Introduction
Colorectal cancer (CRC) is the third most common
type of cancer worldwide and poses a threat to human life (1). Current advances in the treatment of
CRC have not successfully decreased the high incidence of CRC
(2). The pathogenesis of CRC is not
completely understood; therefore, the identification of novel
targets to effectively improved the severe pathological conditions
of patients with CRC is important (3).
Vasohibin 1 (VASH1) is an endogenous inhibitor of
angiogenesis induced by VEGF and fibroblast growth factor
2(4). It has been reported that
angiogenesis is inhibited by a negative feedback mechanism under
physiological conditions (4). A
study revealed that VASH1 was upregulated in several types of tumor
tissues, and exhibited therapeutic efficacy by preventing tumor
angiogenesis and suppressing tumor growth in a nude mice xenograft
model (5). VASH1 is not only
expressed in the vascular endothelium, but also in tumor cells
(6,7). Another study demonstrated that
microRNA (miR)-143-3p triggered by N6-methyladenosine promoted lung
cancer brain metastasis via regulating VASH1(8). In addition, VASH1 could not cause
damage to healthy blood vessels in mice (9). Angiogenesis in hepatocellular
carcinoma (HCC) could be promoted by the VEGF-mediated enhancer of
zeste 2 polycomb repressive complex 2 subunit/VASH1 signaling
pathway in cancer-related fibroblasts (10). VASH1 is primarily expressed in the
cytoplasm of bladder cancer cells and several vascular endothelial
cells. VASH1 was associated with tumor stage, pathological grade
and distant metastasis, whereas its high expression was
significantly associated with 5-year overall survival (OS) and
progression free survival (PFS) rates (11). In CRC tissues, the expression levels
of VASH1 and VEGFA were positively correlated with microvessel
density. The OS and PFS of patients with high VASH1 expression were
significantly lower compared with those with low VASH1 expression,
which indicated that VASH1 protein expression is an independent
risk factor affecting OS and PFS (7). VASH1 overexpression promoted CRC cell
apoptosis and senescence, inhibited CRC cell proliferation and
colony formation, and suppressed tumor growth in vivo
(12).
miRNAs are non-coding endogenous conserved RNA
molecules that bind to the 3'-untranslated regions of their target
mRNAs (13,14). Emerging evidence has suggested that
miR-1269a is upregulated in CRC (15). A study showed that the expression of
miR-1269a was increased in non-small cell lung cancer (NSCLC)
tissues compared with that in adjacent healthy tissues, whereas
miR-1269a knockdown attenuated NSCLC cell proliferation, colony
formation and cell cycle progression (16). Another study suggested that miR-1269
could serve as an oncogene by promoting tumor metastasis and
forming a positive feedback loop with TGF-β (15). Additionally, miR-1269a
overexpression could enhance the apoptosis of gastric cancer cells
(17). Finally, long non-coding RNA
LINC00261 could attenuate lung cancer tumor growth and metastasis
via the miR-1269a/FOXO1 signaling pathway (18). Therefore, we speculated that
miR-1269a may regulate CRC cells via targeting VASH1.
Circular RNAs (circRNAs/circs) are a class of
endogenous non-coding RNAs with covalently closed continuous loops
(19). A previous study reported
that circASS1 expression is decreased and can inhibit the invasion
and migration of breast cancer cells (20). The potential regulatory association
between miR-1269a and circASS1, and how circASS1 regulates CRC
cells via the miR-1269a/VASH1 axis is not completely
understood.
Therefore, the present study aimed to elucidate the
role of the circASS1/miR-1269a/VASH1 axis in regulating the
proliferation, invasion and migration of CRC cells, as well as the
potential underlying mechanism to identify novel therapeutic
targets for CRC.
Materials and methods
Clinical specimens
Tumor tissue samples and pathologically verified
normal tissue samples taken >2 cm from the tumor were collected
from 10 patients with CRC who were not treated with chemotherapy or
radiotherapy from Huizhou Municipal Central Hospital of Guangdong
Province (Huizhou, China) between May 2018 and May 2019. In total,
seven males and three females were recruited (age, 35-72 years).
The inclusion criteria were patients with only primary colon
cancer. Patients who had received preoperative chemotherapy or
radiotherapy were excluded. Samples were snap frozen in liquid
nitrogen immediately after collection and stored at -80˚C until
further analysis. All patients provided written informed consent
prior to enrollment in the present study. The present study was
approved by the Huizhou Municipal Central Hospital of Guangdong
Province.
Cell culture
CRC cell lines (HCT116, Caco2, LoVo, SW480 and HT29)
were obtained from Procell Life Science & Technology Co., Ltd.
The normal colon epithelial cell line (HIEC-6) was obtained from
BioVector Science Lab, Inc. The HT-29 cell line was authenticated
by STR. HIEC-6 cells were cultured in DMEM (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.). CRC cell lines were cultured in DMEM
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS, 100 U/ml penicillin and 100 µg/ml streptomycin (both from
Gibco; Thermo Fisher Scientific, Inc.) All cell lines were cultured
in a humidified incubator at 37˚C in a 5% CO2
atmosphere.
Cell transfection
miR-1269a mimic (100 nM; cat. no. miR10005923-1-5)
negative control (NC) mimic (100 nM; cat. no. miR1N0000001-1-5),
miR-1269a inhibitor (100 nM; cat. no. miR20005923-1-5) and NC
inhibitor (100 nM; cat. no. miR2N0000001-1-5) were purchased from
Guangzhou RiboBio Co., Ltd. pcDNA3.1-circASS1 (Oe-circASS1; 100
nM), pcDNA3.1-NC (Oe-NC; 100 nM), short hairpin RNA (shRNA) against
VASH1 (shRNA-VASH1; 100 nM) and shRNA-NC (100 nM) plasmids were all
constructed by Shanghai GenePharma Co., Ltd. When HT29 cells
reached 60-80% confluence, cell transfection was performed using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 24 h at 37˚C. At 48 h post-transfection,
cells were used for subsequent experiments. Untreated HT29 cells
were used as the control.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells and tissues using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Subsequently, RNA was
reverse transcribed into cDNA using the All-in-One miRNA
First-Strand cDNA Synthesis kit (GeneCopoeia, Inc.) or the
PrimeScript RT Reagent kit with gDNA Eraser (Takara Bio, Inc.) in
accordance with the manufacturer's protocol. qPCR was performed
using the SYBR Green PCR kit (GeneCopoeia, Inc.) on the
StepOnePlus™ Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The thermocycling conditions were as
follows: 95˚C for 30 sec, followed by 40 cycles of 95˚C for 10 sec
and 60˚C for 30 sec. U6 and GAPDH were used as internal controls
for miRNA and mRNA expression, respectively. Additionally, internal
β-actin was used to normalize circRNA expression. The
2-∆∆Cq method was used to determine relative expression
(21). Primer sequences were as
follows: circASS1 forward, 5'-GCCGTATTGACATCGTGGAG-3' and reverse,
5'-TCGAGAATGTCAGGGGTGT-3'; miR1269a forward,
5'-GCTGGACTGAGCCGTGC-3' and reverse, 5'-CAGTGCGTGTCGTGGAGT-3';
VASH1 forward, 5'-AGATCCCCATACCGAGTGTG-3' and reverse,
5'-GCTTCCAGGCATTTGATTGGC-3'; U6 forward, 5'-AGCCCGCACTCAGAACATC-3'
and reverse, 5'-GCCACCAAGACAATCATCC-3'; GAPDH forward,
5'-ACAACTTTGGTATCGTGGAAGG-3' and reverse, 5'-GCCATCACGCCACAGTTTC-3'
and β-actin forward, 5'-CACCTTCTACAATGAGCTGCGTGTG-3' and reverse,
5'-ATAGCACAGCCTGGATAGCAACGTAC-3'.
Western blotting
Total protein was extracted from cells using an RIPA
lysis buffer kit (Boster Biological Technology). Protein
concentrations were quantified using the bicinchoninic acid method
(Thermo Fisher Scientific, Inc.). Following protein separation via
10% SDS-PAGE (40 µg/lane), proteins were transferred onto PVDF
membranes (Beyotime Institute of Biotechnology). Following blocking
with 5% skimmed milk for 2 h at room temperature, the membranes
were then incubated overnight at 4˚C with primary antibodies
targeted against: VASH1 (cat. no. ab199732; 1:1,000; Abcam), Bcl-2
(cat. no. ab32124; 1:1,000; Abcam), Bax (cat. no. ab32503; 1:1,000;
Abcam), cleaved caspase-3 (cat. no. ab32042; 1:500; Abcam),
caspase-3 (cat. no. ab32351; 1:500; Abcam), cleaved poly
(ADP-ribose) polymerase (PARP; cat. no. ab32064; 1:1,000; Abcam),
PARP (cat. no. ab191217; 1:1,000; Abcam) and GAPDH (cat. no.
ab9485; 1:2,500; Abcam). After washing three times with TBST (10%
Tween-20), the membranes were then incubated with the
HRP-conjugated goat anti-rabbit secondary antibody (cat. no.
ab6721; 1:2,000; Abcam) for 1 h at room temperature. The signals
were developed using an enhanced chemiluminescence kit (EMD
Millipore). Densitometric analysis was performed using ImageJ
(version 1.48v; National Institutes of Health).
Colony formation assay
HT29 cells were seeded (2x103 cells/well)
into 96-well plates and cultured in complete medium until colony
formation. After washing three times with PBS, the colonies were
fixed with 100% methanol for 30 min at room temperature and stained
with crystal violet at room temperature for 20 min. Clones
containing ≥50 cells were counted. Cells were observed under a
light microscope (DM1000; Leica Microsystems GmbH) and five fields
of view were randomly selected for manual counting of colonies.
MTT assay
Following transfection, HT29 cells were seeded
(2x104 cells/well) into 96-well plates for 24 h.
Subsequently, culture medium was discarded and serum-free medium
was added. Then, 20 µl MTT reagent (5 mg/ml) was added to each well
and incubated for an additional 4 h. The medium was discarded and
100 µl dimethyl sulfoxide was added to each well to dissolve the
purple formazan. The optical density was measured at a wavelength
of 490 nm using a microplate reader (Bio-Rad Laboratories,
Inc.).
TUNEL assay
Transfected HT29 cells were fixed with 4%
paraformaldehyde for 30 min at room temperature. Subsequently, the
TdT enzyme was added onto the slide and cells were incubated at
37˚C for 2 h. Cells were then incubated for an additional 1 h at
room temperature in the dark in the presence of streptavidin-HRP.
Then, 0.05% DAB was stained at room temperature for 5 min, mounting
medium (Beijing Solarbio Science & Technology, Inc.) was used
to seal the slides, and the cells were observed under a fluorescent
microscope with 5 random fields of view per group. Washing with PBS
was performed between all steps.
Matrigel invasion assays
To evaluate cell migration and invasion abilities, a
Transwell chamber (Corning, Inc.) precoated with Matrigel at 37˚C
overnight was used. Transfected HT29 cells (5x104) were
suspended in 200 µl serum-free DMEM and then added to the upper
chamber, the lower chamber was supplemented with DMEM containing
10% FBS and incubated for 24 h at 37˚C. After washing three times
with PBS, cells were fixed with 4% paraformaldehyde for 30 min at
room temperature and then stained with 0.1% crystal violet solution
for 15 min at room temperature and observed under a light
microscope. Cell migration and invasion rates were determined based
on crystal violet staining.
Wound healing assay
Following transfection, HT29 cells were grown to
100% confluence in 6-well culture plates. Subsequently, a 10-µl
pipette tip was used to scratch the cell monolayer, followed by
washing with PBS washing to remove cell debris. Cells were cultured
in DMEM supplemented with 2% FBS. The wound was observed and
photographed at 0 and 24 h using a light microscope with a digital
camera and quantified using ImageJ (version 1.48v; National
Institutes of Health).
Luciferase reporter assay
The association between miR-1269a, VASH and circASS1
was predicted using the Encyclopedia of RNA Interactomes (ENCORI)
database (starbase.sysu.edu.cn/index.php). The binding sequences
of miR-1269a on circASS1 cDNA and VASH1 and their mutated antisense
sequences were separately subcloned into the psiCHECK-2 vector
(Promega Corporation). Subsequently, HT29 cells (1x105
cells/well) were co-transfected with the reporter plasmids (100 nM)
and miR-1269a mimic (100 nM) or NC mimic (100 nM) using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Following incubation at 37˚C for 48 h, the firefly and
Renilla luciferase activities were measured using the
Renilla-Firefly Luciferase Dual Assay kit (Pierce, Thermo
Fisher Scientific, Inc.).
Statistical analysis
Each experiment was repeated three times.
Statistical analyses were performed using GraphPad Prism 6.0
software (GraphPad Software, Inc.). Comparisons between two groups
were analyzed using paired Student's t test, whereas comparisons
among multiple groups were analyzed using one-way ANOVA followed by
Tukey's post hoc test. Pearson's correlation coefficient was used
to determine correlations. P<0.05 was considered to indicate a
statistically significant difference. All data are presented as
mean ± standard deviation.
Results
circASS1 is downregulated in CRC cells
and tissues
To elucidate the effect of circASS1 on CRC
progression, its expression in CRC cells and tissues was determined
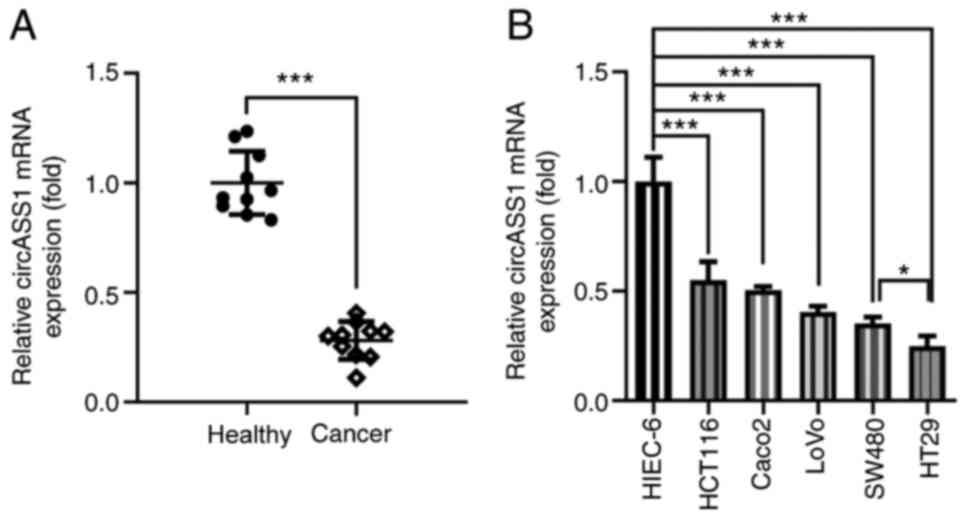
by RT-qPCR. As shown in Fig. 1A,
the expression levels of circASS1 were significantly decreased in
CRC tissues compared with those in adjacent healthy tissues.
Furthermore, circASS1 expression was significantly downregulated in
CRC cells compared with that in HIEC-6 cells, with HT29 cells
displaying the lowest circASS1 expression levels among all CRC cell
lines. Therefore, HT29 cells were selected for subsequent
experiments.
circASS1 overexpression attenuates CRC
cell proliferation, invasion and migration
To further examine the specific effects of circASS1
on CRC development, circASS1 overexpression plasmids were
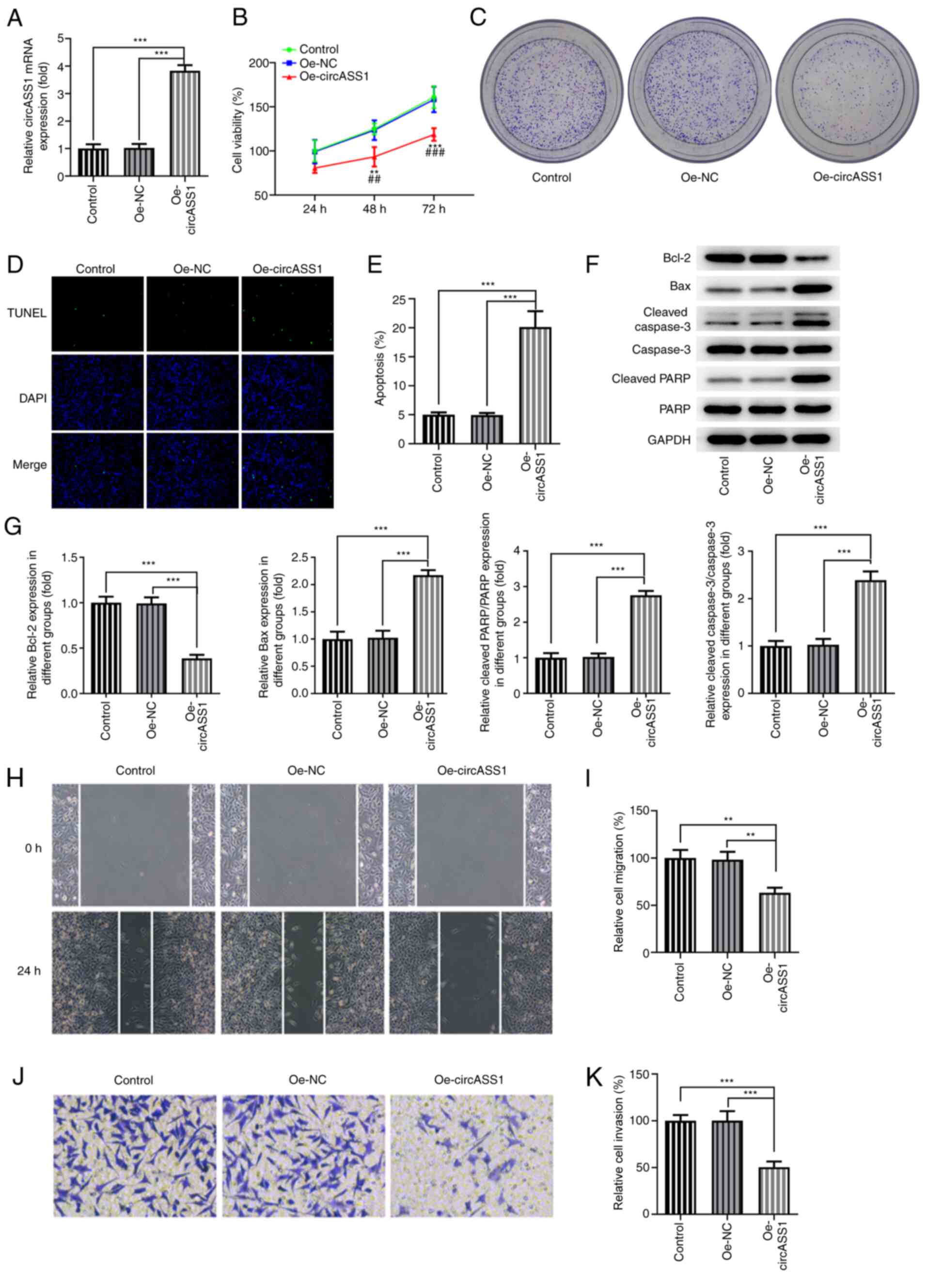
constructed and transfected into HT29 cells (Fig. 2A). The results showed that cell
viability and proliferation were decreased in HT29 cells following
transfection with Oe-circASS1 compared with transfection with Oe-NC
(Fig. 2B and C). Subsequently, TUNEL assays were
performed to assess the effect of circASS1 overexpression on CRC
cell apoptosis. Compared with the Oe-NC group, circASS1
overexpression significantly induced apoptosis in HT29 cells
(Fig. 2D and E). Moreover, compared with the Oe-NC
group, circASS1 overexpression significantly decreased Bcl-2
expression and increased the expression of Bax, cleaved
caspase-3/caspase-3 and cleaved PARP/PARP in HT29 cells (Fig. 2F). The invasion and migration
abilities of HT29 cells were significantly inhibited after circASS1
overexpression in HT29 cells compared with the Oe-NC group
(Fig. 2G-J). Taken together, the
aforementioned findings indicated that circASS1 overexpression
attenuated CRC cell proliferation, invasion and migration.
circASS1 adsorbs and negatively
regulates miR-1269a
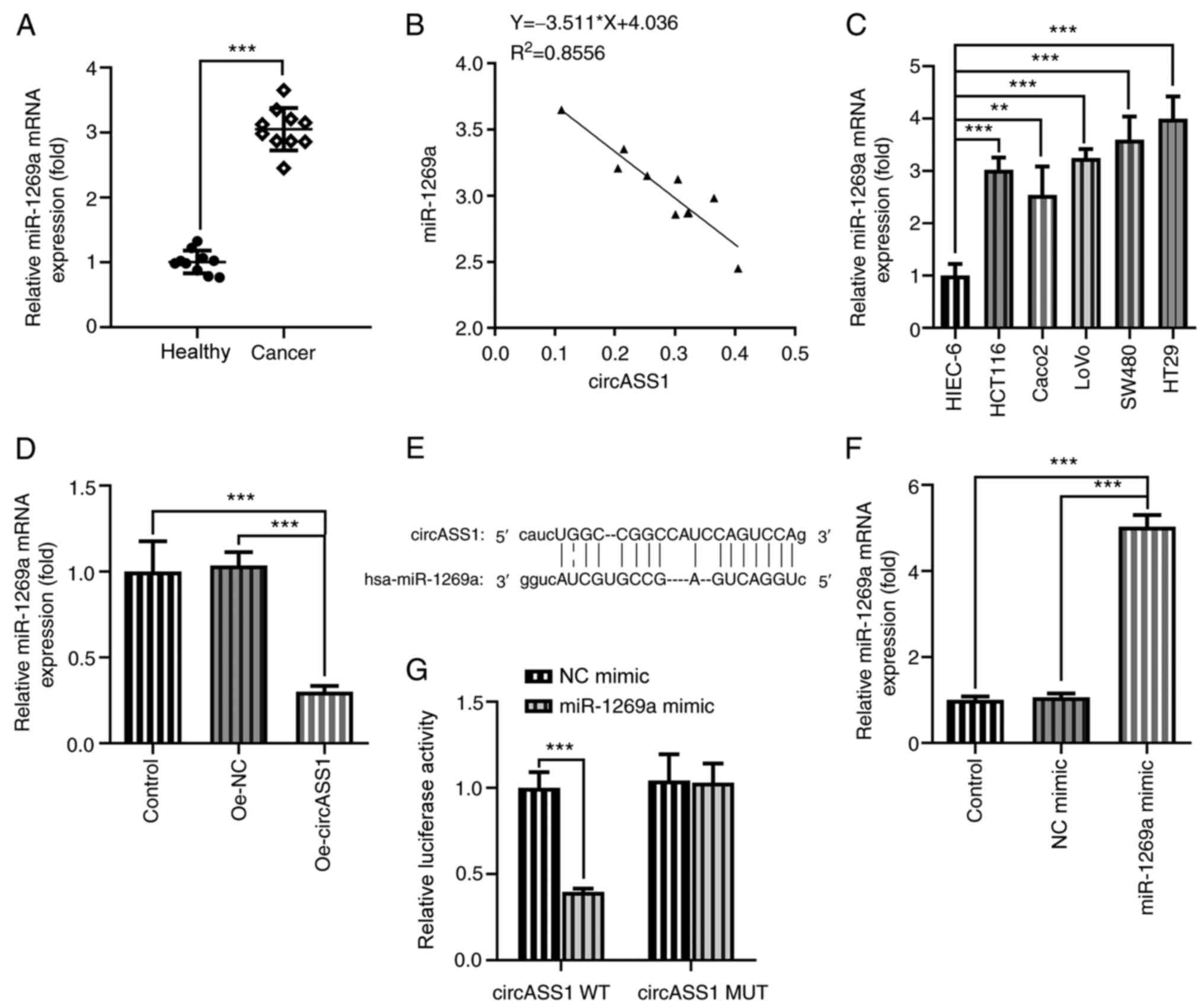
Previous studies have shown that miR-1269a is
upregulated in CRC cells (15,22).
Consistently, miR-1269a expression was significantly increased in
CRC tissue samples compared with that in adjacent healthy tissues
(Fig. 3A). Correlation analysis
confirmed the negative correlation between circASS1 and miR-1269a
expression (Fig. 3B). Subsequently,
miR-1269a expression levels were determined in different CRC cell
lines and HIEC-6 cells. The results demonstrated that miR-1269a
expression was significantly elevated in CRC cells compared with
HIEC-6 cells, with HT29 cells displaying the highest miR-1269a
expression levels (Fig. 3C).
Following circASS1 overexpression in HT29 cells, miR-1269a
expression was significantly downregulated compared with that in
the Oe-NC group (Fig. 3D). The
predicted binding sites between circASS1 and miR-1269a are
presented in Fig. 3E. Subsequently,
HT29 cells were transfected with miR-1269a mimic, and the
association between circASS1 and miR-1269a was verified by
performing the luciferase reporter assay (Fig. 3F and G). The results showed that co-transfection
of circASS1 WT and miR-1269 significantly decreased the luciferase
activity, demonstrating a negative correlation between the two.
This indicated that circASS1 could adsorb and negatively regulate
miR-1269a expression.
VASH1 is directly targeted by
miR-1269a
The expression levels of VASH1 were evaluated by
RT-qPCR and western blotting. As shown in Fig. 4A and B, VASH1 expression levels were
significantly downregulated in CRC tissues compared with those in
adjacent healthy tissues. Correlation coefficient analysis revealed
a negative correlation between miR-1269a and VASH1, and a positive
correlation between circASS1 and VASH1 in CRC tissue samples
(Fig. 4C and D). VASH1 was also expressed at the lowest
levels in HT29 cells among the CRC cell lines (Fig. 4E and F). Compared with NC inhibitor,
transfection with miR-1269a inhibitor significantly decreased the
expression levels of miR-1269a (Fig.
4G) and increased the expression levels of VASH1 (Fig. 4H and I). Following transfection with
Oe-circASS1, the expression of VASH1 was significantly upregulated
compared with that in the Oe-NC group (Fig. 4J and K). Furthermore, a binding association
between miR-1269a and VASH1 was predicted using ENCORI (Fig. 4L), and luciferase results showed
that co-transfection of VASH1 WT and miR-1269 significantly
decreased luciferase activity (Fig.
4M), confirming the binding association between miR-1269a and
VASH1. The results suggested that VASH1 was directly targeted by
miR-1269a.
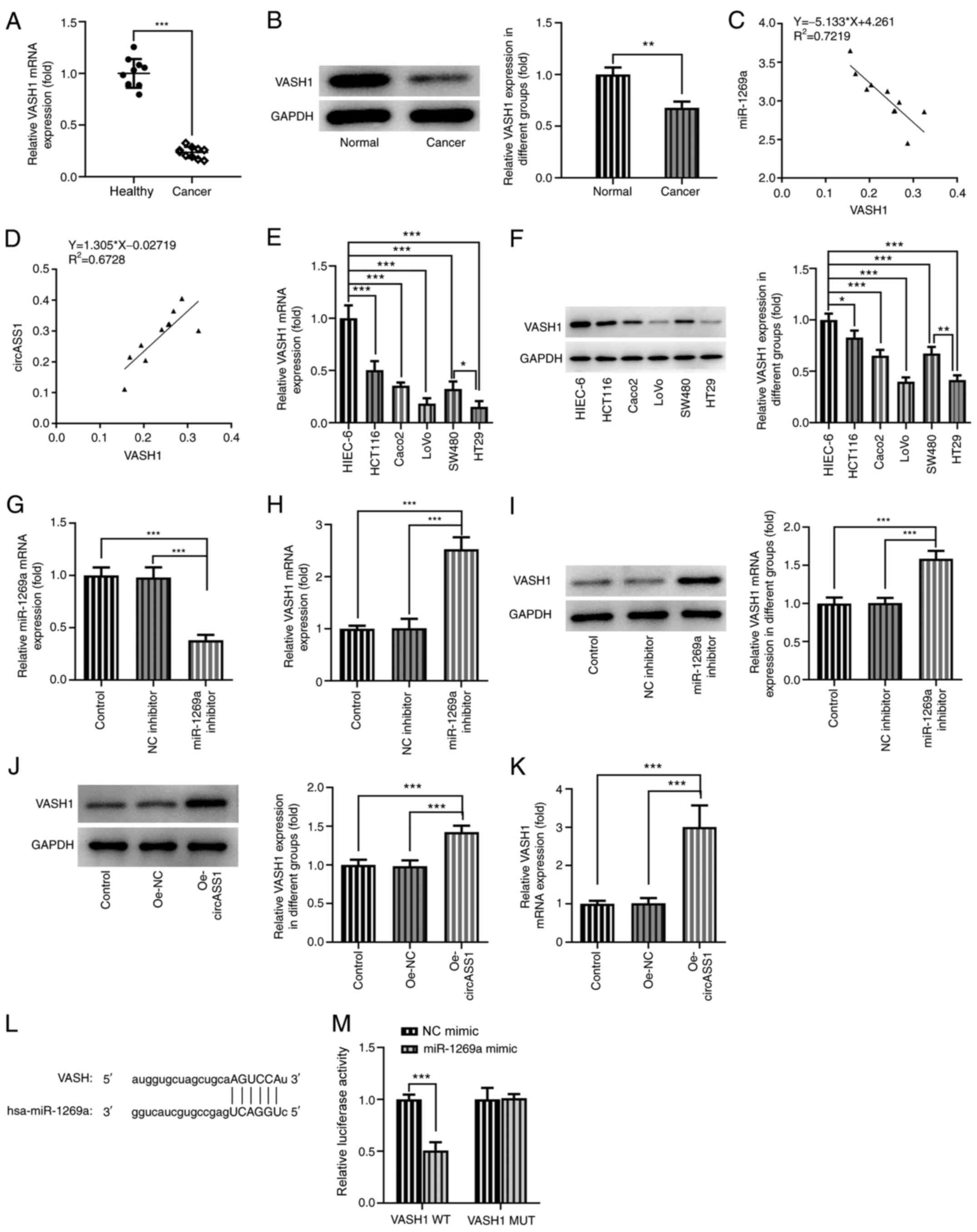 | Figure 4VASH1 is a target protein of
miR-1269a. VASH1 (A) mRNA and (B) protein expression levels in CRC
tissues and adjacent healthy tissues. Correlation between (C)
miR-1269a and VASH1, and (D) circASS1 and VASH1 in CRC tissues.
VASH1 (E) mRNA and (F) protein expression levels in CRC cell lines.
(G) Transfection efficiency of miR-1269a inhibitor. VASH1 (H) mRNA
and (I) protein expression levels in miR-1269a
inhibitor-transfected HT29 cells. VASH1 (J) mRNA and (K) protein
expression levels in Oe-circASS1-transfected HT29 cells. (J-K) The
mRNA and protein levels of VASH1 in HT29 cells treated with
Oe-circASS1. Binding sites between miR-1269a and VASH1 were (L)
predicted and (M) confirmed. *P<0.05,
**P<0.01 and ***P<0.001. VASH1,
vasohibin 1; miR, microRNA; CRC, colorectal cancer; Oe,
overexpression; circASS1, circular RNA argininosuccinate synthase
1; NC, negative control; WT, wild-type; MUT, mutant. |
circASS1/miR-1269a/VASH1 axis enhances
CRC cell proliferation
To investigate whether circASS1 could affect the
characteristics of CRC cells via regulating the miR-1269a/VASH1
axis, VASH1 was knocked down for subsequent experiments (Fig. 5A and B). As shown in Fig. 5C and D, compared with the control group,
circASS1 overexpression attenuated HT29 cell viability and
proliferation, which was abrogated by co-transfection with
miR-1269a mimic or shRNA-VASH1. circASS1 overexpression-mediated
HT29 cell apoptosis was significantly inhibited following
co-transfection with miR-1269a mimic or shRNA-VASH1 (Fig. 5E and F). Compared with the control group,
circASS1 overexpression significantly decreased Bcl-2 expression
and increased the expression of Bax, cleaved caspase-3/caspase-3
and cleaved PARP/PARP in HT29 cells, which was significantly
inhibited by co-transfection with miR-1269a mimic or shRNA-VASH1
(Fig. 5G).
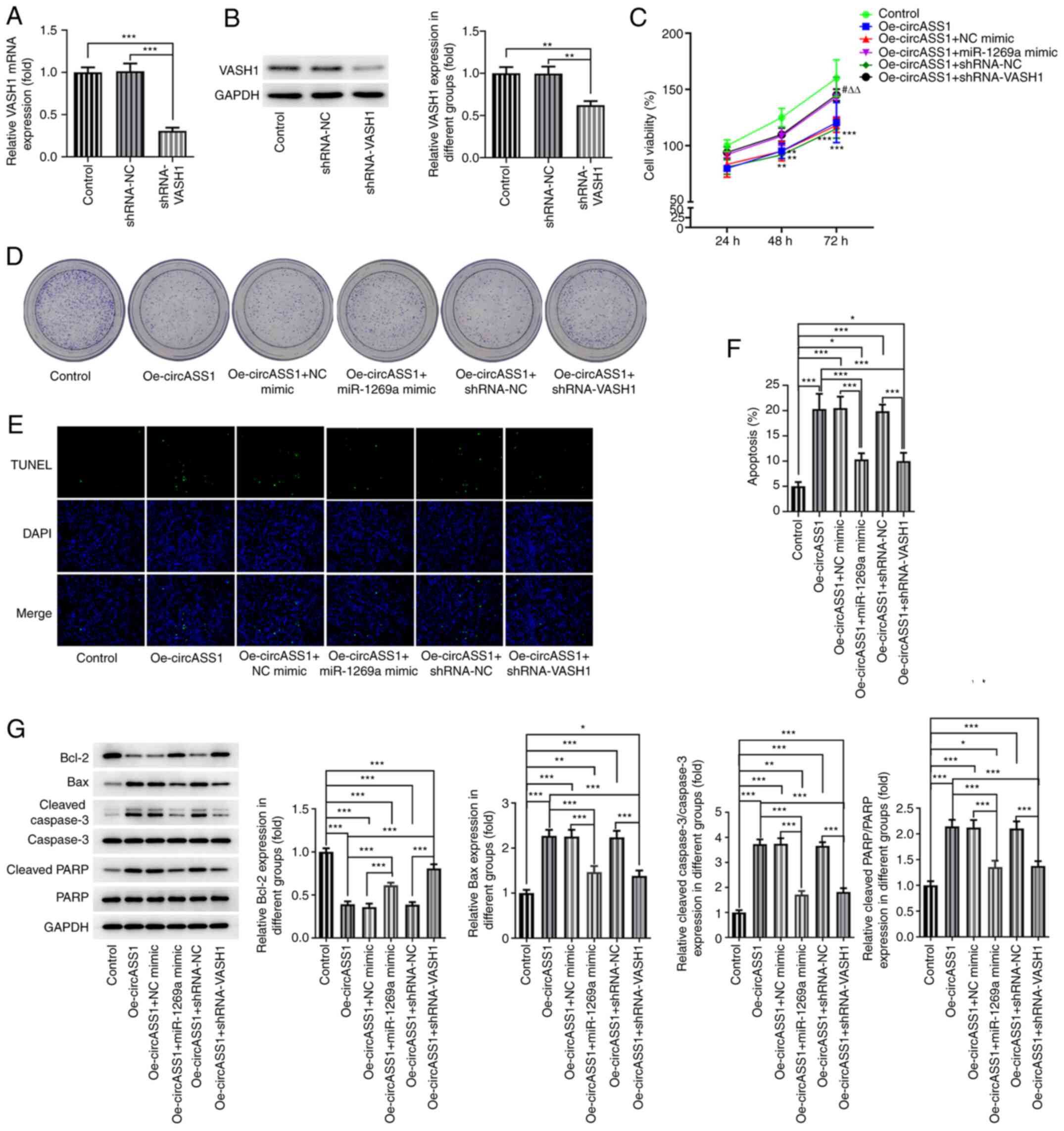 | Figure 5circASS1/miR-1269a/VASH1 axis
enhances colorectal cancer cell proliferation. VASH1 (A) mRNA and
(B) protein expression levels in shRNA-VASH1-transfected HT29
cells. (C) Cell viability and (D) colony formation of
Oe-circASS1-transfected HT29 cells co-transfected with miR-1269a
mimic or shRNA-VASH1. Cell apoptosis was (E) detected by performing
TUNEL assays and (F) quantified. Magnification, x100. (G)
Apoptosis-related protein expression levels were determined by
western blotting. *P<0.05, **P<0.01 and
***P<0.001 vs. Control; #P<0.05,
Oe-circASS1+miR-1296a mimic vs. Oe-circASS1+NC mimic;
∆∆P<0.01, Oe-circASS1+shRNA-VASH1 vs.
Oe-circASS1+shRNA-NC. circASS1, circular RNA argininosuccinate
synthase 1; miR, microRNA; VASH1, vasohibin 1; shRNA, short hairpin
RNA; Oe, overexpression; NC, negative control; PARP,
poly(ADP-ribose) polymerase. |
circASS1/miR-1269a/VASH1 axis enhances
CRC cell invasion and migration
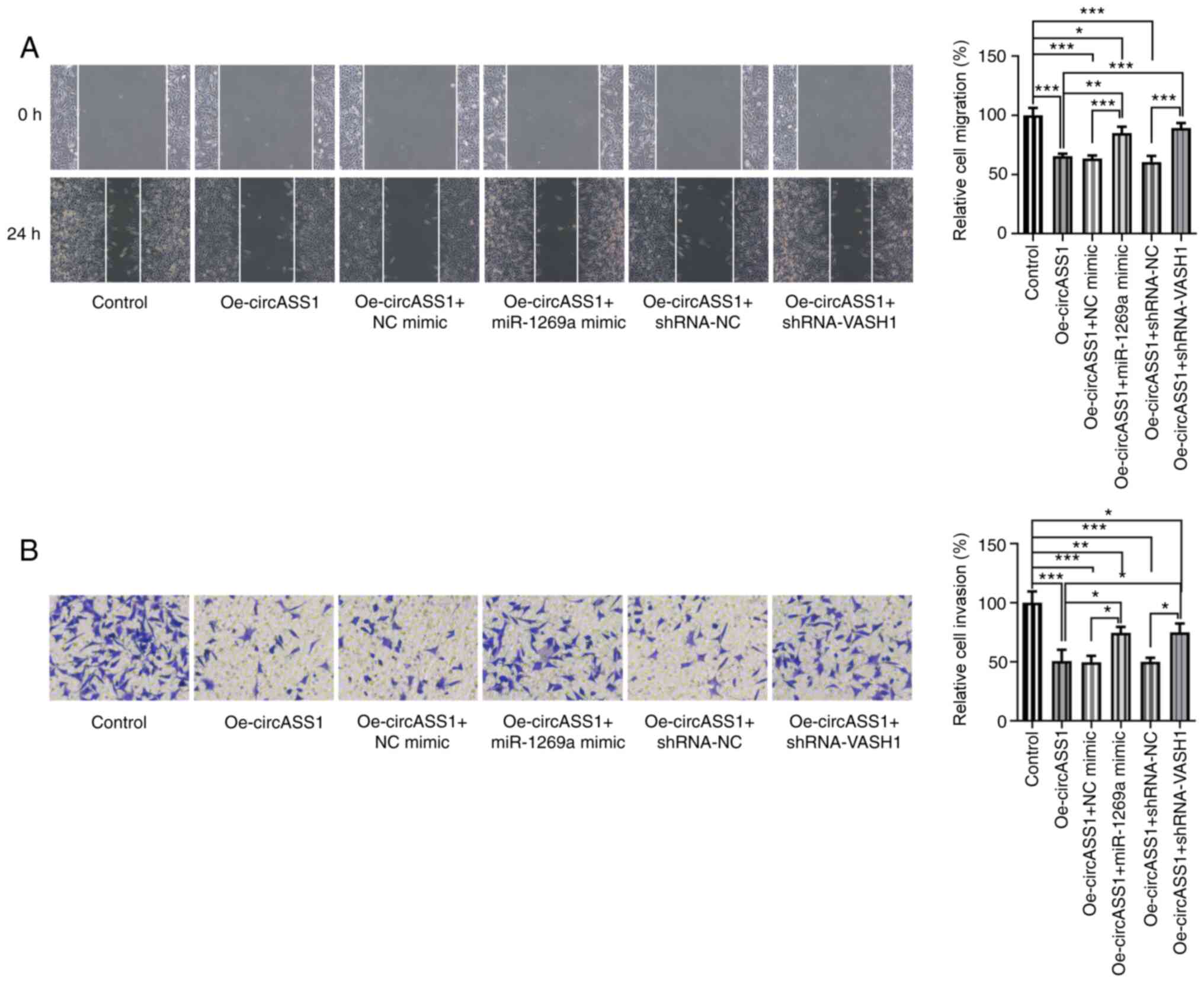
Subsequently, the present study investigated the
invasion and migration abilities of HT29 cells. Compared with the
control group, circASS1 overexpression significantly decreased the
migration and invasion abilities of HT29 cells, whereas
co-transfection with miR-1269a mimic or shRNA-VASH1 significantly
suppressed these effects (Fig. 6A
and B). Therefore, these findings
suggested that the circASS1/miR-1269a/VASH1 axis enhanced CRC cell
proliferation, invasion and migration.
Discussion
The functions of circRNAs have been well documented
(23). It has been reported that
circRNAs interact with RNA binding proteins, serve as miRNA sponges
and are involved in several biological processes, including
transcriptional regulation and alternative splicing, indicating
that circRNAs may participate in the development of various
diseases (24,25). Specifically, the role of circRNAs
has been extensively investigated in multiple types of cancer
(26-28).
The unique characteristics of circRNAs highlight their potential as
markers for the treatment of cancer (29). The tumor suppressive role of
circASS1 in breast cancer has been previously reported (20). Therefore, the present study
hypothesized that circASS1 may also inhibit the progression of CRC
via a certain mechanism. To the best of our knowledge, the present
study was the first to demonstrate that circASS1 was downregulated
in CRC tissues and cells. Furthermore, circASS1 overexpression
inhibited CRC cell proliferation, invasion and migration, but
enhanced cell apoptosis.
A plethora of genes have been identified as direct
targets of miR-1269a. For instance, miR-1269a and its variant could
affect the susceptibility and progression of HCC via targeting
spermatogenesis associated serine rich 2 like and LDL receptor
related protein 6(30). miR-1269a
also promoted NSCLC by regulating SOX6(31). In the present study, miR-1269a was
predicted to interact with circASS1. miR-1269a was reported to be
expressed at abnormal levels in 14 types of cancer, suggesting that
it could serve as a potential diagnostic marker for cancer
treatment (32). A previous study
demonstrated that the expression level of miR-1269a was increased
in CRC and promoted CRC metastasis, which was further verified in
the present study (15). The
present study also indicated a negative correlation between
miR-1269a and circASS1 expression.
VASH1, which was first recognized as a negative
feedback modulator of angiogenesis, can regulate endothelial cell
functions and further inhibit angiogenesis (33). VASH1 is not only expressed in
endothelial cells but is also expressed in tumor cells (34). Therefore, further studies
investigated whether this critical inhibitor of angiogenesis could
serve as a key factor in tumor progression (7,8,12). The
potential value of VASH1 in suppressing cancer cell proliferation
and invasion has been confirmed by several studies, and
antiangiogenic therapy is currently used for several types of
cancer (35-37).
Data have also demonstrated that VASH1 expression may inhibit
tumorigenesis and metastasis in a human colon cancer model
(12). Further associations between
the clinicopathological features and VASH1 expression in patients
with different types of cancer supported the potential use of VASH1
as a prognostic marker for cancer treatment (38,39).
The present study also indicated that VASH1 may prevent the
progression of CRC by altering the cellular functions of CRC cells.
VASH1 was expressed at relatively low levels in CRC tissue
specimens compared with healthy tissue samples. Correlation
analyses verified the negative correlation between miR-1269a and
VASH1 expression, and a positive correlation between circASS1 and
VASH1. Further assays demonstrated that miR-1269a inhibitor
upregulated VASH1 in HT29 cells, and HT29 cells transfected with
the circASS1 overexpression plasmid also displayed increased VASH1
expression. As the predicted direct association between miR-1269a
and VASH1 was confirmed, the circASS1/miR-1269a/VASH1 axis was
subsequently investigated to assess the potential mechanism
underlying the effect of circASS1 on the biological functions of
CRC cells. The results showed that circASS1 overexpression
attenuated cell proliferation, invasion and migration, and enhanced
the apoptosis of HT29 cells, and these effects were inhibited by
co-transfection with miR-1269a mimic or shRNA-VASH1.
In conclusion, the present study demonstrated that
circASS1 overexpression inhibited CRC cell proliferation, invasion
and migration by regulating the miR-1269a/VASH1 axis, thus
providing a potential novel molecular mechanism involved in the
pathogenesis of CRC.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Medical Research
Foundation of Guangdong Province (grant no. A2018240).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XHG collected clinical tissue samples. XY designed
the experiments. HLX, XHZ, XHG and HJL performed the experiments.
HLX and XHZ analyzed the data and wrote the manuscript. All authors
read and approved the final manuscript. HLX and XY confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
All patients provided written informed consent prior
to enrollment in the present study. The present study was approved
by the Huizhou Municipal Central Hospital of Guangdong
Province.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Fedewa SA, Anderson WF, Miller
KD, Ma J, Rosenberg PS and Jemal A: Colorectal cancer incidence
patterns in the United States, 1974-2013. J Natl Cancer Inst 109,
2017.
|
|
3
|
Zha F, Qu X, Tang B, Li J, Wang Y, Zheng
P, Ji T, Zhu C and Bai S: Long non-coding RNA MEG3 promotes
fibrosis and inflammatory response in diabetic nephropathy via
miR-181a/Egr-1/TLR4 axis. Aging (Albany NY). 11:3716–3730.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sato Y: The vasohibin family: A novel
family for angiogenesis regulation. J Biochem. 153:5–11.
2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Li D, Zhou K, Wang S, Shi Z and Yang Z:
Recombinant adenovirus encoding vasohibin prevents tumor
angiogenesis and inhibits tumor growth. Cancer Sci. 101:448–452.
2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhao G, Yang Y, Tang Y, Han R and Sun Y:
Reduced expression of vasohibin-1 is associated with
clinicopathological features in renal cell carcinoma. Med Oncol.
29:3325–3334. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yan Y, Shen Z, Ye Y, Jiang K, Zhang H,
Shen C, Mustonen H, Puolakkainen P and Wang S: A novel molecular
marker of prognosis in colorectal cancer: Vasohibin-1. Med Oncol.
31(816)2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen
Z, Dinglin X, Ma S, Li D, Wu Y, et al: N6-methyladenosine induced
miR-143-3p promotes the brain metastasis of lung cancer via
regulation of VASH1. Mol Cancer. 18(181)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Heishi T, Hosaka T, Suzuki Y, Miyashita H,
Oike Y, Takahashi T, Nakamura T, Arioka S, Mitsuda Y, Takakura T,
et al: Endogenous angiogenesis inhibitor vasohibin1 exhibits
broad-spectrum antilymphangiogenic activity and suppresses lymph
node metastasis. Am J Pathol. 176:1950–1958. 2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Huang B, Huang M and Li Q:
Cancer-associated fibroblasts promote angiogenesis of
hepatocellular carcinoma by VEGF-Mediated EZH2/VASH1 pathway.
Technol Cancer Res Treat. 18(1533033819879905)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang B, Wu Z, Xie W, Tian D, Chen F, Qin
C, Du Z, Tang G, Gao Q, Qiu X, et al: The expression of vasohibin-1
and its prognostic significance in bladder cancer. Exp Ther Med.
14:3477–3484. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu S, Han B, Zhang Q, Dou J, Wang F, Lin
W, Sun Y and Peng G: Vasohibin-1 suppresses colon cancer.
Oncotarget. 6:7880–7898. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
14
|
Zhang Y, Wang Q, Luo N, Liu J, Ren H, Shao
X, Zhang L and Yu Y: MicroRNA-1269a promotes proliferation and
arrest of apoptosis of glioma cells by directly targeting ATRX.
Front Oncol. 10(563901)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bu P, Wang L, Chen KY, Rakhilin N, Sun J,
Closa A, Tung KL, King S, Kristine Varanko A, Xu Y, et al: miR-1269
promotes metastasis and forms a positive feedback loop with TGF-β.
Nat Commun. 6(6879)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jin RH, Yu DJ and Zhong M: MiR-1269a acts
as an onco-miRNA in non-small cell lung cancer via down-regulating
SOX6. Eur Rev Med Pharmacol Sci. 22:4888–4897. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li W, Zhang H, Min P, Zhu J, Xu D, Jiang
W, Ma Y, Qiu J, Xu W, Chen J, et al: Downregulated miRNA-1269a
variant (rs73239138) decreases the susceptibility to gastric cancer
via targeting ZNF70. Oncol Lett. 14:6345–6354. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Guo C, Shi H, Shang Y, Zhang Y, Cui J and
Yu H: LncRNA LINC00261 overexpression suppresses the growth and
metastasis of lung cancer via regulating miR-1269a/FOXO1 axis.
Cancer Cell Int. 20(275)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hou JC, Xu Z, Zhong SL, Zhang HD, Jiang
LH, Chen X, Zhu LP, Li J, Zhou SY, Yang SJ, et al: Circular RNA
circASS1 is downregulated in breast cancer cells MDA-MB-231 and
suppressed invasion and migration. Epigenomics. 11:199–213.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2002.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang J, Luo X, Li H, Deng L and Wang Y:
Genome-wide uncovering of STAT3-mediated miRNA expression profiles
in colorectal cancer cell lines. Biomed Res Int.
2014(187105)2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL and
Yang Y: RNA sequencing reveals the expression profiles of circRNA
and indicates that circDDX17 acts as a tumor suppressor in
colorectal cancer. J Exp Clin Cancer Res. 37(325)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liu J, Liu T, Wang X and He A: Circles
reshaping the RNA world: From waste to treasure. Mol Cancer.
16(58)2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang T, Wu DM, Deng SH, Han R, Liu T, Li
J and Xu Y: RNAseq profiling of circRNA expression in
radiation-treated A549 cells and bioinformatics analysis of
radiation-related circRNA-miRNA networks. Oncol Lett. 20:1557–1566.
2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Patop IL and Kadener S: circRNAs in
Cancer. Curr Opin Genet Dev. 48:121–127. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xu H, Liu Y, Cheng P, Wang C, Liu Y, Zhou
W, Xu Y and Ji G: CircRNA_0000392 promotes colorectal cancer
progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin
Cancer Res. 39(283)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Min P, Li W, Zeng D, Ma Y, Xu D, Zheng W,
Tang F, Chen J, Shi J, Hu H, et al: A single nucleotide variant in
microRNA-1269a promotes the occurrence and process of
hepatocellular carcinoma by targeting to oncogenes SPATS2L and
LRP6. Bull Cancer. 104:311–320. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu J and Dong YJ: Myocardial protective
effect and mechanism of dexmedetomidine on myocardial
ischemia-reperfusion injury in rats. Journal of Clinical and
Experimental Medicine. 17:2476–2479. 2018.(In Chinese).
|
|
32
|
Hu Y, Dingerdissen H, Gupta S, Kahsay R,
Shanker V, Wan Q, Yan C and Mazumder R: Identification of key
differentially expressed MicroRNAs in cancer patients through
pan-cancer analysis. Comput Biol Med. 103:183–197. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Miyashita H, Watanabe T, Hayashi H, Suzuki
Y, Nakamura T, Ito S, Ono M, Hoshikawa Y, Okada Y, Kondo T and Sato
Y: Angiogenesis inhibitor vasohibin-1 enhances stress resistance of
endothelial cells via induction of SOD2 and SIRT1. PLoS One.
7(e46459)2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Shimizu K, Watanabe K, Yamashita H, Abe M,
Yoshimatsu H, Ohta H, Sonoda H and Sato Y: Gene regulation of a
novel angiogenesis inhibitor, vasohibin, in endothelial cells.
Biochem Biophys Res Commun. 327:700–706. 2005.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Takahashi Y, Saga Y, Koyanagi T, Takei Y,
Machida S, Taneichi A, Mizukami H, Sato Y, Matsubara S and Fujiwara
H: The angiogenesis regulator vasohibin-1 inhibits ovarian cancer
growth and peritoneal dissemination and prolongs host survival. Int
J Oncol. 47:2057–2063. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhao G, Na R, Li L, Xiao H, Ding N, Sun Y
and Han R: Vasohibin-1 inhibits angiogenesis and suppresses tumor
growth in renal cell carcinoma. Oncol Rep. 38:1021–1028.
2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhang T, Jing L, Li H, Ding L, Ai D, Lyu J
and Zhong L: MicroRNA-4530 promotes angiogenesis by targeting VASH1
in breast carcinoma cells. Oncol Lett. 14:111–118. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tamaki K, Moriya T, Sato Y, Ishida T,
Maruo Y, Yoshinaga K, Ohuchi N and Sasano H: Vasohibin-1 in human
breast carcinoma: A potential negative feedback regulator of
angiogenesis. Cancer Sci. 100:88–94. 2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kanomata N, Sato Y, Miyaji Y, Nagai A and
Moriya T: Vasohibin-1 is a new predictor of disease-free survival
in operated patients with renal cell carcinoma. J Clin Pathol.
66:613–619. 2013.PubMed/NCBI View Article : Google Scholar
|