Introduction
The incidence of benign esophageal stricture (BES)
as a result of esophageal fibrosis has increased in recent years
and is caused by a variety of esophageal injuries and inflammation,
including partial esophagectomy, gastroesophageal reflux, caustic
esophageal injury and radiotherapy (1-3).
Screening programs of early detection of esophageal carcinoma have
been implemented, which have led to earlier diagnosis and improved
long-term success in patients with esophageal carcinoma. An
increasing number of post-operative esophageal stricture cases
require to be treated. The frequency of post-operative esophageal
stricture ranges from 10 to 43% after esophagectomy (4,5). In
addition, the number of cases of BES after endoscopic submucosal
dissection (ESD) has markedly increased over the past decade
(4,5). Although ESD is gradually becoming a
standard treatment for early esophageal cancer, the post-operative
complication of esophageal stricture cannot be ignored. The rate of
post-operative esophageal stricture is 88-100% if the lesion
measures >3/4 of a circumference (6). In the process of esophageal repair
after ESD, fibroblasts produce extracellular matrix (ECM) and
transform into myofibroblasts from 1 week after the resection. The
latter causes wound contraction, which leads to scar formation and
fibrosis. Thus, it is important to clarify the changes in the wound
microenvironment ~1 week after ESD (7,8).
However, the prevention or treatment of BES remains difficult. The
mechanisms of esophageal fibrosis and BES have remained largely
elusive. Therefore, investigating the mechanism of esophageal
fibrosis after ESD may provide approaches to develop novel
treatments.
MicroRNAs (miRNAs/miRs) are a series of small
non-coding RNAs with a length of 20-24 nucleotides. It has been
reported that miRNAs regulate >60% of protein-coding genes by
acting on mRNAs and then inhibiting or reducing their translation
(9). Multiple miRNAs and mRNAs
interact with each other (10). Of
note, dysregulation of certain miRNAs and their target genes may
contribute to the process of fibrosis. Identifying novel treatment
strategies for BES prevention and treatment is essential. In
postoperative epidural fibrosis tissue, mitomycin C (MMC) was able
to reduce fibrosis by regulating miR-200b expression and targeting
RhoE (11). In a study by Wang
et al (12), microarray
analysis of laminectomy scar tissue indicated that the expression
levels of miR-34a, miR-146a and miR-200 were significantly
increased, while the levels of miR-16, miR-221 and miR-378a were
significantly decreased. Thus, it was speculated that various
dysregulated miRNAs and their genes have a role in the process of
esophageal fibrosis.
Through performing an integrated analysis, the
present study aimed to identify the core miRNAs, differently
expressed (DE) genes (DEGs) and their network, as well as to
explore pathways associated with the pathogenesis of esophageal
fibrosis.
Materials and methods
Participants and tissue
collection
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Soochow University
and The First People's Hospital of Changzhou (Third Affiliated
Hospital of Soochow University, Suzhou, China; approval no.
2020-100). All participants provided written informed consent. The
samples were collected from January 2018 to December 2018. In the
normal esophageal (NE) group, the remaining submucosal tissue at
the edge of the lesion immediately after the ESD procedure was
collected. In the post-operative esophageal (PE) group, the tissue
was collected at the edge of the lesion 7 days after ESD. For each
group, a total of five cases were enrolled in the study. The
inclusion criteria were as follows: i) Patients diagnosed with
esophageal high-grade intraepithelial neoplasia or early esophageal
carcinoma prior to ESD and by post-operative pathology; and ii) the
wound margin and base were both negative. The exclusion criteria
were as follows: i) Patients <18 or >80 years of age; and ii)
patients with coagulation dysfunction, abdominal aortic aneurysm
and those who were not able to tolerate an endoscopic procedure.
The characteristics of the participants are displayed in Table I. The tissue was obtained from the
patients and immediately stored at -80˚C for further RNA
extraction.
 | Table ICharacteristics of the
participants. |
Table I
Characteristics of the
participants.
| Item | NE group (n=5) | PE group (n=5) | P-value |
|---|
| Male sex | 3 | 2 | ns |
| Age (years) | 66 (51-76) | 66 (57-70) | ns |
| Postoperative
pathology | | | |
|
High-grade
intraepithelial neoplasia | 3 | 4 | ns |
|
Early
esophageal carcinoma | 2 | 1 | ns |
Microarray analysis of miRNAs
Total RNA was isolated from the tissue by using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
concentration and purity of total RNA were determined using
NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Inc.).
RNA integrity was accessed by agarose gel electrophoresis. The
microarray with the Agilent Human miRNA Microarray kit, Release
21.0, 8x60K (design ID: 070156) and data analysis of the 10 samples
from esophageal submucosal tissues were performed by OE
Biotechnology Co., Ltd. The miRNA array contained 2,570 probes.
miRNA expression was evaluated with an Agilent Bioanalyzer 2100
(Agilent Technologies, Inc.). Total RNA was dephosphorylated,
ligated with pCpCy3 and then hybridized to the microarray samples,
which were then washed. Using an Agilent Scanner G2505C (Agilent
Technologies, Inc.), the array was analyzed.
Data analysis
Raw data and array images were analyzed using
Feature Extraction software (version 10.7.1.1; Agilent
Technologies, Inc.) and GeneSpring software (version 14.8; Agilent
Technologies, Inc.). The genes detected in all samples were further
analyzed. DE miRNAs were selected based on fold change and P-value
according to Student's t-test. Significant DEGs were considered
those exhibiting a fold change ≥2.0 and a P≤0.05. The target genes
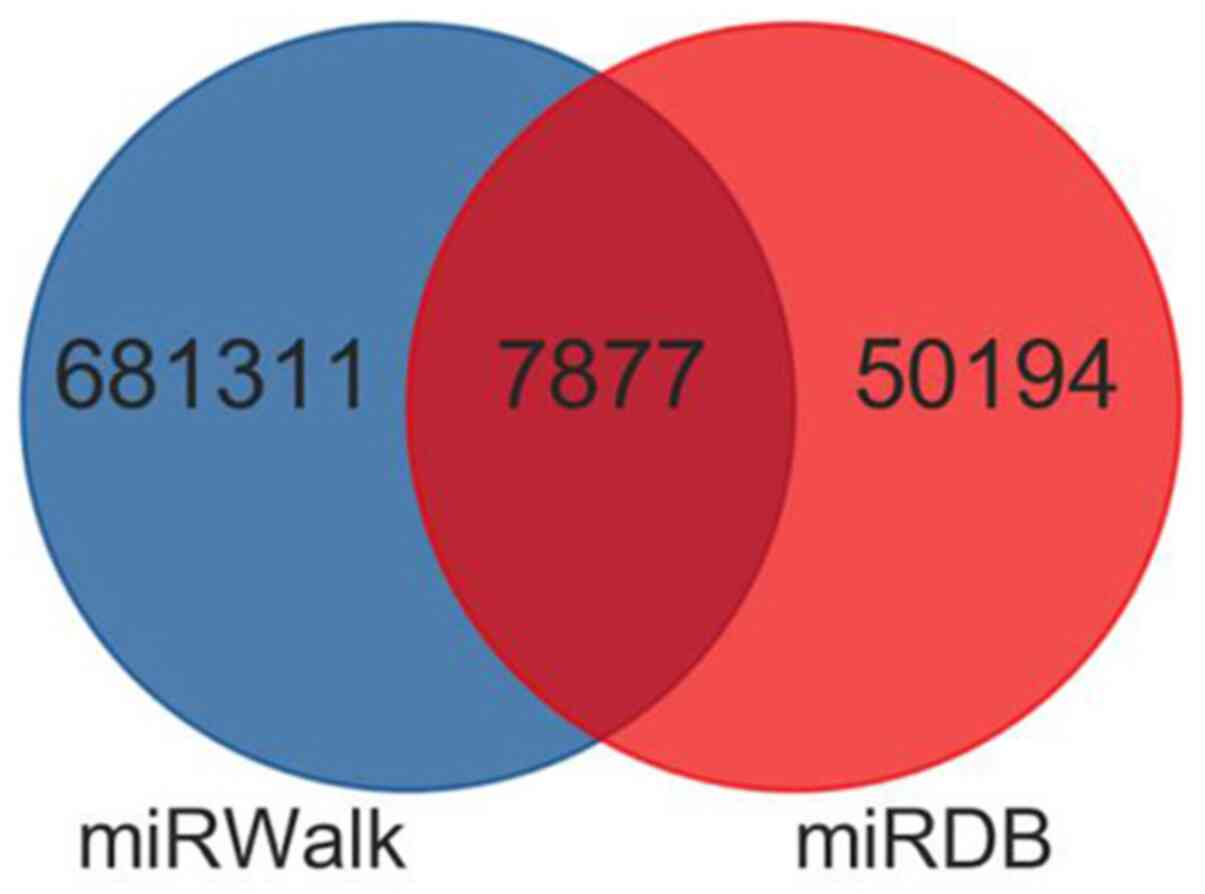
of these dysregulated miRNAs were predicted with two databases
miRDB (http://www.mirdb.org/) and miRWalk
(http://mirwalk.umm.uni-heidelberg.de/). Hierarchical
clustering was performed to confirm the dysregulated miRNAs among
samples between the two groups (NE and PE group).
miRNA-target prediction
In order to identify the key miRNA-mRNA interactions
in esophageal fibrosis, target genes of the DE miRNAs were
predicted using two databases (miRWalk and miRDB). The intersection
of the two databases was considered. Those target genes that were
predicted by ≥4 algorithms were included in the study. miRNAs that
reduced the target gene expression at the post-transcriptional
level and the inversely correlated genes were selected as miRNA
targets. The interaction network was built using Cytoscape software
3.7.2 (http://www.cytoscape.org/).
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analyses
To elucidate the functional roles of miRNAs and
their target genes, GO (molecular function, biological process and
cellular component) and KEGG pathway enrichment analyses were
performed. A false discovery rate (FDR) <0.05 was considered to
indicate a statistically significant difference.
Reverse transcription-quantitative PCR
(RT-qPCR) validation of DE miRNAs and DEGs
RT-qPCR analysis was performed to verify the
expression of the integrated analysis. Total RNA was isolated from
the tissue which was the same as the microarray. Total RNA (400 ng)
was converted to cDNA using TransScript All-in-One First-Strand
cDNA Synthesis SuperMIX (cat. no. AT341-01; Beijing Transgen
Biotech Co., Ltd.) following the manufacturer's instructions. The
reverse transcription temperature protocols were 42˚C for 15 min,
85˚C for 5 sec and maintained at 4˚C. The final products were
stored at -20˚C. qPCR was conducted with Takara SYBR Premix Ex Taq™
(Tli RNaseH Plus; Takara Bio, Inc.) in a 7900HT Fast Real-Time PCR
system (Thermo Fisher Scientific, Inc.). The primer sequences are
showed in Table II. The reaction
procedure consists of 40 cycles: Initial denaturation at 94˚C for
30 sec, followed by 40 cycles of 94˚C for 5 sec and 60˚C for 30
sec. U6 was used as endogenous control for miRNAs and β-actin was
for mRNAs. The relative expression values were calculated using the
2-ΔΔCq method (13).
 | Table IIPrimers used in reverse
transcription-quantitative PCR. |
Table II
Primers used in reverse
transcription-quantitative PCR.
| Name | Primer sequences
(5'-3') |
|---|
| miR-223-3p | F:
GCCGAGACCCCAUAAACUG |
| | R:
CAGTGCGTGTCGTGGAGT |
| miR-142-5p | F:
CATAAAGTAGAAAGCACTACT |
| | R:
GCGAGCACAGAATTAATACGAC |
| miR-582-5p | F:
GCGGTTACAGTTGTTCAACC |
| | R:
CTCAACTGGTGTCGTGGA |
| miR-21-3p | F:
ACGTCAACAGCAGTCGATGG |
| | R:
TATGGTTGTTCTGCTCTCTCTGTCTC |
| miR-218-5p | F:
TGCGGCGGCCCCACGCACCAG |
| | R:
CCAGTGCAGGGTCCGAGGT |
| FOXO1 | F:
AATCCAGAGGGTGGCAAGAGCG |
| | R:
GGCGAAATGTACTCCAGTTATCAA |
| PAX6 | F:
CCCATGCAGATGCAAAAGTC |
| | R:
GCCAGTCTCGTAATACCTGC |
| PIK3CA | F:
CCACGACCATCATCAGGTGAA |
| | R:
CCTCACGGAGATTCTAAAGT |
| ADRB1 | F:
GCTGCAGACGCTCACCA |
| | R:
GCGAGGTAGCGGTCCAG |
| U6 | F:
CTCGCTTCGGCAGCACA |
| | R:
AACGCTTCACGAATTTGCGT |
| β-actin | F:
TGGCACCCAGCACAATGAA |
| | R:
CTAAGTCATAGTCCGCCTAGAA |
Statistical analysis
Statistical analysis was performed using SPSS 19.0
software (SPSS, Inc.). Values are expressed as the mean ± standard
deviation (relative expression of RNAs) or median (range) age. The
unpaired Student's t-test was used to compare parameter variables
(relative expression of RNAs, age). Fisher's exact test was used to
compare categorical variables (male sex, postoperative pathology).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Differential expression analysis of
genes in esophageal fibrosis
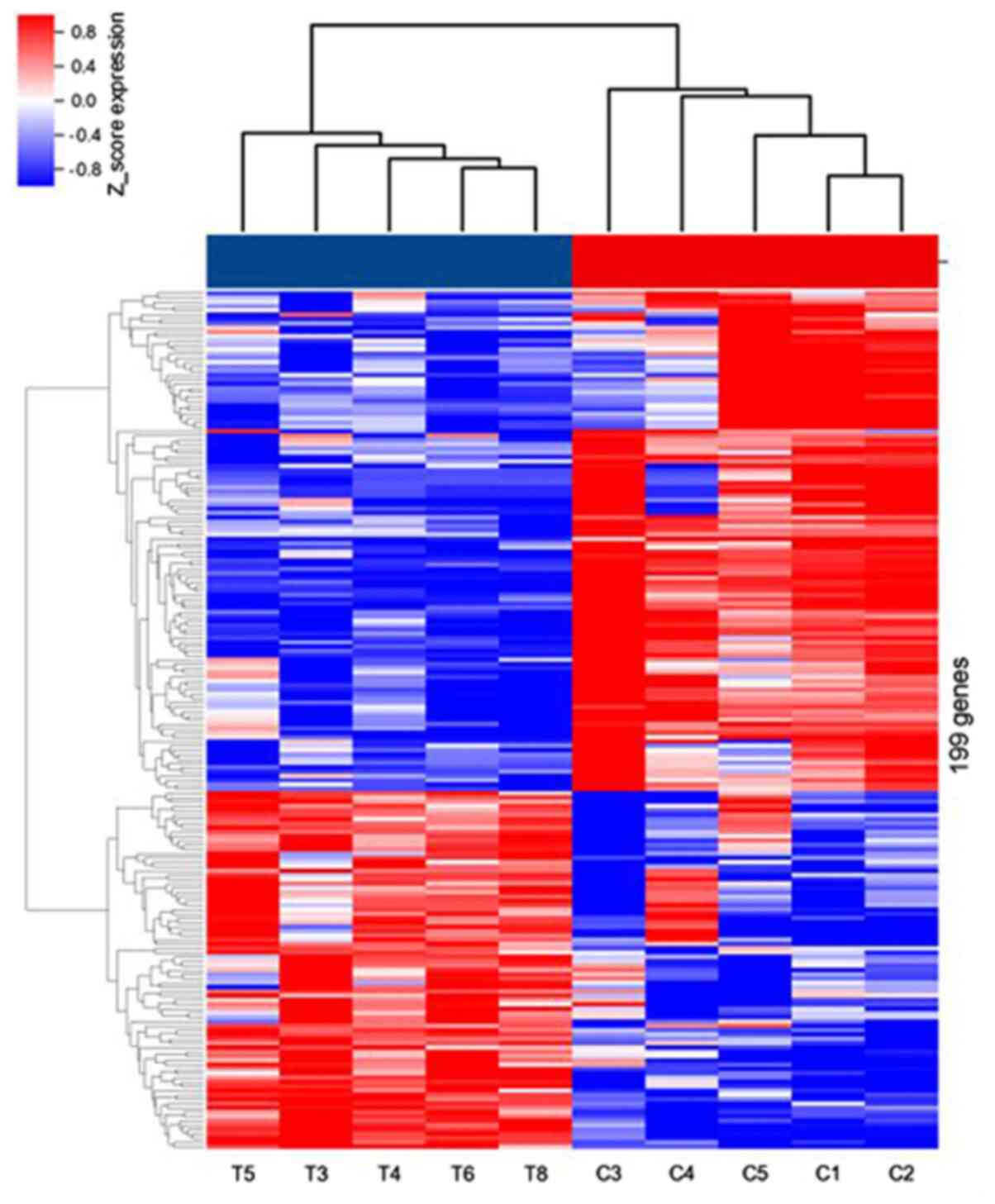
Using miRNA microarray analysis, numerous
transcripts were detected in PE and NE tissues. In total, 199
miRNAs were significantly dysregulated with fold change >2.0,
P<0.05 and FDR <0.05. Among them, 83 miRNAs were upregulated,
while 116 miRNAs were downregulated. The dysregulated miRNAs are
displayed in a heat-map (Fig. 1).
The top 10 upregulated and downregulated miRNAs are listed in
Table III.
 | Table IIITop 10 overexpressed and top 10
downregulated miRNAs in esophageal samples taken at 1 week
post-operatively vs. normal esophageal samples. |
Table III
Top 10 overexpressed and top 10
downregulated miRNAs in esophageal samples taken at 1 week
post-operatively vs. normal esophageal samples.
| Probe ID | P-value | FC | log2FC | Direction of
regulation |
|---|
| hsa-miR-223-3p |
2.75x10-5 | 64.46 | 6.01 | Up |
|
hsa-miR-3663-5p |
1.94x10-5 | 48.76 | 5.61 | Up |
| hsa-miR-223-5p |
1.84x10-4 | 36.43 | 5.19 | Up |
| hsa-miR-142-5p |
4.40x10-4 | 35.13 | 5.13 | Up |
| hsa-miR-31-3p | 0.022 | 30.50 | 4.93 | Up |
| hsa-miR-605-5p |
2.38x10-4 | 27.57 | 4.79 | Up |
| hsa-miR-4451 |
2.81x10-6 | 24.63 | 4.62 | Up |
| hsa-miR-1246 |
1.56x10-7 | 16.45 | 4.04 | Up |
| hsa-miR-582-5p |
9.05x10-3 | 13.71 | 3.78 | Up |
| hsa-miR-21-3p |
4.53x10-4 | 13.17 | 3.72 | Up |
| hsa-miR-4647 |
7.84x10-5 | -58.61 | -5.87 | Down |
| hsa-miR-6129 |
3.89x10-6 | -46.88 | -5.55 | Down |
| hsa-miR-4522 |
4.10x10-5 | -43.70 | -5.45 | Down |
| hsa-miR-1273c |
3.93x10-7 | -40.00 | -5.32 | Down |
|
hsa-miR-4446-3p |
9.54x10-7 | -38.85 | -5.28 | Down |
| hsa-miR-218-5p |
5.03x10-3 | -36.99 | -5.21 | Down |
|
hsa-miR-5699-5p |
1.62x10-4 | -34.97 | -5.13 | Down |
| hsa-miR-424-3p |
4.28x10-7 | -33.47 | -5.06 | Down |
| hsa-miR-345-5p |
6.97x10-6 | -31.75 | -4.99 | Down |
|
hsa-miR-6857-5p |
9.26x10-7 | -26.64 | -4.748 | Down |
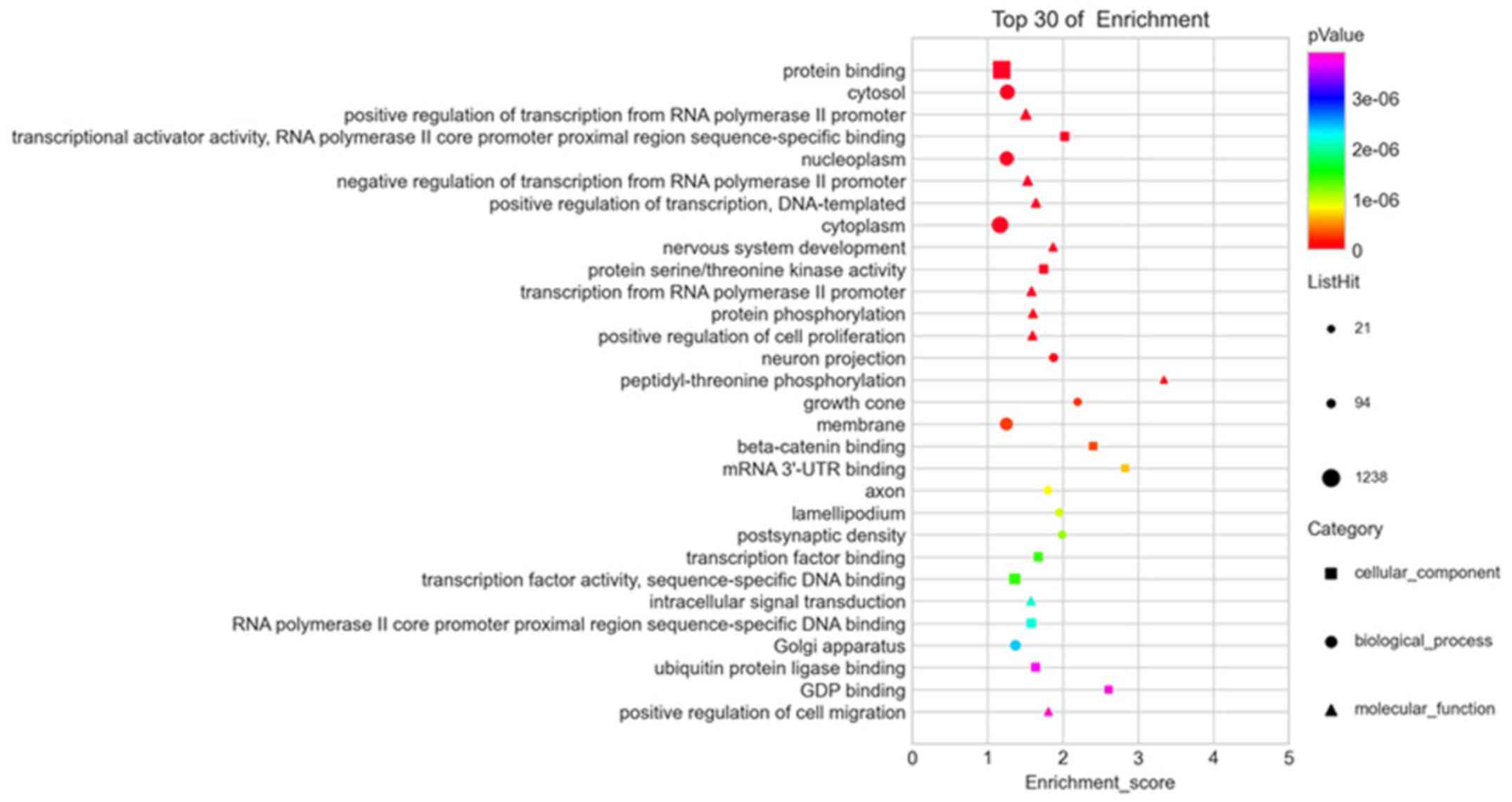
GO and KEGG enrichment analyses of
dysregulated miRNAs
GO and KEGG pathway enrichment analyses were
performed to explore the pathogenesis of esophageal fibrosis. In
the GO enrichment analysis, the main affected biological processes
included positive regulation of transcription mediated by the RNA
polymerase II promoter, negative regulation of transcription
mediated by the RNA polymerase II promoter, positive regulation of
transcription (DNA template), nervous system development and
transcription mediated by the RNA polymerase II promoter. The
cellular component terms included cytosol, nucleoplasm, cytoplasm,
neuron projection and growth cone. Molecular functions mainly
involved protein binding, transcriptional activator activity, RNA
polymerase II core promoter proximal region sequence-specific
binding, protein serine/threonine kinase activity, β-catenin
binding and mRNA 3'-untranslated region (UTR) binding (Fig. 2).
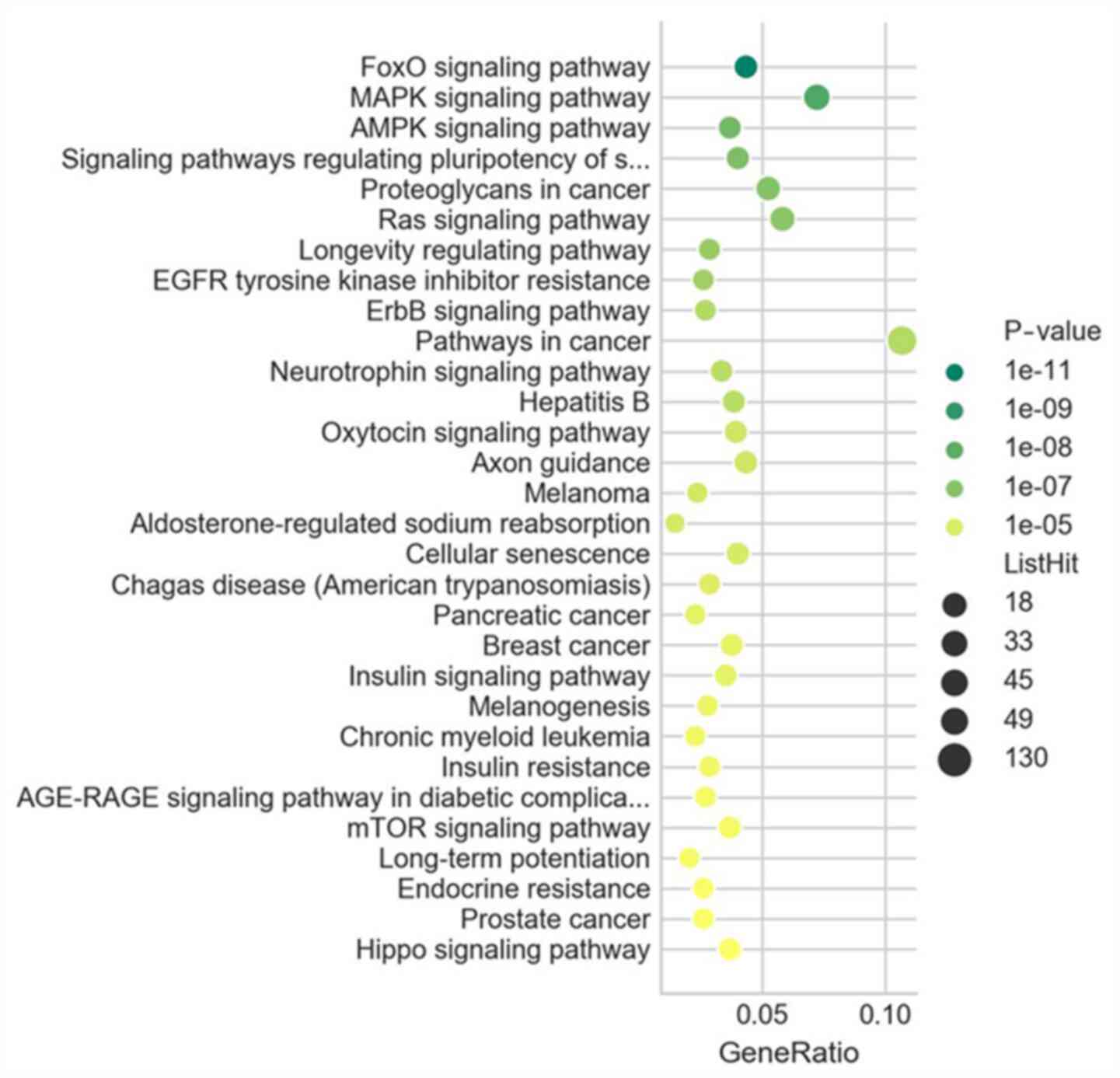
The results of the KEGG pathway enrichment analysis
revealed the core pathways involved in esophageal fibrosis,
including the forkhead box O (FOXO), MAPK and AMP-activated protein
kinase (AMPK) signaling pathways, as well as signaling pathways
regulating the pluripotency of stem cells and proteoglycans in
cancer (Fig. 3).
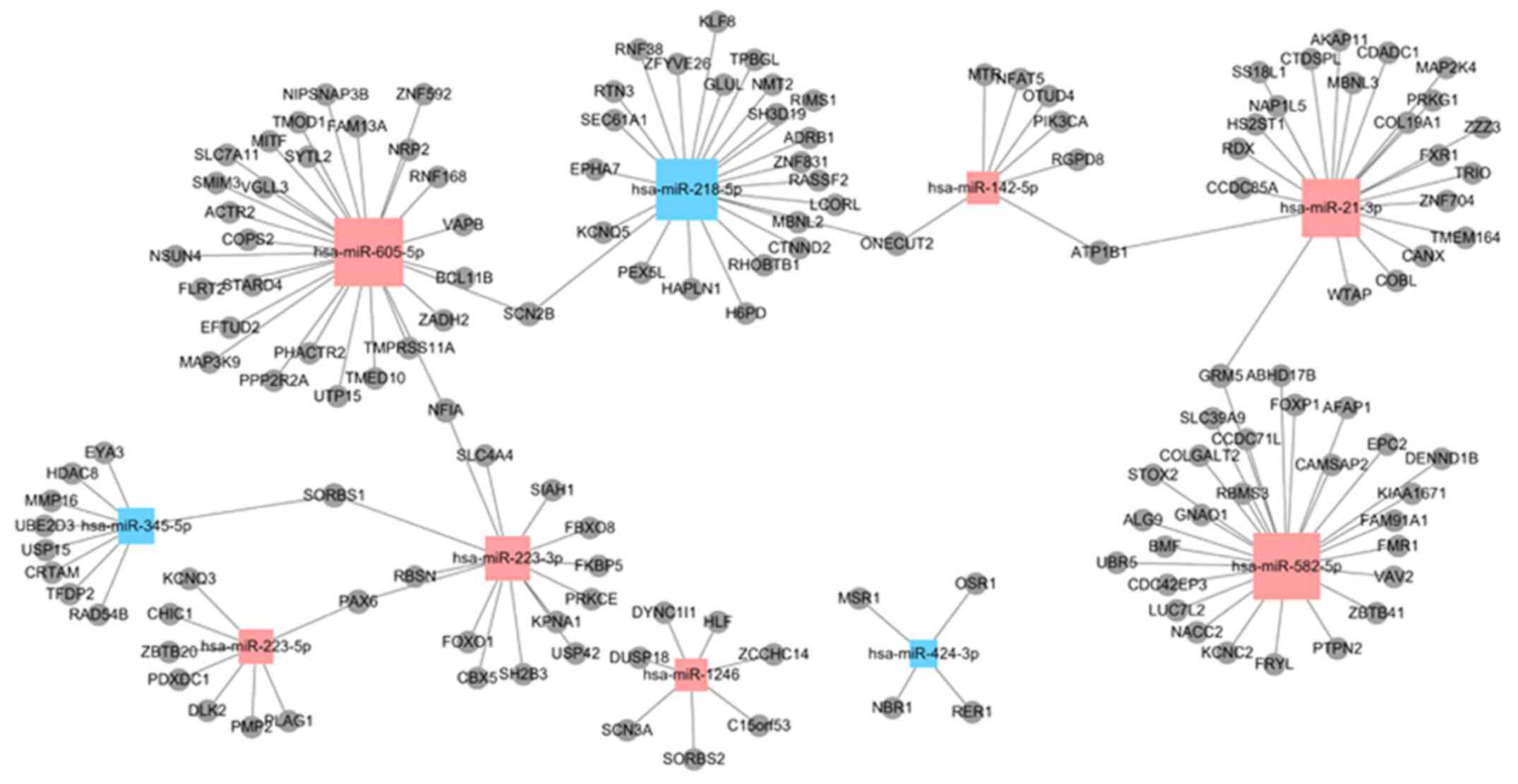
Target genes predicted by DE miRNAs
and miRNA-mRNA interaction network
A total of 7,878 miRNA-mRNA interaction pairs were
obtained, of which 2,745 miRNA-mRNA pairs were upregulated and
5,133 miRNA-mRNA pairs downregulated (Fig. 4). According to these pairs, the
miRNA-mRNA interaction network was built. The network consisted of
the top 10 related dysregulated miRNAs (miR-218-5p, miR-345-5p,
miR-424-3p, miR-1246, miR-142-5p, miR-21-3p, miR-223-3p,
miR-223-5p, miR-582-5p and miR-605-5p) and 143 predicted DEGs
(Fig. 5). In total, five miRNAs
(miR-223-3p, miR-142-5p, miR-582-5p, miR-21-3p and miR-218-5p) were
selected as core DE miRNAs that interacted with the majority of
DEGs according to the interaction network analysis, and the GO and
KEGG analyses. According to the DE miRNAs obtained, four predicted
DEGs [FOXO1, paired box 6 (PAX6),
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
alpha (PIK3CA) and adrenoceptor β1 (ADRB1)] were selected, which
were also involved in the process of fibrosis.
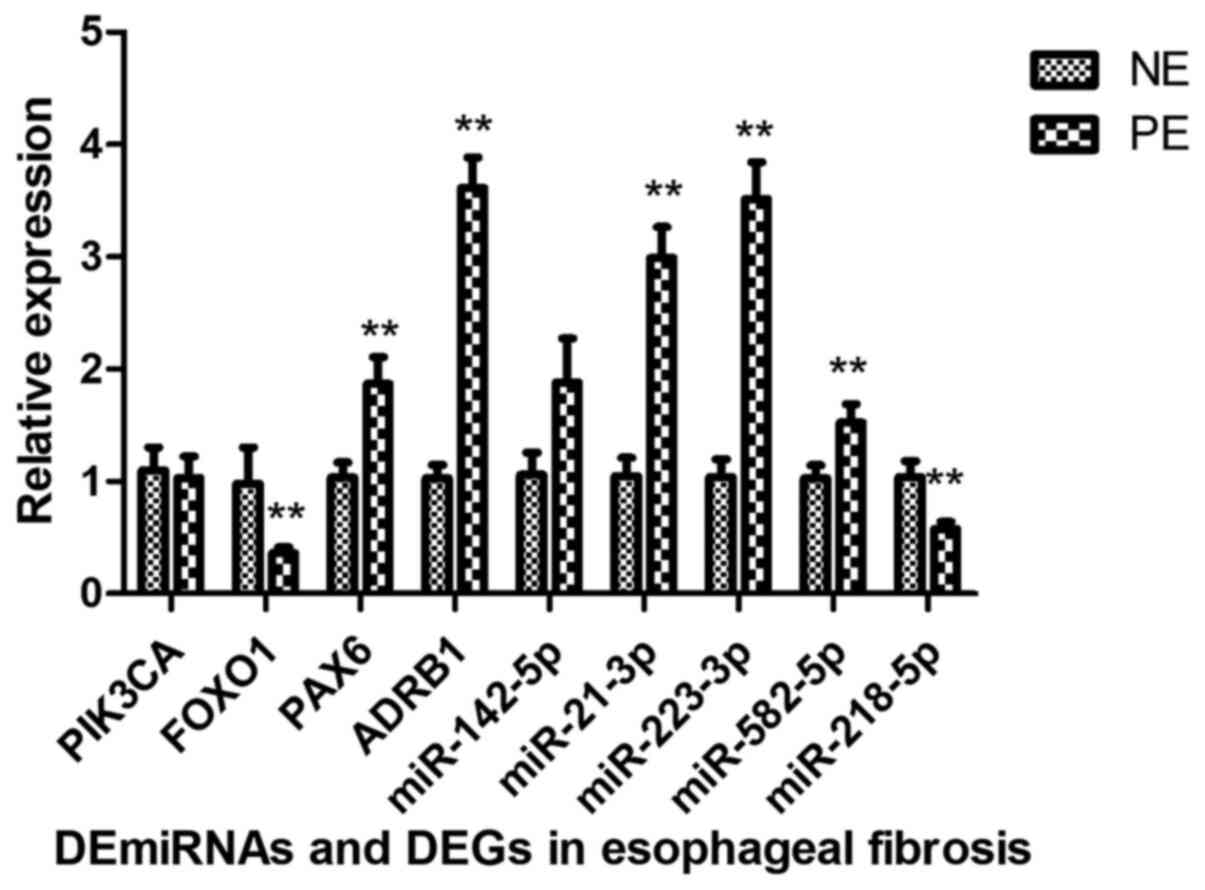
Verification of expression changes in
miRNAs and mRNAs
To confirm whether the genes were consistent with
the integrated analysis, five DE miRNAs (miR-223-3p, miR-142-5p,
miR-582-5p, miR-21-3p and miR-218-5p) and four targets (FOXO1,
PAX6, PIK3CA and ADRB1) were selected. The results revealed that
FOXO1 and miR-218-5p were downregulated, while miR-223-3p,
miR-582-5p, miR-21-3p, PAX6 and ADRB1 were upregulated. However,
the expression of PIK3CA and miR-142-5p was inconsistent with the
results of the integrated analysis (Fig. 6).
Discussion
Studies have indicated that miRNAs have a vital role
in the process of fibrosis, including myocardial, pulmonary and
liver fibrosis (10-12).
However, to the best of our knowledge, no previous studies have
reported on the function of miRNAs in the pathobiology of
esophageal fibrosis and subsequent BES. In the clinic, the
prevention or treatment of BES remain challenging and identifying
novel strategies for the prevention or treatment of esophageal
fibrosis is of great importance. In the present study, miRNA
microarray analysis was performed for evaluating DE miRNA
expression associated with esophageal fibrosis. Compared with those
in the NE control group, 83 overexpressed miRNAs and 116
downregulated miRNAs were identified in the patients 1 week after
the esophageal ESD procedure. According to the results of GO and
KEGG analyses, as well as the interaction network, four DEGs
(FOXO1, PAX6, PIK3CA and ADRB1) and five DE miRNAs (miR-223-3p,
miR-142-5p, miR-582-5p, miR-21-3p and miR-218-5p) were associated
with esophageal fibrosis.
Regarding the collected tissue, the process of
esophageal fibrosis after ESD, in which the mucosa and majority of
the submucosal layer were dissected, were evaluated (6). The majority of fibroblasts and
fibrocytes are located in the connective tissue (submucosal or
serosa layer) of the esophagus. Thus, after ESD, the surrounding
fibroblasts are activated and participate in the process of
esophageal fibrosis mainly in the remaining submucosal layer
(7). In addition, in the process of
esophageal repair after ESD, fibroblasts produce ECM and transform
into myofibroblasts from 1 week after the resection. Thus, the
remaining submucosal tissue after the ESD procedure was harvested
immediately and 7 days thereafter.
FOXO1, a potential target gene of miR-223-3P, which
usually has a key role in cell function, is a member of the FOX
transcription factor family and was reported to participate in cell
metabolism, proliferation, migration, apoptosis and angiogenesis
progression (14). In a study by
Han et al (15), miR-223-3p
was indicated to be highly expressed in neuroblastoma cell lines as
compared with its expression in normal cell lines, and it was able
to bind to the 3'-UTR of FOXO1, thus resulting in a reduction in
FOXO1 expression. The results revealed that miR-223-3p has an
oncogenic role by promoting cell proliferation and invasion, and
inhibiting apoptosis via targeting FOXO1. In other studies, miR-223
was also dysregulated in skeletal muscle and cardiac fibroblasts
and acted as a fibrogenic factor (16,17).
In the present study, miR-223-3p and miR-223-5p were highly
expressed and may serve as therapeutic targets. However, whether
they are associated only with inflammation or with the process of
esophageal fibrosis was not confirmed. The precise role of miR-223
requires further investigation.
PAX6 is a member of the PAX family of transcription
factors and has been indicated to have essential roles in multiple
organs and cell lines, including the eyes, central nervous system,
pancreatic islet cells and olfactory system. It has been reported
that PAX6 acted as an anti-fibrotic agent for cardiac fibroblast
differentiation and cystic fibrosis and repressed the expression of
the pro-fibrotic factor TGF-β1(18). Interference against PAX6 also
inhibited the activation and proliferation of hepatic stellate
cells (HSCs). Thus, PAX6 upregulation may induce HSC activation and
proliferation, resulting in liver fibrosis (19). Based on these results, future
studies should explore the fibrogenic effect of PAX6 on esophageal
fibrosis.
ADRB1 is expressed in the apical membrane of human
bronchial ciliated epithelia and participates in the process of
pulmonary cystic fibrosis (20).
Kiriazis et al (21)
investigated the effect of ADRB1 on cardiac hypertrophic growth and
fibrosis. The results also revealed the key role of β-adrenoceptor
signaling and that ADRB1 may be an alternative therapeutic target
for cardiac hypertrophy and fibrosis. The study by Ito et al
(22) suggested that DRB1 has an
important role in myocyte apoptosis and fibrosis in cardiac muscle
and skeletal muscle. In the present study, ADRB1 was one of the
target mRNAs of miR-218-5p, which may also be a regulator of the
process of fibrosis.
Steiner et al (23) indicated that PIK3CA mutation leads
to abnormal scarring involving the PI3K-Akt-mTOR pathway. Yang
et al (24) reported that
PIK3CA regulated myocardial fibrosis and oxidative stress via the
PI3K/Akt signaling pathway. Wegner et al (25) indicated that PIK3CA and TGF-β1
contributed to prostatic stromal hypertrophy and collagen
accumulation. However, in the present study, the RT-qPCR results
for PIK3CA revealed no significant difference between the two
groups, whilst PIK3CA was negatively correlated with miR-142-5p,
which was predicted with two databases miRDB and miRWalk. In other
studies, PIK3CA was also negatively regulated by miR-142-5p in
multiple tumors (26,27). It may be speculated that this
inconsistency may result from the small size of the samples and
their heterogeneity. Thus, whether PIK3CA regulates esophageal
fibroblast activation requires further investigation.
In cardiac fibrosis after myocardial infarction,
miR-142 contributed to cardiac fibroblast activation by targeting
the adenomatous polyposis coli and the subsequent Wnt signaling
pathway (28). In idiopathic
pulmonary fibrosis, miR-142-3p had a protective role against the
pro-fibrotic effect of TGF-β1(29).
In the process of non-small cell lung cancer, upregulated
miR-142-5p suppressed cell proliferation by regulating PIK3CA
(27). Together with the present
results, the above suggested that miR-142 may have key roles in
inflammation and fibrosis by regulating the expression of PIK3CA.
However, the RT-qPCR results for miR-142-5p indicated no
significant difference. In the microarray results, the miR-142
expression in the majority of patients with PE was higher than that
in the NE group. However, there was excessive heterogeneity among
different samples. In addition, a previous study suggested that
miR-142-5p regulates PIK3CA and has a role in fibrosis (27). Thus, whether miR-142-5p and PIK3CA
are involved in the process of esophageal fibrosis requires further
evaluation.
miR-582-5p has also been revealed to be closely
associated with inflammatory and immune response signaling in
previous studies. It was reported to have a key role in the
development of chronic inflammation of the airway and subsequent
airway remodeling (30). In the
cirrhotic liver, hepatic cytochrome P4503A (CYP3A) activity was
determined to be decreased and miR-582-5p was highly expressed,
which suggested that miR-582-5p expression may regulate hepatic
CYP3A activity and liver cirrhosis (31).
High expression of miR-21 has been determined in
multiple fibrotic tissues. As reported by Yang et al
(32), miR-21 contributed to the
development of hepatic fibrosis via the TGF-β/Smad7 signaling
pathway. miR-21 also regulated myofibroblast transformation and
myocardial fibrosis. TGF-β promoted cardiac fibroblast to
myofibroblast transformation and myocardial fibrosis by
upregulating miR-21, which in turn sponged the target mRNA
Jagged1(33). Furthermore, miR-21
was also reported to be a fibrosis-associated miRNA that promoted
inflammation in ligamentum flavum tissue by activating IL-6
expression, leading to ligamentum flavum fibrosis and hypertrophy
(34). In the present study, the
expression level of miR-21-3p was significantly increased in the PE
group, which is consistent with other studies (32-34).
miR-218-5p was reported to be significantly
downregulated in smokers with chronic obstructive pulmonary disease
compared with its expression in never-smokers. It was mostly highly
expressed in the bronchial airway epithelium and was associated
with airway obstruction (35). Li
et al (36) and Brkić et
al (37) indicated that
miR-218-5p regulated the proliferation, migration and EMT of human
glioma cells. Pterygium is a chronic ocular inflammatory disease
that may result in various ocular surface symptoms. Upregulated
miR-218-5p significantly suppressed the expression of epidermal
growth factor receptor via the PI3K/Akt/mTOR pathway in epithelial
cells affected by pterygium (38).
As the results of the present study, miR-218-5p was downregulated
in the PE group, suggesting that its overexpression may counteract
the fibrotic process.
According to the KEGG enrichment analysis, the
PIK3CA, FOXO, MAPK and AMPK pathway was among the top signaling
pathways. These pathways have all been reported as key signaling
pathways in the activation of fibrosis, which supports the present
results (23-25,15,39,40).
In the clinic, certain drugs may be used to prevent
or treat esophageal stricture, including topical use of
corticosteroids and mitomycin C (MMC) (41,42).
In a previous study by our group, topical injection of MMC was able
to release the post-ESD esophageal stricture (42). However, the mechanisms of esophageal
fibrosis have remained elusive, to the best of our knowledge. Only
MMC was reported to reduce epidural fibrosis by regulating miR-200b
expression and the target gene RhoE (11). In a study by Wang et al
(12), after MMC treatment of
laminectomy scar tissue, the expression levels of miR-34a, miR-146a
and miR-200 were significantly increased, while the levels of
miR-16, miR-221 and miR-378a were significantly decreased. However,
whether these genes are differentially expressed during the process
of esophageal fibrosis remains unclear. Thus, future studies should
aim to validate the associations of the aforementioned drugs and
these DE miRNAs and DE mRNAs.
The present study has several limitations. First,
the post-ESD esophageal stricture model is not suitable for other
types of esophageal fibrosis such as esophagectomy, as the mucosa
and majority of submucosal layer were dissected during the ESD
procedure, but the lamina propria and serosa remained intact.
However, to the best of our knowledge, there is no animal model or
study referring to the mechanism of esophageal stricture resulting
from esophagectomy. However, all BES types are the result of
esophageal fibrosis due to myofibroblast contraction and
substantial deposition of ECM components. Thus, it may be partially
confirmed which factors determine the process of esophageal
fibrosis. In addition, only five cases were recruited in each
group. Furthermore, the tissue was collected by endoscopic biopsy,
which features broad heterogeneity. Thus, further studies with
larger sample sizes are required.
In conclusion, using high-throughput sequencing, DE
miRNAs and their DEGs were detected in the process of esophageal
fibrosis. According to the RT-qPCR results, the expression levels
of FOXO1, PAX6, ADRB1, miR-223-3p, miR-582-5p, miR-21-3p and
miR-218-5p were consistent with the integrated analysis, but PIK3CA
and miR-142-5p could not be confirmed. These reasons may include
the large heterogeneity of patients and the false positive chip
results. Furthermore, miR-223-3p may participate in esophageal
fibrosis by targeting FOXO1 and PAX6. miR-218-5p also has a
promoting role by targeting ADRB1. miR-142-5p may facilitate
fibrosis by regulating PIK3CA. In addition, miR-582-5p and
miR-21-3p also had roles in esophageal fibrosis. Further studies
are required to functionally characterize the role of these
candidate RNAs. The current results may enhance the understanding
of the microenvironmental changes in the process of esophageal
fibrosis and may provide targets for novel treatment strategies for
esophageal fibrosis and BES.
Acknowledgements
Not applicable.
Funding
Funding: The study was partially funded by the National Natural
Science Foundation of China (grant no. 81700575), the Young Talent
Development Plan of Changzhou Health Commission (grant no.
CZQM2020017), the Natural Science Foundation of the Jiangsu Higher
Education Institutions of China (grant no. 18KJB320018) and the
Primary Research & Social Development Plan of Jiangsu Province
(grant no. BE2018659).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
(accession no. GSE169354) repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169354).
Authors' contributions
CX conceived and designed the study, authenticated
the raw data and performed manuscript review. YZ developed the
methodology, performed the experiments, analyzed the data and wrote
the manuscript. Both authors read and approved the final version of
this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Soochow University
and The First People's Hospital of Changzhou (Third Affiliated
Hospital of Soochow University, Suzhou, China; approval no.
2020-100). All participants provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siersema PD: How to approach a patient
with refractory or recurrent benign esophageal stricture.
Gastroenterology. 156:7–10. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yan X, Nie D, Zhang Y, Chang H and Huang
Y: Effectiveness of an orally administered steroid gel at
preventing restenosis after endoscopic balloon dilation of benign
esophageal stricture. Medicine (Baltimore).
98(e14565)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fuccio L, Hassan C, Frazzoni L, Miglio R
and Repici A: Clinical outcomes following stent placement in
refractory benign esophageal stricture: A systematic review and
meta-analysis. Endoscopy. 48:141–148. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
van Halsema EE, Noordzij IC, van Berge
Henegouwen MI, Fockens P, Bergman JJ and van Hooft JE: Endoscopic
dilation of benign esophageal anastomotic strictures over 16 mm has
a longer lasting effect. Surg Endosc. 31:1871–1881. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Haverkamp L, van der Sluis PC, Verhage RJ,
Siersema PD, Ruurda JP and van Hillegersberg R: End-to-end cervical
esophagogastric anastomoses are associated with a higher number of
strictures compared with end-to-side anastomoses. J Gastrointest
Surg. 17:872–876. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liao Z, Liao G, Yang X, Peng X, Zhang X,
Xie X, Zhao X, Yang S, Fan C and Bai J: Transplantation of
autologous esophageal mucosa to prevent stricture after
circumferential endoscopic submucosal dissection of early
esophageal cancer (with video). Gastrointest Endosc. 88:543–546.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rockey DC, Bell PD and Hill JA: Fibrosis -
a common pathway to organ injury and failure. N Engl J Med.
372:1138–1149. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Honda M, Nakamura T, Hori Y, Shionoya Y,
Nakada A, Sato T, Yamamoto K, Kobayashi T, Shimada H, Kida N, et
al: Process of healing of mucosal defects in the esophagus after
endoscopic mucosal resection: Histological evaluation in a dog
model. Endoscopy. 42:1092–1095. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Blaya D, Coll M, Rodrigo-Torres D,
Vila-Casadesús M, Altamirano J, Llopis M, Graupera I, Perea L,
Aguilar-Bravo B, Díaz A, et al: Integrative microRNA profiling in
alcoholic hepatitis reveals a role for microRNA-182 in liver injury
and inflammation. Gut. 65:1535–1545. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sun Y, Ge Y, Fu Y, Yan L, Cai J, Shi K,
Cao X and Lu C: Mitomycin C induces fibroblasts apoptosis and
reduces epidural fibrosis by regulating miR-200b and its targeting
of RhoE. Eur J Pharmacol. 765:198–208. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang BB, Xie H, Wu T, Xie N, Wu J, Gu Y,
Tang F and Liu J: Controlled-release mitomycin C-polylactic acid
film prevents epidural scar hyperplasia after laminectomy by
inducing fibroblast autophagy and regulating the expression of
miRNAs. Eur Rev Med Pharmacol Sci. 21:2526–2537. 2017.PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M,
Zhu Y, Zhao Q, Dong YW, Shao K, et al: MicroRNA-223 regulates FOXO1
expression and cell proliferation. FEBS Lett. 586:1038–1043.
2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Han LL, Zhou XJ, Li FJ, Hao XW, Jiang Z,
Dong Q and Chen X: MiR-223-3p promotes the growth and invasion of
neuroblastoma cell via targeting FOXO1. Eur Rev Med Pharmacol Sci.
23:8984–8990. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Cheng N, Liu C, Li Y, Gao S, Han YC, Wang
X, Du J and Zhang C: MicroRNA-223-3p promotes skeletal muscle
regeneration by regulating inflammation in mice. J Biol Chem.
295:10212–10223. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu X, Xu Y, Deng Y and Li H: MicroRNA-223
regulates cardiac fibrosis after myocardial infarction by targeting
RASA1. Cell Physiol Biochem. 46:1439–1454. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen YT, Chen FY, Vijmasi T, Stephens DN,
Gallup M and McNamara NA: Pax6 downregulation mediates abnormal
lineage commitment of the ocular surface epithelium in
aqueous-deficient dry eye disease. PLoS One.
8(e77286)2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Feng Y, Li M, Wang S, Cong W, Hu G, Song
Y, Xiao H, Dong E and Zhang Y: Paired box 6 inhibits cardiac
fibroblast differentiation. Biochem Biophys Res Commun.
528:561–566. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li C, Tan YH, Sun J, Deng FM and Liu YL:
PAX6 contributes to the activation and proliferation of hepatic
stellate cells via activating Hedgehog/GLI1 pathway. Biochem
Biophys Res Commun. 526:314–320. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kiriazis H, Wang K, Xu Q, Gao XM, Ming Z,
Su Y, Moore XL, Lambert G, Gibbs ME, Dart AM, et al: Knockout of
beta(1)- and beta(2)-adrenoceptors attenuates pressure
overload-induced cardiac hypertrophy and fibrosis. Br J Pharmacol.
153:684–692. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ito A, Ohnuki Y, Suita K, Ishikawa M,
Mototani Y, Shiozawa K, Kawamura N, Yagisawa Y, Nariyama M, Umeki
D, et al: Role of β-adrenergic signaling in masseter muscle. PLoS
One. 14(e0215539)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Steiner JE, Cottrell CE, Streicher JL,
Jensen JN, King DM, Burrows PE, Siegel DH, Tollefson MM, Drolet BA
and Püttgen KB: Scarring in patients With PIK3CA-related overgrowth
syndromes. JAMA Dermatol. 154:452–455. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yang X, Li X, Lin Q and Xu Q:
Up-regulation of microRNA-203 inhibits myocardial fibrosis and
oxidative stress in mice with diabetic cardiomyopathy through the
inhibition of PI3K/Akt signaling pathway via PIK3CA. Gene.
715(143995)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wegner KA, Mueller BR, Unterberger CJ,
Avila EJ, Ruetten H, Turco AE, Oakes SR, Girardi NM, Halberg RB,
Swanson SM, et al: Prostate epithelial-specific expression of
activated PI3K drives stromal collagen production and accumulation.
J Pathol. 250:231–242. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhu J, Zhou L, Wei B, Qian Z, Wang J, Hui
H and Sun Y: miR-142-5p inhibits pancreatic cancer cell migration
and invasion by targeting PIK3CA. Mol Med Rep. 22:2085–2092.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cai L, Chao G, Li W, Zhu J, Li F, Qi B,
Wei Y, Chen S, Zhou G, Lu X, et al: Activated CD4+ T
cells-derived exosomal miR-142-3p boosts post-ischemic ventricular
remodeling by activating myofibroblast. Aging (Albany NY).
12:7380–7396. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Guiot J, Cambier M, Boeckx A, Henket M,
Nivelles O, Gester F, Louis E, Malaise M, Dequiedt F, Louis R, et
al: Macrophage-derived exosomes attenuate fibrosis in airway
epithelial cells through delivery of antifibrotic miR-142-3p.
Thorax. 75:870–881. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang Z, Liu Z, Fang X and Yang H:
MiR-142-5p suppresses tumorigenesis by targeting PIK3CA in
non-small cell lung cancer. Cell Physiol Biochem. 43:2505–2515.
2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Mei J, Zhang Y, Lu S and Wang J: Long
non-coding RNA NNT-AS1 regulates proliferation, apoptosis,
inflammation and airway remodeling of chronic obstructive pulmonary
disease via targeting miR-582-5p/FBXO11 axis. Biomed Pharmacother.
129(110326)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Vuppalanchi R, Liang T, Goswami CP,
Nalamasu R, Li L, Jones D, Wei R, Liu W, Sarasani V, Janga SC, et
al: Relationship between differential hepatic microRNA expression
and decreased hepatic cytochrome P450 3A activity in cirrhosis.
PLoS One. 8(e74471)2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang F, Luo L, Zhu ZD, Zhou X, Wang Y, Xue
J, Zhang J, Cai X, Chen ZL, Ma Q, et al: Chlorogenic acid inhibits
liver fibrosis by blocking the miR-21-regulated TGF-β1/Smad7
signaling pathway in vitro and in vivo. Front Pharmacol.
8(929)2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhou XL, Xu H, Liu ZB, Wu QC, Zhu RR and
Liu JC: miR-21 promotes cardiac fibroblast-to-myofibroblast
transformation and myocardial fibrosis by targeting Jagged1. J Cell
Mol Med. 22:3816–3824. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sun C, Tian J, Liu X and Guan G: MiR-21
promotes fibrosis and hypertrophy of ligamentum flavum in lumbar
spinal canal stenosis by activating IL-6 expression. Biochem
Biophys Res Commun. 490:1106–1111. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Conickx G, Mestdagh P, Avila Cobos F,
Verhamme FM, Maes T, Vanaudenaerde BM, Seys LJ, Lahousse L, Kim RY,
Hsu AC, et al: MicroRNA profiling reveals a role for
MicroRNA-218-5p in the pathogenesis of chronic obstructive
pulmonary disease. Am J Respir Crit Care Med. 195:43–56.
2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li Z, Qian R, Zhang J and Shi X:
MiR-218-5p targets LHFPL3 to regulate proliferation, migration, and
epithelial-mesenchymal transitions of human glioma cells. Biosci
Rep. 39(BSR20180879)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Brkić J, Dunk C, O'Brien J, Fu G, Nadeem
L, Wang YL, Rosman D, Salem M, Shynlova O, Yougbaré I, et al:
MicroRNA-218-5p promotes endovascular trophoblast differentiation
and spiral artery remodeling. Mol Ther. 26:2189–2205.
2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Han S, Chen Y, Gao Y, Sun B and Kong Y:
MicroRNA-218-5p inhibit the migration and proliferation of
pterygium epithelial cells by targeting EGFR via PI3K/Akt/mTOR
signaling pathway. Exp Eye Res. 178:37–45. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Baker Frost D, Savchenko A, Ogunleye A,
Armstrong M and Feghali-Bostwick C: Elucidating the cellular
mechanism for E2-induced dermal fibrosis. Arthritis Res Ther.
23(68)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang Z, Zhu D, Zhang X, Liu Y, Wang J and
Yan L: Tanshinone IIA regulates fibroblast proliferation and
migration and post-surgery arthrofibrosis through the
autophagy-mediated PI3K and AMPK-mTOR signaling pathway. Am J
Transl Res. 13:565–584. 2021.PubMed/NCBI
|
|
41
|
Qi L, He W, Yang J, Gao Y and Chen J:
Endoscopic balloon dilation and submucosal injection of
triamcinolone acetonide in the treatment of esophageal stricture: A
single-center retrospective study. Exp Ther Med. 16:5248–5252.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhang Y, Wang X, Liu L, Chen JP and Fan
ZN: Intramuscular injection of mitomycin C combined with endoscopic
dilation for benign esophageal strictures. J Dig Dis. 16:370–376.
2015.PubMed/NCBI View Article : Google Scholar
|