Introduction
Colorectal cancer (CRC) is a malignant tumor of the
gastrointestinal tract and was a leading cause of cancer-associated
mortality worldwide in 2019(1).
Patients with CRC who are diagnosed at an early stage typically
have a favorable prognosis, with a 5-year survival rate of 70-90%
(2). However, the majority of
patients with CRC are diagnosed at advanced or metastatic stages,
displaying an unfavorable 5-year survival rate at <30% (3). Therefore, identifying novel targets
and developing appropriate treatment strategies for preventing the
progression of CRC is important.
Mex-3 RNA binding family member A (MEX3A) was
initially identified as a translational regulator in
Caenorhabditis elegans, and is typically distributed in
early embryos (4). In addition,
MEX3A has been characterized as a phosphoprotein that can bind with
RNA (5). MEX3A can also regulate
target protein ubiquitination via its ring finger domain, which
results in the regulation of the target protein subcellular
localization and stability (6-8).
As a member of the MEX3 family, MEX3A was also reported to serve a
role in the modulation of mRNA expression, which resulted in
regulation of the progression of numerous types of disease,
including malignant tumors (9,10). To
date, MEX3A has been reported to participate in the progression of
gastric cancer and nephroblastoma (10,11).
However, to the best of our knowledge, the role of MEX3A in CRC is
not completely understood. Therefore, the present study aimed to
investigate the function of MEX3A in CRC to identify novel targets
for CRC treatment. On the other hand, it has been reported that
MEX3A could regulate the cell cycle distribution in multiple types
of cancer, including liver and cervical cancer (7,11). In
addition, CDK4, CDK6 and CDK2 are known to be important mediators
of the G1 phase of the cell cycle (12,13).
However, the association between MEX3A and the CDK family in CRC is
not completely understood. Therefore, the present study also aimed
to investigate the function of MEX3A in these three proteins.
Materials and methods
Cell lines and culture
Human normal intestinal epithelial cells (HIEC-6)
and CRC cell lines (SW480, HCT116 and HT29) were purchased from
American Type Culture Collection. Cells were cultured in RPMI-1640
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin (Thermo
Fisher Scientific, Inc.) and 10% streptomycin (Thermo Fisher
Scientific, Inc.) at 37˚C with 5% CO2.
Bioinformatics analysis
The expression levels of MEX3A in CRC and adjacent
healthy tissues were obtained from The Cancer Genome Atlas (TCGA).
Data from TCGA were analyzed using the Gene Expression Profiling
Interactive Analysis database (GEPIA; http://gepia.cancer-pku.cn/), as previously described
(14).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from CRC cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using
PrimeScript RT Reagent Kit [ELK (Wuhan) Biotechnology Co., Ltd.]
according to the manufacturer's protocol. Subsequently, qPCR was
performed on an ABI 7500 Real-Time PCR Detection system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using SYBR-Green
(Beyotime Institute of Biotechnology). The following thermocycling
conditions were used for qPCR: Initial denaturation for 2 min at
94˚C; followed by 35 cycles for 30 sec at 94˚C and 45 sec at 55˚C.
The following primers were designed by Shanghai GenePharma Co.,
Ltd. and used for qPCR: MEX3A forward,
5'-AGCAGTGTAAGGGAGTTGGAGTC-3' and reverse,
5'-GGAGGGAAAGGAAAGAGTTGAG-3'; and β-actin forward,
5'-GTCCACCGCAAATGCTTCTA-3' and reverse, 5'-TGCTGTCACCTTCACCGTTC-3'.
mRNA expression levels were quantified using the 2-∆∆Cq
method (15) and normalized to the
internal reference gene β-actin.
Cell transfection
Small interfering RNAs (siRNAs/sis) targeting MEX3A
(si-MEX3A-1, si-MEX3A-2 and si-MEX3A-3; 10 nM) and a negative
control siRNA (siRNA-ctrl; 10 nM) were purchased from Guangzhou
RiboBio Co., Ltd. The CDK2 overexpression plasmid (pcDNA3.1-CDK2; 1
µg/μl) and empty vector (pcDNA3.1; 1 µg/μl) were obtained from
Shanghai GenePharma Co., Ltd. The sequences for the siRNAs were as
follows: siRNA-ctrl, 5'-GCAGAATTGGTACCGCCAA-3'; si-MEX3A-1,
5'-CAAGATCCTCGAGTACAACAATGAA-3'; si-MEX3A-2,
5'-CAGCAGCAAACCAACACATACATTA-3'; and si-MEX3A-3,
5'-GCCTAGTCTAGTGGTATCTGGA ATA-3'. CRC cells (5x103
cells/well) were transfected with siRNAs or plasmids using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37˚C. At 48 h post-transfection, subsequent
experiments were performed. Blank refers to cells without
transfection.
Cell Counting Kit-8 (CCK-8) assay
HCT-116 or SW480 cells were plated (5x103
cells/well) into 96-well plates and transfected with siRNA-ctrl or
si-MEX3A-1 for 48 h. Subsequently, 10 µl CCK-8 reagent (Beyotime
Institute of Biotechnology) was added to each well and incubated
for a further 2 h at 37˚C. The absorbance was measured at a
wavelength of 450 nm using a microplate reader (Thermo Fisher
Scientific, Inc.).
Western blotting
Total protein was extracted from CRC cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology) and
quantified using a BCA protein assay kit (Beyotime Institute of
Biotechnology). Proteins (40 µg per lane) were separated via 10%
SDS-PAGE, transferred onto PVDF membranes (Bio-Rad Laboratories,
Inc.) and blocked with 5% skimmed milk at room temperature for 1 h.
The membranes were then incubated at 4˚C overnight with the
following primary antibodies: Anti-CDK2 (1:1,000; cat. no. ab32147;
Abcam), anti-CDK4 (1:1,000; cat. no. ab32147; Abcam), anti-CDK6
(1:1,000; cat. no. ab124821; Abcam) and anti-β-actin (1:1,000; cat.
no. ab8226; Abcam). Following the primary antibody incubation, the
membranes were incubated with a HRP-conjugated goat anti-rabbit IgG
secondary antibody (1:5,000; cat. no. ab7090; Abcam) at room
temperature for 1 h. Protein bands were visualized using an ECL kit
(Thermo Fisher Scientific, Inc.). β-actin was used as the loading
control. ImageJ software (version 6.0; National Institutes of
Health) was used for densitometry.
Flow cytometric analysis of
apoptosis
HCT116 or SW480 cells were trypsinized, washed with
PBS and resuspended in Annexin V binding buffer (BD Biosciences).
Subsequently, cells were stained with 5 µl Annexin V-FITC (20
µg/ml) and 5 µl propidium (PI; 50 µg/ml) in 100 µl Annexin V
binding buffer for 15 min at 4˚C in the dark. The stained cells
were analyzed using a BD flow cytometer (BD Biosciences). The
proportions of apoptotic cells (Annexin
V+/PI- and Annexin
V+/PI+) were estimated using
Fluorescence-activated Cell Sorting (FACSLyric™; BD Biosciences)
and FlowJo software (version 10.6.2; BD Biosciences).
Measurement of mitochondrial membrane
potential (MMP)
CRC cell loss of MMP was measured using the
MitoProbe assay (Molecular Probes; Thermo Fisher Scientific, Inc.).
Cells transfected with siRNA-ctrl or si-MEX3A-1 were seeded
(2x104) into a 6-well plate. Subsequently, JC-1 dye (5
µM) was added for 20 min at 37˚C. Cells were washed with PBS for
three times. Moreover, following fixing with 4% formaldehyde for 10
min at room temperature, cells were stained with Hoechst 33258 at
4˚C for 2 h to stain living cells. Subsequently, cells were
immediately analyzed using a Zeiss 4.4.0 Axiovert 200 inverted
fluorescence microscope with a 100 W mercury lamp under the
following conditions: 330-385 nm excitation filter (excf), 400 nm
dichroic mirror (dm) and 420 nm barrier filter (bf) for Hoechst
33258; and 450-480 nm excf, 500 nm dm and 515 nm bf for JC-1. Red
fluorescence represents the polymer, and green fluorescence
represents the monomer.
Transwell assay
The upper chambers of Transwell plates were
pretreated with 50 µl Matrigel (BD Biosciences) for 4 h at 37˚C.
Subsequently, CRC cells (1x106 cells/ml) were seeded
into the upper chamber with serum-free medium. The lower chamber
was filled with RPMI-1640 supplemented with 1% FBS. Following
incubation for 24 h at 37˚C, cells in the lower chamber were fixed
with 95% alcohol for 10 min at room temperature and stained with
0.1% crystal violet for 5 min at room temperature. Invasive cells
were observed under a light microscope (magnification, x400).
Cell cycle distribution analysis
CRC cells (5x105) were fixed with 75%
ethanol on ice for 20 min, permeabilized with 0.25% Triton X-100
and stained with PI/RNase (BD Pharmingen; BD Biosciences).
Following incubation at 4˚C for 15 min, cells were analyzed using a
flow cytometer (BD FACSAria III; BD Biosciences) and ModFit
(version 3.0; Verity Software House, Inc.). The data were
quantified using FlowJo software (version 3.0; FlowJo, LLC).
Statistical analysis
All experiments were performed in triplicate. Data
are presented as the mean ± SD. Comparisons between two groups were
analyzed using a paired Student's t-test, whereas one-way ANOVA
followed by Tukey's post hoc test was used to analyze comparisons
among multiple groups (using GraphPad Prism 7; GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
MEX3A knockdown suppresses CRC cell
viability
To investigate the role of MEX3A in CRC, TCGA
database was used. As shown in Fig.
1A, MEX3A expression levels were significantly upregulated in
CRC tissues compared with those in adjacent healthy tissues.
Similarly, the expression levels of MEX3A in SW480 or HCT116 cells
were also significantly upregulated compared with those in HIEC-6
cells (Fig. 1B). Conversely, MEX3A
expression levels were significantly downregulated in CRC cells
transfected with si-MEX3As, compared with the Blank (Fig. 1C and D). As the expression levels of MEX3A were
downregulated to the greatest extent in SW480 and HCT116 cells
following transfection with si-MEX3A-1 compared with the two other
siRNAs, si-MEX3A-1 was selected for use in subsequent experiments
(further labelled as si-MEX3A). MEX3A knockdown significantly
inhibited CRC cell viability (Fig.
1E and F). These results
suggested that MEX3A knockdown may decrease CRC cell viability.
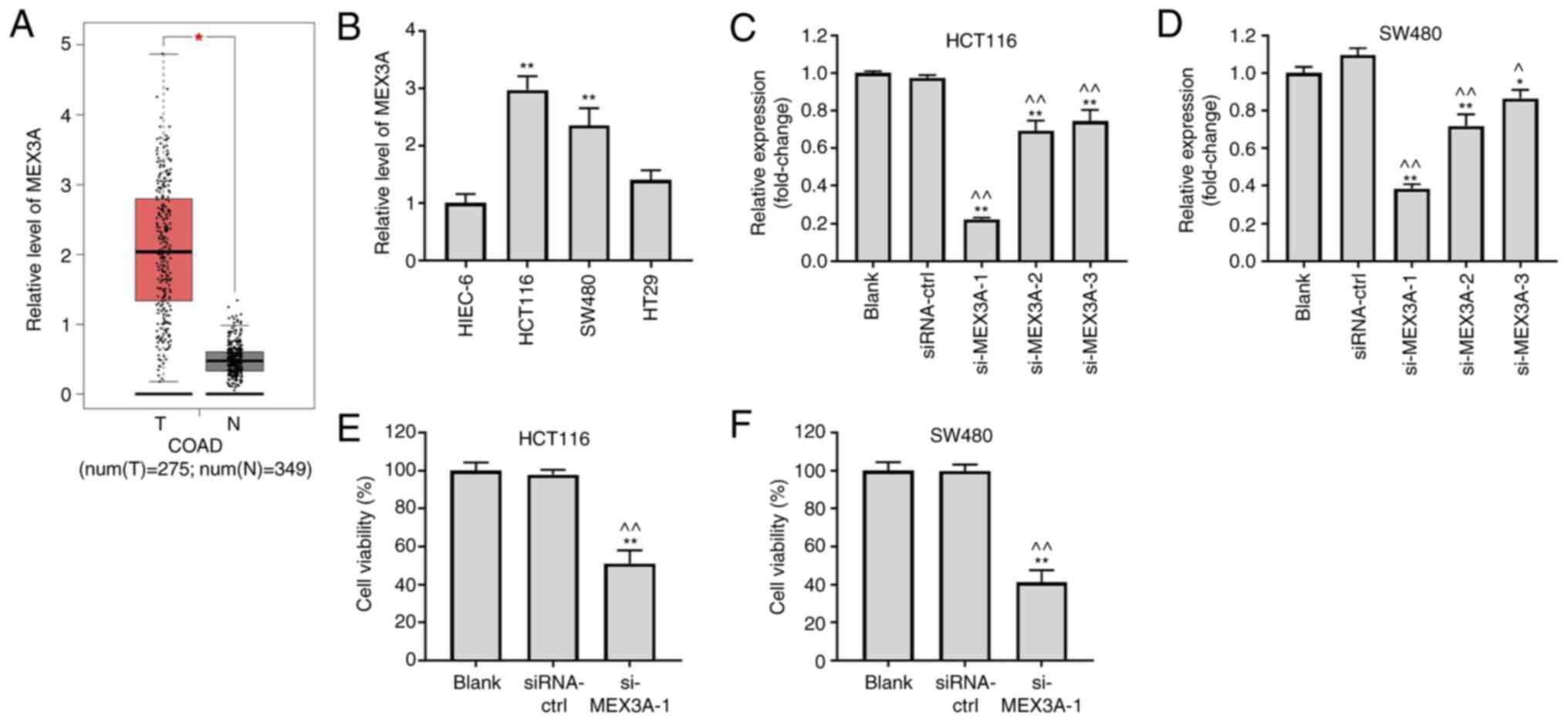 | Figure 1MEX3A knockdown significantly inhibits
CRC cell viability. (A) Expression levels of MEX3A in CRC and
adjacent healthy tissues were analyzed using data from The Cancer
Genome Atlas. (B) Expression levels of MEX3A in HIEC-6, HCT116,
SW480 and HT29 cell lines were analyzed by RT-qPCR. HCT116 and
SW480 cells were transfected with siRNA-ctrl, si-MEX3A-1,
si-MEX3A-2 or si-MEX3A-3. Expression levels of MEX3A in transfected
(C) HCT116 and (D) SW480 cells were analyzed using RT-qPCR. (E)
HCT116 and (F) SW480 cell viability was measured using a Cell
Counting Kit-8 assay. *P<0.05 and
**P<0.01 vs. HIEC-6 or Blank; ^P<0.05
and ^^P<0.01 vs. siRNA-ctrl. MEX3A, mex-3 RNA binding
family member A; CRC, colorectal cancer; RT-qPCR, reverse
transcription-quantitative PCR; siRNA, small interfering RNA; ctrl,
control; N, normal; T, tumor. |
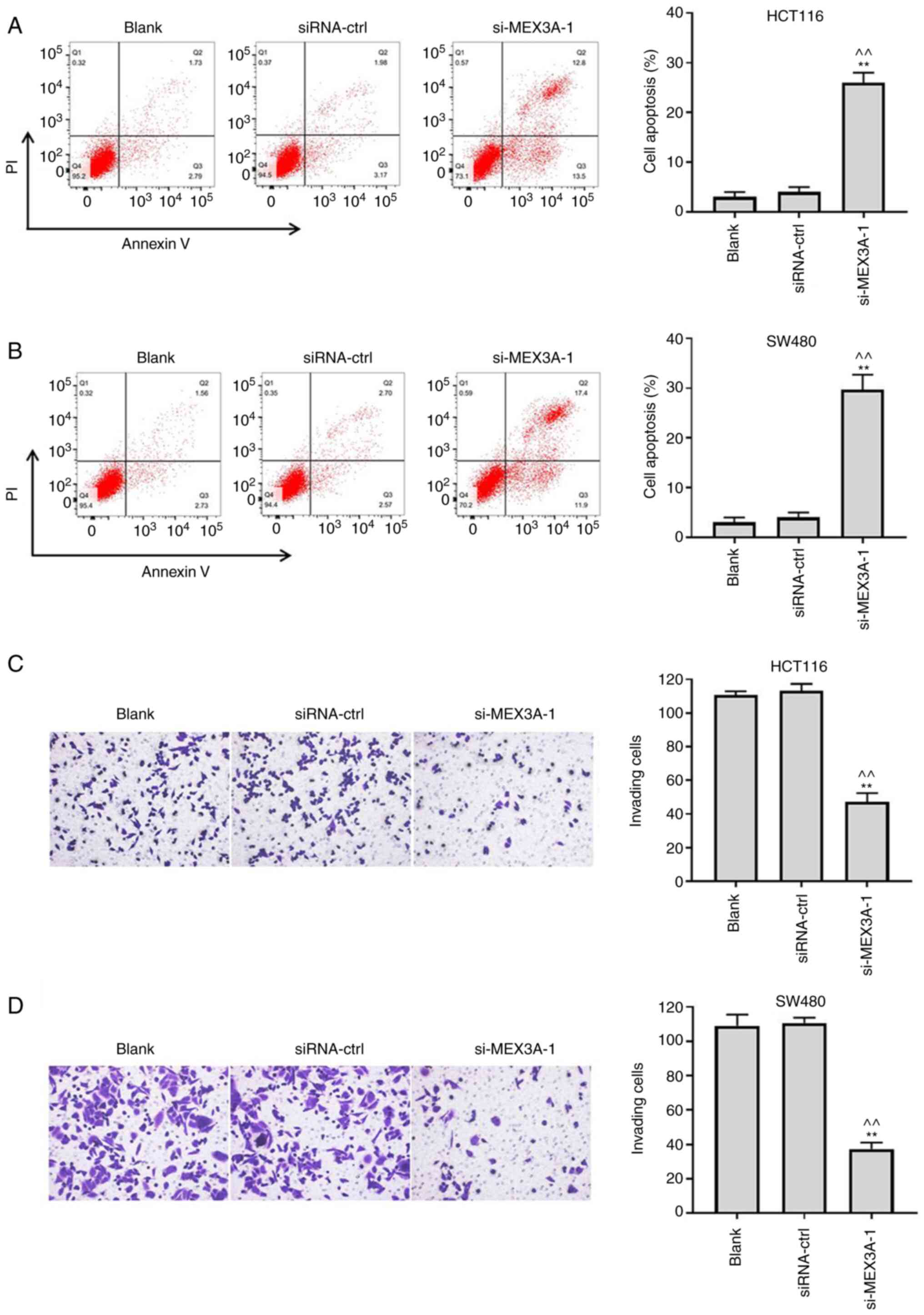
MEX3A knockdown induces the apoptosis
and suppresses the invasion of CRC cells
To investigate the effect of si-MEX3A on cell
apoptosis, flow cytometry was performed. As shown in Fig. 2A and B, si-MEX3A significantly induced CRC cell
apoptosis, compared with the Blank. In addition, the invasive
ability of CRC cells was significantly suppressed following
transfection with si-MEX3A, compared with the Blank (Fig. 2C and D). These findings suggested that MEX3A
knockdown may induce the apoptosis and decrease the invasion of CRC
cells.
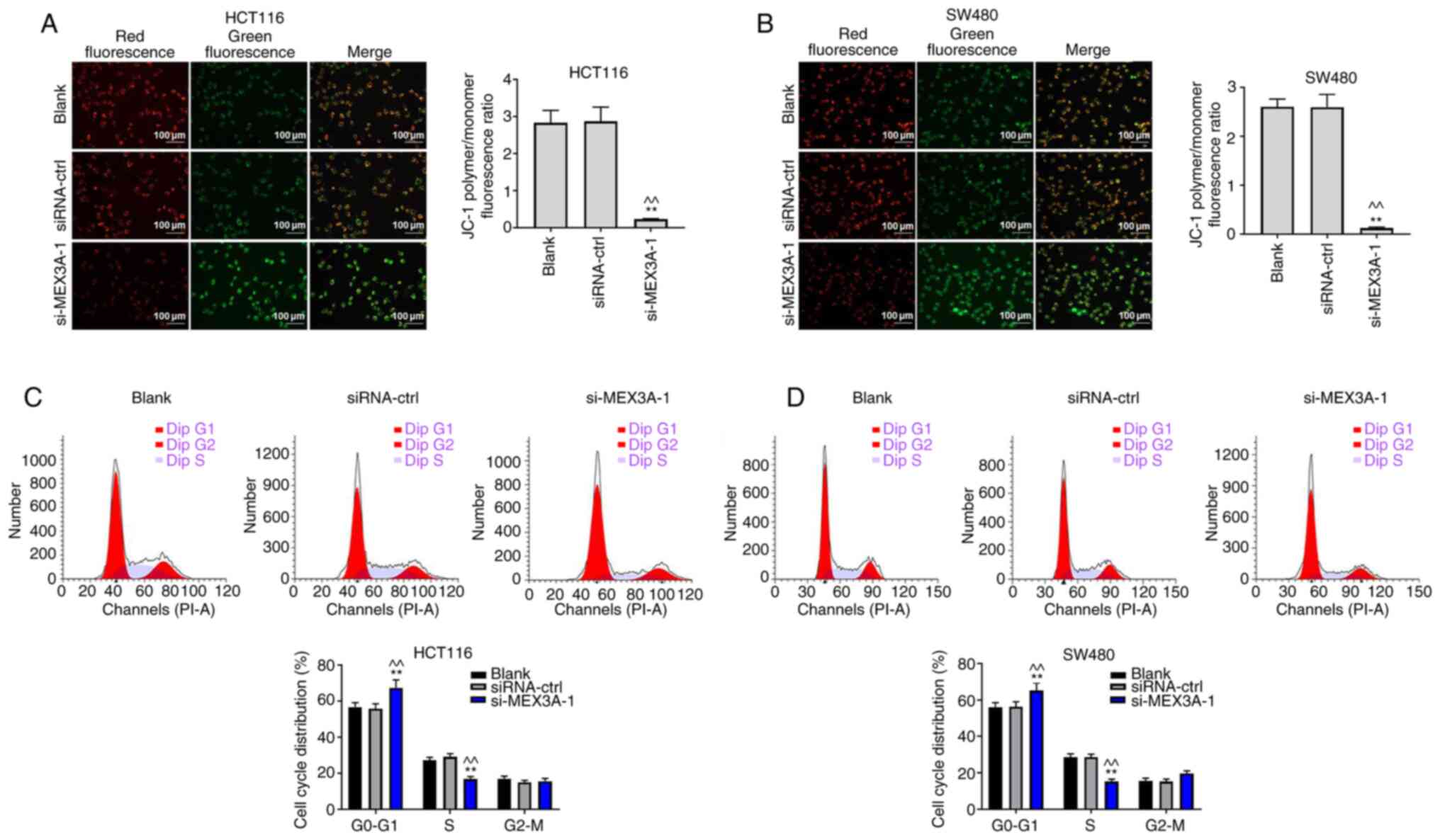
MEX3A knockdown suppresses CRC cell
proliferation by inducing mitochondrial injury
To further verify the function of MEX3A in CRC, JC-1
staining was performed. As shown in Fig. 3A and B, the ratio of polymer/monomer
fluorescence was significantly decreased in CRC cells following
transfection with si-MEX3A, compared with the Blank. In addition,
si-MEX3A significantly induced G1 cell cycle arrest in
CRC cells, compared with the Blank (Fig. 3C and D). These results indicated that MEX3A
knockdown may suppress CRC cell proliferation by inducing
mitochondrial injury.
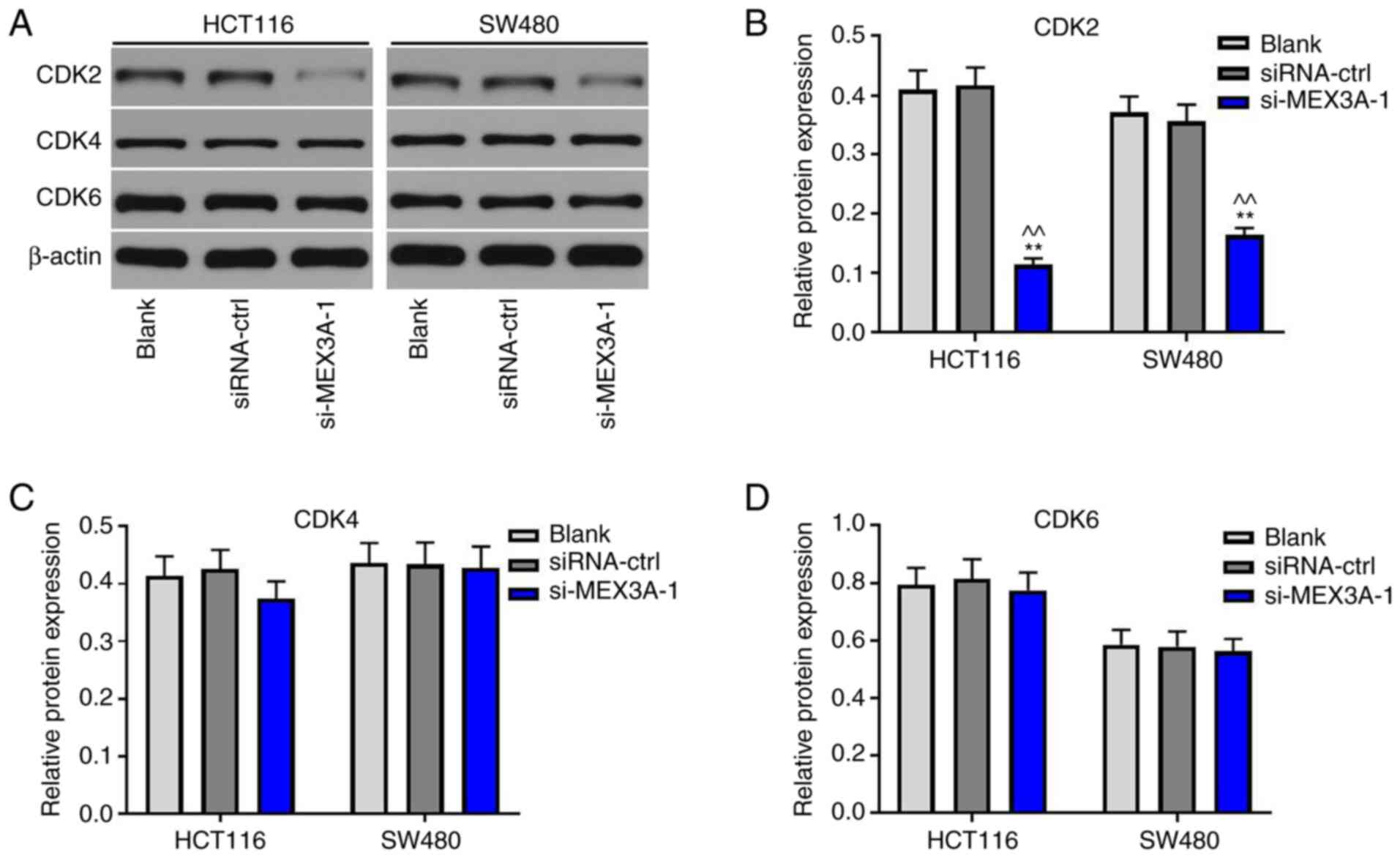
MEX3A knockdown suppresses the cycle
progression of CRC cells via inactivation of CDK2
To investigate the mechanism underlying
MEX3A-mediated tumorigenesis of CRC, western blotting was
performed. CDK2 expression levels were significantly downregulated
in CRC cells following MEX3A knockdown, compared with the Blank
(Fig. 4A and B). However, si-MEX3A exerted very limited
effects on CDK4 and CDK6 expression levels (Fig. 4A, C
and D). These results suggested
that si-MEX3A may suppress the progression of CRC cells by
downregulating CDK2 expression.
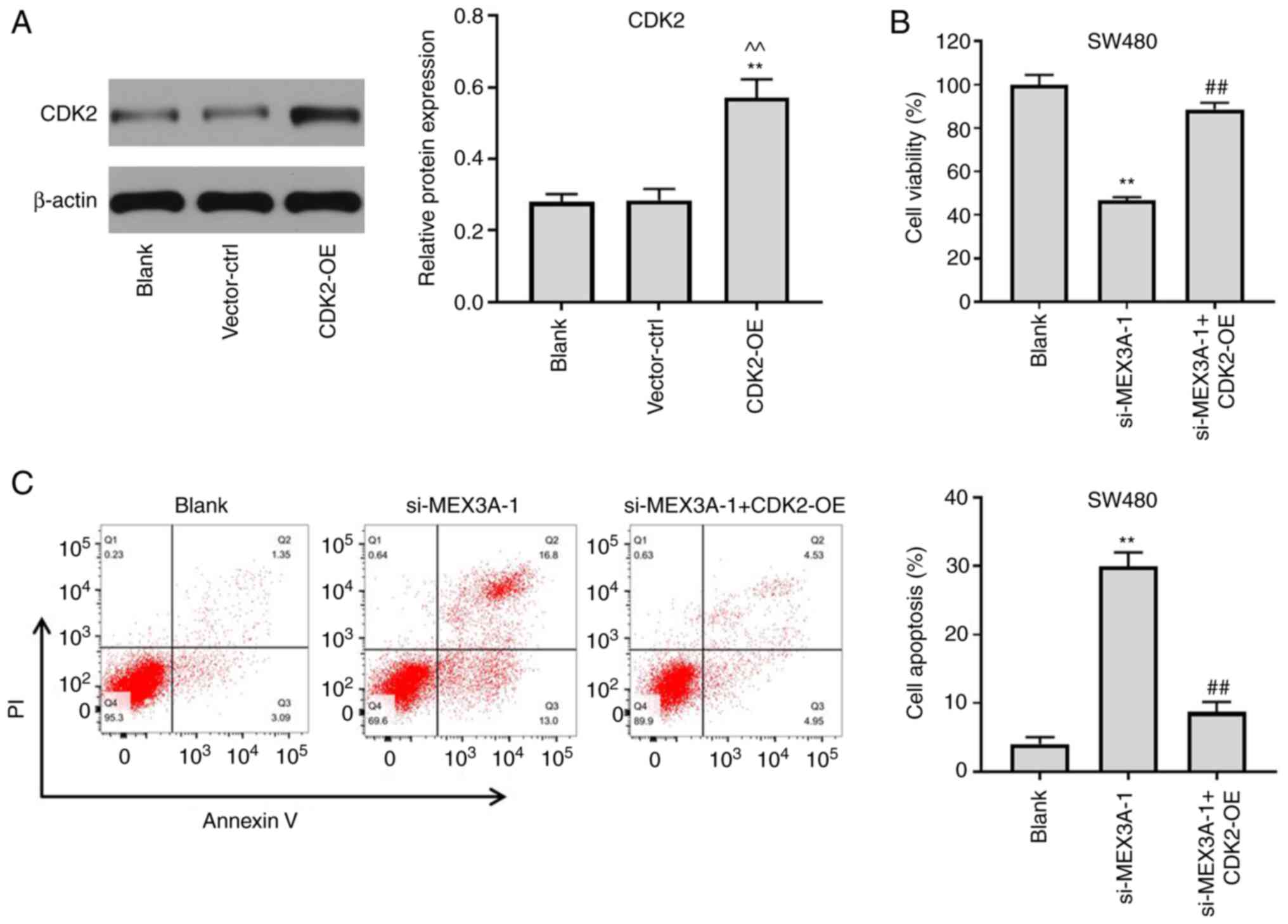
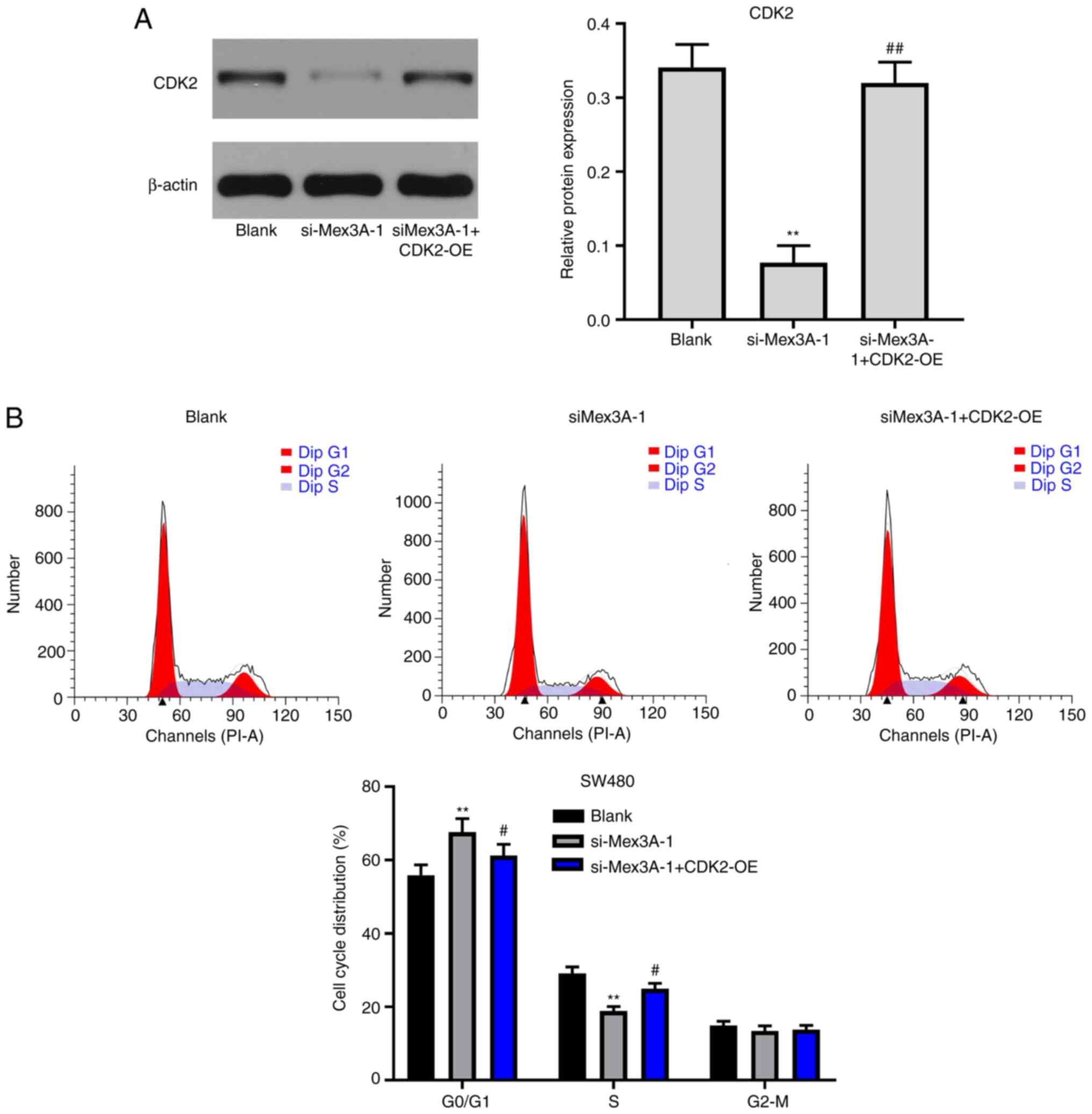
CDK2 overexpression partially reverses
the antitumor effect of si-MEX3A in CRC
To further confirm whether MEX3A inhibited the
tumorigenesis of CRC cells via mediating CDK2 expression, CDK2 was
overexpressed in CRC cells. The transfection efficiency of CDK2
overexpression was analyzed using western blotting. As shown in
Fig. 5A, the expression levels of
CDK2 were significantly upregulated in CRC cells following
transfection with pcDNA3.1-CDK2, compared with the Blank. Notably,
CDK2 overexpression partially reversed si-MEX3A-induced decreases
in cell viability (Fig. 5B). In
addition, si-MEX3A-induced CRC cell apoptosis was reversed by CDK2
overexpression (Fig. 5C). These
findings suggested that CDK2 overexpression may partially reverse
the antitumor effects of MEX3A in CRC cells.
CDK2 overexpression reverses
si-MEX3A-induced G1 cell cycle arrest in CRC cells
To further validate the mechanism underlying
MEX3A-mediated CRC cell proliferation, western blotting was
performed. The results revealed that MEX3A knockdown-induced
downregulation of CDK2 expression was significantly reversed
following CDK2 overexpression (Fig.
6A). In addition, CDK2 overexpression partially reversed the
effects of si-MEX3A on the cell cycle distribution (Fig. 6B). Taken together, these results
suggested that CDK2 may reverse si-MEX3A-induced G1 cell
cycle arrest in CRC cells.
Discussion
MEX3A regulates gene expression and serves a role in
numerous types of cancer. For example, a previous study found that
MEX3A knockdown in gastric cancer cells attenuated cancer cell
proliferation, suggesting that MEX3A may regulate cellular
transformation (11). MEX3A
knockdown could also significantly inhibit gastric cancer cell
invasion (11). Consistent with
these findings, the data from TCGA demonstrated that MEX3A
expression was higher in CRC tissues compared with that in adjacent
healthy tissues, and MEX3A was able to modulate CRC cell cycle
progression. Pereira et al (16) demonstrated that MEX3A participated
in CDX2 regulation by downregulating its expression levels, and
reversed intestinal cell differentiation, indicating that MEX3A may
serve as an oncogene in CRC. The present study also investigated
the function of MEX3A in CRC, indicating that MEX3A may serve as an
oncogene in CRC.
Further experiments demonstrated that MEX3A
knockdown suppressed CRC cell proliferation and invasion. In
addition, the transfection of CRC cells with si-MEX3A exerted
antitumor effects via regulating the expression of CDK2. Similarly,
Li et al (17) indicated
that MEX3A promoted oncogenesis through the RAP1/MAPK signaling
pathway in CRC (17). MAPK
signaling also promoted the tumorigenesis of CRC, and CDK2 was
known to be a promoter in cancer cell growth (18,19).
As important mediators of the cell cycle, CDK2, CDK4
and CDK6 belong to the cell division cycle 20-related kinase family
(20,21). Previous studies have shown that the
expression levels of CDK2, CDK4 and CDK6 are often upregulated in
various types of cancer, and are associated with tumorigenesis by
interacting with other proteins (22,23).
For example, Qu et al (24)
reported that CDK2 served a key role in circular RNA
(circ)_0084927/microRNA (miR)-1179 signaling axis-mediated cervical
cancer development. Zhao et al (25) found that CDK6 knockdown suppressed
gastric cancer cell proliferation via regulating the cell cycle. In
non-small cell lung cancer, CDK4 was identified as a crucial
mediator of the competing endogenous RNA mechanism underlying the
hsa_circ_0014235/miR-520-5p signaling axis (26). Based on the aforementioned findings
and the results obtained in the present study, it was suggested
that MEX3A knockdown may inhibit the tumorigenesis of CRC via
mediating the expression of CDK2.
The present study had a number of limitations. To
further verify the function of MEX3A in CRC, animal studies need to
be performed and lentivirus transfection of MEX3A should be
conducted. Moreover, the mechanism underlying MEX3A-mediated
regulation of CDK2 expression is not completely understood. In
addition, whether CDK2 overexpression can rescue MEX3A
siRNA-induced mitochondrial injury requires further
investigation.
In conclusion, the results of the present study
suggested that MEX3A knockdown may inhibit the tumorigenesis of CRC
via regulating CDK2 expression. Thus, MEX3A may serve as a
potential target for the treatment of CRC. However, the role of
other mRNAs regulated by MEX3A in CRC requires further
investigation. Furthermore, to validate the role of MEX3A in CRC,
the expression levels of epithelial-mesenchymal transition markers
should be investigated in future studies.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81702422), the Natural
Science Research Project of Jiangsu Higher Education Institutions
(grant no. 17KJB320024) and Jiangsu Provincial Health and Family
Planning Commission (grant no. H2017086).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SL and YQ conceived and supervised the study. XZ and
YQ designed the study. XZ, TM, JZ, HL, XL, FL, BJ, MZ and ZL
performed the experiments and analyzed the data. SL and YQ
confirmed the authenticity of the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Parine NR, Azzam NA, Shaik J, Aljebreen
AM, Alharbi O, Almadi MA, Alanazi M and Khan Z: Genetic variants in
the WNT signaling pathway are protectively associated with
colorectal cancer in a Saudi population. Saudi J Biol Sci.
26:286–293. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Feroldi F, Verlaan M, Knaus H, Davidoiu V,
Vugts DJ, van Dongen GA, Molthoff CF and de Boer JF: High
resolution combined molecular and structural optical imaging of
colorectal cancer in a xenograft mouse model. Biomed Opt Express.
9:6186–6204. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gala de Pablo J, Armistead FJ, Peyman SA,
Bonthron D, Lones M, Smith S and Evans SD: Biochemical fingerprint
of colorectal cancer cell lines using label-free live single-cell
Raman spectroscopy. J Raman Spectrosc. 49:1323–1332.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Panzeri V, Manni I, Capone A, Naro C,
Sacconi A, Di Agostino S, de Latouliere L, Montori A, Pilozzi E,
Piaggio G, et al: The RNA-binding protein MEX3A is a prognostic
factor and regulator of resistance to gemcitabine in pancreatic
ductal adenocarcinoma. Mol Oncol. 15:579–595. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Naef V, De Sarlo M, Testa G, Corsinovi D,
Azzarelli R, Borello U and Ori M: The stemness gene Mex3A is a key
regulator of neuroblast proliferation during neurogenesis. Front
Cell Dev Biol. 8(549533)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liang J, Li H, Han J, Jiang J, Wang J, Li
Y, Feng Z, Zhao R, Sun Z, Lv B, et al: Mex3a interacts with LAMA2
to promote lung adenocarcinoma metastasis via PI3K/AKT pathway.
Cell Death Dis. 11(614)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jiang S, Meng L, Chen X, Liu H, Zhang J,
Chen F, Zheng J, Liu H, Wang F, Hu J, et al: MEX3A promotes triple
negative breast cancer proliferation and migration via the PI3K/AKT
signaling pathway. Exp Cell Res. 395(112191)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang X, Shan YQ, Tan QQ, Tan CL, Zhang H,
Liu JH, Ke NW, Chen YH and Liu XB: MEX3A knockdown inhibits the
development of pancreatic ductal adenocarcinoma. Cancer Cell Int.
20(63)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bufalieri F, Caimano M, Lospinoso Severini
L, Basili I, Paglia F, Sampirisi L, Loricchio E, Petroni M,
Canettieri G, Santoro A, et al: The RNA-binding ubiquitin ligase
MEX3A affects glioblastoma tumorigenesis by inducing ubiquitylation
and degradation of RIG-I. Cancers (Basel). 12(12)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yang D, Jiao Y, Li Y and Fang X: Clinical
characteristics and prognostic value of MEX3A mRNA in liver cancer.
PeerJ. 8(e8252)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jiang H, Zhang X, Luo J, Dong C, Xue J,
Wei W, Chen J, Zhou J, Gao Y and Yang C: Knockdown of hMex-3A by
small RNA interference suppresses cell proliferation and migration
in human gastric cancer cells. Mol Med Rep. 6:575–580.
2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yang C, Zhan H, Zhao Y, Wu Y, Li L and
Wang H: MEX3A contributes to development and progression of glioma
through regulating cell proliferation and cell migration and
targeting CCL2. Cell Death Dis. 12(14)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Jin X, Ge LP, Li DQ, Shao ZM, Di GH, Xu XE
and Jiang YZ: LncRNA TROJAN promotes proliferation and resistance
to CDK4/6 inhibitor via CDK2 transcriptional activation in
ER+ breast cancer. Mol Cancer. 19(87)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Krepischi ACV, Maschietto M, Ferreira EN,
Silva AG, Costa SS, da Cunha IW, Barros BDF, Grundy PE, Rosenberg C
and Carraro DM: Genomic imbalances pinpoint potential oncogenes and
tumor suppressors in Wilms tumors. Mol Cytogenet.
9(20)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Pereira B, Sousa S, Barros R, Carreto L,
Oliveira P, Oliveira C, Chartier NT, Plateroti M, Rouault JP,
Freund JN, et al: CDX2 regulation by the RNA-binding protein MEX3A:
Impact on intestinal differentiation and stemness. Nucleic Acids
Res. 41:3986–3999. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li H, Liang J, Wang J, Han J, Li S, Huang
K and Liu C: Mex3a promotes oncogenesis through the RAP1/MAPK
signaling pathway in colorectal cancer and is inhibited by
hsa-miR-6887-3p. Cancer Commun (Lond). 41:472–491. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Peng W, Li J, Chen R, Gu Q, Yang P, Qian
W, Ji D, Wang Q, Zhang Z, Tang J, et al: Upregulated METTL3
promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK
signaling pathway. J Exp Clin Cancer Res. 38(393)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
He X, Li S, Yu B, Kuang G, Wu Y, Zhang M,
He Y, Ou C and Cao P: LINC00467Up-regulation of promotes the
tumourigenesis in colorectal cancer. J Cancer. 10:6405–6413.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wells N, Quigley J, Pascua J, Pinkowski N,
Almaiman L, Brasser SM and Hong MY: Effects of low-to-moderate
ethanol consumption on colonic growth and gene expression in young
adult and middle-aged male rats. PLoS One.
15(e0243499)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kumarasamy V, Vail P, Nambiar R,
Witkiewicz AK and Knudsen ES: Functional determinants of cell-cycle
plasticity and sensitivity to CDK4/6 inhibition. Cancer Res.
81:1347–1360. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shih LJ, Wang JY, Jheng JY, Siao AC, Lin
YY, Tsuei YW, Kuo YC, Chuu CP and Kao YH: Betel nut arecoline
induces different phases of growth arrest between normal and
cancerous prostate cells through the reactive oxygen species
pathway. Int J Mol Sci. 21(21)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Pandey K, Park N, Park KS, Hur J, Cho YB,
Kang M, An HJ, Kim S, Hwang S and Moon YW: Combined CDK2 and CDK4/6
inhibition overcomes palbociclib resistance in breast cancer by
enhancing senescence. Cancers (Basel). 12(12)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Qu X, Zhu L, Song L and Liu S:
circ_0084927 promotes cervical carcinogenesis by sponging miR-1179
that suppresses CDK2, a cell cycle-related gene. Cancer Cell Int.
20(333)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao Y, He J, Li Y, Xu M, Peng X, Mao J,
Xu B and Cui H: PHF14 promotes cell proliferation and migration
through the AKT and ERK1/2 pathways in gastric cancer cells. BioMed
Res Int. 2020(6507510)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xu X, Tao R, Sun L and Ji X:
Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance
and deteriorates the development of non-small cell lung cancer by
mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int.
20(552)2020.PubMed/NCBI View Article : Google Scholar
|