNN | 1,006 patients with
RRMS |
| La Rosa et
al (50) | 2020 | DSC 0.62 | FCNN architecture
based on the 3D U-Net | 90 patients with
MS |
| Gessert et
al (51) | 2020 | DSC up-to
0.656 | CNN | 89 patients with
MS |
| Essa et al
(52) | 2020 | VD down to 0.451,
TPR 0.681, FPR 0.632 | Region-based
CNN | 45 patients with MS
from MICCAI 2008 |
| Barquero et
al (53) | 2020 | DSC 0.835 | CNN (RimNet) | 124 patients with
MS |
 | Table IISummary and performance metrics of
the reviewed publications relevant to classification tasks. |
Table II
Summary and performance metrics of
the reviewed publications relevant to classification tasks.
| Author (Ref.) | Year | Performance | Architecture | Dataset, #patients
(training/test) |
|---|
| Yoo et al
(55) | 2018 | ACC 0.879, SN
0.873, SP 0.886, AUC 0.88 | Unsupervised
four-layer DBN | 55 patients with
RRMS and 44 HC |
| Wang et al
(56) | 2018 | ACC 0.988, SN
0.988, SP 0.988 | 14-layer CNN
combined with batch normalization, dropout and stochastic pooling
techniques | 38 patients with MS
from eHealth laboratory and 26 HC |
| Zhang et al
(57) | 2018 | ACC 0.982, SN
0.982, SP 0.982 | 10-layer deep
convolutional neural network used in this study was consisted of 7
convolution layers and 3 fully-connected layers | 676 MS brain slices
and 681 HC slices |
| Talo et al
(58) | 2019 | Stage-1: ACC 0.979,
SN 0.778, Stage-2: ACC 0.979, SN 0.74 Stage-3: ACC 1, SN 1 | CNN based
ResNet34 | 42 subjects (2 HC
and 40 patients with cerebrovascular, neoplastic, degenerative and
inflammatory disease types) |
| Lu et al
(59) | 2019 | ACC 1 | AlexNet structure
and stochastic gradient descent with momentum transfer learning
technique | MR images from 38
HC and 177 pathological, including pathologies such as Alzheimer's,
glioma, Huntington's, AIDS dementia, MS and Pick's |
| McKinley et
al (60) | 2019 | External validation
sets: Zurich: ACC 0.75, SN 0.60 Munich: ACC 0.85, SN 0.72 | DeepSCAN MS
classifier, a fully-convolutional neural network | Bern (train-test),
Zurich and Munich datasets |
| Marzullo et
al (61) | 2019 | F1 0.74, PR 0.76,
RC 0.75 | Graph CNN | 90 patients with MS
(12 CIS, 30 RRMS, 28 SPMS and 20 PPMS) and 24 HC |
| Eitel et al
(62) | 2019 | ACC 0.87, AUC
0.96 | 3D CNN and a
LRP | 921 patients with
Alzheimer's disease for training, validation in 76 MS and 71
HC |
| Narayana et
al (63) | 2020 | Slice: AUC 0.82, SN
0.78, SP 0.73 Patient: AUC 0.75, SN 0.72, SP 0.70 | CNN (fully
connected) | 1,008 participants
with MS |
| Maggi et al
(64) | 2020 | ACC 0.91, SN 0.89,
SP 0.92 Vesselness: ACC 0.69, SN 0.61, SP 0.77 | 3D CNN
(‘CVSnet’) | 42 MS, 33 MS mimics
and 5 patients with uncertain diagnosis |
| Wang et al
(65) | 2020 | ACC 0.75, SN 0.707,
SP 0.759 | CNN | 41 patients with
NMOSD and 47 patients with MS |
| Roca et al
(66) | 2020 | MSE 3 | CNN | 1,446 MS (971
training, 475 testing) |
| Lopatina et
al (67) | 2020 | ACC up-to 0.95
(echo 1) | CNN | 66 MS and 66
healthy controls |
 | Table IIISummary and performance metrics of
the reviewed publications relevant to imaging protocol related
tasks. |
Table III
Summary and performance metrics of
the reviewed publications relevant to imaging protocol related
tasks.
| Sub-category | Author (Ref.) | Year | Performance mean
(standard deviation) | Architecture | Dataset, #patients
(training/test) |
|---|
| Image quality
evaluation | Sreekumari et
al (68) | 2019 | Not applicable | CNN with seven
convolutional layers, four max pooling layers and three batch
normalization layers | MS and stroke
patients |
| Image quality
evaluation | Sujit et al
(69) | 2019 | ABIDE: AUC 0.90,
ACC 0.84, SN 0.77, SP 0.85 CombiRx: AUC 0.71, ACC 0.73, SN 0.41, SP
0.84 | DCNN | 1,064 images from
patients with autism from ABIDE database (60% training, 20%
validation and 20% test), and 110 patients with MS from the CombiRx
dataset |
| Enhance image | Zhao et al
(71) | 2019 | Not applicable | A DL method called
SMORE, Enhanced Deep Residual Network | Brain white matter
lesions from patients with MS quality |
| Generate synthetic
images | Wei et al
(73) | 2019 | MSE 918.07 (41.70)
SSIM 0.860 (0.031) | 3D FCNN | 20 patients with MS
and 4 HC |
| Generate synthetic
images | Salem et al
(77) | 2019 | T1w: MSE 0.03, SSIM
0.96 FLAIR: MSE 0.02, SSIM 0.98 | A two-input
two-output FCNN for MS lesion synthesis, in which the lesion
information was encoded as discrete binary intensities level
masks | 15 HC and 65
patients with CIS or RRMS, and the ISBI2015 dataset consisted of
five training and 14 testing subjects with four to five follow-up
studies for every subject |
| Generate synthetic
images | Wei et al
(78) | 2019 | MSE: 0.0083, PSNR
30.044 | Sketcher-refiner
GANs | 18 patients with MS
and 10 HC |
| Generate synthetic
images | Finck et al
(79) | 2020 | Not applicable | A diamond shape
topology generative adversarial NN (DiamondGAN) | 110 patients with
MS |
| Computational
analysis | Yoon et al
(80) | 2018 | pSNR 42.6 (0.61)
NRMSE 54.3 (2.5) HFEN 51.8 (2.5) SSIM 0.90 (0.01) | DL network (QSMnet)
with a modified U-net structure | 12 HC, a
microbleed, a MS and a haemorrhage patient |
| Computational
analysis | Bollman et
al (81) | 2019 | NRMSE 41.29 HFEN
23.69 SDSIM 0.21 | FCNN, DeepQSM | Synthetic data, and
a clinical dataset from a patient with MS |
| Computational
analysis | Dewey et al
(82) | 2019 | Not applicable | U-Net | 10 MS and two HC
were scanned twice, longitudinal retrospective data from 45
patients with RRMS |
| Computational
analysis | Liu et al
(83) | 2020 | Myelin water
fraction 0.039 (0.037) | NN | MWI data acquired
by a 32-echo 3D gradient and spin echo sequence, 6 healthy patients
and one MS |
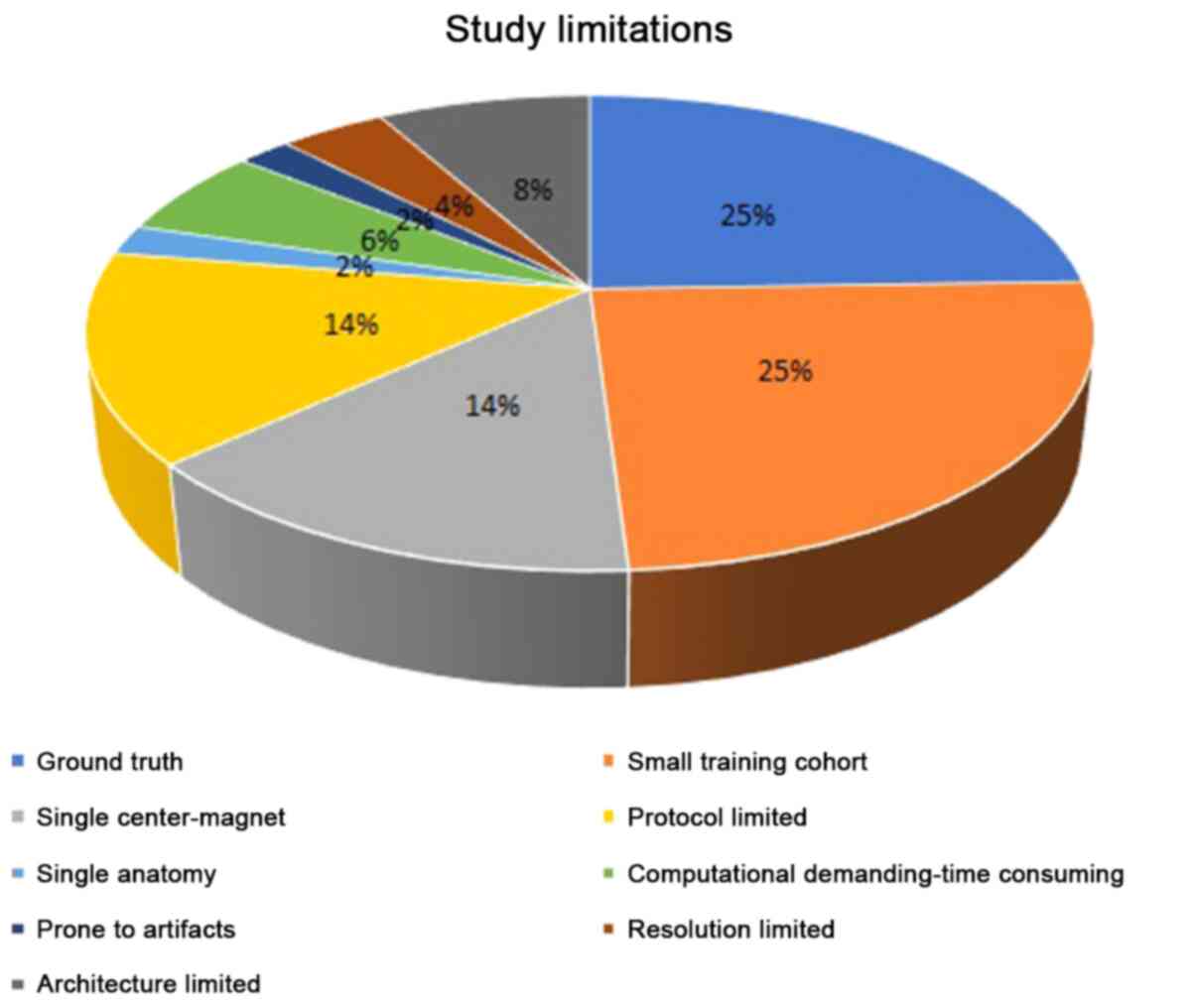
Concerning the limitations of the reviewed papers,
the most pronounced are associated with the absence of ground
truth, the small available cohorts, and the lack of generalization
of the results in multi-center data from diverse vendors, in
Fig. 4 presents the limitations of
the reviewed literature grouped in a chart pie. Finally, another
noticeable limitation of DL techniques is the lack of
explainability, which is particularly critical for the clinical
translation of such methods to clinical practice as well as for
adding precision in MS and CIS diagnosis and disease
management.
Considering the segmentation procedure, the
fundamental MRI protocols, Τ1-w, T2-w and PD can be characterized
based on their advantages concluding that, T1-w images enclose
detailed anatomical information and thus facilitate image
registration tasks involved in e.g. development of brain atlases.
On the contrary, T2-w images are beneficial for highlighting WM
lesions which appear as bright areas, thus assisting lesion
detection tasks. However, the main drawback of these techniques is
the insufficient differentiation of CSF, GM and lesions in terms of
image contrast. Finally, PD-w images are beneficial in revealing MS
lesions as areas of increased contrast compared to WM, even better
than T2-w, but still the relative image contrast between MS lesions
and CSF are in the same range of intensities. A major limitation
for accurate image segmentation is pixel misclassification due to
the partial volume effect generated from the low spatial resolution
of the MR protocols which renders difficult the accurate
delineation of brain regions and decreases the accuracy of
volumetric studies. Finally, inter-observer variability is a major
problem when trying to generate ground truth label data, while some
of the factors that may hamper this task include image quality,
different levels of user expertise and domain knowledge. Concerning
the reviewed research studies, in Narayana et al (41) segmentation results were assessed by
two experts, while in the study from Brown et al (46) results were evaluated with manual
segmentations stemming from only one expert.
Considering the classification related task,
initially it can be reported that fundamentally this is not a
binary problem. CIS diagnosis adds another level of complexity to
the classification problem, which can be treated as multiple binary
comparisons, i.e., controls vs. MS, controls vs CIS, CIS vs MS,
while for each problem a different classifier can be developed.
Furthermore, considering the constantly increasing imaging
databases, especially those including longitudinal data, research
studies may shift to the development of CAD based systems for early
detection of the MS disease. In addition, classification techniques
can be grouped in class-based methods in which MS lesions are
separated in WM, GM and CSF lesions, and outlier-based methods in
which GM, WM, and CSF are distinct classes from MS lesions.
Furthermore, images stemming from different centers,
scanners and protocols, provide a basis in order to build robust
algorithms that can generalize the aforementioned tasks. However,
in the small dataset setting, transfer learning techniques are
methods used for alleviating the limited availability of training
samples by tuning parameters of pre-trained networks with less data
in order to adapt existing models in new domains and achieve higher
level of generalization. In domain adaptation techniques, optimal
transfer is established through a source and a target domain in
order to deploy a model on the target site that was not included in
the training process, and this is a promising research field for
providing integrated solutions.
Moreover, according to best practices that must be
followed in order to build a robust DL methodology, some research
studies mentioned in this review article might not satisfy these
prerequisites. More specific, Ackaouy et al (47) and Talo et al (58) methodology did not follow the
universally accepted best practices in DL training, considering the
data splitting. Data should be split into training set, validation
set and testing set in order to ensure that the model will not be
over-fitted. External validation set is necessary in order to
enhance reproducibility and generalizability of a prediction model
to new patients. In the reviewed research articles, only a few used
external validation set in order to check reproducibility and
enhance generalizability (29,34,40,60,71).
Finally, data imbalance problem is very common in MS
machine learning applications and can lead to a training network
with sufficient high prediction but low recall, thus biased to the
class with the most data. Techniques such as two-step training,
sample re-weighting, balance sampling and similarity loss functions
are promising for bypassing such problems, while they can, also,
provide computational techniques suited to highly unbalanced
problems.
Future DL research work on MS considering also the
aforementioned limitations, should focus on three different
directions. The first direction regards the formation of large and
more diverse datasets, with optimized protocols, for providing a
ground truth validation framework of existing and future
techniques. The second direction concerns the need for MS imaging
data harmonization techniques for reducing the inherent information
heterogeneity of multi-centric data due to different vendors and
protocols and enabling the development of more robust and accurate
AI models. Finally, the development of CAD systems for early
detection of MS disease is of great importance based on the
increased availability of larger datasets. Such systems offering
cloud-based AI services, can provide an enhanced diagnostic
experience to clinicians and offer better diagnostic opportunities
especially in rural areas that usually lack access to specialists.
All these directions regarding the future of DL in MS have to
promote explainability and trustworthiness, and not only target
increased performance, in order to realize the vision of real-world
use of AI algorithms in clinical practice.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
EEK, EP and KM conceived the designed study. EEK,
EP, ET and KM researched the literature, performed analysis and
interpretation of data and drafted the manuscript. EEK, EP, ET,
TGM, PS, GZP, AT, DAS, AK and KM critically revised the article for
important intellectual content. All authors have read and approved
the final manuscript. Data sharing is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief of the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision for this article.
All the authors declare that they have no competing interests.
References
|
1
|
Ortiz GG, Pacheco-Moisés FP, Macías-Islas
MÁ, Flores-Alvarado LJ, Mireles-Ramírez MA, González-Renovato ED,
Hernández-Navarro VE, Sánchez-López AL and Alatorre-Jiménez MA:
Role of the blood-brain barrier in multiple sclerosis. Arch Med
Res. 45:687–697. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lopes Pinheiro MA, Kooij G, Mizee MR,
Kamermans A, Enzmann G, Lyck R, Schwaninger M, Engelhardt B and de
Vries HE: Immune cell trafficking across the barriers of the
central nervous system in multiple sclerosis and stroke. Biochim
Biophys Acta. 1862:461–471. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Miller DH, Chard DT and Ciccarelli O:
Clinically isolated syndromes. Lancet Neurol. 11:157–169.
2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kappos L, Polman CH, Freedman MS, Edan G,
Hartung HP, Miller DH, Montalban X, Barkhof F, Bauer L, Jakobs P,
et al: Treatment with interferon beta-1b delays conversion to
clinically definite and McDonald MS in patients with clinically
isolated syndromes. Neurology. 67:1242–1249. 2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fu Y, Talavage TM and Cheng JX: New
imaging techniques in the diagnosis of multiple sclerosis. Expert
Opin Med Diagn. 2:1055–1065. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Horsfield MA, Rovaris M, Rocca MA, Rossi
P, Benedict RH, Filippi M and Bakshi R: Whole-brain atrophy in
multiple sclerosis measured by two segmentation processes from
various MRI sequences. J Neurol Sci. 216:169–177. 2003.PubMed/NCBI View Article : Google Scholar
|
|
7
|
van Walderveen MA, Kamphorst W, Scheltens
P, van Waesberghe JH, Ravid R, Valk J, Polman CH and Barkhof F:
Histopathologic correlate of hypointense lesions on T1-weighted
spin-echo MRI in multiple sclerosis. Neurology. 50:1282–1288.
1998.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bakshi R, Ariyaratana S, Benedict RH and
Jacobs L: Fluid-attenuated inversion recovery magnetic resonance
imaging detects cortical and juxtacortical multiple sclerosis
lesions. Arch Neurol. 58:742–748. 2001.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Polman CH, Reingold SC, Edan G, Filippi M,
Hartung HP, Kappos L, Lublin FD, Metz LM, McFarland HF, O'Connor
PW, et al: Diagnostic criteria for multiple sclerosis: 2005
revisions to the ‘McDonald Criteria’. Ann Neurol. 58:840–846.
2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Richards TL: Proton MR spectroscopy in
multiple sclerosis: Value in establishing diagnosis, monitoring
progression, and evaluating therapy. AJR Am J Roentgenol.
157:1073–1078. 1991.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pike GB, De Stefano N, Narayanan S,
Worsley KJ, Pelletier D, Francis GS, Antel JP and Arnold DL:
Multiple sclerosis: Magnetization transfer MR imaging of white
matter before lesion appearance on T2-weighted images. Radiology.
215:824–830. 2000.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Filippi M and Rocca MA: Magnetization
transfer magnetic resonance imaging in the assessment of
neurological diseases. J Neuroimaging. 14:303–313. 2004.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Rovaris M, Gass A, Bammer R, Hickman SJ,
Ciccarelli O, Miller DH and Filippi M: Diffusion MRI in multiple
sclerosis. Neurology. 65:1526–1532. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lapointe E, Li DKB, Traboulsee AL and
Rauscher A: What Have We Learned from Perfusion MRI in Multiple
Sclerosis? AJNR Am J Neuroradiol. 39:994–1000. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kotsiantis SB, Zaharakis I and Pintelas P:
Supervised machine learning: A review of classification techniques.
Emerging artificial intelligence applications in computer
engineering. 160:3–24. 2007.
|
|
16
|
Mortazavi D, Kouzani AZ and
Soltanian-Zadeh H: Segmentation of multiple sclerosis lesions in MR
images: A review. Neuroradiology. 54:299–320. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Uddin M, Wang Y and Woodbury-Smith M:
Artificial intelligence for precision medicine in
neurodevelopmental disorders. NPJ Digit Med. 2(112)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bergsland N, Horakova D, Dwyer MG, Uher T,
Vaneckova M, Tyblova M, Seidl Z, Krasensky J, Havrdova E and
Zivadinov R: Gray matter atrophy patterns in multiple sclerosis: A
10-year source-based morphometry study. Neuroimage Clin.
17:444–451. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Pontillo G, Petracca M, Cocozza S and
Brunetti A: The Development of Subcortical Gray Matter Atrophy in
Multiple Sclerosis: One Size Does Not Fit All. AJNR Am J
Neuroradiol. 41:E80–E81. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Almutairi AH, Hassan HA, Suppiah S,
Alomair OI, Alshoaibi A, Almutairi H and Mahmud R: Lesion load
assessment among multiple sclerosis patient using DIR, FLAIR, and
T2WI sequences. Egypt J Radiol Nucl Med. 51(209)2020.
|
|
21
|
Thompson AJ, Banwell BL, Barkhof F,
Carroll WM, Coetzee T, Comi G, Correale J, Fazekas F, Filippi M,
Freedman MS, et al: Diagnosis of multiple sclerosis: 2017 revisions
of the McDonald criteria. Lancet Neurol. 17:162–173.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Valverde S, Cabezas M, Roura E,
González-Villà S, Pareto D, Vilanova JC, Ramió-Torrentà L, Rovira
À, Oliver A and Lladó X: Improving automated multiple sclerosis
lesion segmentation with a cascaded 3D convolutional neural network
approach. Neuroimage. 155:159–168. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Birenbaum A and Greenspan H: Multi-view
longitudinal CNN for multiple sclerosis lesion segmentation. Eng
Appl Artif Intell. 65:111–118. 2017.
|
|
24
|
Carass A, Roy S, Jog A, Cuzzocreo JL,
Magrath E, Gherman A, Button J, Nguyen J, Bazin PL, Calabresi PA,
et al: Longitudinal multiple sclerosis lesion segmentation data
resource. Data Brief. 12:346–350. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gros C, De Leener B, Badji A, Maranzano J,
Eden D, Dupont SM, Talbott J, Zhuoquiong R, Liu Y, Granberg T, et
al: Automatic segmentation of the spinal cord and intramedullary
multiple sclerosis lesions with convolutional neural networks.
Neuroimage. 184:901–915. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Milletari F, Navab N and Ahmadi SA: V-Net:
Fully Convolutional Neural Networks for Volumetric Medical Image
Segmentation. In: Proceedings of the 2016 Fourth International
Conference on 3D Vision (3DV). IEEE, Stanford, CA, pp565-571,
2016.
|
|
27
|
De Leener B, Kadoury S and Cohen-Adad J:
Robust, accurate and fast automatic segmentation of the spinal
cord. Neuroimage. 98:528–536. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Aslani S, Dayan M, Storelli L, Filippi M,
Murino V, Rocca MA and Sona D: Multi-branch convolutional neural
network for multiple sclerosis lesion segmentation. Neuroimage.
196:1–15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Sander L, Pezold S, Andermatt S, Amann M,
Meier D, Wendebourg MJ, Sinnecker T, Radue EW, Naegelin Y,
Granziera C, et al: Alzheimer's Disease Neuroimaging Initiative:
Accurate, rapid and reliable, fully automated MRI brainstem
segmentation for application in multiple sclerosis and
neurodegenerative diseases. Hum Brain Mapp. 40:4091–4104.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Iglesias JE, Van Leemput K, Bhatt P,
Casillas C, Dutt S, Schuff N, Truran-Sacrey D, Boxer A and Fischl
B: Alzheimer's Disease Neuroimaging Initiative. Bayesian
segmentation of brainstem structures in MRI. Neuroimage.
113:184–195. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hashemi SR, Salehi SSM, Erdogmus D, Prabhu
SP, Warfield SK and Gholipour A: Asymmetric Loss Functions and Deep
Densely Connected Networks for Highly Imbalanced Medical Image
Segmentation: Application to Multiple Sclerosis Lesion Detection.
IEEE Access. 7:721–1735. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Commowick O, Cervenansky F and Ameli R:
MSSEG challenge proceedings: multiple sclerosis lesions
segmentation challenge using a data management and processing
infrastructure. MICCAI, Athens, 2016.
|
|
33
|
Carass A, Roy S, Jog A, Cuzzocreo JL,
Magrath E, Gherman A, Button J, Nguyen J, Prados F, Sudre CH, et
al: Longitudinal multiple sclerosis lesion segmentation: Resource
and challenge. Neuroimage. 148:77–102. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Gabr RE, Coronado I, Robinson M, Sujit SJ,
Datta S, Sun X, Allen WJ, Lublin FD, Wolinsky JS and Narayana PA:
Brain and lesion segmentation in multiple sclerosis using fully
convolutional neural networks: A large-scale study. Mult Scler.
26:1217–1226. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Weeda MM, Brouwer I, de Vos ML, de Vries
MS, Barkhof F, Pouwels PJW and Vrenken H: Comparing lesion
segmentation methods in multiple sclerosis: Input from one manually
delineated subject is sufficient for accurate lesion segmentation.
Neuroimage Clin. 24(102074)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Valverde S, Salem M, Cabezas M, Pareto D,
Vilanova JC, Ramió-Torrentà L, Rovira À, Salvi J, Oliver A and
Lladó X: One-shot domain adaptation in multiple sclerosis lesion
segmentation using convolutional neural networks. Neuroimage Clin.
21(101638)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Shiee N, Bazin PL, Ozturk A, Reich DS,
Calabresi PA and Pham DL: A topology-preserving approach to the
segmentation of brain images with multiple sclerosis lesions.
Neuroimage. 49:1524–1535. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Schmidt P: Bayesian Inference for
Structured Additive Regression Models for Large-Scale Problems with
Applications to Medical Imaging (unpublished PhD thesis).
Ludwig-Maximilians-Universität München, 2016.
|
|
39
|
Griffanti L, Zamboni G, Khan A, Li L,
Bonifacio G, Sundaresan V, Schulz UG, Kuker W, Battaglini M,
Rothwell PM, et al: BIANCA (Brain Intensity AbNormality
Classification Algorithm): A new tool for automated segmentation of
white matter hyperintensities. Neuroimage. 141:191–205.
2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
McKinley R, Wepfer R, Aschwanden F,
Grunder L, Muri R, Rummel C, Verma R, Weisstanner C, Reyes M,
Salmen A, et al: Simultaneous lesion and neuroanatomy segmentation
in multiple sclerosis using deep neural networks.
arXiv:1901.07419.
|
|
41
|
Narayana PA, Coronado I, Sujit SJ,
Wolinsky JS, Lublin FD and Gabr RE: Deep-Learning-Based Neural
Tissue Segmentation of MRI in Multiple Sclerosis: Effect of
Training Set Size. J Magn Reson Imaging. 51:1487–1496.
2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Nair T, Precup D, Arnold DL and Arbel T:
Exploring uncertainty measures in deep networks for Multiple
sclerosis lesion detection and segmentation. Med Image Anal.
59(101557)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
McKinley R, Wepfer R, Aschwanden F,
Grunder L, Muri R, Rummel C, Verma R, Weisstanner C, Reyes M,
Salmen A, et al: Simultaneous lesion and brain segmentation in
multiple sclerosis using deep neural networks. Sci Rep.
11(1087)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Narayana PA, Coronado I, Sujit SJ, Sun X,
Wolinsky JS and Gabr RE: Are multi-contrast magnetic resonance
images necessary for segmenting multiple sclerosis brains? A large
cohort study based on deep learning. Magn Reson Imaging. 65:8–14.
2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Salem M, Valverde S, Cabezas M, Pareto D,
Oliver A, Salvi J, Rovira À and Lladó X: A fully convolutional
neural network for new T2-w lesion detection in multiple sclerosis.
Neuroimage Clin. 25(102149)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Brown RA, Fetco D, Fratila R, Fadda G,
Jiang S, Alkhawajah NM, Yeh EA, Banwell B, Bar-Or A and Arnold DL:
Canadian Pediatric Demyelinating Disease Network. Deep learning
segmentation of orbital fat to calibrate conventional MRI for
longitudinal studies. Neuroimage. 208(116442)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ackaouy A, Courty N, Vallée E, Commowick
O, Barillot C and Galassi F: Unsupervised Domain Adaptation With
Optimal Transport in Multi-Site Segmentation of Multiple Sclerosis
Lesions From MRI Data. Front Comput Neurosci. 14(19)2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Commowick O, Istace A, Kain M, Laurent B,
Leray F, Simon M, Pop SC, Girard P, Améli R, Ferré JC, et al:
Objective Evaluation of Multiple Sclerosis Lesion Segmentation
using a Data Management and Processing Infrastructure. Sci Rep.
8(13650)2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Coronado I, Gabr RE and Narayana PA: Deep
learning segmentation of gadolinium-enhancing lesions in multiple
sclerosis. Mult Scler. 27:219–527. 2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
La Rosa F, Abdulkadir A, Fartaria MJ,
Rahmanzadeh R, Lu PJ, Galbusera R, Barakovic M, Thiran JP,
Granziera C and Cuadra MB: Multiple sclerosis cortical and WM
lesion segmentation at 3T MRI: A deep learning method based on
FLAIR and MP2RAGE. Neuroimage Clin. 27(102335)2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Gessert N, Krüger J, Opfer R, Ostwaldt AC,
Manogaran P, Kitzler HH, Schippling S and Schlaefer A: Multiple
sclerosis lesion activity segmentation with attention-guided
two-path CNNs. Comput Med Imaging Graph. 84(101772)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Essa E, Aldesouky D, Hussein SE and Rashad
MZ: Neuro-fuzzy patch-wise R-CNN for multiple sclerosis
segmentation. Med Biol Eng Comput. 58:2161–2175. 2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Barquero G, La Rosa F, Kebiri H, Lu PJ,
Rahmanzadeh R, Weigel M, Fartaria MJ, Kober T, Théaudin M, Du
Pasquier R, et al: RimNet: A deep 3D multimodal MRI architecture
for paramagnetic rim lesion assessment in multiple sclerosis.
Neuroimage Clin. 28(102412)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Gautam R and Sharma M: Prevalence and
Diagnosis of Neurological Disorders Using Different Deep Learning
Techniques: A Meta-Analysis. J Med Syst. 44(49)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yoo Y, Tang LYW, Brosch T, Li DKB, Kolind
S, Vavasour I, Rauscher A, MacKay AL, Traboulsee A and Tam RC: Deep
learning of joint myelin and T1w MRI features in normal-appearing
brain tissue to distinguish between multiple sclerosis patients and
healthy controls. Neuroimage Clin. 17:169–178. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Wang SH, Tang C, Sun J, Yang J, Huang C,
Phillips P and Zhang YD: Multiple Sclerosis Identification by
14-Layer Convolutional Neural Network With Batch Normalization,
Dropout, and Stochastic Pooling. Front Neurosci.
12(818)2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhang YD, Pan C, Sun J and Tang C:
Multiple sclerosis identification by convolutional neural network
with dropout and parametric ReLU. J Comput Sci. 28(818)2018.
|
|
58
|
Talo M, Baloglu UB, Yıldırım Ö and Acharya
UR: Application of deep transfer learning for automated brain
abnormality classification using MR images. Cogn Syst Res.
54:176–188. 2019.
|
|
59
|
Lu S, Lu Z and Zhang YD: Pathological
brain detection based on AlexNet and transfer learning. J Comput
Sci. 30:41–47. 2019.
|
|
60
|
McKinley R, Wepfer R, Grunder L,
Aschwanden F, Fischer T, Friedli C, Muri R, Rummel C, Verma R,
Weisstanner C, et al: Automatic detection of lesion load change in
Multiple Sclerosis using convolutional neural networks with
segmentation confidence. Neuroimage Clin. 25(102104)2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Marzullo A, Kocevar G, Stamile C,
Durand-Dubief F, Terracina G, Calimeri F and Sappey-Marinier D:
Classification of Multiple Sclerosis Clinical Profiles via Graph
Convolutional Neural Networks. Front Neurosci.
13(594)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Eitel F, Soehler E, Bellmann-Strobl J,
Brandt AU, Ruprecht K, Giess RM, Kuchling J, Asseyer S, Weygandt M,
Haynes JD, et al: Uncovering convolutional neural network decisions
for diagnosing multiple sclerosis on conventional MRI using
layer-wise relevance propagation. Neuroimage Clin.
24(102003)2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Narayana PA, Coronado I, Sujit SJ,
Wolinsky JS, Lublin FD and Gabr RE: Deep Learning for Predicting
Enhancing Lesions in Multiple Sclerosis from Noncontrast MRI.
Radiology. 294:398–404. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Maggi P, Fartaria MJ, Jorge J, La Rosa F,
Absinta M, Sati P, Meuli R, Du Pasquier R, Reich DS, Cuadra MB, et
al: CVSnet: A machine learning approach for automated central vein
sign assessment in multiple sclerosis. NMR Biomed.
33(e4283)2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wang Z, Yu Z, Wang Y, Zhang H, Luo Y, Shi
L, Wang Y and Guo C: 3D Compressed Convolutional Neural Network
Differentiates Neuromyelitis Optical Spectrum Disorders From
Multiple Sclerosis Using Automated White Matter Hyperintensities
Segmentations. Front Physiol. 11(612928)2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Roca P, Attye A, Colas L, Tucholka A,
Rubini P, Cackowski S, Ding J, Budzik JF, Renard F, Doyle S, et al:
OFSEP Investigators; Steering Committee; Investigators; Imaging
group: Artificial intelligence to predict clinical disability in
patients with multiple sclerosis using FLAIR MRI. Diagn Interv
Imaging. 101:795–802. 2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Lopatina A, Ropele S, Sibgatulin R,
Reichenbach JR and Güllmar D: Investigation of Deep-Learning-Driven
Identification of Multiple Sclerosis Patients Based on
Susceptibility-Weighted Images Using Relevance Analysis. Front
Neurosci. 14(609468)2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Sreekumari A, Shanbhag D, Yeo D, Foo T,
Pilitsis J, Polzin J, Patil U, Coblentz A, Kapadia A, Khinda J, et
al: A Deep Learning-Based Approach to Reduce Rescan and Recall
Rates in Clinical MRI Examinations. AJNR Am J Neuroradiol.
40:217–223. 2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sujit SJ, Coronado I, Kamali A, Narayana
PA and Gabr RE: Automated image quality evaluation of structural
brain MRI using an ensemble of deep learning networks. J Magn Reson
Imaging. 50:1260–1267. 2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Lublin FD, Cofield SS, Cutter GR, Conwit
R, Narayana PA, Nelson F, Salter AR, Gustafson T and Wolinsky JS:
CombiRx Investigators. Randomized study combining interferon and
glatiramer acetate in multiple sclerosis. Ann Neurol. 73:327–340.
2013.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Zhao C, Shao M, Carass A, Li H, Dewey BE,
Ellingsen LM, Woo J, Guttman MA, Blitz AM, Stone M, et al:
Applications of a deep learning method for anti-aliasing and
super-resolution in MRI. Magn Reson Imaging. 64:132–141.
2019.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Jog A, Carass A and Prince JL: Self
Super-resolution for Magnetic Resonance Images. Med Image Comput
Comput Assist Interv. 9902:553–560. 2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Wei W, Poirion E, Bodini B, Durrleman S,
Colliot O, Stankoff B and Ayache N: Fluid-attenuated inversion
recovery MRI synthesis from multisequence MRI using
three-dimensional fully convolutional networks for multiple
sclerosis. J Med Imaging (Bellingham). 6(014005)2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Ye DH, Zikic D, Glocker B, Criminisi A and
Konukoglu E: Modality propagation: coherent synthesis of
subject-specific scans with data-driven regularization. Med Image
Comput Comput Assist Interv. 13:606–613. 2013.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Jog A, Carass A, Pham DL and Prince JL:
RANDOM FOREST FLAIR RECONSTRUCTION FROM T1,
T2, AND PD-WEIGHTED MRI. Proc
IEEE Int Symp Biomed Imaging. 2014:1079–1082. 2014.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Ronneberger O, Fischer P and Brox T:
U-Net: Convolutional Networks for Biomedical Image Segmentation.
Lect Notes Comput Sci. 9351:234–241. 2015.
|
|
77
|
Salem M, Valverde S, Cabezas M, Pareto D,
Oliver A, Salvi J, Rovira À and Lladó X: Multiple sclerosis lesion
synthesis in MRI using an encoder-decoder U-NET. IEEE Access.
7:25171–25184. 2019.
|
|
78
|
Wei W, Poirion E, Bodini B, Durrleman S,
Ayache N, Stankoff B and Colliot O: Predicting PET-derived
demyelination from multimodal MRI using sketcher-refiner
adversarial training for multiple sclerosis. Med Image Anal.
58(101546)2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Finck T, Li H, Grundl L, Eichinger P,
Bussas M, Mühlau M, Menze B and Wiestler B: Deep-Learning Generated
Synthetic Double Inversion Recovery Images Improve Multiple
Sclerosis Lesion Detection. Invest Radiol. 55:318–323.
2020.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Yoon J, Gong E, Chatnuntawech I, Bilgic B,
Lee J, Jung W, Ko J, Jung H, Setsompop K, Zaharchuk G, et al:
Quantitative susceptibility mapping using deep neural network:
QSMnet. Neuroimage. 179:199–206. 2018.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Bollmann S, Rasmussen KGB, Kristensen M,
Blendal RG, Østergaard LR, Plocharski M, O'Brien K, Langkammer C,
Janke A and Barth M: DeepQSM-using deep learning to solve the
dipole inversion for quantitative susceptibility mapping.
Neuroimage. 195:373–383. 2019.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Dewey BE, Zhao C, Reinhold JC, Carass A,
Fitzgerald KC, Sotirchos ES, Saidha S, Oh J, Pham DL, Calabresi PA,
et al: DeepHarmony: A deep learning approach to contrast
harmonization across scanner changes. Magn Reson Imaging.
64:160–170. 2019.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Liu H, Xiang QS, Tam R, Dvorak AV, MacKay
AL, Kolind SH, Traboulsee A, Vavasour IM, Li DKB, Kramer JK, et al:
Myelin water imaging data analysis in less than one minute.
Neuroimage. 210(116551)2020.PubMed/NCBI View Article : Google Scholar
|