Introduction
Cataracts can occur due to various factors,
including aging, a familial history, immune and metabolic
abnormalities, trauma to the eye and exposure of the eye to poison
or radiation (1,2). Cataracts can lead to lens protein
degeneration and opacity (3). At
present, age-related cataracts is the most common type of cataracts
(4-6).
Cataracts commonly develop in middle-aged and elderly individuals
aged >50, where the incidence increases with age (7). Age-related cataracts are associated
with the natural degeneration of the lens, which occurs over time
during old age (8). Clinically,
age-related cataracts can be divided into three subtypes, namely
cortical, nuclear and posterior subcapsular cataracts (9,10).
There is currently no strict distinction between these various
subtypes of age-related cataracts, but all can progress into the
total calcification of the lens (11). Cortical cataracts are the most
common type of age-related cataracts, accounting for 65-70% of all
age-related cataract cases, followed by nuclear cataracts,
accounting for 25-35% of cases and finally subcapsular opacity
cataracts, which are relatively rare and only account for 5% of all
age-related cataracts (12,13). Age-related cataracts is one of the
main causes of blindness, and the incidence rate increases with
age. Worldwide, patients aged 43-54 years have an incidence rate of
8.3%, compared with an incidence as high as 70.5% in patients
>75 years-old (14,15). It was previously discovered that
oxidative stress serves an important role in the pathogenesis of
cataracts. Under oxidative stress, apoptosis is induced in lens
epithelial cells (LECs), which was found to promote the
opacification of the lens and accelerate the development of
cataracts (16,17). Therefore, the present study used
human LECs to study the pathogenesis of age-related cataracts.
Long non-coding RNAs (lncRNAs) are a type of
non-coding RNA that are >200 nucleotides in length but lack
protein coding ability (18,19).
Although lncRNAs do not generally encode protein, they participate
in the regulation of protein-coding gene expression at multiple
levels, including epigenetic, transcriptional and
post-transcriptional regulation (20,21).
MicroRNAs (miRNAs/miRs) are a type of endogenous non-coding small
RNA that are 21-25 nucleotides in length and exist in both animals
and plants (22,23). The tissue specificity of miRNA and
the time at which they are expressed determines their functional
specificity in tissues and cells (24). This suggests that miRNAs can serve
important roles in the regulation of cell proliferation, in
addition to having a key role in the regulation of
post-transcriptional gene expression. It was previously reported
that the dysregulated expression levels of lncRNAs and miRNAs were
associated with the occurrence of cataracts (25-27).
For example, Chen et al (25) demonstrated that increased expression
of miR-26a and miR-26b inhibited lens fibrosis and cataract
formation by regulating the Jagged-1/Notch signaling pathway. Zhang
et al (26) previously found
that downregulation of miRNA-133b suppressed apoptosis of LECs by
upregulating BCL2L2 in age-related cataracts. In addition, the
expression of TUG1 in the anterior lens capsules of age-related
cataract were revealed to be significantly higher compared with
normal anterior lens capsules, where TUG1 promoted ultraviolet
radiation-induced apoptosis by downregulating the expression of
miR-421(27).
miR-196a-5p has been studied in various diseases,
including cancer, pre-eclampsia and postmenopausal osteoporosis
(28-30).
miR-196a-5p serves a key role in the regulation of tumor cell
apoptosis and proliferation (31,32).
However, the effects of miR-196a-5p on age-related cataracts and in
lens epithelial cells remain unclear.
The present study aimed to investigate the effects
of TUG1 in oxidative stress-induced apoptosis in age-related
cataracts and to determine its underlying mechanism of action, with
a focus on providing novel insights into potential therapeutic
targets for cataracts.
Materials and methods
Cell culture and establishment of
oxidative stress model
SRA01/04 cells, a human LEC line, were obtained from
the American Type Culture Collection (ATCC). SRA01/04 cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin/streptomycin, which were maintained at 37˚C
with 5% CO2.
To establish an in vitro oxidative stress
model, SRA01/04 cells were exposed to 200 µM hydrogen peroxide
(H2O2) at 37˚C for 24 h (33).
Cell transfection
SRA01/04 cells were cultured in six-well plates at
37˚C for 24 h and then transfected with 200 pmol/l control small
interfering RNA (siRNA; sense, 5'-UUCUCCGAACGUGUCACGUTT-3';
antisense, 3'-ACGUGACACGUUCGGAGAATT-5'; Shanghai GenePharma Co.,
Ltd.), 200 pmol/l TUG1-siRNA (sense, 5'-CCAUCUCACAAGGCUUCAATT-3';
antisense, 3'-TTGGUAGAGUGUUCCGAAGUU-5'; Shanghai GenePharma Co.,
Ltd.), 50 nM inhibitor control (5'-CAGUACUUUUGUGUAGUACAA-3';
Shanghai GenePharma Co., Ltd.), 50 nM miR-196a-5p inhibitor
(5'-CCCAACAACAUGAAACUACCUA-3'; Shanghai GenePharma Co., Ltd.), 100
nM mimic control (sense, 5'-UUCUCCGAACGUGUCACGUTT-3'; antisense,
5'-ACGUGACACGUUCGGAGAATT-3'; Shanghai GenePharma Co., Ltd.) or 100
nM miR-196a-5p mimic (sense, 5'-UAGGUAGUUUCAUGUUGUUGGG-3';
antisense, 5'-CAACAACAUGAAACUACCUAUU-3'; Shanghai GenePharma Co.,
Ltd.) for 24 h using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Following
transfection, the cells were collected to determine the
transfection efficiencies by reverse transcription-quantitative PCR
(RT-qPCR).
To determine the effect of TUG1-siRNA in SRA01/04
cells exposed to 200 µM H2O2, cells were
divided into the following groups: i) Control group, with cells
without any treatment; ii) H2O2 group, where
the SRA01/04 cells were exposed to 200 µM
H2O2 at 37˚C for 24 h; iii)
H2O2 + control-siRNA group, where SRA01/04
cells were transfected with control-siRNA for 24 h and then exposed
to 200 µM H2O2 at 37˚C for 24 h; iv)
H2O2 + TUG1-siRNA group, where SRA01/04 cells
were transfected with TUG1-siRNA for 24 h and then exposed to 200
µM H2O2 at 37˚C for 24 h; v)
H2O2 + TUG1-siRNA + inhibitor control group,
where SRA01/04 cells were co-transfected with the TUG1-siRNA +
inhibitor control for 24 h and then exposed to 200 µM
H2O2 at 37˚C for 24 h; and vi)
H2O2 + TUG1-siRNA + miR-196a-5p inhibitor
group, where SRA01/04 cells were co-transfected with TUG1-siRNA +
miR-196a-5p inhibitor for 24 h and then exposed to 200 µM
H2O2 at 37˚C for 24 h.
To determine the effect of the miR-196a-5p mimic on
SRA01/04 cells following exposure to 200 µM
H2O2, cells were divided into the following
groups: i) Control group, which consists of cells without any
treatment; ii) H2O2 group, where SRA01/04
cells were exposed to 200 µM H2O2 at 37˚C for
24 h; iii) H2O2 + mimic group, where SRA01/04
cells were transfected with the mimic control for 24 h and then
exposed to 200 µM H2O2 at 37˚C for 24 h; and
iv) H2O2 + miR-196a-5p mimic group, where
SRA01/04 cells were transfected with miR-196a-5p mimic for 24 h and
then exposed to 200 µM H2O2 at 37˚C for 24
h.
miRNA target analysis and
dual-luciferase reporter assay
The direct binding site between TUG1 and miR-196a-5p
was identified using StarBase version 2.0 (http://starbase.sysu.edu.cn/). The 3'-untranslated
region (UTR) sequences of TUG1 [TUG1-wild-type (WT),
5'-AUCGUCAAUUUUCUACUACCUU-3'], which included the target sequence
for miR-196a-5p, or the mutated (MUT) target site (TUG1-MUT,
5'-AUGGUGUUUUAUCUUGAUGGAU-3') were obtained by PCR using a
Transcriptor First Strand cDNA Synthesis kit (cat. no. 04896866001;
Roche Diagnostics GmbH). The thermocycling conditions were as
follows: Incubation for 5 min at 25˚C, followed by 60 min at 42˚C.
The 3'-UTR products were cloned into the pmirGLO vector (Promega
Corporation) to construct the TUG1-WT reporter vector. In addition,
a TUG1-MUT reporter vector was also generated. 293 cells were
obtained from the ATCC and cultured in Eagle's Minimum Essential
Medium (ATCC) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C with 5% CO2. Briefly, 293
cells were cultured for 24 h before being co-transfected with 1 ng
TUG1-WT or 1 ng TUG1-MUT luciferase reporter gene plasmid and 100
nM miR-196a-5p mimic or 100 nM mimic control using
Lipofectamine® 2000 reagent for 48 h. The relative
luciferase activity was measured using a Dual Luciferase Reporter
assay system (Promega Corporation), according to the manufacturer's
protocol. The results were normalized to Renilla luciferase
activity.
RT-qPCR
Total RNA was extracted from cells using the
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific
Inc.). Total RNA was reverse transcribed into cDNA using a Maxima
First Strand cDNA synthesis kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The reaction conditions for RT-PCR were as
follows: 70˚C for 5 min, 37˚C for 5 min and 42˚C for 60 min. qPCR
was performed in an ABI Prism 7000 Real-Time PCR Detection system
(Applied Biosciences; Thermo Fisher Scientific, Inc.) using a
SYBR™-Green qPCR Master mix (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The primers used for the
qPCR were synthesized by Genscript and primer sequences were listed
as the following: GAPDH forward, 5'-CTTTGGTATCGTGGAAGGACTC-3' and
reverse, 5'-GTAGAGGCAGGGATGATGTTCT-3'; U6 forward,
5'-GCTTCGGCAGCACATATACTAAAAT-3' and reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3'; TUG1 forward:
5'-GACCGTCCAATGACCTTCCT-3' and reverse, 5'-TGGCTGAATGCTTCTTGGGT-3'
and miR-196a-5p forward, 5'-CCGACGTAGGTAGTTTCATGTT-3' and reverse,
5'-GTGCAGGGTCCGAGGTATTC-3'. The following thermocycling conditions
were used for the qPCR: Initial denaturation for 5 min at 95˚C;
followed by 40 cycles for 10 sec at 95˚C and 30 sec at 60˚C. GAPDH
or U6 were used as the internal controls for TUG1 and miR-196a-5p,
respectively. The relative mRNA expression levels of TUG1 and
miR-196a-5p were calculated using the 2-ΔΔCq method
(34).
MTT assay
MTT assay was performed to evaluate cell viability.
Briefly, 24 h after cell transfection, SRA01/04 cells were exposed
to 200 µM H2O2 at 37˚C for another 24 h,
before the cells were seeded into a 96-well plate (1x104
cells per well). They were then treated with 10 µl 5 mg/ml MTT
solution (Beyotime Institute of Technology) per well and incubated
at 37˚C for an additional 4 h. Following incubation, the medium was
removed and 100 µl DMSO was added to each well to dissolve the
formazan product. The absorbance was measured at a wavelength of
570 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Flow cytometry analysis of
apoptosis
Flow cytometry analysis was used to detect cell
apoptosis. Briefly, following transfection, the cells were
harvested by trypsinization and resuspended in 1X buffer (Annexin
V-FITC/PI apoptosis detection kit; Beyotime Biotechnology). In
total, 100 µl of this cell suspension (1x106 cells) was
incubated with 5 µl Annexin V-FITC and propidium iodide at 4˚C in
the dark for 15 min. The stained cells were analyzed using a BD
FACSCalibur™ flow cytometer (BD Biosciences) and FlowJo software
(version 7.2.4; FlowJo LLC).
Western blotting
Total protein was extracted from SRA01/04 cells
using RIPA lysis buffer (Beyotime Institute of Biotechnology) and
the lysate was centrifuged at 4˚C at 10,000 x g for 15 min to
obtain the total protein. Total protein (40 µg per lane) was
quantified using a BCA protein assay kit (Bio-Rad Laboratories,
Inc.) and separated by 10% SDS-PAGE. The separated proteins were
subsequently transferred onto PVDF membranes and blocked with 5%
non-fat milk diluted in PBS-0.1% Tween-20 (PBST) solution at room
temperature for 1 h. The membranes were then incubated with the
following primary antibodies overnight at 4˚C: Anti-cleaved
caspase-3 (cat. no. ab32042; 1:1,000; Abcam), anti-caspase-3 (cat.
no. ab32351; 1:1,000; Abcam) and anti-GAPDH (cat. no. ab9485;
1:1,000; Abcam). Following primary antibody incubation, the
membranes were washed three times with PBST and incubated with a
goat anti-rabbit IgG H&L (HRP) pre-adsorbed (cat. no. ab97080;
1:2,000; Abcam) for 1 h at room temperature. Protein bands were
visualized using an ECL substrate (Cytiva), according to the
manufacturer's protocol on an ImageQuant800 western blotting
imaging system (Amersham; Cytiva).
Statistical analysis
Data are presented as the mean ± SD from three
independent experiments. Statistical differences among groups were
determined using an unpaired Student's t-test or one-way ANOVA
followed by Tukey's post-hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
TUG1 is a direct target gene of
miR-196a-5p
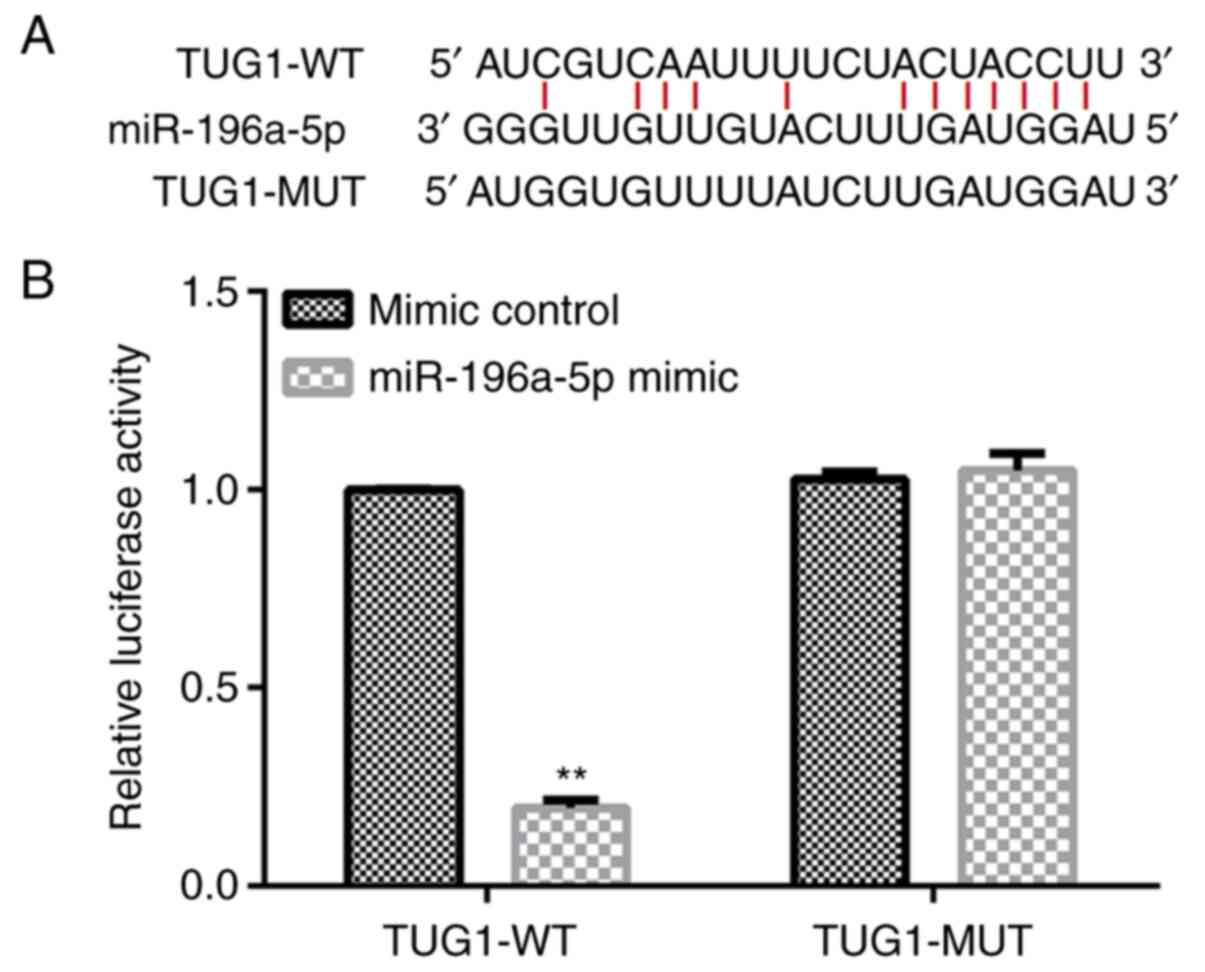
Analysis using the StarBase database identified a
binding site between TUG1 and miR-196a-5p (Fig. 1A), where this binding between TUG1
and miR-196a-5p was validated using a dual luciferase reporter
assay (Fig. 1B). Compared with that
in cells co-transfected with the mimic control and TUG1-WT, the
luciferase activity of cells co-transfected with miR-196a-5p mimic
and TUG1-WT was significantly reduced (Fig. 1B). By contrast, the luciferase
activity of cells co-transfected with miR-196a-5p mimic and
TUG1-MUT demonstrated no significant changes compared with the
luciferase activity of cells co-transfected with mimic control and
TUG1-MUT (Fig. 1B).
Expression levels of TUG1 and
miR-196a-5p in SRA01/04 cells after H2O2
treatment
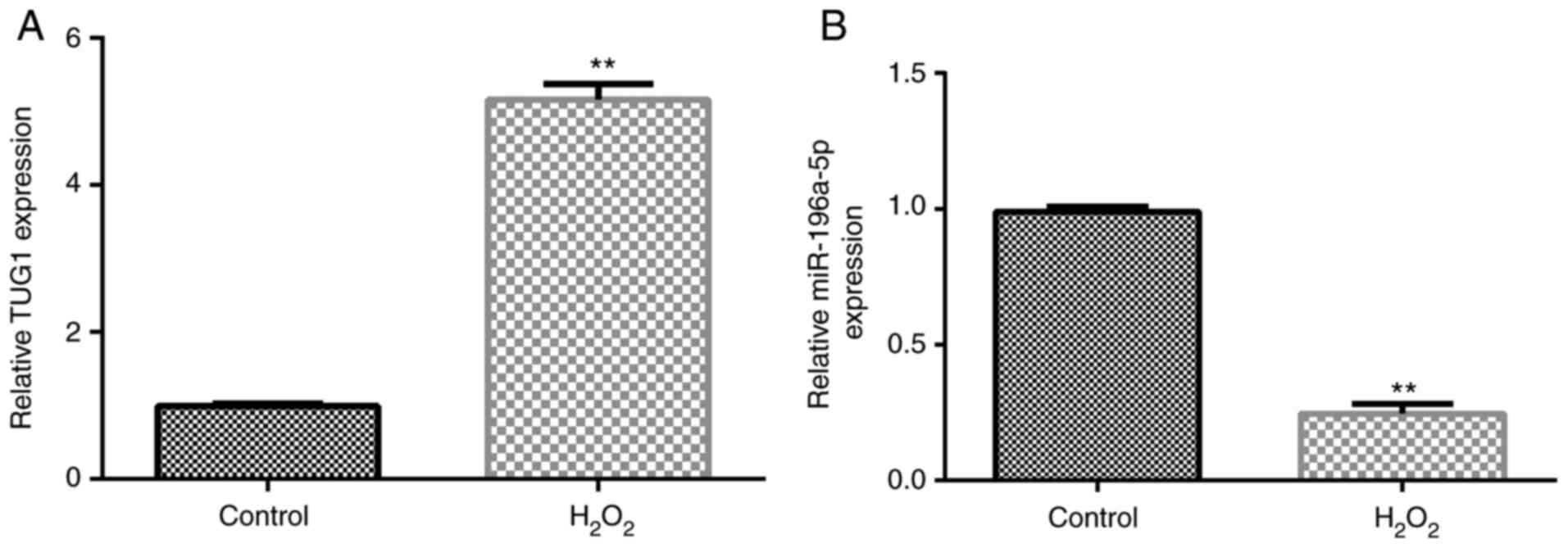
The expression levels of TUG1 and miR-196a-5p in
SRA01/04 cells were analyzed using RT-qPCR after cells were exposed
to H2O2 for 24 h. As shown in Fig. 2A and B, compared with those in the control
group, the expression levels of TUG1 were significantly upregulated
in the H2O2 group, whilst the expression
levels of miR-196a-5p were significantly downregulated.
TUG1 negatively regulates the
expression of miR-196a-5p in SRA01/04 cells
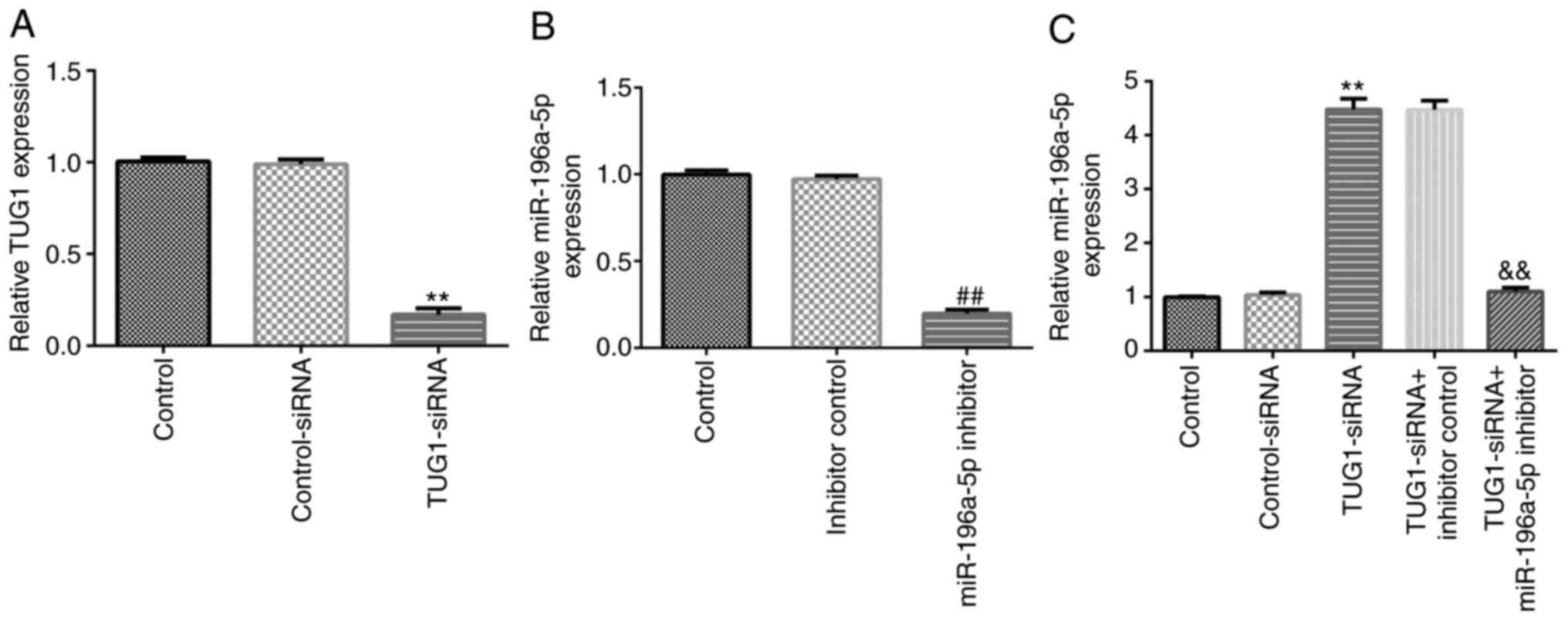
SRA01/04 cells were transfected with TUG1-siRNA or
the miR-196a-5p inhibitor for 24 h before RT-qPCR was performed to
determine the transfection efficiency. Compared with those in the
control-siRNA or inhibitor control groups, transfection with
TUG1-siRNA or the miR-196a-5p inhibitor significantly downregulated
the expression levels of TUG1 and miR-196a-5p in SRA01/04 cells,
respectively (Fig. 3A and B). In addition, compared with those in the
control-siRNA group, transfection with TUG1-siRNA significantly
upregulated the expression levels of miR-196a-5p in SRA01/04 cells,
which was significantly reversed following co-transfection with the
miR-196a-5p inhibitor (Fig.
3C).
miR-196a-5p inhibitor reverses the
effects of TUG1-siRNA on H2O2-induced
oxidative damage of SRA01/04 cells
SRA01/04 cells were exposed to
H2O2 following transfection for 24 h and were
subsequently divided into the following six groups: i) Control
group; ii) H2O2 group; iii)
H2O2 + control-siRNA group; iv)
H2O2 + TUG1-siRNA group; v)
H2O2 + TUG1-siRNA + inhibitor control group;
and vi) H2O2 + TUG1-siRNA + miR-196a-5p
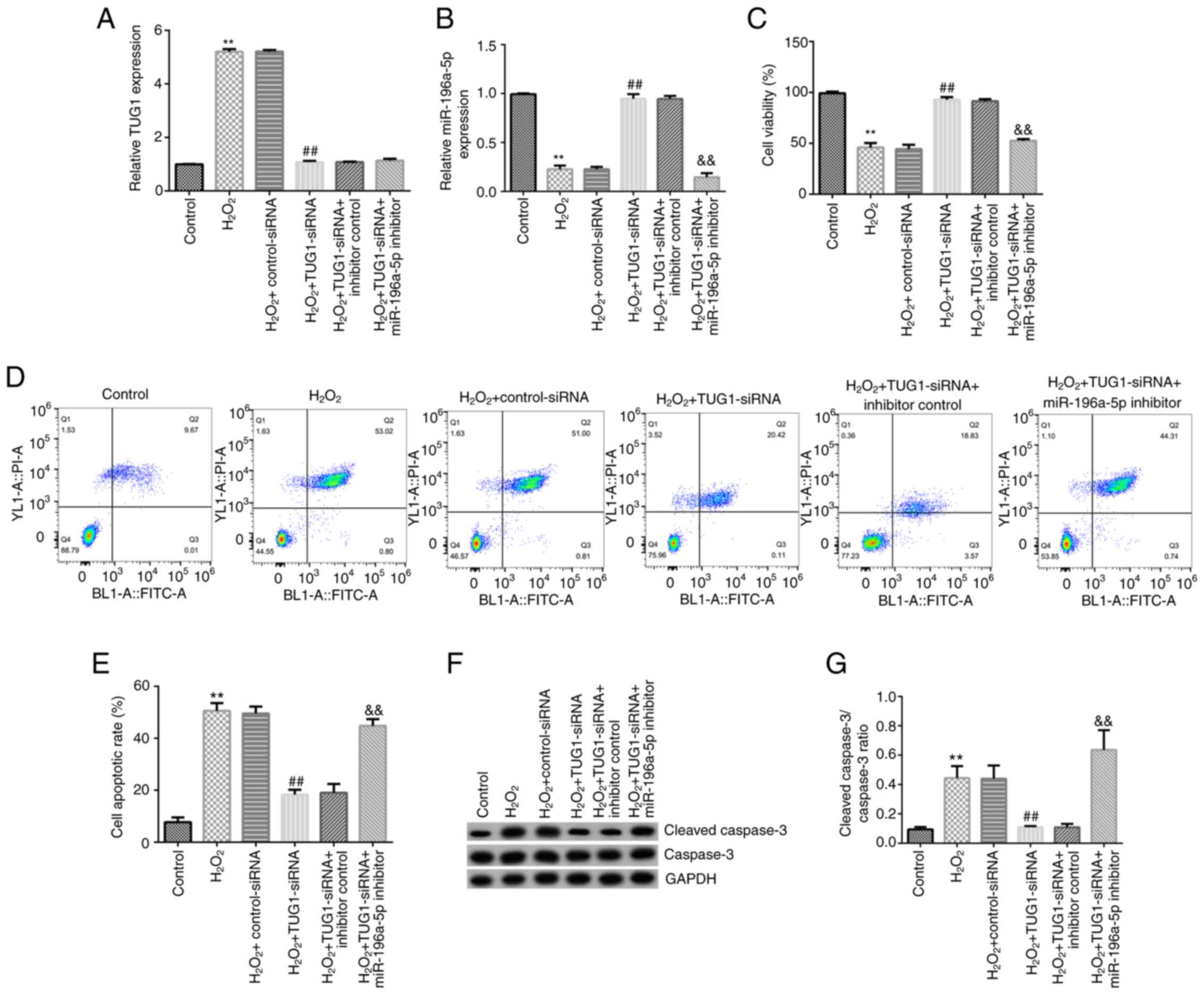
inhibitor group. Compared with those in the control group, TUG1
expression levels were significantly upregulated in the
H2O2 group, whilst the expression levels of
miR-196a-5p were significantly downregulated (Fig. 4A and B). Compared with those in the
H2O2 + control-siRNA group, TUG1 expression
levels were significantly downregulated in the
H2O2 + TUG1-siRNA group (Fig. 4A). In addition, compared with those
in the H2O2 + control-siRNA group,
miR-196a-5p expression levels were found to be significantly
upregulated in the H2O2 + TUG1-siRNA group
(Fig. 4B). Notably, this effect was
reversed following co-transfection with the miR-196a-5p inhibitor
(Fig. 4B).
In addition, the viability, apoptosis, and
expression levels of cleaved caspase-3 and caspase-3 were analyzed
in SRA01/04 cells. Compared with that in the control group, the
viability of cells in the H2O2 group was
significantly reduced (Fig. 4C),
whilst the cell apoptotic rate, protein expression levels of
cleaved caspase-3 and the cleaved caspase-3/caspase-3 ratio were
significantly increased in the H2O2 group
(Fig. 4D and E). By contrast, compared with that in the
H2O2 + control-siRNA group, the viability of
cells in the H2O2 + TUG1-siRNA group was
significantly increased (Fig. 4C),
whilst the cell apoptotic rate, protein expression levels of
cleaved caspase-3 and the cleaved caspase-3/caspase-3 ratio were
significantly reduced (Fig. 4D and
E). All of these effects
aforementioned were found to be significantly reversed following
co-transfection with the miR-196a-5p inhibitor.
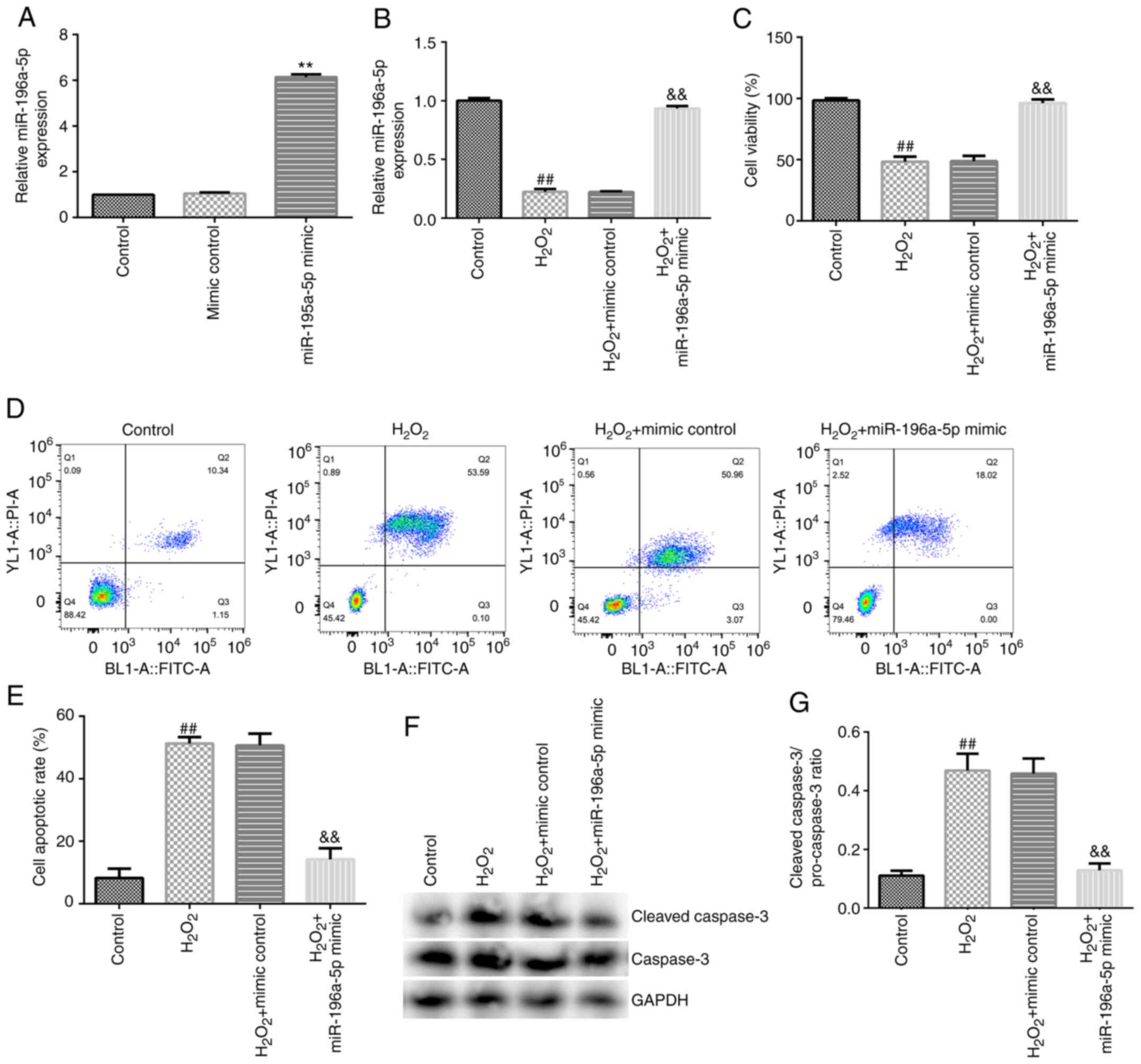
miR-196a-5p attenuates
H2O2-induced oxidative damage in SRA01/04
cells
SRA01/04 cells were exposed to
H2O2 following 24 h of transfection and
subsequently divided into the following four groups: i) Control
group; ii) H2O2 group; iii)
H2O2 + mimic control group; and iv)
H2O2 + miR-196a-5p mimic group. Transfection
with the miR-196a-5p mimic significantly upregulated the expression
levels of miR-196a-5p in SRA01/04 cells (Fig. 5A), suggesting the successful
transfection of the miR-196a-5p mimic. Subsequently, the underlying
molecular mechanism of miR-196a-5p in SRA01/04 cells was
investigated. Compared with those in the control group, miR-196a-5p
expression levels and cell viability were significantly reduced in
the H2O2 group (Fig. 5B and C), whilst the apoptosis levels, protein
expression levels of cleaved caspase-3 and the cleaved
caspase-3/caspase-3 ratio were all significantly increased
(Fig. 5D-G). Conversely, compared
with those in the H2O2 + mimic control group,
miR-196a-5p expression levels and cell viability were significantly
increased in the H2O2 + miR-196a-5p mimic
group (Fig. 5B and C), whilst the levels of apoptosis, protein
expression levels of cleaved caspase-3 and the cleaved
caspase-3/caspase-3 ratio were significantly reduced (Fig. 5D-G).
Discussion
Age-related cataracts is a type of degenerative
disease as a complication of aging, where its pathogenesis is
closely associated with cellular senescence and decreased metabolic
function in the lens (35). The
incidence of age-related cataracts increases with age;
43-54-year-old patients have an incidence of 8.3%, compared with an
incidence as high as 70.5% in patients over 75 (14,15).
Oxidative stress occurs when the oxidative and antioxidant
mechanisms in the body become unbalanced, such that an enhanced
oxidative state is favored (36,37).
This then promotes inflammatory infiltration by neutrophils,
increased secretion of proteases and the production of large
quantities of reactive oxidative intermediate products (36,37).
Oxidative stress is an adverse effect that is caused by the
production and accumulation of free radicals in the body, which is
considered to be an important contributing factor to aging and
disease (38). It was previously
reported that oxidative stress serves an important role in the
pathogenesis of various types of cataracts (39,40).
Age-related cataracts are mainly caused by oxygen free
radical-induced damage to LECs, which prompts conformational
changes in important proteins, such as E3 ubiquitin-protein ligase
Mdm2 and Rho-associated protein kinase 1 in LECs (41,42).
To study the role of lncRNA TUG1 in age-related cataracts in
vitro, the present study established an in vitro
oxidative stress model by exposing the LEC line, SRA01/04, to 200
µM H2O2 for 24 h.
LncRNA TUG1 is expressed in the retina and brain and
was discovered to serve an important role in numerous cancer types,
including colorectal, esophageal and bladder cancer (43-45).
However, to the best of our knowledge, the underlying mechanism of
action of TUG1 in age-related cataracts remains to be determined.
To investigate the underlying mechanisms of TUG1 in age-related
cataracts, the present study predicted and verified the binding
site between TUG1 and miR-196a-5p. Through bioinformatics software
analysis, it was found that there may be a binding site between
miR-196a-5p and TUG1. Therefore, TUG1 may regulate the
proliferation and apoptosis of lens epithelial cells by regulating
the expression of miR-196a-5p, thereby participating in the
occurrence of age-related cataracts. In addition, the expression
levels of TUG1 were found to be upregulated, whilst miR-196a-5p
expression levels were downregulated, in SRA01/04 cells induced by
H2O2. Subsequent transfection experiments
revealed that TUG1 negatively regulated miR-196a-5p expression in
SRA01/04 cells. However, whether the overexpression of TUG1 has a
significant inhibiting effect on miR-196a-5p was not studied in the
present study and is a limitation.
To determine the effects of TUG1 on
H2O2-induced oxidative damage in SRA01/04
cells and miR-196a-5p expression, cell function experiments were
performed in SRA01/04 cells following TUG1 knockdown. Results from
the present study revealed that transfection with TUG1-siRNA
reduced the H2O2-induced oxidative damage,
which was evidenced by the increased cell viability, reduced cell
apoptosis, cleaved-caspase3 protein expression and reduced
cleaved-caspase3/caspase3 ratios in SRA01/04 cells. By contrast,
co-transfection with the miR-196a-5p inhibitor reversed these
effects aforementioned. In addition, the overexpression of
miR-196a-5p attenuated H2O2-induced oxidative
damage in SRA01/04 cells. It was worth mentioning that the
apoptosis rate of H2O2 + TUG1-siRNA +
miR-196a-5p inhibitor group was similar to that in the
H2O2 and H2O2 +
control-siRNA group. However, the ratio of
cleaved-caspase3/caspase3 in the H2O2 +
TUG1-siRNA + miR-196a-5p inhibitor group, was higher compared with
that in the H2O2 and
H2O2 + control-siRNA group. The reason for
this difference between the apoptosis rate and the
cleaved-caspase3/caspase3 ratio remain unclear, which require
further study.
In conclusion, the findings of the present study
revealed that knockdown of lncRNA TUG1 expression protected LECs
from oxidative stress-induced apoptosis by increasing miR-196a-5p
expression. These results suggest that targeting TUG1 and
miR-196a-5p may provide a new therapeutic strategy for patients
with age-related cataracts.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or generated during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
QS contributed to study design, data collection,
statistical analysis, data interpretation and manuscript
preparation. TZ contributed to data collection, statistical
analysis and manuscript preparation. QS and TZ confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Thompson J and Lakhani N: Cataracts. Prim
Care. 42:409–423. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Shiels A and Hejtmancik JF: Biology of
inherited cataracts and opportunities for treatment. Annu Rev Vis
Sci. 5:123–149. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Vrensen GF: Early cortical lens opacities:
A short overview. Acta Ophthalmol. 87:602–610. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ten Berge JC, Fazil Z, van den Born I,
Wolfs RCW, Schreurs MWJ, Dik WA and Rothova A: Intraocular cytokine
profile and autoimmune reactions in retinitis pigmentosa,
age-related macular degeneration, glaucoma and cataract. Acta
Ophthalmol. 97:185–192. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lee CM and Afshari NA: The global state of
cataract blindness. Curr Opin Ophthalmol. 28:98–103.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Olson RJ, Braga-Mele R, Chen SH, Miller
KM, Pineda R II, Tweeten JP and Musch DC: Cataract in the adult eye
preferred practice pattern®. Ophthalmology. 124:P1–P119.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yuan XB, Zhang DY, Chen SJ, Wu PC and
Zhang WF: Prevalence of cataract among the population aged 50 years
and over at different altitudes in Gansu Province. Zhonghua Yan Ke
Za Zhi. 55:589–594. 2019.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
8
|
Asbell PA, Dualan I, Mindel J, Brocks D,
Ahmad M and Epstein S: Age-related cataract. Lancet. 365:599–609.
2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shiels A and Hejtmancik JF: Mutations and
mechanisms in congenital and age-related cataracts. Exp Eye Res.
156:95–102. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liu YC, Wilkins M, Kim T, Malyugin B and
Mehta JS: Cataracts. Lancet. 390:600–612. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Keel S and He M: Risk factors for
age-related cataract. Clin Exp Ophthalmol. 46:327–328.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hashemi H, Pakzad R, Yekta A, Aghamirsalim
M, Pakbin M, Ramin S and Khabazkhoob M: Global and regional
prevalence of age-related cataract: A comprehensive systematic
review and meta-analysis. Eye (Lond). 34:1357–1370. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
National Institute for Health and Care
Excellence (UK): Cataracts in adults: Management. National
Institute for Health and Care Excellence, London, 2017.
|
|
14
|
Truscott RJW and Friedrich MG: Molecular
processes implicated in human age-related nuclear cataract. Invest
Ophthalmol Vis Sci. 60:5007–5021. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Klein BE, Klein R and Lee KE: Incidence of
age-related cataract: The beaver dam eye study. Arch Ophthalmol.
116:219–225. 1998.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lu B, Christensen IT, Yu T, Wang C, Yan Q
and Wang X: SUMOylation evoked by oxidative stress reduced lens
epithelial cell antioxidant functions by increasing the stability
and transcription of TP53INP1 in age-related cataracts. Oxid Med
Cell Longev. 2019(7898069)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yang H, Cui Y, Tang Y, Tang X, Yu X, Zhou
J, Yin Q and Shentu X: Cytoprotective role of humanin in lens
epithelial cell oxidative stress-induced injury. Mol Med Rep.
22:1467–1479. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Panni S, Lovering RC, Porras P and Orchard
S: Non-coding RNA regulatory networks. Biochim Biophys Acta Gene
Regul Mech. 1863(194417)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Puvvula PK: LncRNAs regulatory networks in
cellular senescence. Int J Mol Sci. 20(2615)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
He Z, Yang D, Fan X, Zhang M, Li Y, Gu X
and Yang M: The roles and mechanisms of lncRNAs in liver fibrosis.
Int J Mol Sci. 21(1482)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang J and Cen S: Roles of lncRNAs in
influenza virus infection. Emerg Microbes Infect. 9:1407–1414.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Correia de Sousa M, Gjorgjieva M, Dolicka
D, Sobolewski C and Foti M: Deciphering miRNAs' action through
miRNA editing. Int J Mol Sci. 20(6249)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ghafouri-Fard S, Shoorei H and Taheri M:
miRNA profile in ovarian cancer. Exp Mol Pathol.
113(104381)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Permenter MG, McDyre BC, Ippolito DL and
Stallings JD: Alterations in tissue microRNA after heat stress in
the conscious rat: Potential biomarkers of organ-specific injury.
BMC Genomics. 20(141)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chen X, Xiao W, Chen W, Liu X, Wu M, Bo Q,
Luo Y, Ye S, Cao Y and Liu Y: MicroRNA-26a and -26b inhibit lens
fibrosis and cataract by negatively regulating Jagged-1/Notch
signaling pathway. Cell Death Differ. 24:1431–1442. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang F, Meng W and Tong B:
Down-regulation of MicroRNA-133b suppresses apoptosis of lens
epithelial cell by up-regulating BCL2L2 in age-related cataracts.
Med Sci Monit. 22:4139–4145. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li G, Song H, Chen L, Yang W, Nan K and Lu
P: TUG1 promotes lens epithelial cell apoptosis by regulating
miR-421/caspase-3 axis in age-related cataract. Exp Cell Res.
356:20–27. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Xin H, Wang C and Liu Z: miR-196a-5p
promotes metastasis of colorectal cancer via targeting IκBα. BMC
Cancer. 19(30)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Mi C, Ye B, Gao Z, Du J, Li R and Huang D:
BHLHE40 plays a pathological role in pre-eclampsia through
upregulating SNX16 by transcriptional inhibition of miR-196a-5p.
Mol Hum Reprod. 26:532–548. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang L, Xie H and Li S: LncRNA LOXL1-AS1
controls osteogenic and adipocytic differentiation of bone marrow
mesenchymal stem cells in postmenopausal osteoporosis through
regulating the miR-196a-5p/Hmga2 axis. J Bone Miner Metab.
38:794–805. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wang L, Wei Y, Yan Y, Wang H, Yang J,
Zheng Z, Zha J, Bo P, Tang Y, Guo X, et al: CircDOCK1 suppresses
cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in
OSCC. Oncol Rep. 39:951–966. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang JP, Yang JK, Li C, Cui ZQ, Liu HJ,
Sun XF, Geng SM, Lu SK, Song J, Guo CY and Jiao BH: Downregulation
of ZMYND11 induced by miR-196a-5p promotes the progression and
growth of GBM. Biochem Biophys Res Commun. 494:674–680.
2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tu Y, Li L, Qin B, Wu J, Cheng T, Kang L
and Guan H: Long noncoding RNA glutathione peroxidase 3-antisense
inhibits lens epithelial cell apoptosis by upregulating glutathione
peroxidase 3 expression in age-related cataract. Mol Vis.
25:734–744. 2019.PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Fukuoka H and Afshari NA: The impact of
age-related cataract on measures of frailty in an aging global
population. Curr Opin Ophthalmol. 28:93–97. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chainy GBN and Sahoo DK: Hormones and
oxidative stress: An overview. Free Radic Res. 54:1–26.
2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chirumbolo S: Oxidative stress, nutrition
and cancer: Friends or foes? World J Mens Health. 39:19–30.
2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Shao A, Lin D, Wang L, Tu S, Lenahan C and
Zhang J: Oxidative stress at the crossroads of aging, stroke and
depression. Aging Dis. 11:1537–1566. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wu C, Liu Z, Ma L, Pei C, Qin L, Gao N, Li
J and Yin Y: MiRNAs regulate oxidative stress related genes via
binding to the 3' UTR and TATA-box regions: A new hypothesis for
cataract pathogenesis. BMC Ophthalmol. 17(142)2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Liu XF, Hao JL, Xie T, Malik TH, Lu CB,
Liu C, Shu C, Lu CW and Zhou DD: Nrf2 as a target for prevention of
age-related and diabetic cataracts by against oxidative stress.
Aging Cell. 16:934–942. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang Z, Su D, Sun Z, Liu S, Sun L, Li Q,
Guan L, Liu Y, Ma X and Hu S: MDM2 phosphorylation mediates
H2O2-induced lens epithelial cells apoptosis
and age-related cataract. Biochem Biophys Res Commun. 528:112–119.
2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hu S, Su D, Sun L, Wang Z, Guan L, Liu S,
Zhao B, Liu Y, Shi C, Yu J and Ma X: High-expression of ROCK1
modulates the apoptosis of lens epithelial cells in age-related
cataracts by targeting p53 gene. Mol Med. 26(124)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yan Z, Bi M, Zhang Q, Song Y and Hong S:
LncRNA TUG1 promotes the progression of colorectal cancer via the
miR-138-5p/ZEB2 axis. Biosci Rep. 40(BSR20201025)2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zong M, Feng W, Wan L, Yu X and Yu W:
LncRNA TUG1 promotes esophageal cancer development through
regulating PLK1 expression by sponging miR-1294. Biotechnol Lett.
42:2537–2549. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yu G, Zhou H, Yao W, Meng L and Lang B:
lncRNA TUG1 promotes cisplatin resistance by regulating CCND2 via
epigenetically silencing miR-194-5p in bladder cancer. Mol Ther
Nucleic Acids. 16:257–271. 2019.PubMed/NCBI View Article : Google Scholar
|