Introduction
Ovarian cancer (OC) is the third commonest
gynecological malignancy worldwide (1). Patients are frequently diagnosed with
OC at an advanced stage and the 5-year survival rate is only ~35%
(2). Despite advances in
chemotherapy and surgery, the prognosis of patients with OC remains
unsatisfactory (3). High recurrence
rates and poor outcomes due to OC metastasis pose serious
challenges (4). Therefore, it is
necessary to explore the molecular mechanisms of OC progression to
discover new therapeutic strategies.
Long non-coding (lnc) RNAs serve multiple roles in
the occurrence and development of OC. LncRNA PVT1 regulates EZH2 to
downregulate microRNA (miRNA/miR)-214, resulting in the suppression
of OC progression (5). LncRNA
LINC00319 contributes to OC progression by inhibiting miR-423-5p
and enhancing nucleus accumbens-associated 1 expression (6). LncRNA PCAT6 restrains PTEN expression
to accelerate the initiation and progression of OC (7). Numerous studies have shown that lncRNA
miR155HG (miR155HG) serves critical roles in diverse tumors
(8-10).
miR155HG promotes pancreatic cancer cell growth and inhibits
apoptosis by suppressing miR-802 expression (8). miR155HG deficiency attenuates
glioblastoma tumorigenesis by downregulating Annexin A2 expression
via an increase in miR-185 expression (9). Notably, the expression of miR155HG is
enhanced in OC tissues and cells (10).
miRNAs have been shown to participate in different
gynecological malignancies, including OC. miR-126-3p modulates
PLXNB2 expression to attenuate OC progression (11). miR-603 suppresses the malignancy of
OC cells by targeting hexokinase-2(12). miR-122 inhibits OC cell growth by
repressing prolyl 4-hydroxylase subunit α1(13). miR-155-5p has been regarded as a
pivotal regulator of multiple cancers. miR-155-5p elevation impedes
gastric cancer cell proliferation and triggers apoptosis (14). miR-155-5p regulates IGF2 through the
PI3K pathway to exert tumor-repressing roles in Wilms tumors
(15). Notably, miR-155-5p reduces
OC cell viability by inhibiting HIF1α (16).
Tyrosinase-related protein (TYRP) 1 is a melanogenic
enzyme and protein (17). Previous
research has reported on the relationship between melanogenic
proteins and cancers, such as TYRP1 in breast cancer (18) and TYRP2 in retinoblastoma (19). TYRP1 exerts tumor-promoting
functions in different types of cancer. Increased TYRP1 expression
in lymph node metastases from melanoma patients is related to
unfavorable prognosis (20). TYRP1
has emerged as an oncogene in colon cancer and high levels of TYRP1
are associated with decreased overall survival rates (21). Notably, El Hajj et al
(22) reported that TYRP1 is a
target of miR-155.
Despite the aforementioned studies, the molecular
mechanisms of miR155HG, miR-155-5p and TYRP1 in OC progression
remain unclear. The present study assessed the expression and roles
of miR155HG in OC. In addition, the relationships between miR155HG,
miR-155-5p and TYRP1 in OC were determined. The present study aimed
to reveal the molecular mechanism of miR155HG in OC cell viability,
migration, invasion and apoptosis.
Materials and methods
Ethics statement
The study was approved by the ethics committee of
the Affiliated Hospital of North Sichuan Medical College [approval
no. 2020ER (A) 066)]. Written informed consent was obtained from
all participants. The study was conducted in accordance with the
principles of the Declaration of Helsinki.
Clinical samples
Patients with OC (n=55) who underwent ovariectomy
between September 2017 and October 2019 at the Affiliated Hospital
of North Sichuan Medical College (Nanchong, China) were enrolled in
this study. Among them, 41 cases were the serous subtype, 8 cases
were the endometrioid subtype and 6 cases were other subtypes. The
histopathological diagnosis of the cases was in accordance with the
diagnostic categories of the World Health Organization
2020(23). OC tissues (n=55; tumor
group) and paired adjacent non-tumor tissues (tissue which were 1-2
cm away from the tumor tissues; adjacent group) were obtained the
from patients with OC who underwent ovariectomy. Prior to
ovariectomy, radiotherapy or chemotherapy was not administered to
the patients.
Cell culture
The OVCAR3 and SK-OV-3 OC cell lines and normal
IOSE80 ovarian cell line (Chinese Academy of Sciences) were
cultured in Roswell Park Memorial Institute (RPMI)-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
exosome-free fetal bovine serum (FBS; Invitrogen; Thermo Fisher
Scientific, Inc.) at 37˚C with 5% CO2.
Cell transfection
For cell transfection, OVCAR3 and SK-OV-3 cells
grown to 85% confluence were transfected with 100 nM of negative
control (NC) small interfering (si) RNA (si-NC;
5'-UUCUCCGAACGUGUCACGU-3'), 100 nM of miR155HG siRNA
(si-miR155HG-1; 5'-CUGGGAUGUUCAACCUUAA-3'; si-miR155HG-2;
5'-UCUUAAAGGGAAACUGAAA-3'), 100 nM of mimics NC
(5'-UCACAACCUCCUAGAAAGAGUAGA-3'), 100 nM of miR-155-5p mimics
(5'-UUAAUGCUAAUCGUCAUAGGGGU-3'), 100 nM of inhibitor NC
(5'-CAGUACUUUUGUGUAGUACAA-3'), 100 nM of miR-155-5p inhibitor
(5'-ACCCCUAUCACGAUUAGCAUUAA-3'), 1 µg of empty plasmid
(pcDNA3.1-NC) and 1 µg of TYRP1 overexpression plasmid
(pcDNA3.1-TYRP1), or co-transfected with 100 nM of si-miR155HG-1
and miR-155-5p inhibitor, 100 nM of si-miR155HG-1 and 1 µg of
pcDNA3.1-TYRP1 using Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) for 6 h at 37˚C. All
oligonucleotides or plasmids were purchased from Shanghai
GenePharma Co., Ltd. At 48 h post-transfection, the cells were
harvested and reverse transcription-quantitative (RT-q) PCR was
conducted to determine the transfection efficiency.
RT-qPCR
RT-qPCR procedures were performed according to the
corresponding manufacturer's protocols. In brief, total RNA was
extracted from tissues and cells using TRIzol® reagent
(Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into
complementary DNA using the Prime Script RT reagent kit (Takara
Biotechnology Co., Ltd.). The SYBR-Green PCR kit (Takara
Biotechnology Co., Ltd.) and TaqMan MicroRNA Assay Kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) were used for qPCR
analysis. The following thermocycling conditions were used for the
qPCR: Initial denaturation at 95˚C for 3 min; followed by 40 cycles
at 95˚C for 15 sec, annealing at 60˚C for 30 sec, elongation at
72˚C for 1 min; and a final extension at 72˚C for 5 min. GAPDH, U6
and β-actin were used for the normalization of miR155HG, miR-155-5p
and TYRP1, respectively (8,24,25).
Relative expression was calculated using the 2-ΔΔCq
method (26). The primers used are
presented in Table I.
 | Table IPrimers sequences. |
Table I
Primers sequences.
| Name of primer | Sequences
(5'-3') |
|---|
| miR155HG-F |
CCCAAATCTAGGTTCAAGTTC |
| miR155HG-R |
CATCTAAGCCTCACAACAAC |
| GAPDH-F |
AGGTGAAGGTCGGAGTCAACG |
| GAPDH-R |
AGGGGTCATTGATGGCAACA |
| miR-155-5p-F |
GTGCAGGGTCCGAGGTATT |
| miR-155-5p-R |
GCCGCTTAATGCTAATCGTGATAG |
| U6-F |
GCTTCGGCAGCACATATACTAAAAT |
| U6-R |
CGCTTCACGAATTTGCGTGTCAT |
| TYRP1-F |
GCTCAGTGCTTGGAAGTTGGT |
| TYRP1-R |
AGTTTGTCCTCCAGTTCCGTTTAG |
| β-actin-F |
GACCCTGCCATCTGTGC |
| β-actin-R |
CGGGTGGAGGAGTTTCA |
Western blot analysis
The transfected OVCAR3 and/or SK-OV-3 cells were
lysed with RIPA buffer (Beyotime Institute of Biotechnology) to
extract total protein. The protein concentration was detected via
the BCA Protein Assay kit. Subsequently, a total of 50 µg
protein/lane was separated by 10% SDS-PAGE and then transferred
onto PVDF membranes. Following blocking with 5% skimmed milk for 2
h at 25˚C, the membranes were incubated overnight at 4˚C with
primary antibodies, including anti-TYRP1 (1:1,000; ab235447;
Abcam), anti-Bax (1:1,000; ab32503; Abcam), anti-Bcl-2 (1:2,000;
ab182858; Abcam) and anti-β-actin (1:1,000; ab265588; Abcam).
Thereafter, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibodies (1:5,000; ab97080;
Abcam) at 25˚C for 1 h. The proteins bound by their respective
antibodies on the immunoblots were measured using an enhanced ECL
kit (Thermo Fisher Scientific, Inc.) and quantified using ImageLab
software (version 2.3; Bio-Rad Laboratories, Inc.).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
OVCAR3 and SK-OV-3 cells (2x103/well)
were seeded into 96-well plates and incubated at 37˚C with 5%
CO2. At each time point (0, 24, 48 and 72 h
post-transfection), cell viability was determined using the MTT
cell viability assay kit (Sigma-Aldrich; Merck KGaA) under an
inverted light microscope (magnification x400; Olympus
Corporation).
Wound healing assay
OVCAR3 and SK-OV-3 cells (1x106/well)
were incubated in 6-well plates. The cell monolayer was then
wounded with a 10-µl pipette tip and cultured in serum-free medium.
Images of the different stages of wound healing were captured by a
light microscopy (magnification, x400; Olympus Corporation) at 0
and 48 h.
Invasion assay
Transwell chambers (24-well; 8 µM pore size; BD
Biosciences) coated with Matrigel at 37˚C for 30 min (BD
Biosciences) were used to evaluate cell invasion. OVCAR3 and
SK-OV-3 cells (1x105) were seeded into the upper chamber
of Transwell plates (Corning, Inc.) in serum-free RPMI-1640 medium.
Exosome-free FBS (10%) RPMI-1640 medium was added to the lower
chamber of the Transwell plates. After 24 h, cells that had invaded
the pores were fixed with methanol and stained with 0.5% crystal
purple at 37˚C for 30 min. Stained cells were imaged using an
inverted light microscope (magnification, x400; Olympus
Corporation).
Dual-luciferase reporter assay
The putative binding sites of miR-155-5p on miR155HG
and the TYRP1 3' untranslated region (UTR) were predicted using
StarBase (version 2.0; http://starbase.sysu.edu.cn) and TargetScan (release
7.2; http://www.targetscan.org/vert_72/), respectively.
miR155HG and TYRP1 sequences were generated with wild-type (WT) or
mutant miR-155-5p binding sites and cloned them into pmirGLO
vectors (Shaanxi Youbio Technology Co., Ltd.). OVCAR3 and SK-OV-3
cells were co-transfected with the luciferase vectors and NC mimics
or miR-155-5p mimics using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h at 37˚C.
Relative luciferase activity was examined using a dual-luciferase
reporter assay system (Promega Corporation). The activity of
firefly luciferase was normalized to that of Renilla
luciferase.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 8.0 software (GraphPad Software, Inc.). Data are
expressed as means ± standard deviations. The miR155HG, miR-155-5p
and TYRP1 expression of OC tissues and paired adjacent non-tumor
tissues were assessed using paired Student's t-test. In addition,
differences between two groups were analyzed using unpaired
Student's t-test. Differences among multiple groups were assessed
by a one-way analysis of variance followed by Tukey's post-hoc
test. In the analysis of clinicopathological features, the age,
diameter, lymph node metastasis, International Federation of
Gynecology and Obstetrics (FIGO) stage (27) and histological grade were analyzed
using the χ2 test. The pathological subtype was analyzed
using the Fisher's exact test. The significance of the correlations
was determined using Pearson's correlation analysis. All
experiments were performed in triplicate, and each experiment was
repeated three times. P<0.05 was considered to indicate a
statistically significant difference.
Results
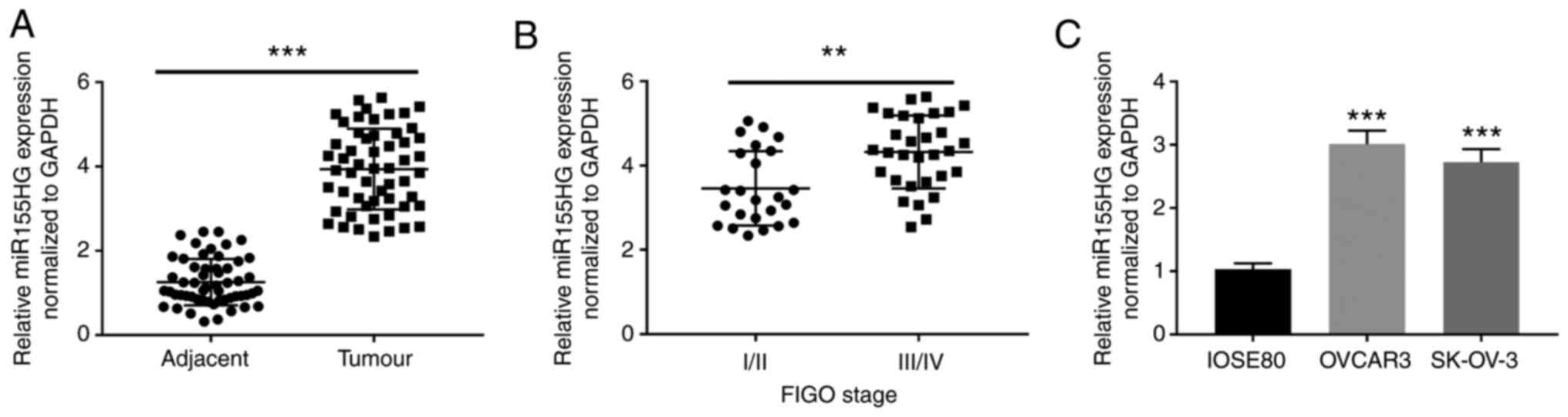
miR155HG is upregulated in OC
To confirm whether miR155HG is differentially
expressed in OC tissues, miR155HG expression was analyzed by
RT-qPCR in 55 patients with OC. The results showed that miR155HG
expression was considerably upregulated in OC tissues compared with
that in adjacent non-tumor tissues (P<0.001; Fig. 1A). Additionally, miR155HG expression
was notably elevated in tumors at FIGO stage III/IV (P<0.01;
Fig. 1B). Furthermore, miR155HG
expression was clearly enhanced in OVCAR3 and SK-OV-3 cells
compared with that in IOSE80 cells (P<0.001; Fig. 1C). According to the analysis of
clinicopathological features shown in Table II, the expression of miR155HG was
not correlated with age, pathological subtype, diameter and lymph
node metastasis (P>0.05) but was closely associated with the
FIGO stage (P<0.05) and histological grade (P<0.01) of
patients with OC. Notably, the number of patients with OC with high
miR155HG expression was greater than the number of patients with OC
with low miR155HG expression in the ‘low-grade’ (grade G3) type of
OC (P<0.01).
 | Table IICorrelation between miR155HG
expression and clinicopathological features in ovarian cancer
patients. |
Table II
Correlation between miR155HG
expression and clinicopathological features in ovarian cancer
patients.
|
Characteristics | n | miR155HG (Low)
27 | miR155HG (High)
28 | P-value |
|---|
| Age | | | | 0.698a |
|
<55
years | 23 | 12 | 11 | |
|
≥55
years | 32 | 15 | 17 | |
| Pathological
subtype | | | | 0.749a |
|
Serous | 41 | 19 | 22 | |
|
Endometrioid | 8 | 5 | 3 | |
|
Others | 6 | 3 | 3 | |
| Diameter | | | | 0.891a |
|
<5
cm | 27 | 13 | 14 | |
|
≥5 cm | 28 | 14 | 14 | |
| Lymph node
metastasis | | | | 0.341a |
|
No | 29 | 16 | 13 | |
|
Yes | 26 | 11 | 15 | |
| FIGO stage | | | | 0.022b |
|
I + II | 24 | 16 | 8 | |
|
III +
IV | 31 | 11 | 20 | |
| Histological
grade | | | | 0.004c |
|
G1-G2 | 25 | 15 | 10 | |
|
G3 | 30 | 12 | 18 | |
miR155HG silencing restrains OC cell
viability, migration and invasion while promoting apoptosis
Loss-of-function experiments were performed to
investigate whether miR155HG knockdown affects OC progression in
vitro. Fig. 2A shows that
miR155HG was effectively silenced following si-miR155HG-1
(P<0.001) and si-miR155HG-2 (P<0.01) transfection in OVCAR3
and SK-OV-3 cells. Si-miR155HG-1 was used for subsequent assays
because of its high silencing efficiency. The MTT assay revealed
that the viability of OVCAR3 and SK-OV-3 cells was markedly reduced
after si-miR155HG-1 transfection (P<0.01; Fig. 2B). The wound healing and invasion
assays revealed that the migration and invasion of OVCAR3 and
SK-OV-3 cells were visibly suppressed by miR155HG deficiency
(P<0.001; Fig. 2C and D). Bax and Bcl-2 are biomarkers of
apoptosis. miR155HG silencing markedly increased the Bax protein
expression level and decreased the Bcl-2 protein expression level
in OVCAR3 and SK-OV-3 cells (P<0.05; Fig. 2E).
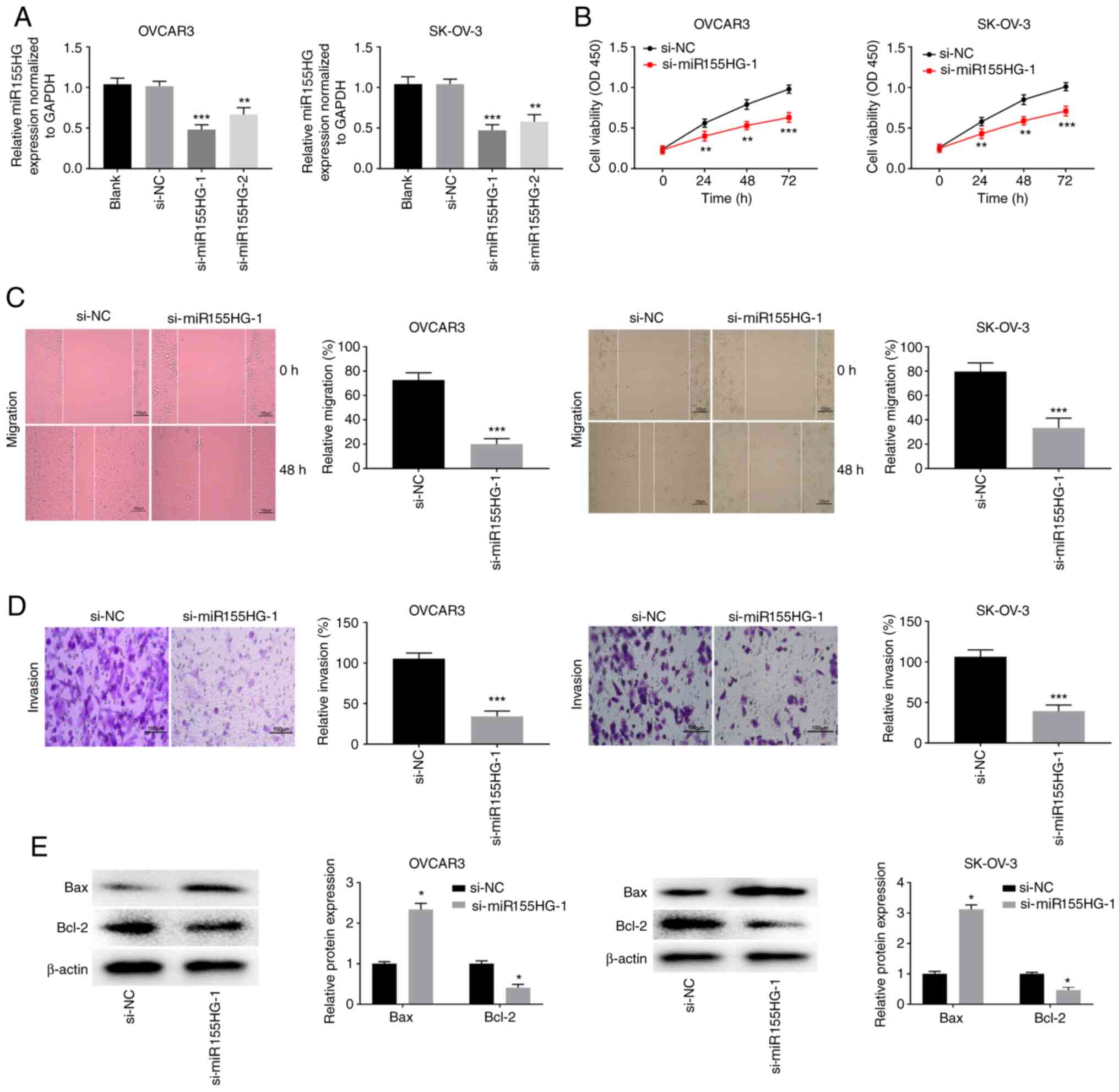 | Figure 2miR155HG silencing restrains the
viability, migration and invasion, while promoting apoptosis of OC
cells. (A) The transfection efficiency of si-NC, si-miR155HG-1 and
si-miR155HG-2 in OVCAR3 and SK-OV-3 cells was measured by reverse
transcription-quantitative PCR. **P<0.01,
***P<0.001 vs. si-NC. (B) The viability of OVCAR3 and
SK-OV-3 cells was assessed by MTT assay. **P<0.01,
***P<0.001 vs. si-NC. The (C) migration and (D)
invasion of OVCAR3 and SK-OV-3 cells were analyzed by wound-healing
assay and invasion assay. ***P<0.001 vs. si-NC. Scale
bar=100 µm; (E) The protein expression of Bax and Bcl-2 in OVCAR3
and SK-OV-3 cells were measured by western blotting.
*P<0.05 vs. si-NC. miR, microRNA; OC, ovarian cancer;
si, short interfering; NC, negative control; OD, optical
density. |
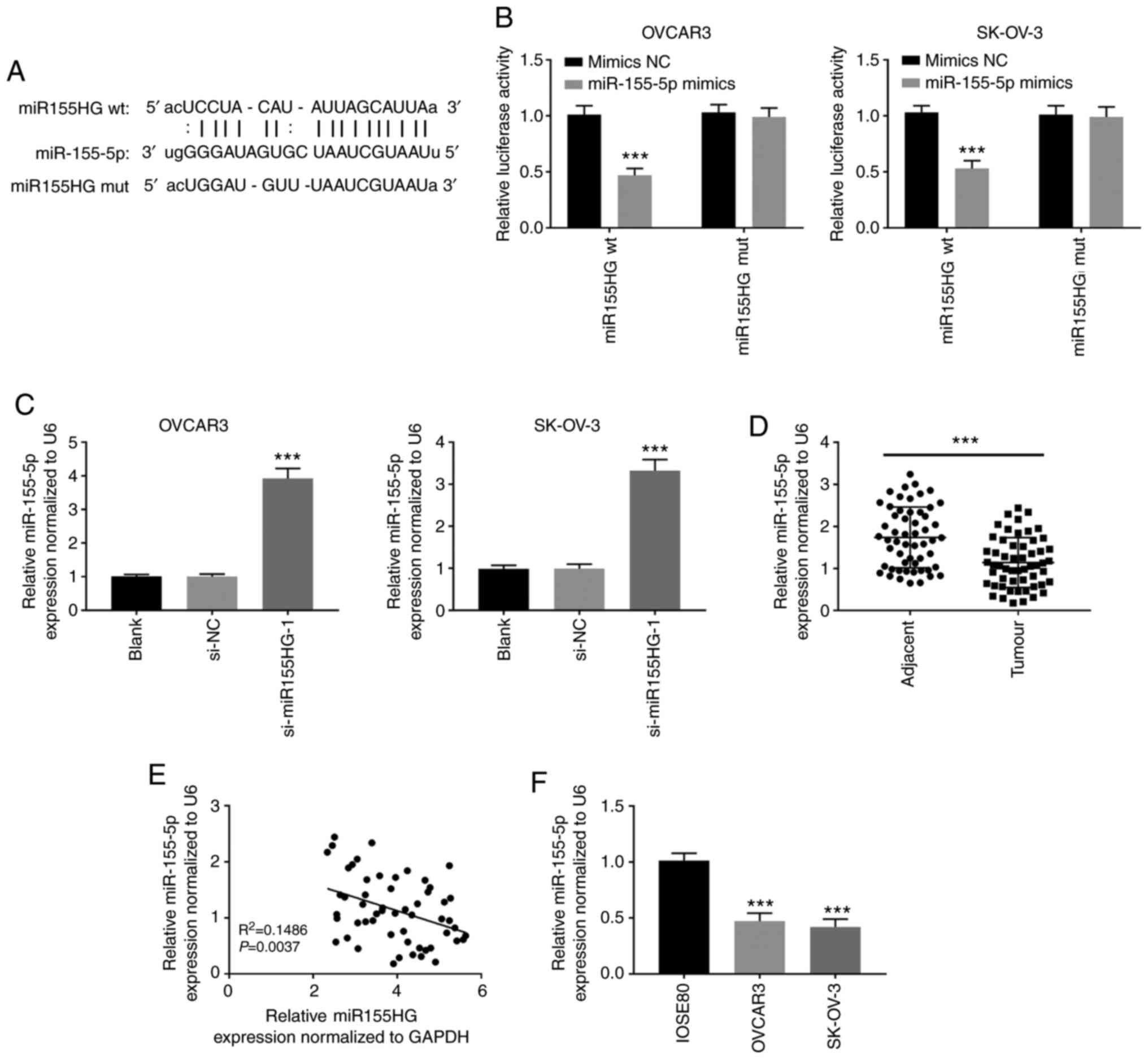
miR155HG directly targets
miR-155-5p
To confirm the miR155HG mechanism of action in OC
development, miR-155-5p was identified as a potential miR155HG
target via StarBase (Fig. 3A). The
dual-luciferase reporter assay showed that miR-155-5p upregulation
evidently attenuated the activity of the WT-miR155HG reporter in
OVCAR3 and SK-OV-3 cells (P<0.001; Fig. 3B). In addition, si-miR155HG-1
transfection markedly enhanced miR-155-5p expression in OVCAR3 and
SK-OV-3 cells (P<0.001; Fig.
3C). In addition, miR-155-5p expression was dramatically
inhibited in OC tissues compared with that in adjacent non-tumor
tissues (P<0.001; Fig. 3D). An
inverse correlation between miR155HG and miR-155-5p expression was
observed in OC tissues (Fig. 3E).
miR-155-5p expression was considerably downregulated in OVCAR3 and
SK-OV-3 cells compared with that in IOSE80 cells (P<0.001,
Fig. 3F).
miR-155-5p elevation impedes OC cell
viability, migration and invasion while facilitating apoptosis
To determine the biological function of miR-155-5p
in OC, miR-155-5p was enhanced or blocked after miR-155-5p mimic
(P<0.001) or miR-155-5p inhibitor (P<0.01) transfection in
OVCAR3 and SK-OV-3 cells (Fig. 4A).
miR-155-5p elevation significantly attenuated the viability of
OVCAR3 and SK-OV-3 cells (P<0.01; Fig. 4B). Furthermore, miR-155-5p elevation
markedly retarded the migration and invasion of OVCAR3 and SK-OV-3
cells (P<0.001; Fig. 4C and
D). Overexpression of miR-155-5p
not only significantly elevated the Bax protein expression level
but also reduced the Bcl-2 protein expression level in OVCAR3 and
SK-OV-3 cells (P<0.05; Fig.
4E).
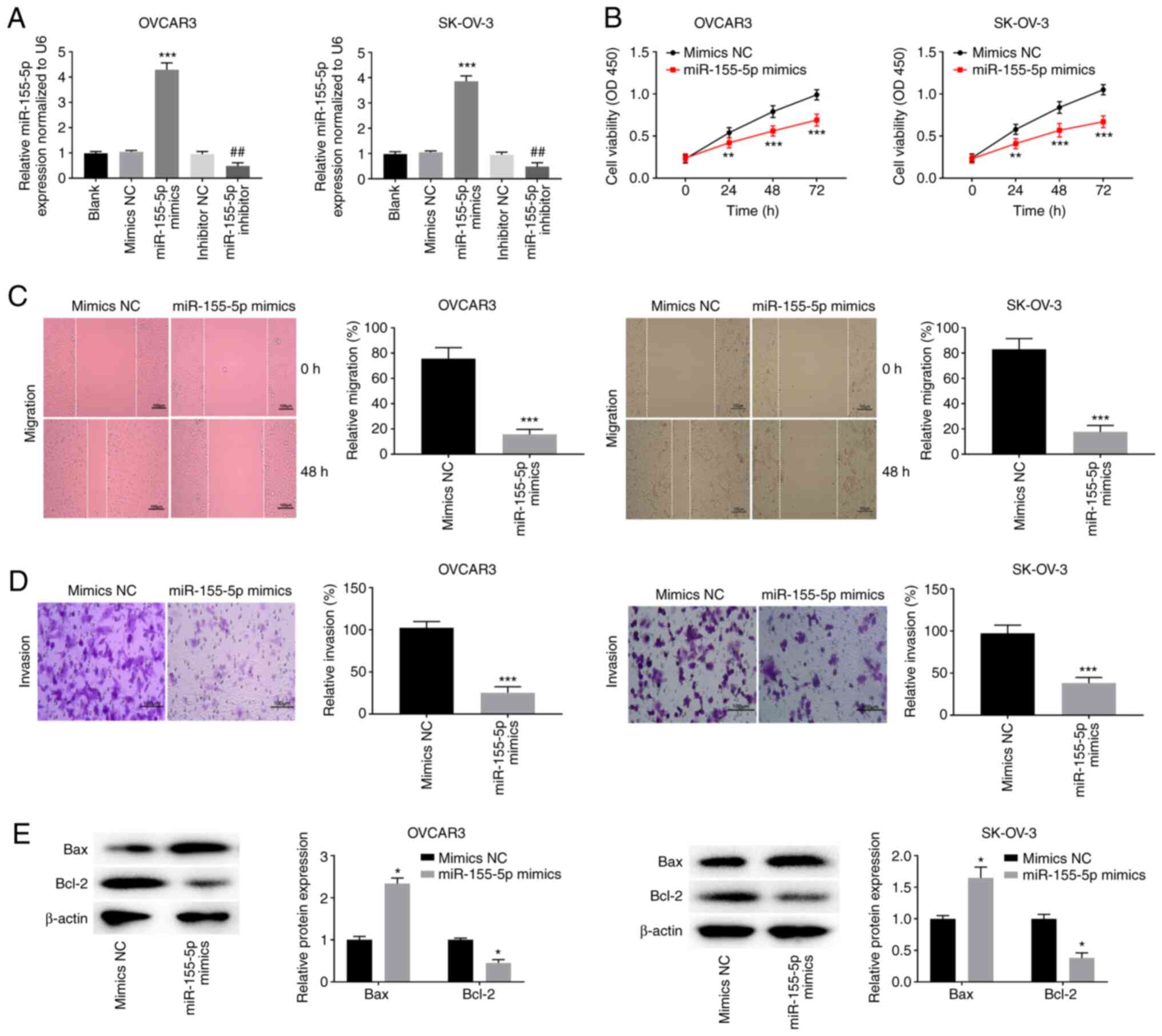 | Figure 4miR-155-5p elevation impedes the
viability, migration and invasion while facilitating apoptosis of
OC cells. (A) The transfection efficiency of mimics NC, miR-155-5p
mimics, inhibitor NC and miR-155-5p inhibitor was evaluated by
reverse transcription-quantitative PCR in OVCAR3 and SK-OV-3 cells.
***P<0.001 vs. mimics NC, ##P<0.01 vs.
inhibitor NC; (B) MTT assay were performed after transfected with
mimics NC or miR-155-5p mimics in OVCAR3 and SK-OV-3 cells.
**P<0.01, ***P<0.001 vs. mimics NC. The
effects of miR-155-5p overexpression on the (C) migration and (D)
invasion of OVCAR3 and SK-OV-3 cells were assessed.
***P<0.001 vs. mimics NC. Scale bar=100 µm; (E)
Western blotting was performed to detect the protein expression of
Bax and Bcl-2 in OVCAR3 and SK-OV-3 cells. *P<0.05
vs. mimics NC. miR, microRNA; OC, ovarian cancer; NC, negative
control; si, short interfering; OD, optical density. |
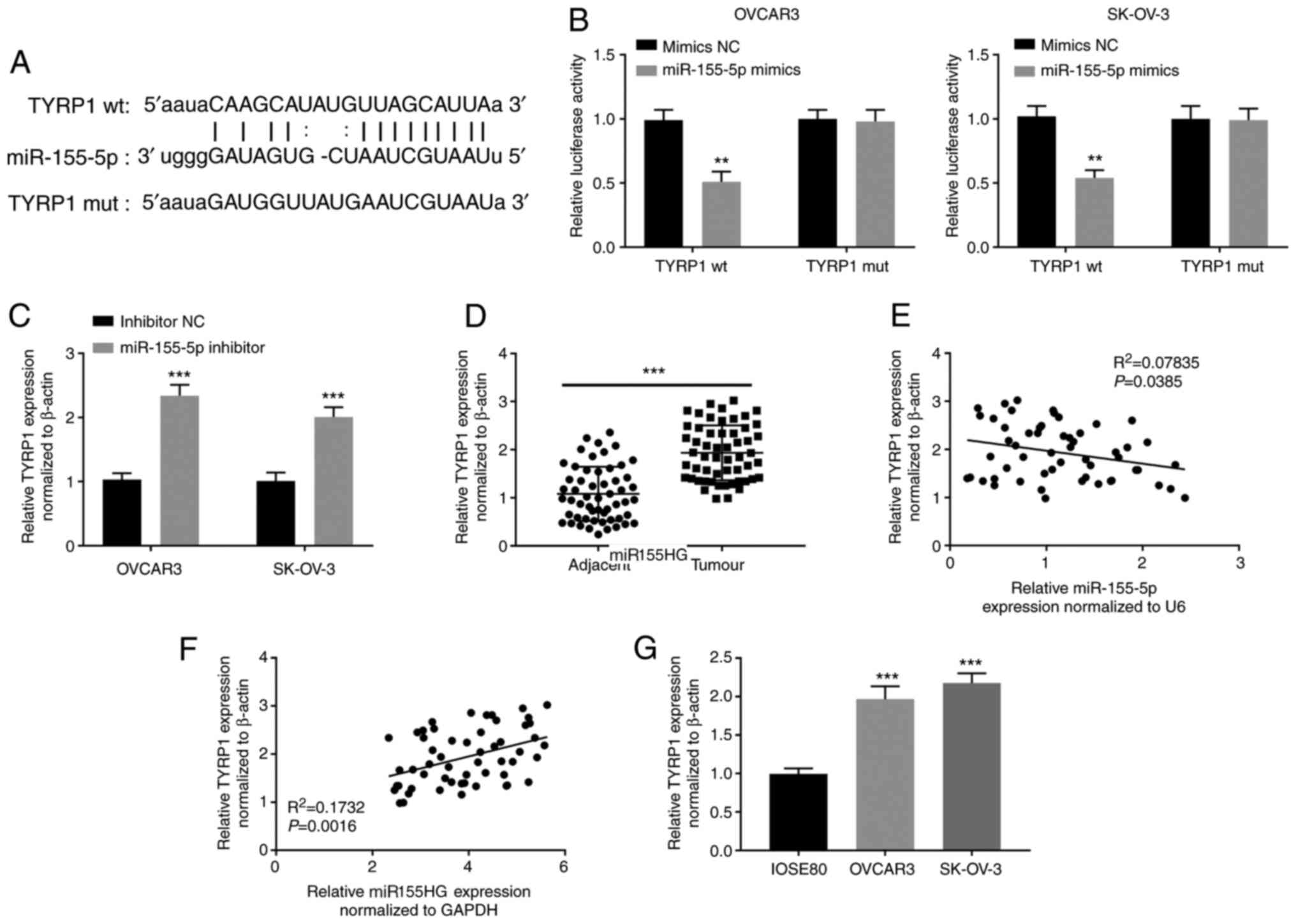
TYRP1 is a target of miR-155-5p
To demonstrate whether TYRP1 is a direct target of
miR-155-5p in OC, TargetScan was used to predict the binding site
for miR-155-5p on the 3'-UTR of TYRP1 (Fig. 5A). The dual-luciferase reporter
assay showed that miR-155-5p elevation clearly hindered the
activity of the WT-TYRP1 reporter in OVCAR3 and SK-OV-3 cells
(P<0.01; Fig. 5B). Additionally,
miR-155-5p deficiency visibly enhanced TYRP1 expression in OVCAR3
and SK-OV-3 cells (P<0.001; Fig.
5C). Furthermore, TYRP1 expression was upregulated in OC
tissues compared with that in adjacent non-tumor tissues
(P<0.001; Fig. 5D). A negative
correlation between TYRP1 and miR-155-5p expression (Fig. 5E) and a positive correlation between
TYRP1 and miR155HG expression (Fig.
5F) were observed in OC tissues. TYRP1 expression was
upregulated in OVCAR3 and SK-OV-3 cells compared with that in
IOSE80 cells (P<0.001; Fig.
5G).
miR155HG silencing hampers the
malignant biological behavior of OC cells by targeting the
miR-155-5p/TYRP1 axis
To ascertain whether miR155HG modulates TYRP1
expression by affecting miR-155-5p repression activity, feedback
approaches were used, in which the effects of miR155HG silencing
were reversed by TYRP1 overexpression or miR-155-5p inhibition.
OVCAR3 cells were transiently transfected with pcDNA3.1-NC or
pcDNA3.1-TYRP1. The overexpression efficiency of pcDNA3.1-TYRP1 was
high in OVCAR3 cells. TYRP1 expression was effectively enhanced
following transfection with pcDNA3.1-TYRP1 in OVCAR3 cells
(P<0.001; Fig. 6A). In addition,
the protein expression level of TYRP1 was markedly increased in
OVCAR3 cells following transfection with pcDNA3.1-TYRP1 (P<0.01;
Fig. 6B). As shown in Fig. 6C, miR155HG silencing visibly
suppressed TYRP1 expression in OVCAR3 cells (P<0.001), whereas
miR-155-5p deficiency weakened the inhibitory effect of miR155HG
silencing on TYRP1 expression (P<0.05). The feedback approaches
showed that TYRP1 elevation or miR-155-5p deficiency considerably
mitigated the inhibitory effects of miR155HG silencing on OVCAR3
cell viability, migration and invasion (P<0.01; Fig. 6D-G). In addition, TYRP1
overexpression or miR-155-5p inhibition markedly weakened the
promotive effect of miR155HG silencing on the Bax protein
expression level and reversed the reduction effect of miR155HG
silencing on the Bcl-2 protein expression level in OVCAR3 cells
(P<0.05; Fig. 6H).
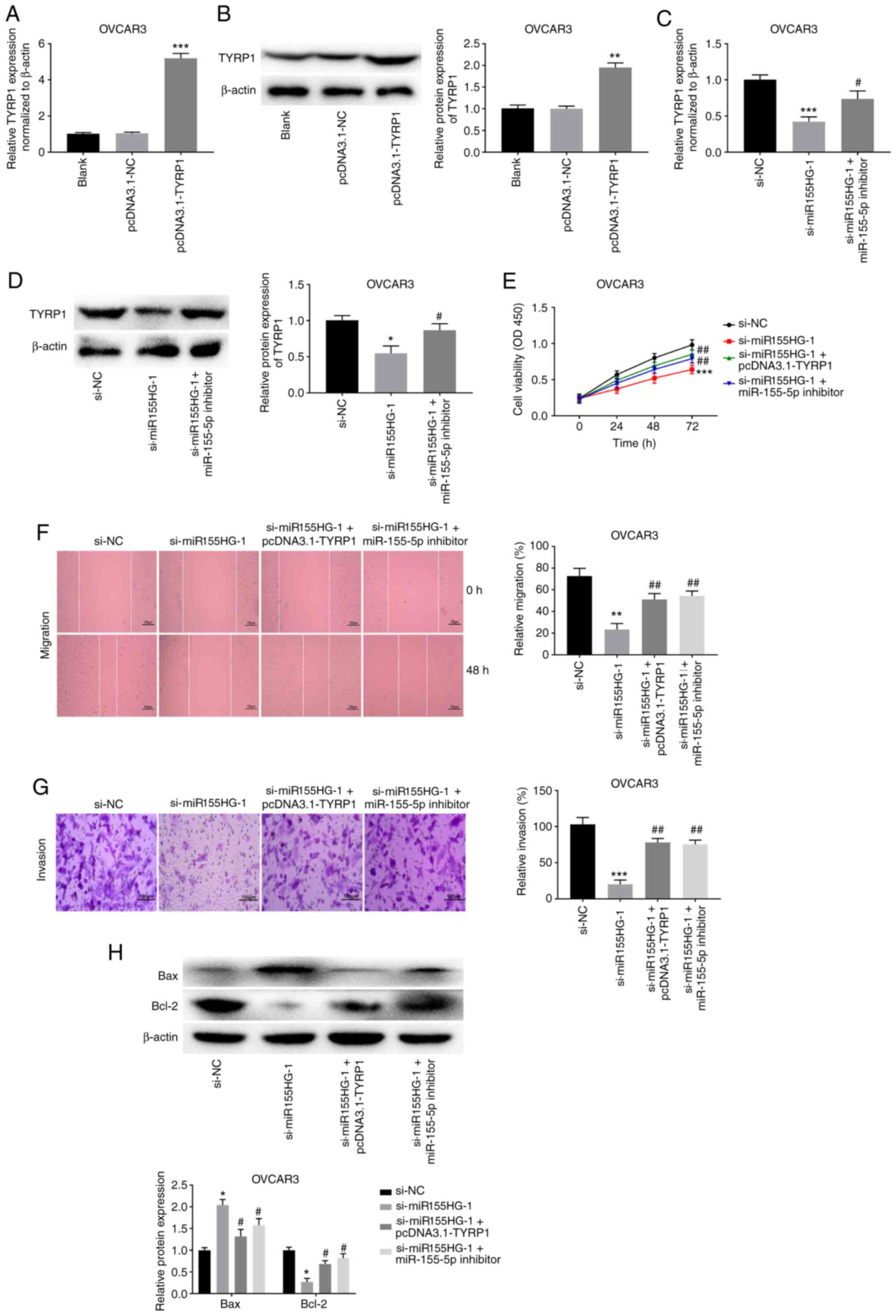 | Figure 6miR155HG silencing hampers the
malignant biological behavior of OC cells by targeting the
miR-155-5p/TYRP1 axis. (A) The transfection efficiency of
pcDNA3.1-NC and pcDNA3.1-TYRP1 was measured by RT-qPCR in OVCAR3
cells. ***P<0.001 vs. pcDNA3.1-NC. (B) The protein
expression of TYRP1 in OVCAR3 cells was detected by western
blotting. **P<0.01 vs. pcDNA3.1-NC. (C) RT-qPCR was
used to detect the expression of TYRP1 in OVCAR3 cells.
***P<0.001 vs. si-NC, #P<0.05 vs.
si-miR155HG-1. (D) Downregulation of miR-155-5p or upregulation of
TYRP1 reversed the inhibitory effects of miR155HG silencing on (E)
viability, (F) migration and (G) invasion of OVCAR3 cells.
*P<0.05, **P<0.01,
***P<0.001 vs. si-NC. #P<0.05,
##P<0.01 vs. si-miR155HG-1. Scale bar=100 µm. (H)
Overexpression of TYRP1 or inhibition of miR-155-5p reversed the
effects of miR155HG silencing on the protein expression of Bax and
Bcl-2 in OVCAR3 cells. *P<0.05 vs. si-NC;
#P<0.05 vs. si-miR155HG-1. miR, microRNA; OC, ovarian
cancer; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; si, short interfering; OD, optical
density. |
Discussion
It has been documented that the expression of
lncRNAs, such as PTAR (28),
TP73-AS1(29) and CCAT1(30), is increased in OC. In the present
study, miR155HG expression was upregulated in OC, suggesting that
miR155HG may be an oncogene. In addition, a high level of miR155HG
was correlated with FIGO stage in patients with OC. Certain lncRNAs
are similar to miR155HG. For instance, high lncRNA PVT1 expression
is related to poor prognosis and advanced FIGO stage in patients
with OC (5). Overexpression of
lncRNA HOTTIP is markedly correlated with advanced FIGO stage in
patients with OC (31). Above all,
the present study suggested that miR155HG expression may be
associated with OC development. Previous studies have demonstrated
that miR155HG participates in the malignant biological behavior of
diverse cancers (24,32,33).
miR155HG silencing diminishes cell viability while facilitating
cell apoptosis by targeting PTBP1 to restrain glioma development
(32). miR155HG knockdown
upregulates miR-155-3p and downregulates TP53INP1, thus retarding
the biological behavior of non-small cell lung cancer (33). miR155HG downregulation hinders
laryngeal squamous cell carcinoma progression by increasing
miR-155-5p and reducing SOX10 expression (24). In the present study, miR155HG
knockdown attenuated OC cell viability, invasion and migration
while promoting OC cell apoptosis, indicating that miR155HG
silencing may inhibit the development of OC.
Some lncRNAs serve as competing endogenous RNAs or
molecular sponges to regulate miRNAs in OC. For instance, lncRNA
EWSAT1 decreases miR-330-5p expression to accelerate OC progression
(34). LncRNA CCAT1 exerts a
tumor-promoting role in OC progression by sponging miR-490-3p
(35). Notably, lncRNA NORAD
induces OC cell proliferation and cell cycle transition by
downregulating miR-155-5p (36). In
the present study, miR-155-5p was a target of miR155HG and
inversely correlated with miR155HG expression, suggesting that
miR155HG may influence OC by regulating miR-155-5p. In fact,
miR-155-5p is downregulated and acts as a target of miR155HG in
some cancers, such as glioma (37),
laryngeal squamous cell carcinoma (24) and clear cell renal cell carcinoma
(38). In the present study,
miR-155-5p was downregulated in OC, indicating that miR-155-5p may
be an anti-oncogene. Previous studies have determined that
miR-155-5p acts as a tumor suppressor in different cancers. For
instance, miR-155-5p decreases CREB1 expression to prevent the
malignant behavior of WT cells (39). miR-155-5p attenuates bladder cancer
progression by regulating MTGR1(40). Notably, miR-155-5p overexpression
weakens the promotive effects of lncRNA NORAD on OC progression
(36). In the present study,
miR-155-5p overexpression inhibited the malignant biological
behavior of OC cells and miR-155-5p inhibition reversed the
anti-tumor effects of miR155HG silencing in OC cells. In summary,
miR155HG knockdown may suppress the malignant biological behavior
of OC cells by upregulating miR-155-5p.
Melanogenic proteins are frequently upregulated in
different tumors, such as TYRP1 in melanoma (41) and TYRP2 in glioma (42). Similarly, TYRP1 expression was
increased in OC, indicating that TYRP1 may be an oncogene. TYRP1
often acts as an oncogene in certain cancers. TYRP1 attenuates the
tumor suppressor activity of miR-16 to facilitate melanoma cell
proliferation (43). CXCL1 promotes
tumor progression in colon cancer by upregulating TYRP1(21). Notably, miR-155 downregulates TYRP1
expression to improve the survival of patients with melanoma
(22). In the present study, TYRP1
was a target of miR-155-5p and negatively correlated with
miR-155-5p. It was hypothesized that miR-155-5p was involved in OC
progression by regulating TYRP1. Furthermore, TYRP1 expression was
positively associated with miR155HG in OC. Considering the
miR155HG/miR-155-5p targeting relationship, it was hypothesized
that miR155HG may enhance TYRP1 expression by inhibiting miR-155-5p
in OC. In the present study, feedback approaches revealed that
TYRP1 upregulation reversed the suppressed OC cell behaviors caused
by miR155HG knockdown. Taken together, the present study suggested
that miR155HG knockdown constrained the malignant biological
behavior of OC cells by regulating the miR-155-5p/TYRP1 axis.
There were some limitations to the present study.
First, the correlation between the expression of miR155HG and the
survival rate of patients with OC was not analyzed; however, more
data about patient survival will be collected in the future.
Second, a normal healthy control group was not set up to compare
with the disease group. Third, the present study did not evaluate
the effect of radiotherapy or chemotherapy on miR155HG-silenced
cells. Fourth, there are a number of other downstream targets of
miR155HG that have not yet been evaluated in OC. Fifth, the present
study was limited to the cellular level and further in vivo
experiments are needed to confirm the role of miR155HG in OC.
In summary, miR155HG expression was enhanced in OC.
In addition, miR155HG knockdown appeared to exert tumor-repressing
roles in OC progression by regulating the miR-155-5p/TYRP1 axis.
Thus, the current study may enhance our understanding of the
mechanism underlying miR155HG in OC progression.
Acknowledgements
Not applicable.
Funding
Funding: The present study was financially supported by the
Government- Universities Specific Cooperative Scientific Research
Project of Nanchong (grant no. 18SXHZ0251).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AW and LL conceived and designed the present study,
analyzed data, performed the experiments and data analyses and
wrote the manuscript. CD and XL contributed significantly to
analysis and manuscript preparation, performed the experiments and
data analyses and revised the manuscript. AW and LL confirm the
authenticity of all the raw data. All authors reviewed and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was conducted after obtaining
ethical approval from the Affiliated Hospital of North Sichuan
Medical College [Nanchong, China; approval no. 2020ER (A)066].
Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nash Z and Menon U: Ovarian cancer
screening: Current status and future directions. Best Pract Res
Clin Obstet Gynaecol. 65:32–45. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nimmagadda S and Penet MF: Ovarian cancer
targeted theranostics. Front Oncol. 9(1537)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chandra A, Pius C, Nabeel M, Nair M,
Vishwanatha JK, Ahmad S and Basha R: Ovarian cancer: Current status
and strategies for improving therapeutic outcomes. Cancer Med.
8:7018–7031. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Stewart C, Ralyea C and Lockwood S:
Ovarian cancer: An integrated review. Semin Oncol Nurs. 35:151–156.
2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chen Y, Du H, Bao L and Liu W: LncRNA PVT1
promotes ovarian cancer progression by silencing miR-214. Cancer
Biol Med. 15:238–250. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Du W, Feng Z and Sun Q: LncRNA LINC00319
accelerates ovarian cancer progression through miR-423-5p/NACC1
pathway. Biochem Biophys Res Commun. 507:198–202. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kong FR, Lv YH, Yao HM, Zhang HY, Zhou Y
and Liu SE: LncRNA PCAT6 promotes occurrence and development of
ovarian cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci.
23:8230–8238. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Qin Y, Liu X, Pan L, Zhou R and Zhang X:
Long noncoding RNA MIR155HG facilitates pancreatic cancer
progression through negative regulation of miR-802. J Cell Biochem.
120:17926–17934. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wu W, Yu T, Wu Y, Tian W, Zhang J and Wang
Y: The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma
growth and progression. J Exp Clin Cancer Res.
38(133)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li N and Zhan X: Identification of
clinical trait-related lncRNA and mRNA biomarkers with weighted
gene co-expression network analysis as useful tool for personalized
medicine in ovarian cancer. EPMA J. 10:273–290. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xiang G and Cheng Y: miR-126-3p inhibits
ovarian cancer proliferation and invasion via targeting PLXNB2.
Reprod Biol. 18:218–224. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lu J, Wang L, Chen W, Wang Y, Zhen S, Chen
H, Cheng J, Zhou Y, Li X and Zhao L: miR-603 targeted hexokinase-2
to inhibit the malignancy of ovarian cancer cells. Arch Biochem
Biophys. 661:1–9. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Duan Y, Dong Y, Dang R, Hu Z, Yang Y, Hu Y
and Cheng J: MiR-122 inhibits epithelial mesenchymal transition by
regulating P4HA1 in ovarian cancer cells. Cell Biol Int.
42:1564–1574. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li S, Zhang T, Zhou X, Du Z, Chen F, Luo J
and Liu Q: The tumor suppressor role of miR-155-5p in gastric
cancer. Oncol Lett. 16:2709–2714. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Luo X, Dong J, He X, Shen L, Long C, Liu
F, Liu X, Lin T, He D and Wei G: MiR-155-5p exerts
tumor-suppressing functions in wilms tumor by targeting IGF2 via
the PI3K signaling pathway. Biomed Pharmacother.
125(109880)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ysrafil Y, Astuti I, Anwar SL, Martien R,
Sumadi FAN, Wardhana T and Haryana SM: MicroRNA-155-5p diminishes
in vitro ovarian cancer cell viability by targeting HIF1α
expression. Adv Pharm Bull. 10:630–637. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lai X, Wichers HJ, Soler-Lopez M and
Dijkstra BW: Structure and function of human tyrosinase and
tyrosinase-related proteins. Chemistry. 24:47–55. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang-Rodriguez J, Urquidi V, Rivard A and
Goodison S: Elevated osteopontin and thrombospondin expression
identifies malignant human breast carcinoma but is not indicative
of metastatic status. Breast Cancer Res. 5(9)2003.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Udono T, Takahashi K, Yasumoto K,
Yoshizawa M, Takeda K, Abe T, Tamai M and Shibahara S: Expression
of tyrosinase-related protein 2/DOPAchrome tautomerase in the
retinoblastoma. Exp Eye Res. 72:225–234. 2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
El Hajj P, Journe F, Wiedig M, Laios I,
Salès F, Galibert MD, Van Kempen LC, Spatz A, Badran B, Larsimont
D, et al: Tyrosinase-related protein 1 mRNA expression in lymph
node metastases predicts overall survival in high-risk melanoma
patients. Br J Cancer. 108:1641–1647. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hsu YL, Chen YJ, Chang WA, Jian SF, Fan
HL, Wang JY and Kuo PL: Interaction between tumor-associated
dendritic cells and colon cancer cells contributes to tumor
progression via CXCL1. Int J Mol Sci. 19(2427)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
El Hajj P, Gilot D, Migault M, Theunis A,
van Kempen LC, Salés F, Fayyad-Kazan H, Badran B, Larsimont D,
Awada A, et al: SNPs at miR-155 binding sites of TYRP1 explain
discrepancy between mRNA and protein and refine TYRP1 prognostic
value in melanoma. Br J Cancer. 113:91–98. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Choi JH and Ro JY: The 2020 WHO
classification of tumors of soft tissue: Selected changes and new
entities. Adv Anat Pathol. 28:44–58. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cui W, Meng W, Zhao L, Cao H, Chi W and
Wang B: TGF-β-induced long non-coding RNA MIR155HG promotes the
progression and EMT of laryngeal squamous cell carcinoma by
regulating the miR-155-5p/SOX10 axis. Int J Oncol. 54:2005–2018.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li N, Liu Y and Cai J: LncRNA miR155HG
regulates M1/M2 macrophage polarization in chronic obstructive
pulmonary disease. Biomed Pharmacother. 117(109015)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Javadi S, Ganeshan DM, Qayyum A, Iyer RB
and Bhosale P: Ovarian cancer, the revised FIGO staging system, and
the role of imaging. AJR AM J Roentgenol. 206:1351–1360.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liang H, Yu T, Han Y, Jiang H, Wang C, You
T, Zhao X, Shan H, Yang R, Yang L, et al: LncRNA PTAR promotes EMT
and invasion-metastasis in serous ovarian cancer by competitively
binding miR-101-3p to regulate ZEB1 expression. Mol Cancer.
17(119)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang X, Yang B, She Y and Ye Y: The lncRNA
TP73-AS1 promotes ovarian cancer cell proliferation and metastasis
via modulation of MMP2 and MMP9. J Cell Biochem. 119:7790–7799.
2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lai XJ and Cheng HF: LncRNA colon
cancer-associated transcript 1 (CCAT1) promotes proliferation and
metastasis of ovarian cancer via miR-1290. Eur Rev Med Pharmacol
Sci. 22:322–328. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zou T, Wang PL, Gao Y and Liang WT: Long
noncoding RNA HOTTIP is a significant indicator of ovarian cancer
prognosis and enhances cell proliferation and invasion. Cancer
Biomark. 25:133–139. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
He X, Sheng J, Yu W, Wang K, Zhu S and Liu
Q: LncRNA MIR155HG promotes temozolomide resistance by activating
the wnt/β-catenin pathway via binding to PTBP1 in glioma. Cell Mol
Neurobiol. 11:1271–1284. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ren XY, Han YD and Lin Q: Long non-coding
RNA MIR155HG knockdown suppresses cell proliferation, migration and
invasion in NSCLC by upregulating TP53INP1 directly targeted by
miR-155-3p and miR-155-5p. Eur Rev Med Pharmacol Sci. 24:4822–4835.
2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fu X, Zhang L, Dan L, Wang K and Xu Y:
LncRNA EWSAT1 promotes ovarian cancer progression through targeting
miR-330-5p expression. Am J Transl Res. 9:4094–4103.
2017.PubMed/NCBI
|
|
35
|
Mu Y, Li N and Cui YL: The lncRNA CCAT1
upregulates TGFβR1 via sponging miR-490-3p to promote TGFβ1-induced
EMT of ovarian cancer cells. Cancer Cell Int.
18(145)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tong L, Ao Y, Zhang H, Wang K, Wang Y and
Ma Q: Long noncoding RNA NORAD is upregulated in epithelial ovarian
cancer and its downregulation suppressed cancer cell functions by
competing with miR-155-5p. Cancer Med. 8:4782–4791. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wu X, Wang Y, Yu T, Nie E, Hu Q, Wu W, Zhi
T, Jiang K, Wang X, Lu X, et al: Blocking MIR155HG/miR-155 axis
inhibits mesenchymal transition in glioma. Neuro Oncol.
19:1195–1205. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tao M, Zhou Y, Jin Y and Pu J: Blocking
lncRNA MIR155HG/miR-155-5p/-3p inhibits proliferation, invasion and
migration of clear cell renal cell carcinoma. Pathol Res Pract.
216(152803)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhao XS, Han B, Zhao JX, Tao N and Dong
CY: miR-155-5p affects Wilms' tumor cell proliferation and
apoptosis via targeting CREB1. Eur Rev Med Pharmacol Sci.
23:1030–1037. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen L, Yang X, Zhao J, Xiong M,
Almaraihah R, Chen Z and Hou T: Circ_0008532 promotes bladder
cancer progression by regulation of the miR-155-5p/miR-330-5p/MTGR1
axis. J Exp Clin Cancer Res. 39(94)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Journe F, Id Boufker H, Van Kempen L,
Galibert MD, Wiedig M, Salès F, Theunis A, Nonclercq D, Frau A,
Laurent G, et al: TYRP1 mRNA expression in melanoma metastases
correlates with clinical outcome. Br J Cancer. 105:1726–1732.
2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu G, Khong HT, Wheeler CJ, Yu JS, Black
KL and Ying H: Molecular and functional analysis of
tyrosinase-related protein (TRP)-2 as a cytotoxic T lymphocyte
target in patients with malignant glioma. J Immunother. 26:301–312.
2003.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Gilot D, Migault M, Bachelot L, Journé F,
Rogiers A, Donnou-Fournet E, Mogha A, Mouchet N, Pinel-Marie ML,
Mari B, et al: A non-coding function of TYRP1 mRNA promotes
melanoma growth. Nat Cell Biol. 19:1348–1357. 2017.PubMed/NCBI View Article : Google Scholar
|