Introduction
Pancreatic adenocarcinoma (PAAD) is a
gastrointestinal malignancy with high mortality. The 5-year
survival rate for patients with pancreatic cancer is <5%
(1,2). It is expected that pancreatic cancer
will rank second in terms of mortality rate among cancers worldwide
in the next 20 years (3). For
patients with pancreatic cancer, the only cure is surgical
resection, but the majority of patients are diagnosed with local
inoperable tumors or distant metastasis; thus, patients with
pancreatic cancer have a poor prognosis (4,5).
Therefore, further research is urgently required to develop
effective prevention measures and early diagnostic methods. In the
past few decades, efforts to study the molecular mechanisms of
pancreatic cancer have provided hope for molecular diagnostics and
molecular targeted therapy for various diseases.
Genes with significantly high expression levels in
pancreatic cancer include KRAS, BRAF serine/threonine kinase (BRAF)
and AKT serine/threonine kinase 2 (AKT2), as previously reported
(6). These genes may be used as
biomarkers for early diagnosis of pancreatic cancer. Gene
expression profile analysis is a high-throughput method for
detecting mRNA expression in tissues or cell samples. For instance,
based on data from Gene Expression Omnibus (GEO), Long et al
(7) screened the differentially
expressed genes (DEGs) in pancreatic cancer and analyzed the copy
number variation in DEGs. Their study indicated that transforming
growth factor β receptor 1 (TGFBR1) and transforming growth factor
β 1 (TGFB1) have an important role in the development of pancreatic
cancer. The Wnt (8) and hedgehog
(9) signaling pathways have been
identified as being of marked significance in pancreatic cancer.
microRNAs (miRNAs or miRs) have attracted widespread attention in
recent years (10). A previous
study indicated that miRNA-27a promotes the proliferation of
pancreatic cancer cells by activating the Wnt/β-catenin signaling
pathway (11). miR-132 has recently
been demonstrated to promote pancreatic cancer cell proliferation
and inhibit apoptosis through the hedgehog signaling pathway
(12). Despite these tremendous
advances, the underlying key mechanisms of pancreatic cancer
require to be further elucidated to screen promising prognostic
biomarkers and potential targets for diagnosis and treatment of
pancreatic cancer. In the present study, genes associated with
pancreatic cancer were determined from datasets obtained from the
online database GEO. GEO2R analysis was performed to identify the
DEGs associated with pancreatic cancer (13). Further enrichment analysis of DEGs
was applied to explore the molecular mechanisms associated with
pancreatic cancer. The core genes in the development of pancreatic
cancer were then explored through the analysis of differential
gene-protein networks and sub-network modules. Overexpression of
integrin subunit α 2 (ITGA2) and ITGB6 was determined to be
associated with poor prognosis. Silencing of ITGB6 inhibited cell
proliferation in pancreatic cancer and produced cell cycle arrest
at G2/M phase.
Materials and methods
Screening of DEGs from GEO
datasets
The GEO database is an international public database
of datasets, including data from single- and dual-channel
determination of mRNA expression and experimental data for genomic
DNA and proteins (14). In the
present study, three expression profiling datasets
[GSE28735(15), GSE16515(16) and GSE15471(17)] were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/). GSE15471
contains 39 tumor and paired adjacent normal tissues; GSE16515
contains 36 tumor and 16 adjacent normal tissues; and GSE28735
contains 45 tumor and paired adjacent normal tissues. GEO2R
(13) was used to screen for DEGs,
and those DEGs shared by the three sets of expression profiles were
further selected by the Venn mapping tool. A log2 fold change
>1.5 and adjusted P<0.05 were considered to indicate a
statistically significant difference.
Database for Annotation, Visualization
and Integrated Discovery database enrichment analysis
Cluster Profiler is an ontology-based R package that
automates the process of biological-term classification and the
enrichment analysis of gene clusters, and provides a visualization
module for displaying the analysis results (18). DEGs were subjected to Gene Ontology
(GO) enrichment analysis [molecular function (MF), biological
process (BP) and cellular component (19,20)]
and signaling pathway Kyoto Encyclopedia of Genes and Genomes
(KEGG) enrichment analysis using Cluster Profiler V3.6.0. The
enrichment analysis and function annotation data were obtained and
displayed in the form of a bubble chart. P<0.05 was considered
to indicate a statistically significant difference.
Protein interaction network
construction and sub-network module analysis
The Search Tool for the Retrieval of Interacting
Genes/proteins (STRING; https://string-db.org/) is an online tool for
searching for gene interactions and protein interactions (21). Selected DEGs were inputted into the
online database STRING to generate a protein network diagram, which
was visualized by Cytoscape v3.7.0 software (https://cytoscape.org/). Degree was used as the
criterion for screening key target genes (where the degree of nodes
indicates the number of proteins that the nodes are able to
interact with). The sub-network modules related to pancreatic
cancer development were analyzed using MCODE plug-in.
Gene Expression Profiling Interactive
Analysis (GEPIA) online survival analysis
GEPIA is a newly developed interactive web server
for analyzing the RNA sequencing expression data of 9,736 tumors
and 8,587 normal samples from The Cancer Genome Atlas (TCGA) and
the GTEx projects using a standard processing pipeline. It is able
to perform survival and correlation analyses of DEGs (22). In the present study, candidate key
genes were incorporated into the GEPIA database to further verify
their expression in normal pancreatic and pancreatic cancer
tissues. To produce the survival curves for key genes, the genes to
be analyzed were searched in the main interface of the GEPIA
database (http://gepia.cancer-pku.cn/index.html). Subsequently,
‘Survival Plots’ was selected in the analysis toolbar, the tumor
type was set to PAAD and the confidence interval was set to 95%.
For the other parameters, the database's default settings were
used.
Screening of cell lines for expression
of ITGb6 by reverse transcription-quantitative (RT-q)PCR
A total of six pancreatic cancer cell lines (BXPC-3,
CFPAC-1, MIA PaCa-2, ASPC-1, PANC-1 and SW1990) were provided by
Shanghai GeneChem Co., Ltd. and cultured in RPMI-1640 basic medium
(Corning, Inc.). All cells were routinely subcultured at 37˚C in
the presence of 5% CO2 in an incubator with saturated
humidity. Total RNA was extracted from the six cell lines using
TRIzol reagent according to the manufacturer's instructions. RNA
was reverse transcribed to complementary DNA using Promega M-MLV at
42˚C (Promega Corp.). The mRNA expression levels of the ITGB6 gene
in different cell lines of interest were detected by quantitative
PCR using a LightCycler 480 II (Roche Molecular Systems, Inc.). The
composition of the reaction mixture was SYBR premix ex taq 6.0 µl,
primer mix 0.3 µl, reverse transcription product 0.6 µl and
RNase-free H2O 5.1 µl. The reaction conditions were as
follows pre-denaturation at 95˚C for 30 sec, followed by
denaturation for 5 sec at 95˚C and annealing for 30 sec at 60˚Cfor
a total of 40 cycles. The primer sequences were as follows: ITGB6
forward, 5'-TGATCTTCGCTGTAACCC-3' and reverse,
5'-CAGACCGCAGTTCTTCATA-3'; GAPDH forward,
5'-TGACTTCAACAGCGACACCCA-3' and reverse,
5'-CACCCTGTTGCTGTAGCCAAA-3'. The experimental results were analyzed
by the 2-∆∆Cq method (23) for relative quantitative
analysis.
Lentivirus (LV) transfection to
silence ITGB6 expression in the ASPC-1 cell line
Negative control (NC) virus CON077 and LV-ITGB6-RNA
interference (RNAi) LV were constructed by Shanghai Jikai. The
ITGB6-small interfering (si)RNA target sequence designed for the
ITGB6 gene sequence was 5'-gcCTCCAAACATTCCCATGAT-3'. The CON077
sequence was 5'-TTCTCCGAACGTGTCACGT-3'. ASPC-1 cells with
relatively high expression were selected for transfection and the
following experimental groups were established: i) Mock group,
normal ITGB6 cells; ii) Short hairpin (sh)NC group, ASPC-1 cells
transfected with recombinant LV carrying NC-siRNA; iii) shITGB6
group, ASPC-1 cells transfected with recombinant LV carrying
ITGB6-siRNA. Total RNA was extracted from cells in the shNC and
shITGB6 groups using Total RNA extraction reagent (Shanghai Pufei)
according to the manufacturer's instructions. and RT-qPCR was used
to detect the expression levels of ITGB6 mRNA in the cells.
Cell proliferation assays
Cells transfected for 24 h were inoculated into
96-well plates and cultured for a further 24 h before MTT (Shanghai
Dingguo Biotechnology Co., Ltd.) solution (20 µl at 5 g/l) was
added to each well. After incubation at 37˚C for 4 h, the
supernatant was discarded and dimethyl sulfoxide was added to each
well (100 µl). The absorbance of each well was determined at a
wavelength of 490 nm.
Cell cycle detection
The cells from the different experimental groups
were digested with trypsin and centrifuged to collect the cells.
After washing with D-Hanks' solution (pH 7.2) pre-cooled at 4˚C,
cells were fixed with 75% ethanol at 4˚C for 1 h. The cell cycle
was detected by flow cytometry, as previously described (24).
Statistical analysis
Values are expressed as the mean ± standard
deviation and all experiments were repeated independently three
times. Statistical analyses were performed with SPSS 24.0
statistical software (IBM Corp.). Graphs and curves were
constructed by GraphPad Prism 7 (GraphPad Software, Inc.). An
independent-samples t-test was used to assess differences between
paired samples. One-way ANOVA was used for comparison between
groups with the least-significant difference method used for
pairwise comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEGs in PAAD
In the present study, the GEO2R analysis platform
was used to preprocess and filter the original data of three
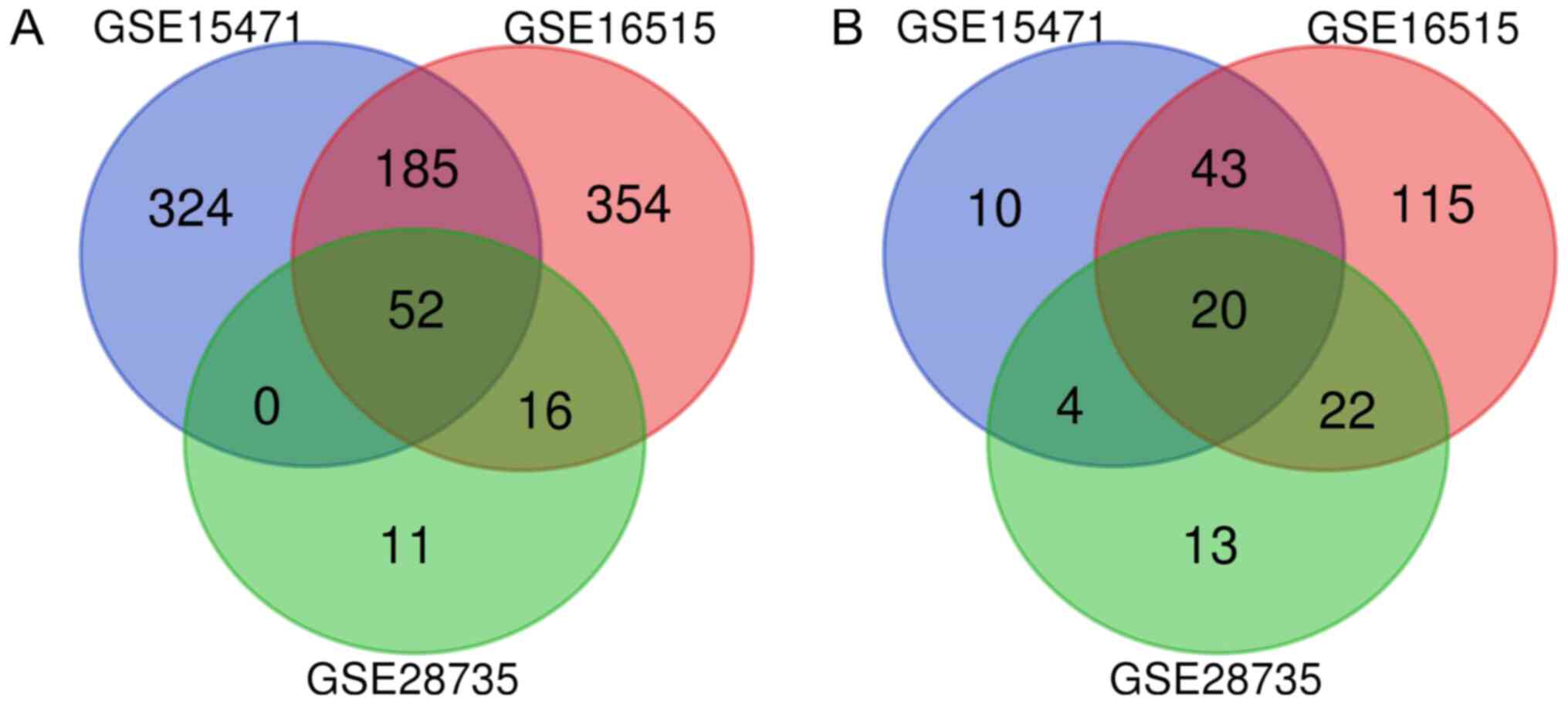
selected datasets (GSE28735, GSE16515 and GSE15471). The Venn
diagram (Fig. 1) contained a total
of 72 genes with an intersection in the three datasets, including
20 downregulated and 52 upregulated genes (Table I).
 | Table IDifferentially expressed genes
(n=72). |
Table I
Differentially expressed genes
(n=72).
| Item | Gene names |
|---|
| Downregulated
genes | ALB, ANPEP, AQP8,
CELA2B, EGF, ERO1B, ERP27, FGL1, GP2, KIAA1324, KLK1, PAIP2B,
PDIA2, PDK4, PNLIPRP1, PNLIPRP2, RBPJL, SERPINI2, TMED6, TRHDE |
| Upregulated
genes | AGR2, AHNAK2, ANLN,
ANTXR1, ANXA10, CEACAM5, CEACAM6, CEMIP, CLDN18, COL10A1, COL11A1,
COL12A1, COL1A1, CP, CST1, CTRL, CTSE, CXCL5, DPCR1, EDIL3, FERMT1,
FN1, FXYD3, GABRP, GATM, INHBA, ITGA2, ITGB6, KRT17, KRT19, KRT7,
LAMB3, LAMC2, MMP11, MMP12, NOX4, NR5A2, PLAC8, POSTN, SDR16C5,
SERPINB5, SLC6A14, SLPI, SULF1, TCN1, TFF1, THBS2, TMC5, TMPRSS4,
TSPAN1, VCAN, VSIG1 |
Differential gene enrichment
analysis
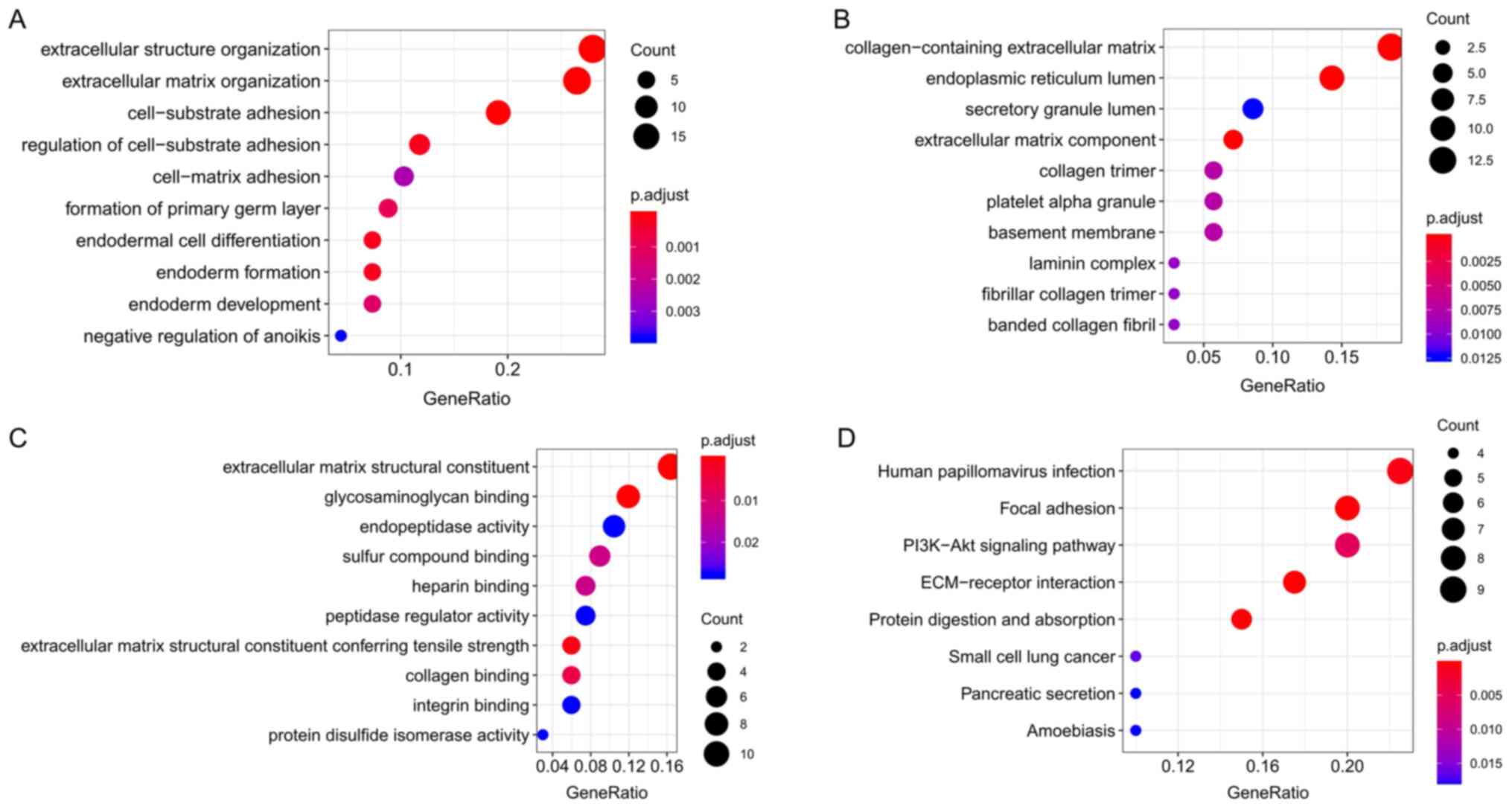
In order to further study the role of DEGs in the
development of pancreatic cancer, Cluster Profiler was used to
subject the DEGs to GO functional enrichment and KEGG pathway
analysis. GO analysis indicated that the DEGs mainly act on the
extracellular matrix (ECM); are involved in BPs such as
cell-substrate adhesion, extracellular structure organization and
ECM organization; and participate in the composition of the
collagen-containing ECM and endoplasmic reticulum lumen. Other MFs
such as ECM structural constituent, endopeptidase activity and
glycosaminoglycan binding were also determined (Table II; Fig.
2A-C). Through KEGG signaling pathway analysis, it was
indicated that the DEGs are mainly involved in human papillomavirus
infection, ECM-receptor interaction, focal adhesion, PI3K-Akt
signaling pathway, protein digestion and absorption, and pathways
in pancreatic secretion (Table
III; Fig. 2D).
 | Table IIGO enrichment analysis of the
differentially expressed genes. |
Table II
GO enrichment analysis of the
differentially expressed genes.
| Category | Term | Count | P-value | Exemplary
genes |
|---|
| BP |
GO:0030198~extracellular matrix
organization | 12 |
3.55x10-10 | LAMB3, ERO1B,
ITGB6 |
| BP | GO:0007155~cell
adhesion | 12 |
2.12x10-6 | LAMB3, ITGB6,
FERMT1 |
| BP |
GO:0035987~endodermal cell
differentiation | 5 |
4.02x10-6 | INHBA, LAMB3,
COL12A1 |
| BP | GO:0030574~collagen
catabolic process | 6 |
5.94x10-6 | COL1A1,MMP12,
COL11A1 |
| BP |
GO:0022617~extracellular matrix
disassembly | 5 |
2.53x10-4 | LAMB3, LAMC2,
FN1 |
| CC |
GO:0005615~extracellular space | 24 |
4.85x10-10 | CXCL5, CST1,
POSTN |
| CC |
GO:0005576~extracellular region | 22 |
3.63x10-7 | PNLIPRP1, PNLIPRP2,
CXCL5 |
| CC |
GO:0070062~extracellular exosome | 27 |
7.40x10-6 | FXYD3, TSPAN1,
KIAA1324 |
| CC |
GO:0031012~extracellular matrix | 9 |
1.74x10-5 | COL12A1, SLPI,
POSTN |
| MF |
GO:0005509~calciumion binding | 10 | 0.001453 | PNLIPRP1, SULF1,
ANXA10 |
| MF |
GO:0004252~serine-type endopeptidase
activity | 6 | 0.002791 | CELA2B, KLK1,
MMP12 |
| MF | GO:0003756~protein
disulfide isomerase activity | 3 | 0.003112 | ERO1B, ERP27,
PDIA2 |
| MF | GO:0005178~integrin
binding | 4 | 0.007401 | ITGB6, ITGA2,
EDIL3, FN1 |
 | Table IIIKyoto Encyclopedia of Genes and
Genomes pathways significantly enriched by the differentially
expressed genes. |
Table III
Kyoto Encyclopedia of Genes and
Genomes pathways significantly enriched by the differentially
expressed genes.
| Term | Count | P-value | Exemplary
genes |
|---|
|
hsa04512:ECM-receptor interaction | 8 |
1.06x10-7 | LAMB3, ITGB6,
ITGA2 |
| hsa04510:Focal
adhesion | 9 |
3.20x10-6 | LAMB3, ITGB6,
ITGA2 |
| hsa04151:PI3K-Akt
signaling pathway | 6 |
1.34x10-4 | LAMB3, ITGB6,
ITGA2 |
| hsa04974:Protein
digestion and absorption | 9 |
4.69x10-5 | COL12A1, CELA2B,
COL1A1 |
| hsa04972:Pancreatic
secretion | 4 | 0.00893 | PNLIPRP2, CELA2B,
CTRL |
Protein network and sub-network module
analysis
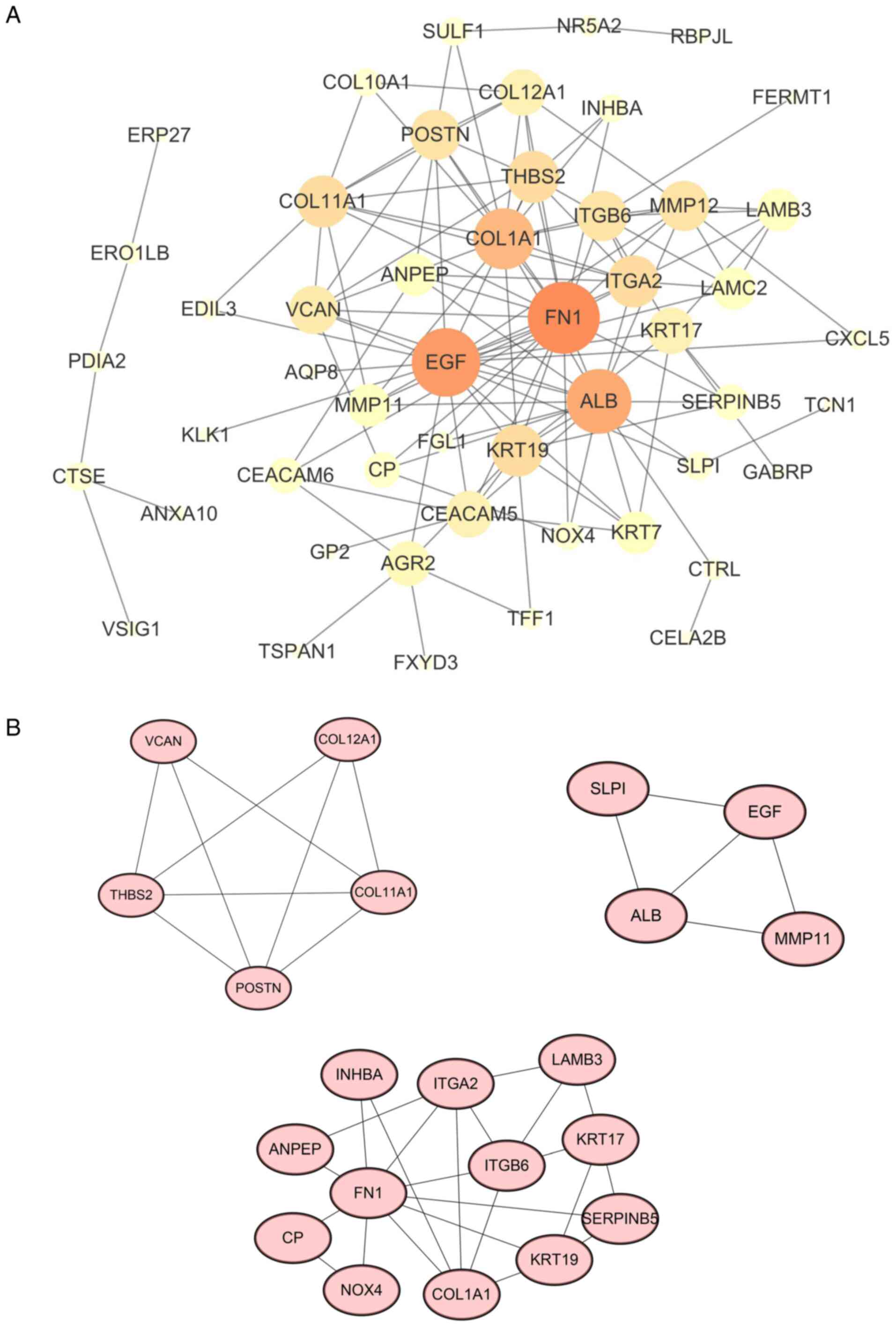
A protein-protein interaction (PPI) network for DEGs
was constructed using the STRING database (Fig. 3A). The first 10 nodes [fibronectin
(FN)1, EGF, albumin (ALB), collagen α1 chain 1 (COL1A1), integrin
subunit α2(ITGA2), keratin 19(KRT19), collagen type XI α 1 chain
(COL11A1), thrombospondin 2 (THBS2), integrin subunit β6 (ITGB6)
and matrix metallopeptidase 12 (MMP12)] with the highest degrees
were screened as hub genes. In order to further explore the
association in the PPI network, the first three modules in the PPI
network were extracted and certain key genes [periostin (POSTN),
matrix metallopeptidase 11 (MMP11) and KRT19] were indicated to
have local regulatory roles in the development and progression of
pancreatic cancer (Fig. 3B). This
provides additional grounds for studying the molecular mechanisms
of pancreatic cancer.
Online survival analysis through
GEPIA
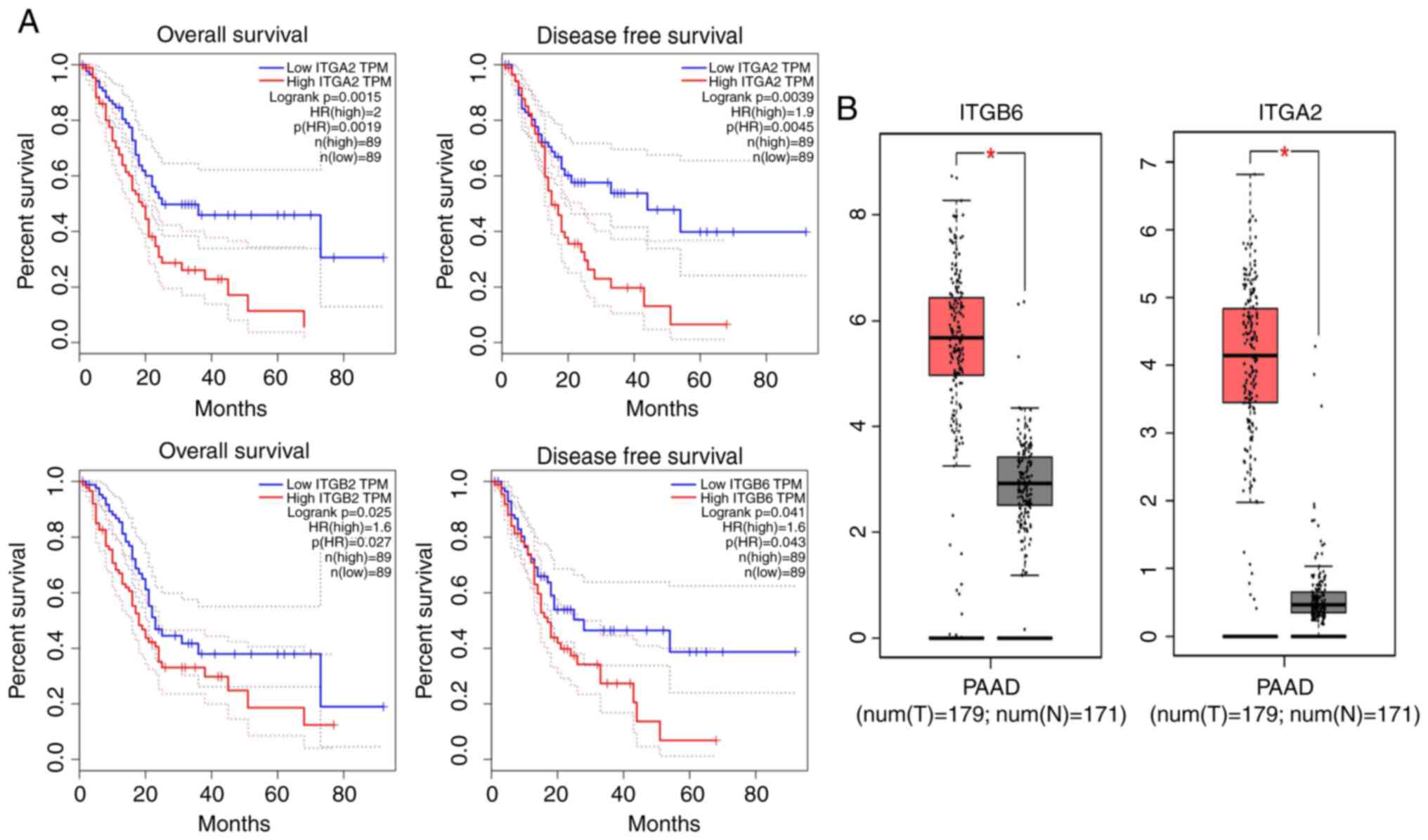
To explore the association between the expression of
key genes and the prognosis of patients with pancreatic cancer, the
key genes were inputted into the GEPIA database for survival
analysis. It was indicated that the expression levels of the ITGB6
and ITGA2 genes are closely associated with the patients' survival
rate (Fig. 4A). This means that
these genes have a negative impact on the survival time of patients
with pancreatic cancer. Finally, using the GEPIA online database,
it was verified that these genes were highly expressed in
pancreatic cancer (Fig. 4B).
Expression of ITGB6 in various cell
lines
As indicated by the RT-qPCR results, the expression
levels of the gene ITGB6 were low in MIA PaCa-2 and PANC-1 cells,
but high in BXPC-3, CFPAC-1, ASPC-1 and SW1990 cells (Fig. 5A). Therefore, ASPC-1 cells were
selected for the subsequent experiments.
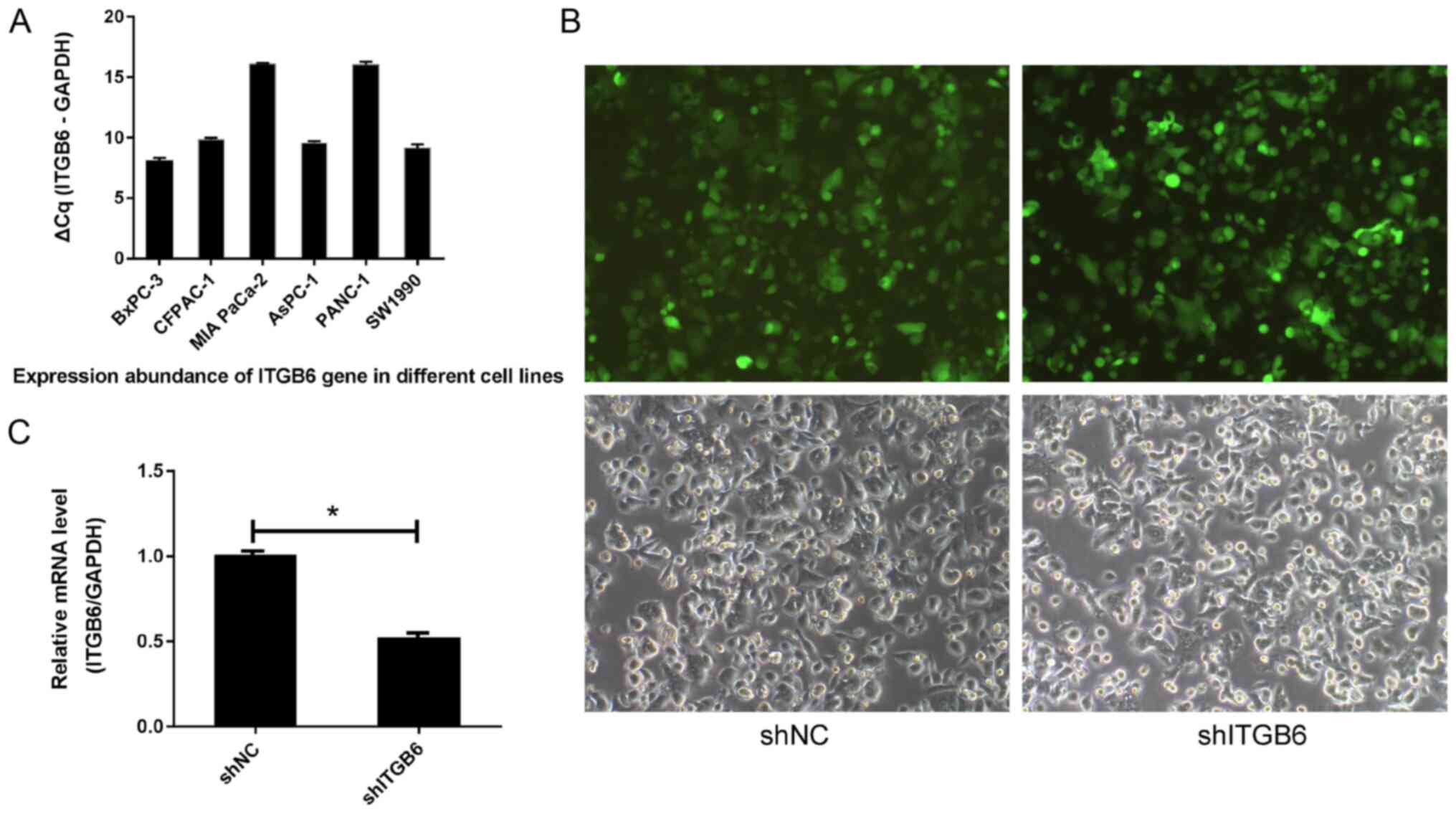 | Figure 5(A) Expression abundance of the ITGB6
gene in different cell lines. When the ∆Cq value was ≤12, the gene
expression abundance in the cell was high; when the ∆Cq value was
≥16, the gene expression abundance in the cell was low. (B)
Recombinant lentivirus infection image in ASPC-1 cells (upper
panels, bright field; lower panels, green fluorescence field;
magnification, x100). (C) Results of the reverse
transcription-quantitative PCR analysis of ITGB6.
*P<0.05. ITGA6, integrin subunit β 6; shNC, negative
control shRNA; shITGB6, shRNA targeting ITGB6; shRNA, short hairpin
RNA; Cq, quantification cycle. |
Results of LV-ITGB6-RNAi lentivirus
infection in AsPC-1 cells
Observation of the green fluorescent protein encoded
in the plasmid with ITGB6 under a fluorescence microscope confirmed
that 24 h after AsPC-1 cells had been infected with the recombinant
virus LV-ITGB6-RNAi, the expression of green fluorescent protein
was observed in both groups of cells (Fig. 5B). The transfection efficiency
detected by flow cytometry was >80%. RT-qPCR analysis
demonstrated that the ITGB6 silencing effect of the plasmid was
evident 72 h after infection (P<0.05; Fig. 5C).
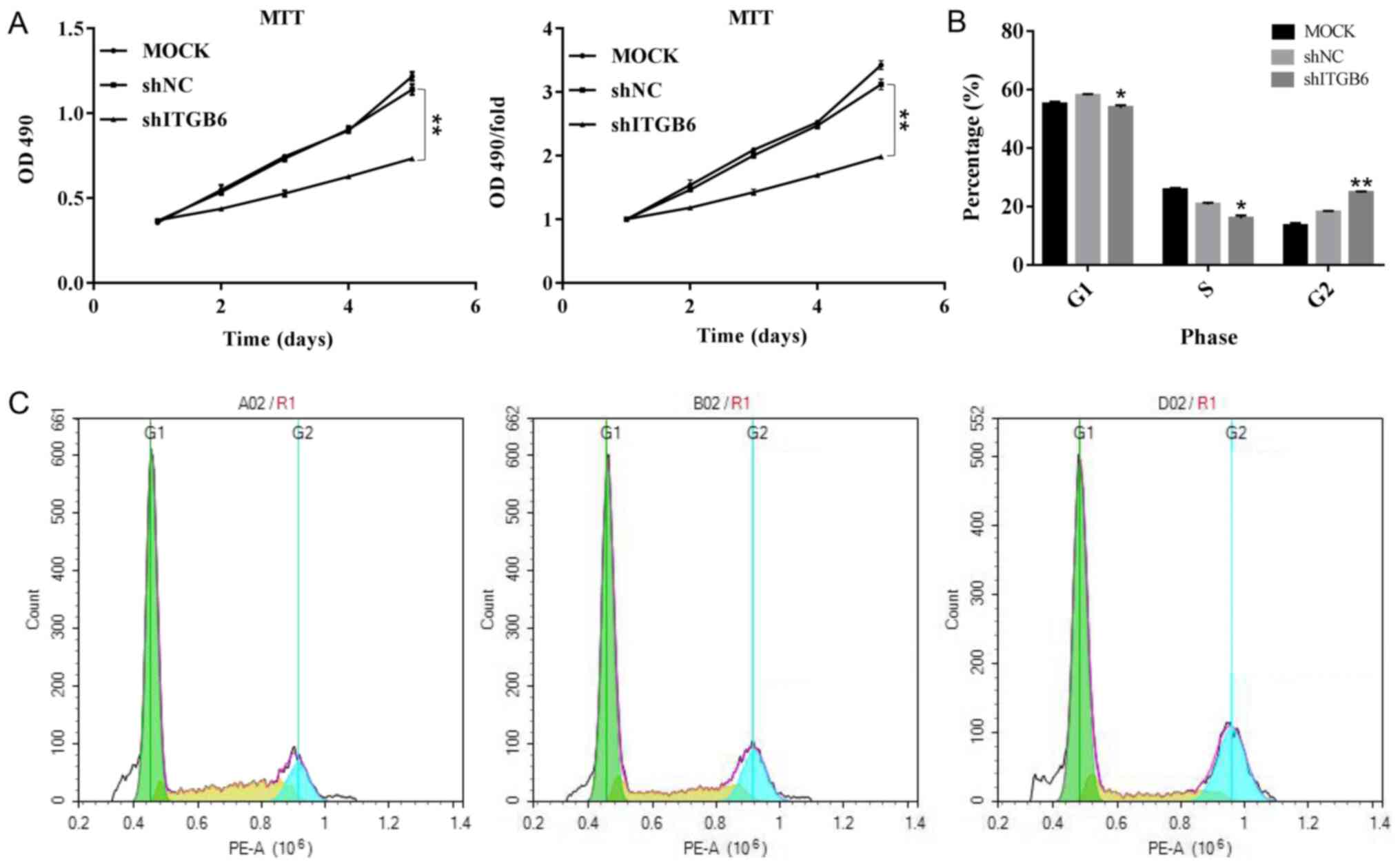
Effect of ITGB6 on the proliferation
of pancreatic cancer ASPC-1 cells
According to the results of the growth curve
analysis, compared with that of the shNC group, the growth curve of
the shITGB6 group was flatter and the overall growth rate of cells
was significantly reduced. From the fourth day onwards, the
proliferation rate of cells in the experimental group was
significantly lower than that of cells in the control group and the
difference was statistically significant (P<0.01). It was
indicated that knockdown of ITGB6 significantly inhibited the
proliferation of ASPC-1 cells (Fig.
6A).
Effect of ITGB6 on the cell cycle of
pancreatic cancer ASPC-1 cells
Flow cytometric analysis of the cell cycle with PI
indicated that, compared with that of the shNC group, the
percentage of cells in G1- and S-phase in the shITGB6 group was
significantly reduced (P<0.05), while the percentage of cells in
G2/M-phase was significantly increased (P<0.01). This suggested
that silencing of ITGB6 gene expression causes cell-cycle arrest in
the G2/M phase, thus significantly inhibiting the cell cycle
(Fig. 6B and C).
Discussion
Pancreatic cancer is a highly malignant tumor type
of the digestive tract (25). Due
to the lack of specific clinical manifestations in the early stage
of the disease, >80% of patients are diagnosed at an advanced
stage, where treatment becomes markedly difficult (26). Revealing the molecular mechanisms
involved in the development of pancreatic cancer will help to
discover novel tumor markers that may allow for an early diagnosis
of pancreatic cancer, develop novel effective treatment strategies,
evaluate prognosis and improve patient survival. In the present
study, datasets of pancreatic cancer-related chips from the GEO
database were analyzed and 72 significant DEGs were mined.
GO functional analysis indicated that the DEGs are
mainly involved in BPs such as cell adhesion and ECM decomposition.
KEGG signaling pathway analysis revealed the involvement of focal
adhesion, ECM-receptor interaction, PI3K-Akt signaling pathway and
protein digestion and absorption. Of these, the PI3K-Akt signaling
pathway is particularly noteworthy, as it is known to be an
important oncogenic signaling pathway involved in tumorigenesis and
resistance to targeted anticancer therapies for various tumor types
(27). The PI3K-Akt signaling
pathway is also an important cause of pancreatic cancer and is
associated with poor prognosis of pancreatic cancer. Abnormal
activation of this pathway involves cellular metabolism and
survival and cell cycle progression. Several inhibitors targeting
Akt, PI3K and mTOR have recently been developed for clinical
research (28).
In the present study, the STRING online tool was
used to analyze the PPIs encoded by the DEGs associated with
pancreatic cancer. It was determined that the interactions between
the proteins encoded by these genes were mainly concentrated in key
node genes such as FN1, EGF, ALB, ITGA2 and ITGB6. FN1 exhibited
the highest connectivity in the PPI network, suggesting its
important role in pancreatic cancer. A recent study (29) revealed that FN1 was abundantly
expressed in the tumor microenvironment of PAAD, which is
consistent with the present study, demonstrating that FN1 is highly
expressed in PAAD, while its expression in normal pancreatic tissue
was low or marginal. Expression of FN1 matrix was associated with
aggressive tumor characteristics, including greater tumor size and
advanced T and N stages.
ALB and EGF were the only downregulated genes among
the hub genes. The ALB gene, which encodes the most abundant
protein in human blood, was a downregulated gene in pancreatic
cancer. The protein regulates plasma colloid osmotic pressure and
serves as a carrier protein for a variety of endogenous molecules
and exogenous drugs. A recent study indicated that the combined
detection of the derived neutrophil-to-lymphocyte ratio and ALB are
able to improve the diagnostic efficiency for pancreatic cancer
(30). Another study demonstrated
the C-reactive protein (CRP)/ALB ratio may serve as a significant
and promising inflammatory prognostic score in pancreatic cancer,
since high CRP/Alb indicates a poor prognosis (31). Combined with the results of the
present study, this suggests that the detection of ALB levels in
patients with pancreatic cancer may improve the sensitivity of
pancreatic cancer diagnosis. EGF was indicated to be significantly
downregulated in the present study, which is inconsistent with the
results of Hao et al (32).
A study with a larger sample of patients is required for further
validation.
Furthermore, the present study detected novel genes
involved in the local regulation of the development of pancreatic
cancer, including POSTN, MMP11 and KRT19. PPI module analysis
indicated the important role of these central genes, which are
involved in key pathways and BPs associated with pancreatic
cancer.
COL1A1 is a key structural component of the ECM. It
occurs in the majority of connective and embryonic tissues, and is
an important member of the collagen family (33). Typically, type I collagen consists
of COL1A1 and COL1A2(34). Abnormal
expression of COL1A1 has been reported in kidney cancer,
hepatocellular carcinoma and melanoma (35-37).
Li et al (38) indicated
that COL1A1 and COL1A2 were overexpressed in gastric cancer and
high COL1A1 may be a monitoring factor for early gastric cancer or
a prognostic factor for predicting overall survival. COL1A1
secreted by pancreatic stellate cells (PSCs) promotes invasion and
migration of pancreatic cancer cells (39). A previous study (40) has indicated that inhibition of
COL1A1 with the novel hydrophilic agent palmatine (PMT) is able to
inhibit the growth of PSCs. The role of collagen family members in
pancreatic cancer deserves further investigation.
Improving the prognosis of patients with pancreatic
cancer is an urgent problem to be solved. In the present study, the
key genes selected from pancreatic cancer samples in TCGA database
were analyzed and the results suggested that high expression of the
ITGA2 and ITGB6 genes is a high-risk factor for poor prognosis in
patients with pancreatic cancer. This evaluation of prognosis has
obvious clinical significance and future studies focusing on these
genes may contribute to the treatment of pancreatic cancer. ITGA2
is an essential collagen receptor on epithelial cells. Gong et
al (41) suggested that changes
in UCA1 (urothelial cancer associated 1) Expression may affect the
expression of ITGA2, further interfering with the progression of
cancer. The adhesion spot pathway was identified as the regulatory
mechanism of ITGA2. The expression profile of the long non-coding
RNA UCA1 was associated with the migration and apoptosis of SW-1990
cells (42). Specifically,
upregulated co-expression of UCA1-ITGA2 promoted the migration and
invasion of pancreatic cancer cells. Thus, ITGA2 may become a novel
potential target for gene therapy.
The mechanisms of ITGB6 in cancer have remained
largely elusive and studying the TGF-β/ITGB6 signaling pathway may
be worthwhile. ITGB6 has a role in signal transmission from the ECM
to cells. TGF-β is an important inflammatory factor produced by
macrophages, stromal cells and tumor cells in the tumor
microenvironment and is involved in the occurrence, development and
metastasis of tumors. A previous study (43) indicated that the TGF-β/ITGB6
signaling pathway has an important role in the invasion and
metastasis of esophageal squamous cell carcinoma and the present
study suggested that this pathway may be inhibited by miR-17/20a. A
recent bioinformatics study on pancreatic cancer used ITGB6 as an
independent risk factor for the prognosis of pancreatic cancer,
which also supports the present results (44). However, the function and mechanism
of ITGB6 in pancreatic cancer are unclear and further studies
should be performed to detect the levels of this gene and confirm
its role in PAAD. The present study suggested that the expression
of ITGB6 mRNA was high in the majority of pancreatic cancer cell
lines evaluated, including BXPC-3, CFPAC-1, ASPC-1 and SW1990. On
the contrary, according to the RT-qPCR results, the expression of
ITGB6 in the MIA PaCa-2 and PANC-1 cell lines was lower than that
in the other four cell lines. Infinite cell proliferation caused by
imbalances in various stages of the cell cycle is closely
associated with formation of tumors (45). In the cellular functional
experiments of the present study, the role of ITGB6 as an oncogene
was confirmed. Through functional analyses with silencing of ITGB6
and determination of the cell proliferation and cell cycle
distribution in ASPC-1 cells, it was indicated that inhibition of
ITGB6 decreased cell proliferation and induced G2/M arrest, which
supports the results of the present bioinformatics analysis. The
effect of silencing the ITGB6 gene on the cell cycle may be a
potential mechanism for inhibition of further progression of
pancreatic tumors. The above results strongly supported the
possibility of ITGB6 as an optimal target for therapeutic
intervention. Detection of the cell cycle, particularly the
expression of G2/M phase-related regulatory proteins, will be the
focus of future studies by our group. At the same time, the study
still has certain limitations, such as the lack of western blot
data. Further research on the impact of ITGB6 on pancreatic cancer
cell invasion and migration, and the potential molecular mechanism
of ITGB6 in pancreatic cancer, require to be further explored.
In conclusion, the present study employed a series
of bioinformatics methods to identify key genes in pancreatic
cancer. ITGA2 and ITGB6 may be used as potential biomarkers for the
diagnosis and treatment of patients with pancreatic cancer. The
DEGs and metabolic pathways revealed in the present study may help
us understand the mechanisms of the molecular development of
pancreatic cancer and provide a theoretical basis for future
research on clinical targeted therapies. Analytical data mining
through bioinformatics analysis is a feasible method to
systematically identify potential biomarkers. However, the
molecular mechanisms of pancreatic cancer require to be further
investigated through biological experiments.
Acknowledgements
Not applicable.
Funding
The present study was supported by Zhenjiang Science and
Technology Committee (grant. no. SH 2019061).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XX and TR were responsible for designing the study,
performing the experiment, collecting the data and writing the
manuscript. XW, XZ and SD were responsible for designing the study,
performing the experiment, and collecting the data and reviewing
the manuscript. SD was responsible for providing experimental ideas
and reviewing the manuscript. XX and TR were responsible for the
confirming the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All animal experimentation protocols were approved
by the Institutional Animal Care and Use Committees of Jiangsu
University and were conducted according to the Regulation on Animal
Experimentation at Jiangsu University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rizzato C, Campa D, Talar-Wojnarowska R,
Halloran C, Kupcinskas J, Butturini G, Mohelníková-Duchoňová B,
Sperti C, Tjaden C, Ghaneh P, et al: Association of genetic
polymorphisms with survival of pancreatic ductal adenocarcinoma
patients. Carcinogenesis. 37:957–964. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Vincent A, Herman J, Schulick R, Hruban RH
and Goggins M: Pancreatic cancer. Lancet. 378:607–620.
2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Halbrook CJ and Lyssiotis CA: Employing
metabolism to improve the diagnosis and treatment of pancreatic
cancer. Cancer Cell. 31:5–19. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Griffin JF, Poruk KE and Wolfgang CL:
Pancreatic cancer surgery: Past, present, and future. Chin J Cancer
Res. 27:332–348. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Maitra A, Kern SE and Hruban RH: Molecular
pathogenesis of pancreatic cancer. Best Pract Res Clin
Gastroenterol. 20:211–226. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Long J, Liu Z, Wu X, Xu Y and Ge C: Gene
expression profile analysis of pancreatic cancer based on
microarray data. Mol Med Rep. 13:3913–3919. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Morris JP IV, Wang SC and Hebrok M: KRAS,
hedgehog, wnt and the twisted developmental biology of pancreatic
ductal adenocarcinoma. Nat Rev Cancer. 10:683–695. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Lauth M and Toftgård R: Hedgehog signaling
and pancreatic tumor development. Adv Cancer Res. 110:1–17.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shigeyasu K, Toden S, Zumwalt TJ, Okugawa
Y and Goel A: Emerging role of MicroRNAs as liquid biopsy
biomarkers in gastrointestinal cancers. Clin Cancer Res.
23:2391–2399. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cui Z, Liu G and Kong D: miRNA27a promotes
the proliferation and inhibits apoptosis of human pancreatic cancer
cells by wnt/β-catenin pathway. Oncol Rep. 39:755–763.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhao DW, Hou YS, Sun FB, Han B and Li SJ:
Effects of miR-132 on proliferation and apoptosis of pancreatic
cancer cells via hedgehog signaling pathway. Eur Rev Med Pharmacol
Sci. 23:1978–1985. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995.
2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhang G, Schetter A, He P, Funamizu N,
Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, et al:
DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity
and predicts clinical outcome in pancreatic ductal adenocarcinoma.
PLoS One. 7(e31507)2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
18
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gene Ontology Consortium. The gene
ontology (GO) project in 2006. Nucleic Acids Res. 34:D322–D326.
2006.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Krishan A: Rapid flow cytofluorometric
analysis of mammalian cell cycle by propidium iodide staining. J
Cell Biol. 66:188–193. 1975.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ilic M and Ilic I: Epidemiology of
pancreatic cancer. World J Gastroenterology. 22:9694–9705.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Courtney KD, Corcoran RB and Engelman JA:
The PI3K pathway as drug target in human cancer. J Clin Oncol.
28:1075–1083. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ebrahimi S, Hosseini M, Shahidsales S,
Maftouh M, Ferns GA, Ghayour-Mobarhan M, Hassanian SM and Avan A:
Targeting the Akt/PI3K signaling pathway as a potential therapeutic
strategy for the treatment of pancreatic cancer. Curr Med Chem.
24:1321–1331. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hu D, Ansari D, Zhou Q, Sasor A,
Hilmersson KS and Andersson R: Stromal fibronectin expression in
patients with resected pancreatic ductal adenocarcinoma. World J
Surg Oncol. 17(29)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liu JX, Li A, Zhou LY, Liu XF, Wei ZH,
Wang XZ and Ying HQ: Significance of combined preoperative serum
Alb and dNLR for diagnosis of pancreatic cancer. Future Oncol.
14:229–239. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu Z, Jin K, Guo M, Long J, Liu L, Liu C,
Xu J, Ni Q, Luo G and Yu X: Prognostic value of the CRP/Alb ratio,
a novel inflammation-based score in pancreatic cancer. Ann Surg
Oncol. 24:561–568. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hao C, Li Z, Zhang X, Zhang H, Shang H,
Bao J and Wang H: Expression and clinical significance of EGF and
TGF-alpha in chronic pancreatitis and pancreatic cancer. Minerva
Endocrinol. 43:253–258. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Cole WG: Collagen genes: Mutations
affecting collagen structure and expression. Prog Nucleic Acid Res
Mol Biol. 47:29–80. 1994.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Exposito JY, Valcourt U, Cluzel C and
Lethias C: The fibrillar collagen family. Int J Mol Sci.
11:407–426. 2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
de Caceres II, Dulaimi E, Hoffman AM,
Al-Saleem T, Uzzo RG and Cairns P: Identification of novel target
genes by an epigenetic reactivation screen of renal cancer. Cancer
Res. 66:5021–5028. 2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hayashi M, Nomoto S, Hishida M, Inokawa Y,
Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S,
et al: Identification of the collagen type 1 α 1 gene (COL1A1) as a
candidate survival-related factor associated with hepatocellular
carcinoma. BMC Cancer. 14(108)2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bonazzi VF, Nancarrow DJ, Stark MS, Moser
RJ, Boyle GM, Aoude LG, Schmidt C and Hayward NK: Cross-platform
array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as
genes frequently silenced by methylation in melanoma. PLoS One.
6(e26121)2011.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li J, Ding Y and Li A: Identification of
COL1A1 and COL1A2 as candidate prognostic factors in gastric
cancer. World J Surg Oncol. 14(297)2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ikenaga N, Ohuchida K, Mizumoto K, Akagawa
S, Fujiwara K, Eguchi D, Kozono S, Ohtsuka T, Takahata S and Tanaka
M: Pancreatic cancer cells enhance the ability of collagen
internalization during epithelial-mesenchymal transition. PLoS One.
7(e40434)2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chakravarthy D, Muñoz AR, Su A, Hwang RF,
Keppler BR, Chan DE, Halff G, Ghosh R and Kumar AP: Palmatine
suppresses glutamine-mediated interaction between pancreatic cancer
and stellate cells through simultaneous inhibition of survivin and
COL1A1. Cancer Lett. 419:103–115. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gong J, Lu X, Xu J, Xiong W, Zhang H and
Yu X: Coexpression of UCA1 and ITGA2 in pancreatic cancer cells
target the expression of miR-107 through focal adhesion pathway. J
Cell Physiol. 234:12884–12896. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen P, Wan D, Zheng D, Zheng Q, Wu F and
Zhi Q: Long non-coding RNA UCA1 promotes the tumorigenesis in
pancreatic cancer. Biomed Pharmacother. 83:1220–1226.
2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Jing C, Ma G, Li X, Wu X, Huang F, Liu K
and Liu Z: MicroRNA-17/20a impedes migration and invasion via
TGF-β/ITGB6 pathway in esophageal squamous cell carcinoma. Am J
Cancer Res. 6:1549–1562. 2016.PubMed/NCBI
|
|
44
|
Wu M, Li X, Zhang T, Liu Z and Zhao Y:
Identification of a nine-gene signature and establishment of a
prognostic nomogram predicting overall survival of pancreatic
cancer. Front Oncol. 9(996)2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu
Y, Qian H and Dai T: LncRNA UCA1 impacts cell proliferation,
invasion, and migration of pancreatic cancer through regulating
miR-96/FOXO3. IUBMB life. 70:276–290. 2018.PubMed/NCBI View Article : Google Scholar
|