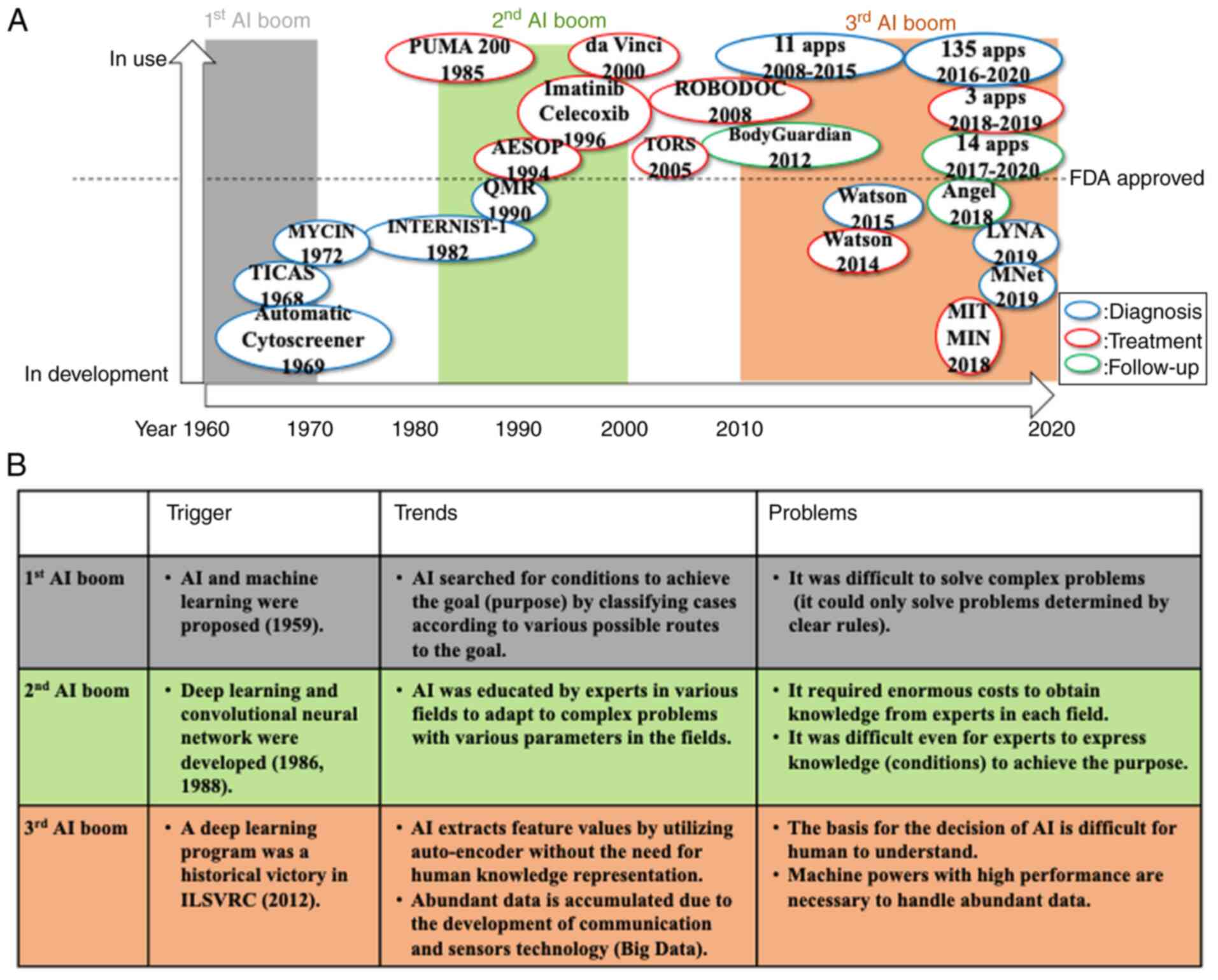
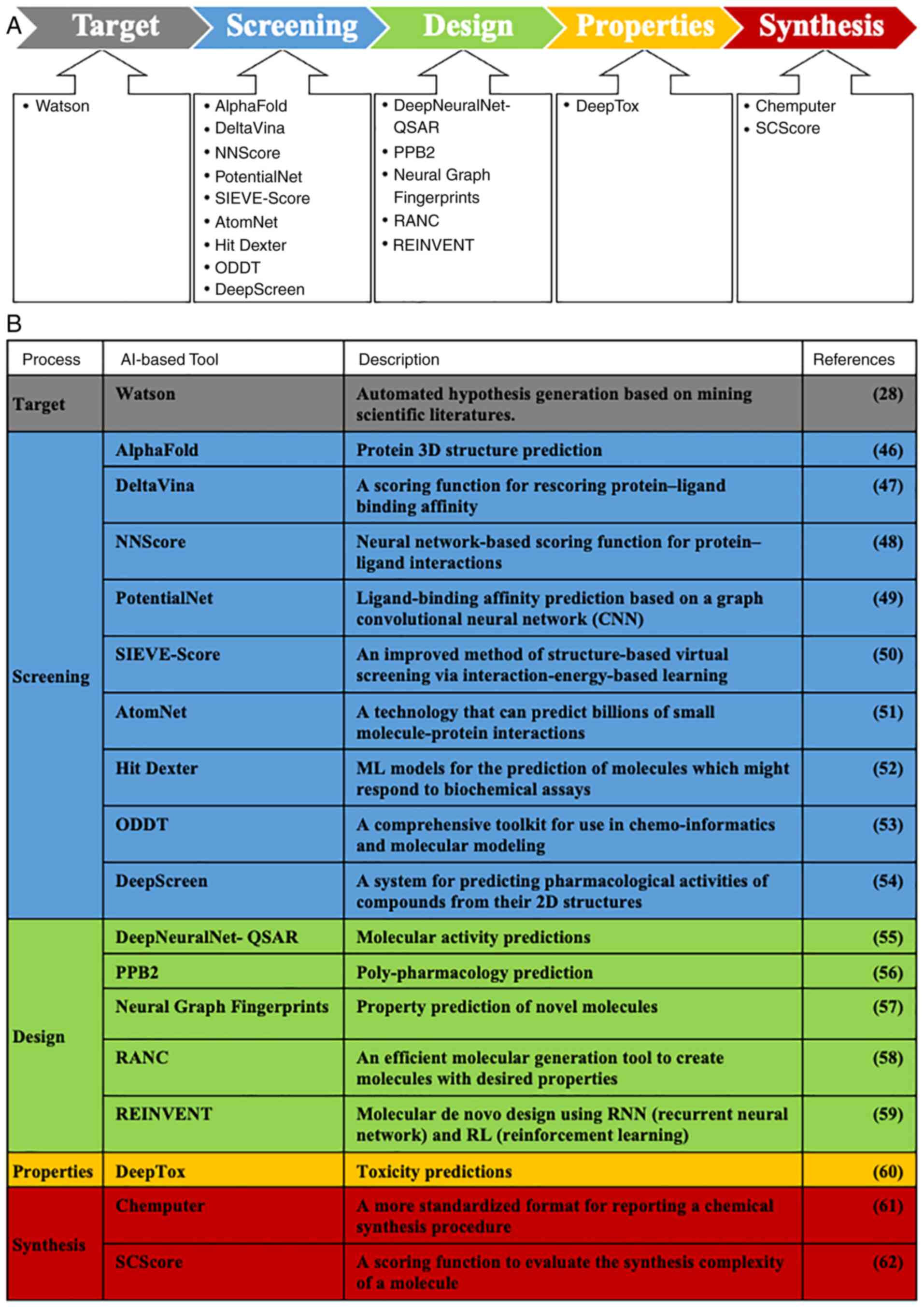
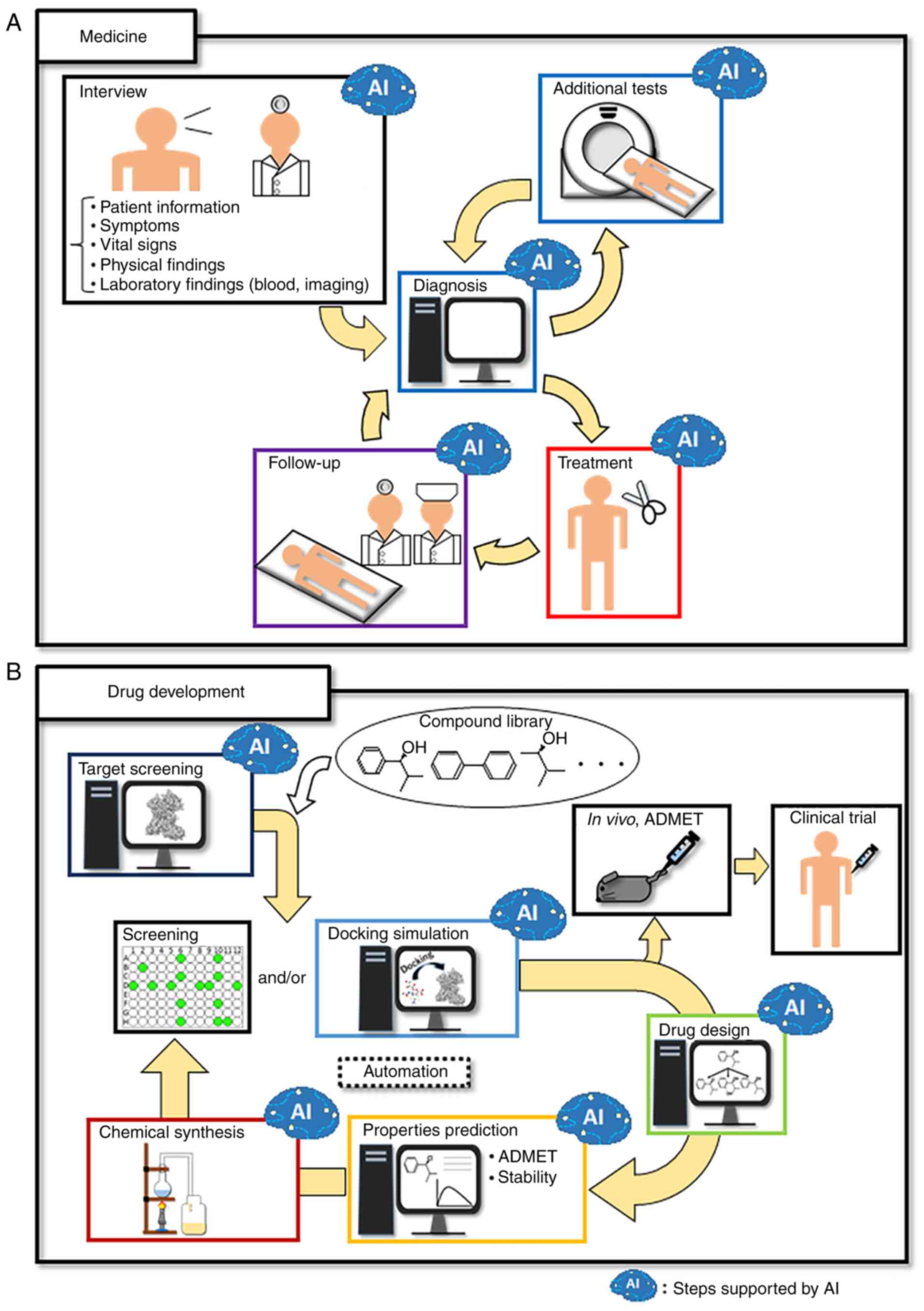
|
1
|
Santos DP and Baeßler B: Big data,
artificial intelligence, and structured reporting. Eur Radiol Exp.
2(42)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Akyazi T, Goti A, Oyarbide A, Alberdi E
and Bayon F: A guide for the food industry to meet the future
skills requirements emerging with industry 4.0. Foods.
9(492)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Favaretto M, Shaw D, De Clercq E, Joda T
and Elger BS: Big data and digitalization in dentistry: A
systematic review of the ethical issues. Int J Environ Res Public
Health. 17(2495)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Etienne H, Hamdi S, Le Roux M, Camuset J,
Khalife-Hocquemiller T, Giol M, Debrosse D and Assouad J:
Artificial intelligence in thoracic surgery: Past, present,
perspective and limits. Eur Respir Rev. 29(200010)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
McCarthy J, Minsky ML, Rochester N and
Shannon CE: A Proposal for the Dartmouth Summer Research Project on
Artificial Intelligence, August 31, 1955. AI Magazine.
27(12)2006.
|
|
6
|
Samuel AL: Some studies in machine
learning using the game of checkers. IBM J Res Dev. 3:210–229.
1959.
|
|
7
|
Wied GL, Bahr GF, Oldfield DG and Bartels
PH: Computer-assisted identification of cells from uterine
adenocarcinoma. A clinical feasibility study with TICAS. I.
Measurements at wavelength 530 nm. Acta Cytol. 12:357–370.
1968.PubMed/NCBI
|
|
8
|
Ishiyama T, Tsubura E, Hirao F, Yamamura
Y, Takeda S, Tateisi K, Yamamoto M, Uemura M, Yaida K, Hayakawa F,
et al: A study of the automation of cytodiagnosis. Med Biol Eng.
7:297–306. 1969.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shortliffe EH, Axline SG, Buchanan BG,
Merigan TC and Cohen SN: An artificial intelligence program to
advise physicians regarding antimicrobial therapy. Comput Biomed
Res. 6:544–560. 1973.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Miller RA, Pople HE and Myers JD:
INTERNIST-1, An experimental computer-based diagnostic consultant
for general internal medicine. N Engl J Med. 307:478–486.
1982.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dechter R: Learning while searching in
constraint-satisfaction-problems. In: Proceedings of the Fifth
National Conference on Artificial Intelligence. American
Association for Artificial Intelligence, Philadelphia, pp178-183,
1986.
|
|
12
|
LeCun Y, Boser B, Denker JS, Henderson D,
Howard RE, Hubbard W and Jackel LD: Backpropagation applied to
handwritten zip code recognition. Neural Comput. 1:541–551.
1989.
|
|
13
|
Kwoh YS, Hou J, Jonckheere EA and Hayati
S: A robot with improved absolute positioning accuracy for CT
guided stereotactic brain surgery. IEEE Trans Biomed Eng.
35:153–160. 1988.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Jacobs LK, Shayani V and Sackier JM:
Determination of the learning curve of the AESOP robot. Surg
Endosc. 11:54–55. 1997.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sung GT and Gill IS: Robotic laparoscopic
surgery: A comparison of the DA Vinci and Zeus systems. Urology.
58:893–898. 2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Weinstein GS, O'malley BW Jr and Hockstein
NG: Transoral robotic surgery: Supraglottic laryngectomy in a
canine model. Laryngoscope. 115:1315–1319. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kurumbail RG, Stevens AM, Gierse JK,
McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM,
Penning TD, Seibert K, et al: Structural basis for selective
inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature.
384:644–648. 1996.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Molnár L and Losonczy H: Tyrosine kinase
inhibitor STI571: New possibility in the treatment of chronic
myeloid leukemia. Orv Hetil. 143:2379–2384. 2002.PubMed/NCBI(In Hu).
|
|
19
|
Lanfranco AR, Castellanos AE, Desai JP and
Meyers WC: Robotic surgery: A current perspective. Ann Surg.
239:14–21. 2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hinton GE, Osindero S and The YW: A fast
learning algorithm for deep belief nets. Neural Comput.
18:1527–1554. 2006.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hinton GE and Salakhutdinov RR: Reducing
the dimensionality of data with neural networks. Science.
313:504–507. 2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Komura D and Ishikawa S: Machine learning
approaches for pathologic diagnosis. Virchows Arch. 475:131–138.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
American College of Radiology. Cleared AI
Algorithms. 2021. https://models.acrdsi.org. Accessed July 1, 2021.
|
|
24
|
U.S. Food and Drug Administration: FDA
allows marketing of first whole slide imaging system for digital
pathology. 2017. https://www.fda.gov/news-events/press-announcements/fda-allows-marketing-first-whole-slide-imaging-system-digital-pathology.
Accessed July 1, 2021.
|
|
25
|
Mills AM, Gradecki SE, Horton BJ,
Blackwell R, Moskaluk CA, Mandell JW, Mills SE and Cathro HP:
Diagnostic efficiency in digital pathology: A comparison of optical
versus digital assessment in 510 surgical pathology cases. Am J
Surg Pathol. 42:53–59. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
The Medical Futurist: FDA-approved
A.I.-based algorithms. 2021. https://medicalfuturist.com/fda-approved-ai-based-algorithms/.
Accessed July 1, 2021.
|
|
27
|
Asai A, Koseki J, Konno M, Nishimura T,
Gotoh N, Satoh T, Doki Y, Mori M and Ishii H: Drug discovery of
anticancer drugs targeting methylenetetrahydrofolate dehydrogenase
2. Heliyon. 4(e01021)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Spangler S, Wilkins AD, Bachman BJ,
Nagarajan M, Dayaram T, Haas P, Regenbogen S, Pickering CR, Comer
A, Myers JN, et al: Automated hypothesis generation based on mining
scientific literature. KDD'14: Proceedings of the 20th ACM SIGKDD
international conference on Knowledge discovery and data mining:
1877-1886, 2014.
|
|
29
|
Doyle-Lindrud S: Watson will see you now:
A supercomputer to help clinicians make informed treatment
decisions. Clin J Oncol Nurs. 19:31–32. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tojo A: Clinical sequencing in leukemia
with the assistance of artificial intelligence. Rinsho Ketsueki.
58:1913–1917. 2017.PubMed/NCBI View Article : Google Scholar : (In Japanese).
|
|
31
|
Esteva A, Kuprel B, Novoa RA, Ko J,
Swetter SM, Blau HM and Thrun S: Dermatologist-level classification
of skin cancer with deep neural networks. Nature. 542:115–118.
2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Toratani M, Konno M, Asai A, Koseki J,
Kawamoto K, Tamari K, Li Z, Sakai D, Kudo T, Satoh T, et al: A
convolutional neural network uses microscopic images to
differentiate between mouse and human cell lines and their
radioresistant clones. Cancer Res. 78:6703–6707. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yanagisawa K, Toratani M, Asai A, Konno M,
Niioka H, Mizushima T, Satoh T, Miyake J, Ogawa K, Vecchione A, et
al: Convolutional neural network can recognize drug resistance of
single cancer cells. Int J Mol Sci. 21(3166)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tanos R, Tosato G, Otandault A, Al Amir
Dache Z, Pique Lasorsa L, Tousch G, El Messaoudi S, Meddeb R, Assaf
MD, Ychou M, et al: Machine learning-assisted evaluation of
circulating DNA quantitative analysis for cancer screening. Adv Sci
(Weinh). 7(2000486)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Im YR, Tsui DWY, Diaz LA Jr and Wan JCM:
Next-generation liquid biopsies: Embracing data science in
oncology. Trends Cancer. 7:283–292. 2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Harada Y and Shimizu T: Impact of a
Commercial Artificial Intelligence-Driven Patient Self-Assessment
Solution on Waiting Times at General Internal Medicine Outpatient
Departments: Retrospective Study. JMIR Med Inform.
8(e21056)2020.PubMed/NCBI View
Article : Google Scholar
|
|
37
|
Litjens G, Kooi T, Bejnordi BE, Setio AAA,
Ciompi F, Ghafoorian M, van Der Laak JA, van Ginneken B and Sánchez
CI: A survey on deep learning in medical image analysis. Med Image
Anal. 42:60–88. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Khosravi P, Kazemi E, Imielinski M,
Elemento O and Hajirasouliha I: Deep convolutional neural networks
enable discrimination of heterogeneous digital pathology images.
EBioMedicine. 27:317–328. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Meyer A, Zverinski D, Pfahringer B,
Kempfert J, Kuehne T, Sündermann SH, Stamm C, Hofmann T, Falk V and
Eickhoff C: Machine learning for real-time prediction of
complications in critical care: A retrospective study. Lancet
Respir Med. 6:905–914. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Saeidi H, Opfermann JD, Kam M, Raghunathan
S, Leonard S and Krieger A: A confidence-based shared control
strategy for the smart tissue autonomous robot (STAR). Rep US.
2018:1268–1275. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Brown N, Cambruzzi J, Cox PJ, Davies M,
Dunbar J, Plumbley D, Sellwood MA, Sim A, Williams-Jones BI,
Zwierzyna M and Sheppard DW: Big data in drug discovery. Prog Med
Chem. 57:277–356. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Vélez-Guerrero MA, Callejas-Cuervo M and
Mazzoleni S: Artificial intelligence-based wearable robotic
exoskeletons for upper limb rehabilitation: A review. Sensors
(Basel). 21(2146)2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Comendador B, Francisco B, Medenilla J,
Nacion S and Serac T: Pharmabot: A pediatric generic medicine
consultant chatbot. J Automat Control Eng. 3:137–140. 2015.
|
|
44
|
Wong V, Rosenbaum S, Sung W, Kaplan RM,
Bott N, Platchek T, Milstein A and Shah NR: Caring for caregivers:
bridging the gap between family caregiving policy and practice.
NEJM Catal Innov Care Deliv. 2:2021.
|
|
45
|
Paul D, Sanap G, Shenoy S, Kalyane D,
Kalia K and Tekade RK: Artificial intelligence in drug discovery
and development. Drug Discov Today. 26:80–93. 2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
AlQuraishi M: AlphaFold at CASP13.
Bioinformatics. 35:4862–4865. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang C and Zhang Y: Improving
scoring-docking-screening powers of protein-ligand scoring
functions using random forest. J Comput Chem. 38:169–177.
2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Durrant JD and McCammon JA: NNScore 2.0: A
neural-network receptor-ligand scoring function. J Chem Inf Model.
51:2897–2903. 2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Feinberg EN, Sur D, Wu Z, Husic BE, Mai H,
Li Y, Sun S, Yang J, Ramsundar B and Pande VS: PotentialNet for
molecular property prediction. ACS Cent Sci. 4:1520–1530.
2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yasuo N and Sekijima M: Improved method of
structure-based virtual screening via interaction-energy-based
learning. J Chem Inf Model. 59:1050–1061. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wallach I, Dzamba M and Heifets A:
AtomNet: A deep convolutional neural network for bioactivity
prediction in structure-based drug discovery. arXiv: 1510.02855,
2015.
|
|
52
|
Stork C, Chen Y, Šícho M and Kirchmair J:
Hit dexter 2.0: Machine-learning models for the prediction of
frequent hitters. J Chem Inf Model. 59:1030–1043. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wójcikowski M, Zielenkiewicz P and
Siedlecki P: Open drug discovery toolkit (ODDT): A new open-source
player in the drug discovery field. J Cheminform.
7(26)2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Rifaioglu AS, Nalbat E, Atalay V, Martin
MJ, Cetin-Atalay R and Doğan T: DEEPScreen: High performance
drug-target interaction prediction with convolutional neural
networks using 2-D structural compound representations. Chem Sci.
11:2531–2557. 2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Xu Y, Ma J, Liaw A, Sheridan RP and
Svetnik V: Demystifying multitask deep neural networks for
quantitative structure-activity relationships. J Chem Inf Model.
57:2490–2504. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Awale M and Reymond JL: Polypharmacology
Browser PPB2: Target prediction combining nearest neighbors with
machine learning. J Chem Inf Model. 59:10–17. 2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Duvenaud DK, Maclaurin D, Iparraguirre J,
Bombarell R, Hirzel T, Aspuru-Guzik A and Adams RP: Convolutional
networks on graphs for learning molecular fingerprints. In:
Advances in neural information processing systems 28. NIPS
Foundation, Montreal. 2:2224–2232. 2015.
|
|
58
|
Putin E, Asadulaev A, Ivanenkov Y,
Aladinskiy V, Sanchez-Lengeling B, Aspuru-Guzik A and Zhavoronkov
A: Reinforced adversarial neural computer for de novo molecular
design. J Chem Inf Model. 58:1194–1204. 2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Olivecrona M, Blaschke T, Engkvist O and
Chen H: Molecular de-novo design through deep reinforcement
learning. J Cheminform. 9(48)2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Mayr A, Klambauer G, Unterithiner T and
Hochreiter S: DeepTox: Toxicity prediction using deep learning.
Front Environ Sci. 3(80)2016.
|
|
61
|
Steiner S, Wolf J, Glatzel S, Andreou A,
Granda JM, Keenan G, Hinkley T, Aragon-Camarasa G, Kitson PJ,
Angelone D and Cronin L: Organic synthesis in a modular robotic
system driven by a chemical programming language. Science.
363(eaav2211)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Coley CW, Rogers L, Green WH and Jensen
KF: SCScore: Synthetic complexity learned from a reaction corpus. J
Chem Inf Model. 58:252–261. 2018.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Antunes DA, Devaurs D and Kavraki LE:
Understanding the challenges of protein flexibility in drug design.
Expert Opin Drug Discov. 10:1301–1313. 2015.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Uehara S and Tanaka S: AutoDock-GIST:
Incorporating thermodynamics of active-site water into scoring
function for accurate protein-ligand docking. Molecules.
21(1604)2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Jamal S, Ali W, Nagpal P, Grover S and
Grover A: Computational models for the prediction of adverse
cardiovascular drug reactions. J Transl Med. 17(171)2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Walonoski J, Kramer M, Nichols J, Quina A,
Moesel C, Hall D, Duffett C, Dube K, Gallagher T and McLachlan S:
Synthea: An approach, method, and software mechanism for generating
synthetic patients and the synthetic electronic health care record.
J Am Med Inform Assoc. 25:230–238. 2018.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Choi E, Biswal S, Malin B, Duke J, Stewart
WF and Sun J: Generating multi-label discrete patient records using
generative adversarial networks. PMLR. 68:286–305. 2017.
|
|
68
|
Linardatos P, Papastefanopoulos V and
Kotsiantis S: Explainable AI: A Review of Machine Learning
Interpretability Methods. Entropy (Basel). 23(18)2021.PubMed/NCBI View Article : Google Scholar
|