Introduction
Pancreatic cancer (PC) results in a relatively low
5-year survival rate (2-9%) and is one of the most fatal cancer
types, ranking as the 7th highest cause of cancer-associated
mortality worldwide (1,2). Moreover, there has been a rapid
increase (1.46 to 7.21 per 100,000) in the occurrence of PC in
China over the past several decades (3,4).
Although the understanding of PC has increased, its prognosis
remains unsatisfactory, mainly due to the lack of specific symptoms
and curative therapies, as well as delayed diagnoses (5,6).
Therefore, the discovery of novel PC biomarkers may be a promising
tool to facilitate the establishment of more accurate and reliable
therapeutic options.
Similar to other types of non-coding RNA molecules
(<22-25 nucleotides), microRNAs (miRNAs/miRs) possess
post-transcriptional functions that can affect the expression
levels of specific genes (7). In
non-small-cell lung cancer (NSCLC), a reduction in miR-361
expression has been observed in tumor cells and tissues, and
overexpression of miR-361 inhibits the proliferative and migratory
abilities of NSCLC cells (8).
miR-361 is considered to directly target the Wilms' tumor 1 (WT1)
gene in NSCLC. Moreover, WT1 knockdown has a similar influence on
miR-361 overexpression in NSCLC cells (8). In esophageal carcinoma tissues, the
expression of GLI family zinc finger 1 (Gli1) is notably increased,
while that of miR-361 is decreased, corresponding with the TNM
stage (9). Additionally, miR-361
reduces Gli1 expression by targeting the 3'-untranslated region
(UTR) of the Gli1 gene (9). With
the transfection of small interfering RNA-Gli1 and/or miR-361
mimics, it has been shown that malignant tumor cell development is
significantly inhibited (9).
Furthermore, miR-361 overexpression and/or Gli1 silencing decreases
the cellular production of N-cadherin, Gli1 and Snail, while
increasing E-cadherin expression, to suppress the
epithelial-mesenchymal transition process and invasive ability of
cancer cells; the opposite effects are achieved via Gli1
overexpression (9). However, the
effect of miR-361 on PC is yet to be elucidated.
The present study assessed the role of miR-361 on PC
progression. The expression of miR-361 was determined in PC cells
and in specimens obtained from patients with PC. Moreover, the
effect of miR-361 overexpression on the proliferation of PC cells,
as well as impaired PC cell migration, were tested in vitro
and in animal models. Therefore, the current study provided
evidence that miR-361 was involved in PC progression in
vitro and in vivo.
Materials and methods
Experimental samples
Patients (n=30; 15 males; 15 females; age range,
40-71 years; median age, 58 years) were selected from
histopathologically and clinically diagnosed patients with PC who
underwent a primary resection at the No. 215 Hospital of the
Shaanxi Nuclear Industry between June 2015 and January 2019. The
inclusion criteria for the current study was patients diagnosed
with PC. Patients who were treated with radiotherapy or
chemotherapy before surgery were excluded. Relevant clinical data
were collected, as presented in Table
I.
 | Table IAssociation of miR-361 expression
with clinicopathologic features of patients with pancreatic
cancer. |
Table I
Association of miR-361 expression
with clinicopathologic features of patients with pancreatic
cancer.
| Characteristic | Low miR-361, n
(%) | High miR-361, n
(%) | P-value |
|---|
| Age, years | | | |
|
≤63 | 8 (26.7) | 10 (33.3) | 0.39 |
|
>63 | 6 (20.0) | 6 (20.0) | |
| Sex | | | |
|
Male | 5 (16.7) | 10 (33.3) | 0.22 |
|
Female | 8 (26.7) | 7 (23.3) | |
| Clinical stage
(TNM) | | | |
|
I-II | 2 (6.7) | 10 (33.3) | 0.04 |
|
III-IV | 15 (50.0) | 3 (10.0) | |
| Lymph node | | | |
|
Negative | 3 (10.0) | 4 (13.3) | 0.66 |
|
Positive | 13 (43.3) | 10 (33.3) | |
| Vascular
infiltration | | | |
|
No | 8 (26.7) | 12 (40.0) | 0.56 |
|
Yes | 6 (20.0) | 4 (13.3) | |
| Neural
infiltration | | | |
|
No | 9 (30.0) | 12 (40.0) | 0.51 |
|
Yes | 3 (10.0) | 6 (20.0) | |
| PanIN | | | |
|
No | 5 (16.7) | 3 (10.0) | 0.27 |
|
Yes | 16 (53.3) | 6 (20.0) | |
| Grading (AJCC) | | | |
|
1-2 | 1 (3.3) | 13 (43.3) | 0.01 |
|
3 | 14 (46.7) | 2 (6.7) | |
| Setting | | | |
|
Metastatic | 5 (16.7) | 15 (50.0) | 0.08 |
|
Adjuvant | 6 (20.0) | 4 (13.3) | |
Healthy non-cancerous pancreatic tissue was obtained
from volunteers at the No. 215 Hospital of the Shaanxi Nuclear
Industry between September 2018 and January 2019 (n=10; 7 males; 3
females; age range, 39-65 years; median age, 53 years, healthy
group) for use in this research. Clinicopathologic classification
and stage of tissues were determined according to the current
National Cancer Center Network (NCCN), Union for International
Cancer Control TNM classification (10). Patients with PC were divided to
three groups, according to the guidelines from the NCCN, as
follows: i) Poor prognosis (n=10); ii) medium prognosis (n=10); and
iii) improved prognosis (n=10) groups. All patients provided
written informed consent. The present research was conducted
according to the Declaration of Helsinki following ethical approval
by the Board of Ethics Review of the No. 215 Hospital of the
Shaanxi Nuclear Industry.
Cell culture, transfection and
lentiviral infection
DMEM (Gibco; Thermo Fisher Scientific, Inc.) was
supplemented with 1% penicillin/streptomycin (Gibco; Thermo Fisher
Scientific, Inc.), 1% glutamine and 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), which was then used to cultivate PC cell line
PANC-1 [American Type Culture Collection (ATCC)] and AsPC-1 (ATCC)
cells. Human pancreatic ductal epithelial cell lines (HPDE6-E6E7
clone 6), immortalized by infection with E6/E7 genes of human
papillomavirus 16, were kindly provided by Dr Haoming-Li (Beijing
Capital University, Beijing, China) and served as normal pancreatic
cells control. Then, 50 nmol/l miR-361 mimic
(5'-UCCCCCAGGUGUGAUUCUGAUUU-3') and a corresponding negative
control mimic (NC group) (5'-TTCTCCGAACGTGTCACGT-3') were purchased
from GenScript Biotech and used to transfect each cell type using
Lipofectamine® RNAiMAX (Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions.
Full-length cDNA for human MAPK/JNK was obtained via
PCR using PrimeSTAR Max DNA polymerase (Takara Biotechnology Co.,
Ltd.). Primer sequences were as follows: Forward,
5'-ATGACTCACGGTCGTAGA-3' and reverse, 5'-GGACCTACGGGTCACTTG-3'.
Thermocycling conditions were as follows: Initial denaturation at
95˚C for 5 min; 35 cycles of 95˚C for 10 sec, 61˚C for 10 sec and
72˚C for 15 sec. Purified PCR products were subsequently cloned
into a pcDNA3 plasmid (GenScript Biotech). The construct was
verified via sequencing by Genscript Biotech and designated as
‘pcDNA3-MAPK/JNK’. PANC-1 and AsPC-1 cells (1x105
cells/well) seeded in a 6-well plate were transfected with 8 µg
pcDNA3-MAPK/JNK or pcDNA3-NC using Lipofectamine® 2000
transfection reagent (Thermo Fisher Scientific, Inc.). At 36 h
post-transfection, subsequent experiments were performed.
MiR-361 mimic oligonucleotides were analyzed and a
scrambled NC sequence served as the NC. The oligonucleotides were
phosphorylated, annealed and cloned into the pLVX-puro vector
(Clontech Laboratories, Inc), designated as ‘pLVX-miR-361’ and
‘pLVX-NC’. Lentiviral-miR-361 and lentiviral-NC particles were
produced via triple transfection of 293T cells (Invitrogen; Thermo
Fisher Scientific, Inc.) using the vectors pLVX-miR-361 and
pLVX-NC, along with 0.4 µg psPAX2 and pMD2.G (GenScript Biotech).
For infection experiments, AsPC-1 cells were incubated with
1x104 cells/ml lentiviral particles (MOI, 8-10) and
polybrene (5 µg/ml) in growth medium. After 6 h, the infection
medium was discarded and the cells were used for subsequent
experimentation.
MTT assay
Cell viability was assessed via MTT assays. Cells
were treated with MTT (0.5 mg/ml; 20 µl; Sigma-Aldrich; Merck
KGaA). The supernatant was removed using a pipette and 150 µl DMSO
was added to the cells. Then, the cell samples were rotated for 10
min to allow the incorporation of formazan dye. An Infinite M200
microplate reader (Tecan Group, Ltd.) was used to measure the
absorbance of the samples at 490 nm.
Apoptosis assay
To characterize apoptotic proportion, PANC-1 and
AsPC-1 cells were collected, washed and resuspended. Following the
addition of 5 µl Annexin V (cat. no. V13242; Thermo Fisher
Scientific, Inc.) and 5 µl propidium iodide (PI; cat. no. V13242;
Thermo Fisher Scientific, Inc.), cells were incubated at room
temperature for 20 min in the dark, washed with PBS and
re-suspended with 300 ml of PBS. Cell apoptosis proportion was
analyzed using a BD FACSCalibur™ flow cytometer (BD Biosciences)
equipped with FACS Diva software (version no. 6.0; BD Biosciences)
to measure the percentages of early and late apoptotic cells.
Transwell invasion assay
After transfection for 24 h, PC cells were
trypsinized and washed with D-Hanks solution (Gibco; Thermo Fisher
Scientific, Inc.). Wells of 24-well plates were divided into an
upper and bottom chamber with Matrigel inserts (pore size, 8 µm).
The cell suspension of PC cells (5x105 cells) in DMEM
was added to the upper chamber. F-12 medium (400 µl) supplemented
with FBS (10%; Thermo Fisher Scientific, Inc.) and hepatocyte
growth factor (20 ng/ml; Thermo Fisher Scientific, Inc.) was added
to the bottom compartment. After incubation for 24 h at 37˚C, cells
that migrated from the upper chamber to the lower one were stained
with crystal violet. Cell migration was evaluated with a light
inverted microscope (Zeiss AG; magnification, x200).
Wound healing assay
Following 36 h of transfection, PANC-1 and AsPC-1
cells at a concentration of 1x106 cells/ml were placed
in 24-well plates and grown to 80% confluence at 37˚C. Using a
sterilized pipette tip, a scratch wound was made. After rinsing
with PBS twice, the floating cells were removed. The original
medium (with 10% FBS) was replaced with fresh medium (with 0% FBS)
and cells were incubated for 0 (control) or 36 h at 37˚C. An
inverted microscope (Nikon Corporation, magnification, x40) and
NIS-Elements (version no. 11.0; Nikon Corporation) was used to
obtain and analyze the images of the cells.
Western blot (WB) analysis
Total protein was extracted from cells using cold
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.). Total protein was quantified using a BCA assay kit (Thermo
Fisher Scientific, Inc.) and 50 µg protein/lane was separated via
12% SDS-PAGE. The resultant protein bands were transferred onto
PVDF membranes and the membranes were blocked with 5% non-fat milk
in Tris-buffered saline with Tween® 20 (Thermo Fisher
Scientific, Inc.) for 2 h at room temperature. After blocking the
membranes, they were incubated with primary and secondary
antibodies for 2 h at room temperature. JNK (1:1,000; cat. no.
ab112501; Abcam), phosphorylated (p-)JNK (1:500; cat. no. ab4821;
Abcam), β-actin (1:5,000; cat. no. ab8227; Abcam), horseradish
peroxidase-conjugated goat anti-rabbit secondary (1:5,000; cat. no.
ab6721; Abcam) antibodies were purchased from Abcam. Protein bands
were visualized using an ECL western blotting kit (Thermo Fisher
Scientific, Inc.) and protein expression was quantified using
Image-Pro Plus software (version no. 6.0; Media Cybernetics,
Inc.).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to obtain total RNA from the
transfected cells or tissues, according to the manufacturer's
protocol. Total RNA (1 µg) was reverse transcribed into cDNA using
the miScript Reverse Transcription kit (Qiagen, Inc.) for
incubation at 37˚C for 30 min and inactivated at 95˚C for 5 min,
according to the manufacturer's protocol. The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 95˚C for 5 min; 40 cycles of 95˚C for 10 sec, 60˚C
for 30 sec and 72˚C for 15 sec. The Roche Light-Cycler 480 RT PCR
System (Roche; Thermo Fisher Scientific, Inc.) and Fast SYBR™ Green
Master Mix (cat. no. 4385617; Thermo Fisher Scientific, Inc.) were
used to determine miR-361 expression. GAPDH was set as the internal
control. SYBR Green PCR Master Mix was used for RT-qPCR. The number
of targets was measured with respect to the calibrator (average
value of the control groups) using the 2-ΔΔCq method
(11). The primers used were as
follows: miR-361 forward, 5'-TTGCATAGTCACAAAAGTGAT-3' and reverse,
5'-CAGTGCGTGTCGTGGAGT-3'; U6 forward, 5'-CTCGCTTCGGCAGCACATATACT-3'
and reverse, 5'-ACGCTTCACGAATTTGCGTGTC-3'; JNK forward,
5'-TTTACTTAGGGGTCATAGGTGAG-3' and reverse,
5'-AGACTCCCGCTCTCCAACAAG-3'; and GAPDH forward,
5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3'.
Target Scan prediction
The prediction algorithm TargetScan was used to
identify miR-361 targets. TargetScan Database predictions (version
no. 7.2; http://www.targetscan.org) are ranked
according to the predicted targeting effectiveness as measured
using the cumulative weighted context + values of the sites
(12). Additionally, conserved
targeting probability can be selected to rank the predictions
(13).
Dual-luciferase reporter assay
(DLRA)
The 3'-UTR luciferase reporter assay was performed
to determine the target gene for miR-361, wherein the mutant and
wild-type (WT) 3'-UTR of MAPK/JNK were inserted in psi-CHECK2
(Genscript). PANC-1 cells were co-transfected with 10 µg miR-361
mimic (or NC mimic) and 4 µg WT-MAPK/JNK-3'UTR plasmid (or
MU-MAPK/JNK-3'UTR plasmid) using Lipofectamine® 2000.
Following incubation for 48 h, reporter luminescence was normalized
using the Renilla luciferase sequence and calibration
luminescence was measured using the firefly luciferase sequence
using a Dual-Luciferase Reporter Assay system (Promega
Corporation).
Animal experiments
The experimental protocol involving animals was
approved by the Animal Ethics Committee of the No. 215 Hospital of
Shaanxi Nuclear Industry. Humane endpoints were set as follows: i)
Tumor burden (combined burden if >1 mass is present) is >15%
body weight; ii) mean tumor diameter ≥20 mm; and iii) tumor
ulceration, infection or necrosis. No mice died during the
experiments. The duration of the experiment was 28 days.
Nude BALB/c-nu mice (n=16; age, 5 weeks; female;
weight, 20-30 g) were purchased from Beijing Vital River Laboratory
Animal Technology Co., Ltd. The mice were housed in groups of four
in the animal facility for the entire experimental period. All mice
were under constant temperature (21˚C) and humidity (45-55%) with
12-h light/dark cycles, with lights on at 7:00 am. Standard food
and filtered water were available ad libitum. AsPC-1 cells
(1x106) were infected with lentiviral-miR-361 or
lentiviral-NC viral particles to generate AsPC-1 cells with
constitutive expression of miR-361 (AsPC-1-miR-361) and control
cells (AsPC-1-NC), respectively. All mice received a subcutaneous
injection (50 µl) containing transfected cells in the right armpit
during administration of inhalant anesthetics isoflurane (2%),
which was used to induce general anesthesia, after adapting to the
environment for 3 days. The tumor size was measured every 3 days
using a vernier caliper. The animals were sacrificed by cervical
dislocation on day 28 after injection. Death was confirmed by the
lack of breathing and corneal reflex. Tumor weights are presented
in g and the formula (LxW2)/2 was used to measure tumor
size. The maximum tumor diameter observed in mice was 1.05 cm.
Statistical analysis
Experiments were performed in triplicate. Data were
analyzed by SPSS Statistics for Windows (version 18.0; SPSS, Inc.).
Data are presented as the mean ± SD. One-way ANOVA followed by a
Tukey's or Dunnett's post-hoc test and the two-tailed Student's
t-test were used to compare the results of multiple group and two
experimental groups, respectively. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-361 expression is decreased in PC
specimens and cells
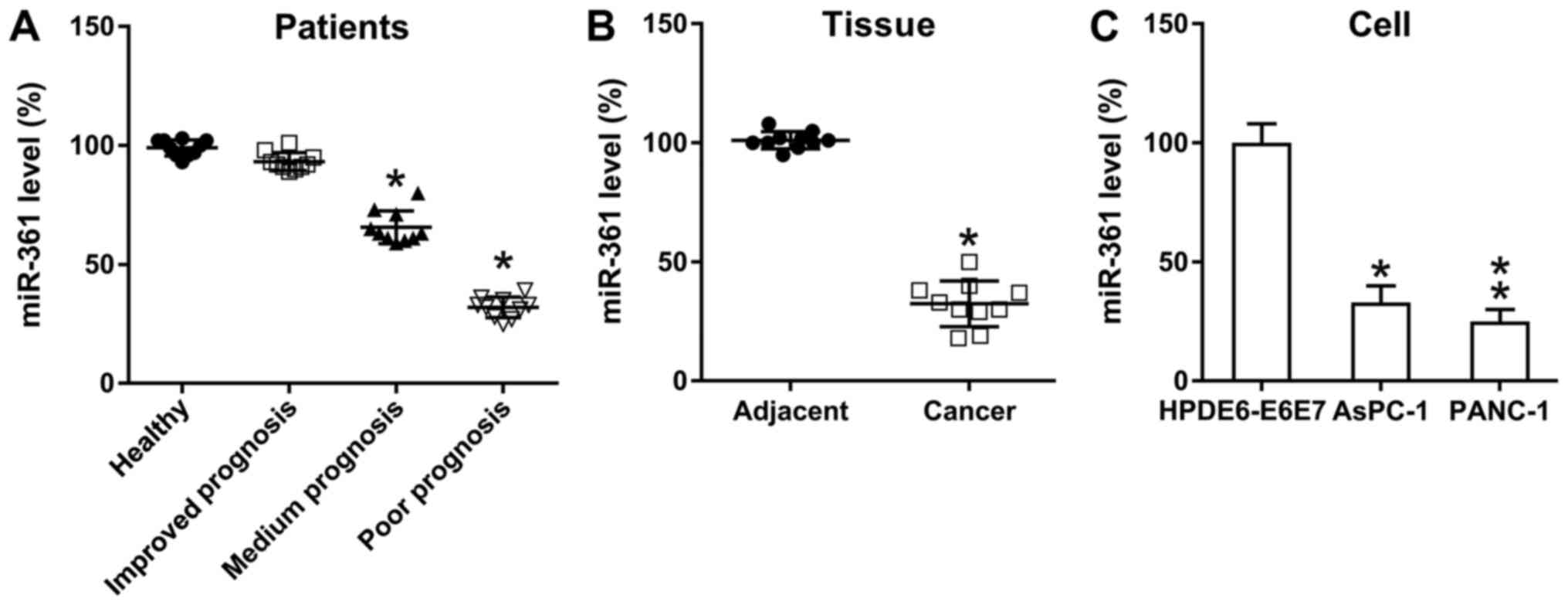
To determine the role of miR-361 on PC development,
miR-361 expression in PC tissues was examined using RT-qPCR. In
accordance with the guidelines from the NCCN, three test subject
groups were established as follows: Poor, medium and improved
prognosis groups. Compared with the control group, the medium and
poor prognosis groups demonstrated a significantly reduced
expression of miR-361 in pancreatic cancer, while the improved
prognosis group showed no significant difference (Fig. 1A). Moreover, compared with samples
from the adjacent healthy pancreas, PC specimens presented a
significant reduction in miR-361 expression (Fig. 1B), while PANC-1 and AsPC-1 cells
also exhibited downregulated miR-361 expression compared with
normal pancreatic cells (Fig. 1C).
Thus, miR-361 expression was found to be reduced in PC, suggesting
a potential role for miR-361 during PC cell proliferation in
vivo.
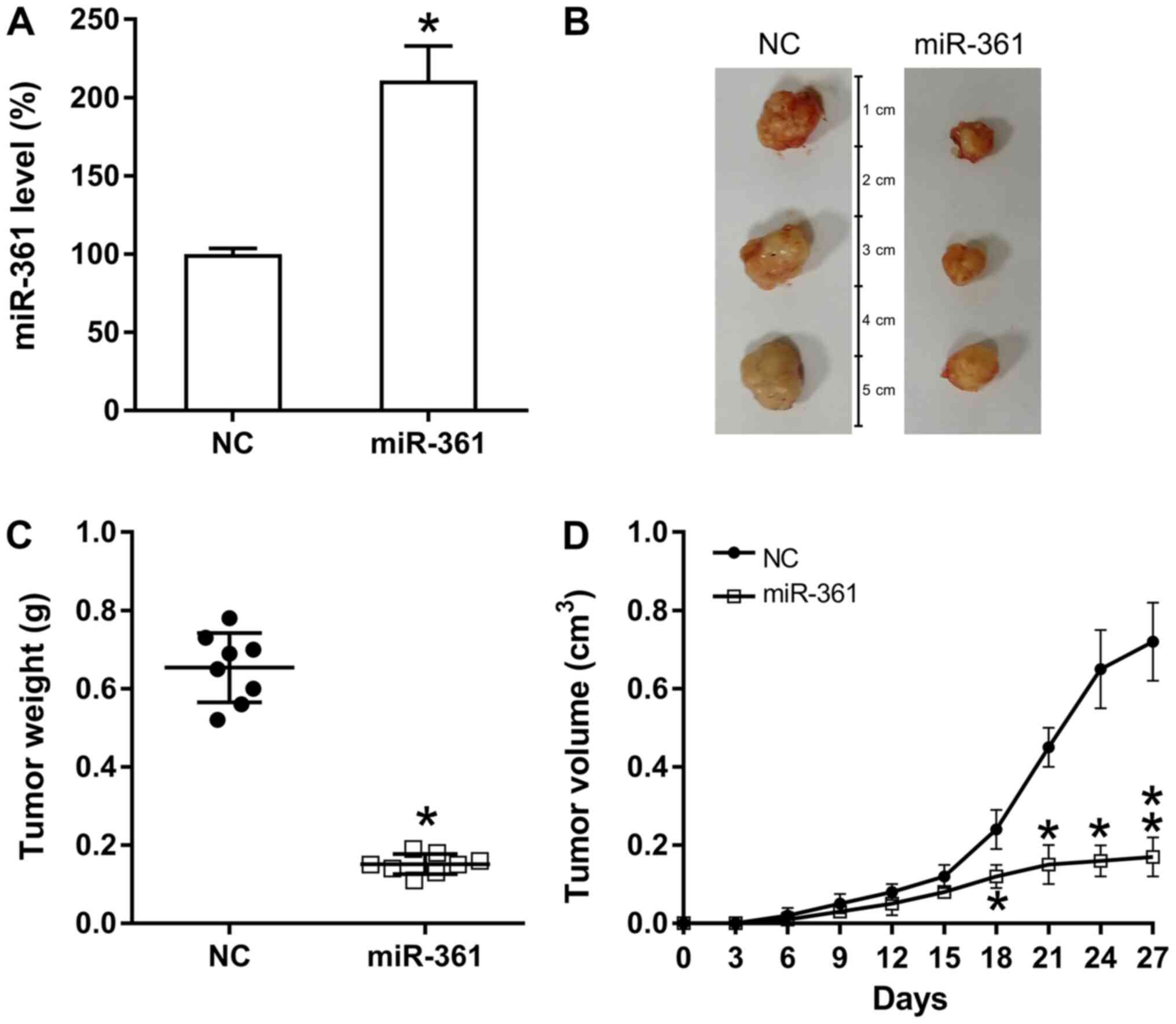
miR-361 suppresses PC cell
proliferation in vivo
To assess the influence of miR-361 on the in
vivo proliferation of xenograft PC tumor cells, nude BALB/c
mice were subcutaneously injected with AsPC-1-miR-361 (stably
expressing miR-361) or AsPC-1-NC cells, and observed every 3 days
for tumor development and overexpression of miR-361. Pancreatic
miR-361 expression was examined in different groups and miR-361
upregulation was identified in the miR-361 group (Fig. 2A). Mice were sacrificed at 28 days
following injection and their pancreatic tissues were removed and
weighed (Fig. 2B). The results
demonstrated that based on the average weight and volume of the
tumors, miR-361-generated tumors developed much slower compared
with those from mice in the NC group and the average neoplasm
volume and weight was significantly lower compared with neoplasms
in mice from the NC groups at day 27 post-injection (Fig. 2C and D).
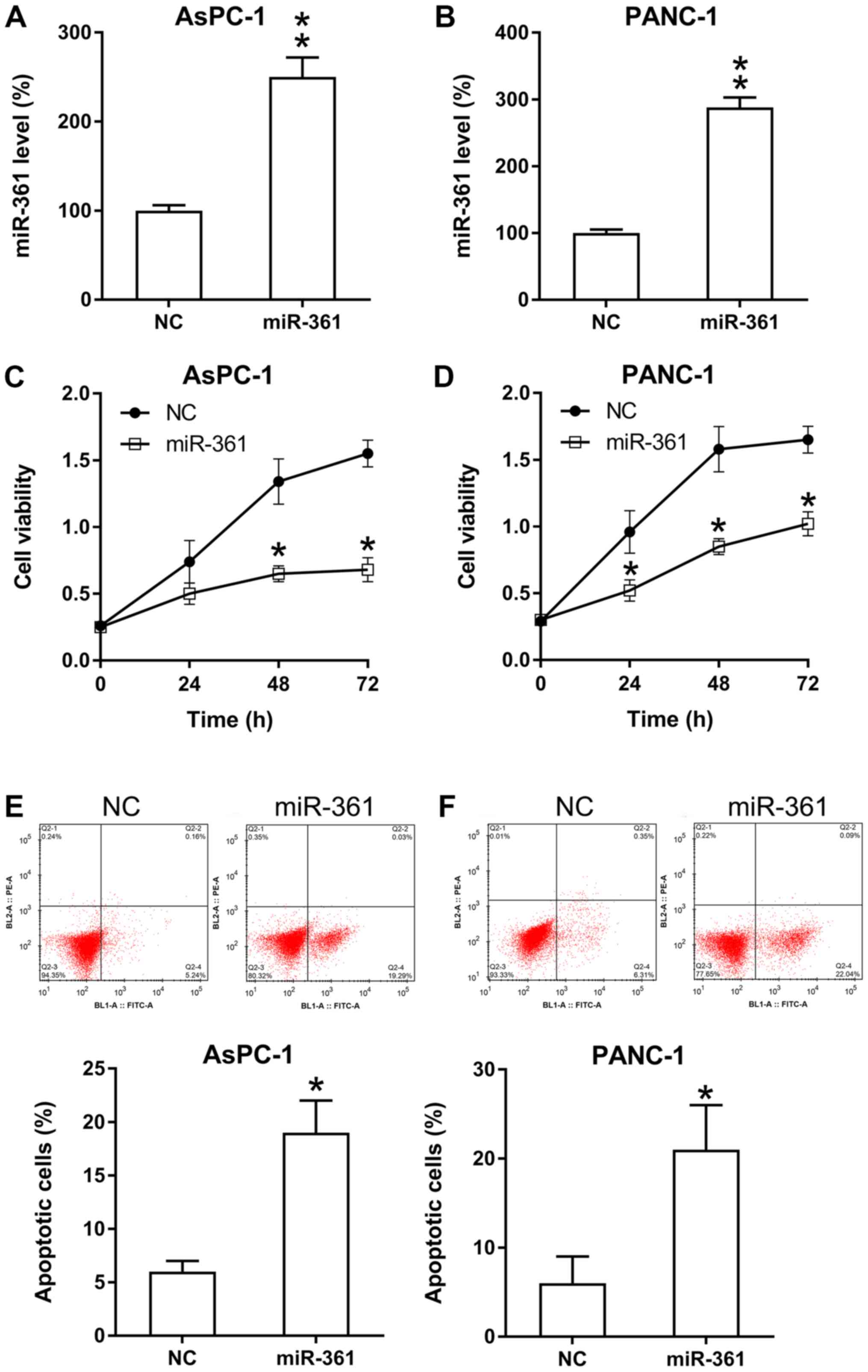
miR-361 inhibits the viability and
triggers apoptosis of PC cells
The influence of miR-361 on PC cell proliferation
was analyzed. miR-361 mimic transfection significantly increased
the expression of miR-361 in PANC-1 and AsPC-1 cells (Fig. 3A and B). Then, based on the MTT assay results,
it was found that ectopic miR-361 overexpression caused a
significant decrease in the rate of proliferation of PANC-1 and
AsPC-1 cells (Fig. 3C and D).
It was also hypothesized that miR-361 may stimulate
apoptosis in these two cell lines. Therefore, both PI flow
cytometry and Annexin V-FITC staining were performed to detect the
number of apoptotic cells in each group. The results identified a
significant increase in the apoptotic rate in PC cells that were
transfected with the miR-361 mimic compared with the NC group
(Fig. 3E and F).
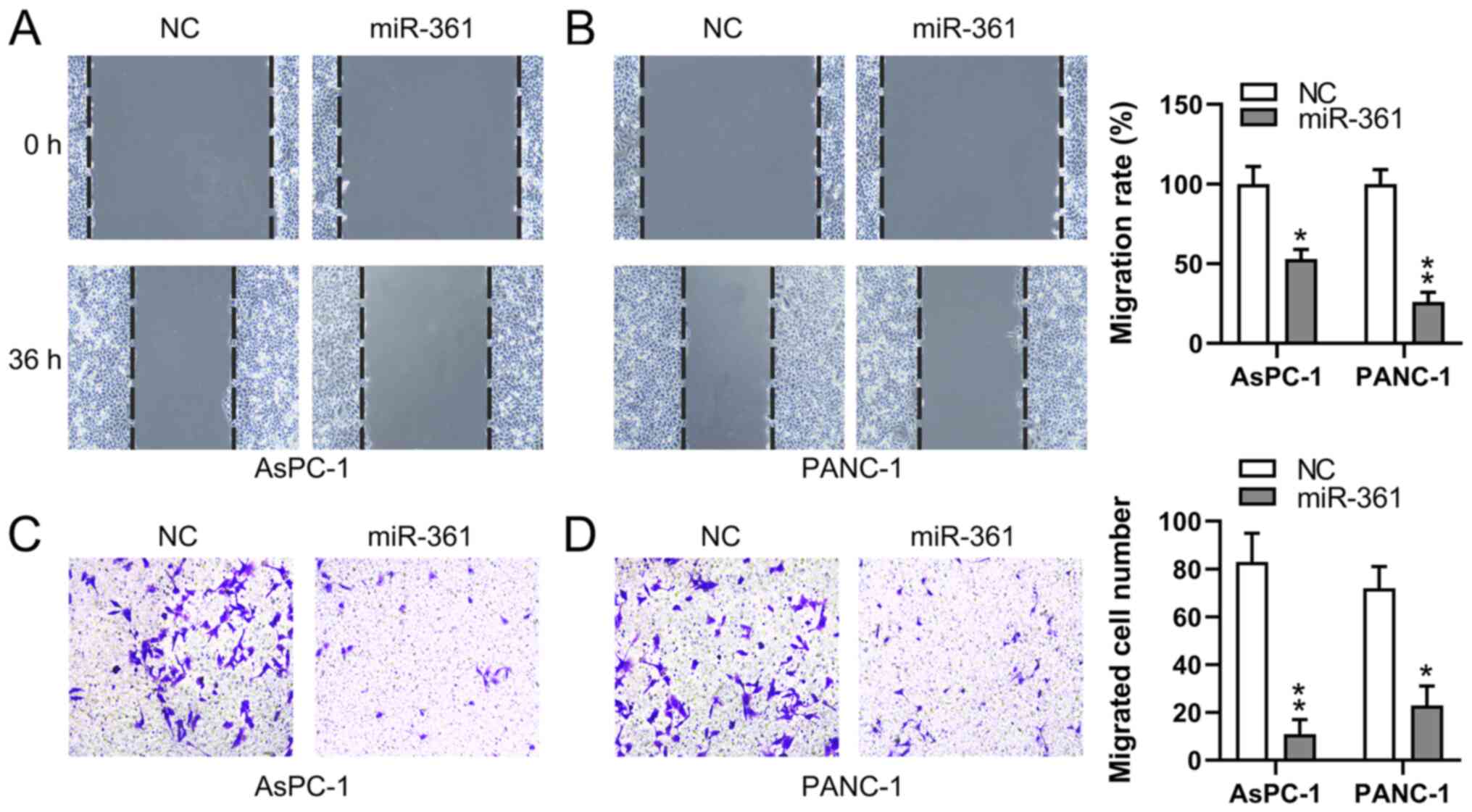
miR-361 inhibits the migratory ability
of PC cells
To assess whether the migratory process of PC cells
is affected by miR-361, wound-healing and Transwell migration
assays were conducted to measure the migration of two PC cell lines
following transfection. miR-361 overexpression demonstrated a
significant inhibitory influence on the migratory capabilities of
both PANC-1 and AsPC-1 cells at 36 h, as determined using
wound-healing assays (Fig. 4A and
B). Additionally, the Transwell
migration assay identified that the miR-361 mimic induced a
significant decrease in PC cell migration (Fig. 4C and D). These findings suggested that miR-361
suppressed the migration of PC cells.
miR-361 targets the 3'-UTR of
MAPK/JNK
MAPK/JNK has been reported to be involved in
apoptosis in various cancer cells (14-16).
Therefore, it was hypothesized that MAPK/JNK may be involved in
miR-361-mediated apoptosis and metastasis of PC cells.
Bioinformatic predictions indicated that miR-361 possesses the
ability to target the 3'-UTR in MAPK/JNK (Fig. 5A). Furthermore, DLRA found a direct
interaction between the WT-MAPK/JNK 3'-UTR and miR-361 (Fig. 5B) and that luciferase function was
inhibited following transfection with the miR-361 mimic, which
bound to the WT-MAPK/JNK 3'-UTR with <50% affinity compared with
the control groups.
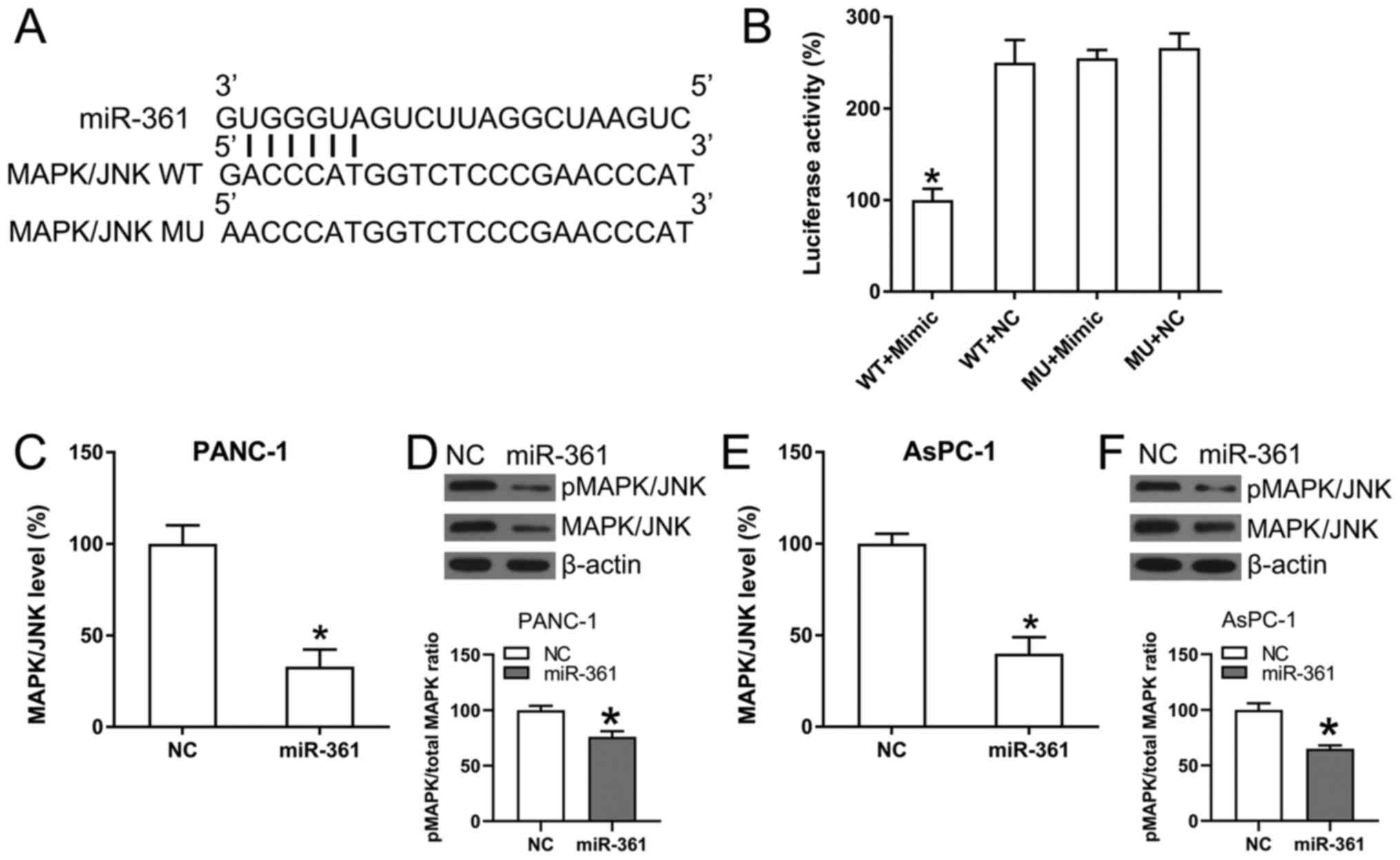 | Figure 5MAPK/JNK is targeted by miR-361. (A)
Conserved miR-361 binding motif on the 3'-UTRs of MAPK. (B)
Following the transfection of the miR-361 mimic, luciferase
activity was assessed using the luciferase reporter construct,
which contains either the MU or WT copy of the human MAPK/JNK
3'-UTRs. The luciferase activity was standardized to the
Renilla luciferase activity. Data were analyzed using
one-way ANOVA. (C) RT-qPCR and (D) WB analyses were performed to
assess the expression and phosphorylation of MAPK/JNK following
transfection of PANC-1 cells using the NC mimic or the miR-361
mimic. (E) RT-qPCR and (F) WB analyses were performed to assess the
expression and phosphorylation of MAPK/JNK following transfection
of AsPC-1 cells using the NC mimic or the miR-361 mimic. Data were
analyzed using unpaired t-test. Data are presented as the mean ±
SD. *P<0.05 vs. NC groups. NC, negative control; miR,
microRNA; WT, wild-type; MU, mutant; UTR, untranslated region;
MAPK, mitogen-activated protein kinase; p, phosphorylated; RT-qPCR,
reverse transcription-quantitative PCR; WB, western blot. |
The effect of ectopic overexpression of miR-361 on
MAPK/JNK expression was also analyzed in AsPC-1 and PANC-1 cells
using RT-qPCR and WB analysis. MAPK/JNK mRNA and protein expression
levels were significantly downregulated as a result of the
transfection with the miR-361 mimic (Fig. 5C-F). Thus, it was found that
MAPK/JNK expression decreased following miR-361 overexpression.
MAPK/JNK restores PC cell viability
and metastasis attenuated by miR-361
To determine whether MAPK/JNK participated in the
effects induced by miR-361 on PC cell viability and metastasis,
MAPK/JNK was overexpressed in the AsPC-1 and PANC-1 cells. WB
analysis results demonstrated the successful overexpression of
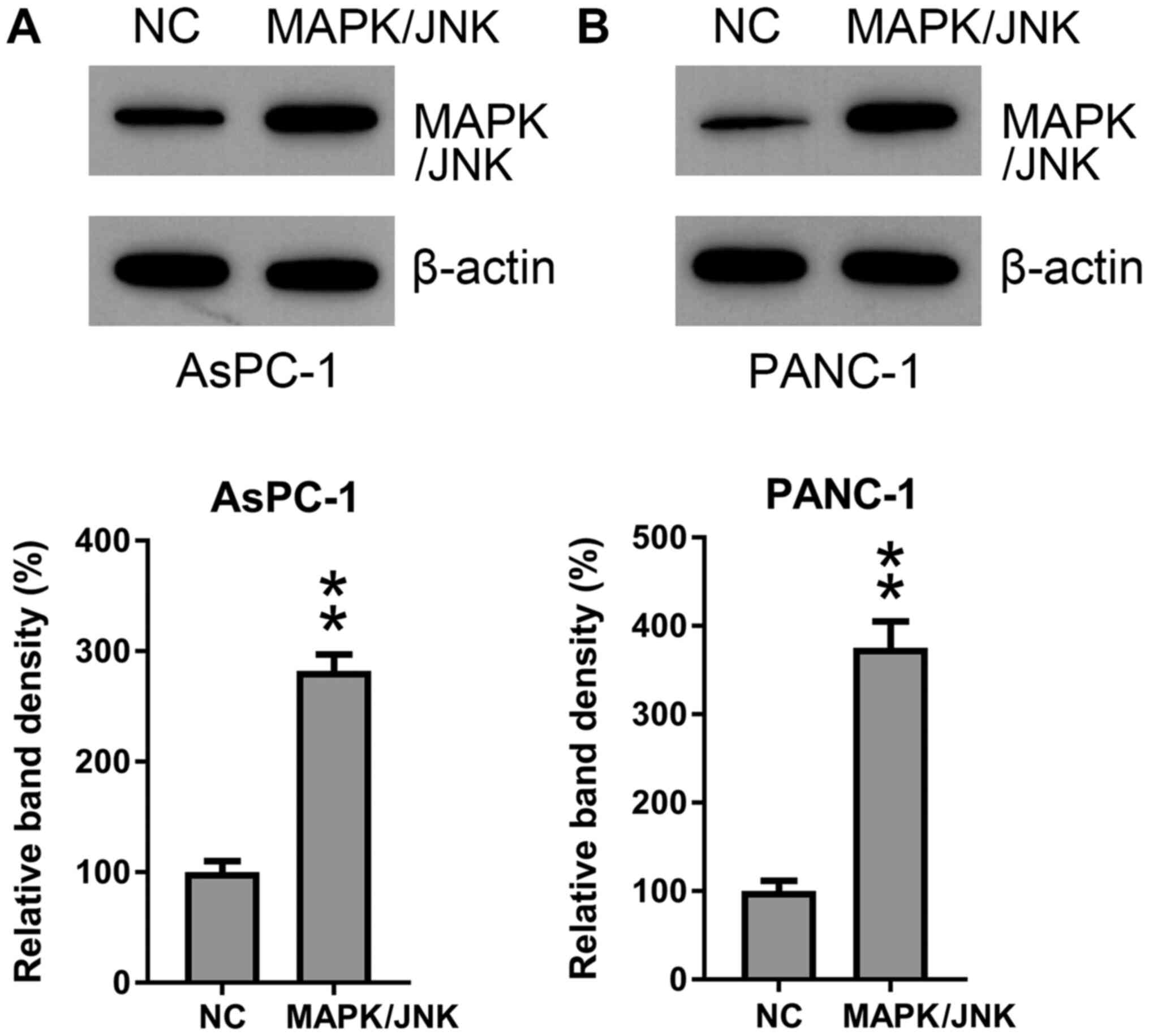
MAPK/JNK (Fig. 6A and B). MAPK/JNK was overexpressed in AsPC-1
and PANC-1 cells that were also co-transfected with the miR-361
mimic and WB analyses were conducted to confirm MAPK/JNK
upregulation in cells (Fig. 7A and
B).
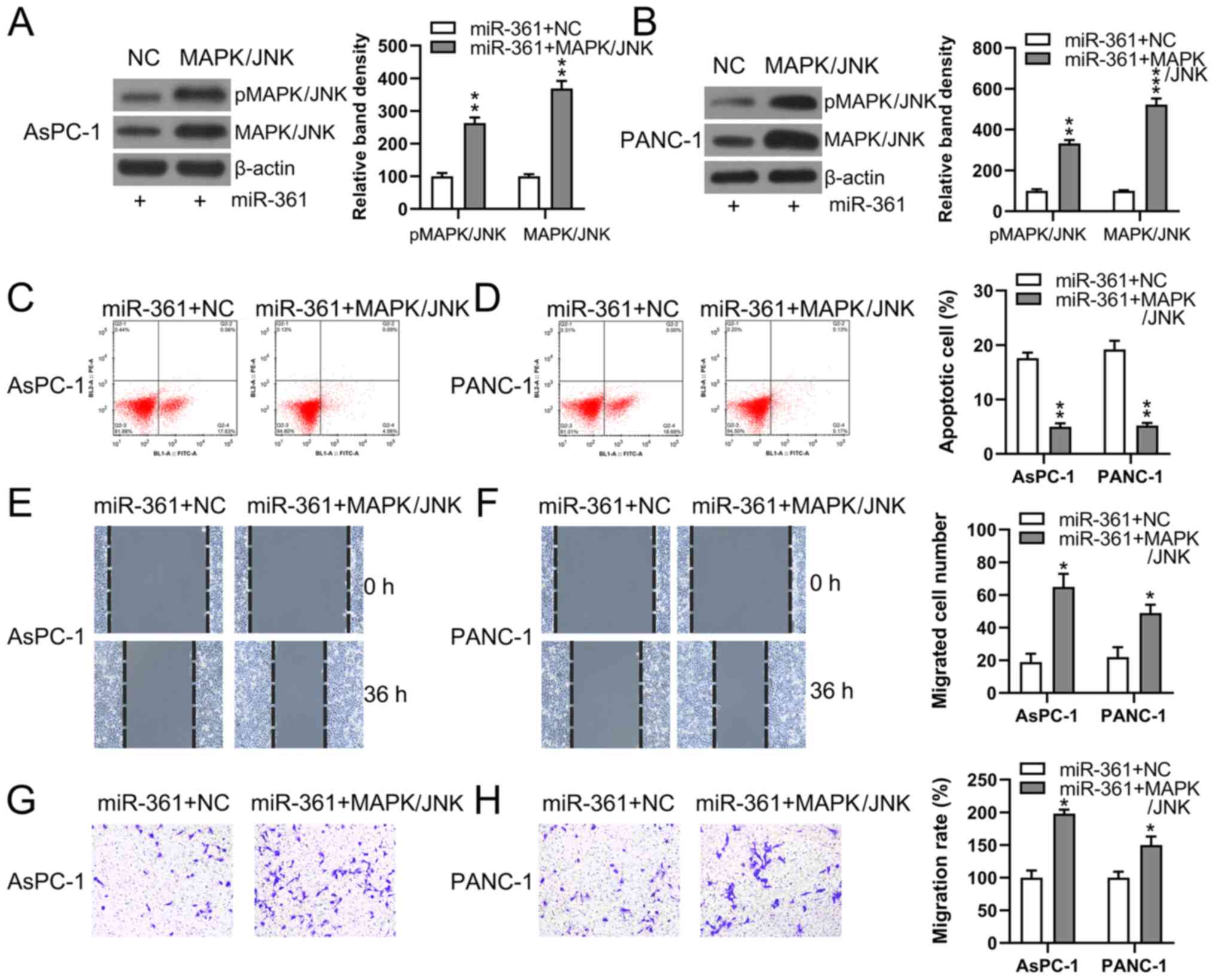
Flow cytometry was used to detect the quantity of
apoptotic cells. While MAPK/JNK was overexpressed and pMAPK/JNK
expression was significantly increased (Fig. 7A and B), the apoptotic process induced by
miR-361 in AsPC-1 and PANC-1 cells was significantly impeded in
cells transfected with MAPK/JNK overexpression vector + miR-361
mimic (Fig. 7C and D).
To evaluate the role of MAPK/JNK in the process of
PC cell metastasis, Transwell migration and would healing assays
were performed on two PC cell lines following their transfection
with different constructs. Cells overexpressing MAPK/JNK + miR-361
demonstrated restored migratory abilities compared with the NC
groups (Fig. 7E-H).
Discussion
PC is a malignant condition that severely affects
the health of patients (16). Only
1/5 patients with PC are viable for surgical therapy, which is
currently the sole reliable curative therapy for PC (16). Therefore, identifying effective
methods of PC inhibition are of great significance. However, the
limitation of the present study was the lack of animal experiments,
which will be performed in future studies.
Previous studies have reported that dysregulated
miRNA expression may cause the development and progression of PC
carcinoma (17-20).
Thus, the discovery of tumor-associated miRNAs, assessment of their
therapeutic effects and identification of their target mRNAs will
be important for finding novel therapeutic targets (20). It has been revealed that miRNAs
perform vital functions with regard to various biological
activities involved in PC progression. For instance, miR-195
expression is decreased in PC cell lines and tissues (21). Furthermore, increased expression of
miR-195 suppresses PC invasion and proliferation by modulating the
Wnt/fatty acid synthase signal pathway (21) The expression of miR-181d is
increased not only in PC cells, but also in human PC (22). It has also been shown that
downregulation of miR-181d induced via transfection with specific
lentiviral constructs inhibits the processes of fluorouracil
resistance, migration and proliferation in PC cells, and prevents
the tumor explant from growing in animal models (23). Additionally, miR-181d can directly
target sodium/potassium transporting ATPase interacting 2 (NKAIN2)
in PC, while NKAIN2 knockdown mediated by small interfering RNA
could reverse the suppression of miR-181d downregulation during PC
development (22). PC utilizes
autophagy as a means of survival to generate fundamental glucose
necessary for glycolysis metabolism and it has been revealed that
miR-7 inhibits autophagy via activating the serine/threonine kinase
11/AMP-activated protein kinase/mTOR signaling pathway in stressful
tumor microenvironments (23).
miR-7 also directly targets specific stages of cellular autophagy
induction and vesicle elongation to decrease glucose provision in
cells involved in glycolysis metabolism (24). Moreover, miR-7 suppresses PC cell
metastasis and proliferation in vitro and in animal models
(24). The present results
demonstrated that the expression of miR-361 was decreased in PC
cells and tissues and that miR-361 overexpression significantly
inhibited the viability and migration of PC cells. In addition,
MAPK/JNK was found to be the direct target of miR-361. Thus, it was
suggested that miR-361 may function as a PC inhibitor and could be
a promising target in PC treatment.
miR-361 is located in Xq21.2, an intron located
between exon 10 and exon 9 in the choroideremia gene (2). A decrease in miR-361 is observed in
various human malignancies, including liver carcinoma (25), gastric tumor (26), colorectal malignancy (26), prostate carcinoma (27), NSCLC (8), esophageal tumors (9) and cutaneous squamous cell carcinoma
(24). Its targets include, but are
not limited to, SH2B adaptor protein 1 in NSCLC and colorectal
cancer (28,29), ERK1/1 in pancreatic ductal
adenocarcinoma (30), hedgehog in
retinoblastoma (31), granzyme B in
cutaneous leishmaniasis (32) and
β-secretase 1 in Alzheimer's disease (33). It has been reported that miR-361 may
regulate C-X-C motif chemokine receptor 6 to inhibit liver tumor
cell proliferation, while another study revealed that miR-361
regulated STAT6 expression by functioning as a tumor inhibition
gene in prostate tumors. Moreover, Ma et al (26) found that miR-361 could decrease the
recurrence rate of gastric cancer via interacting with the CD80
gene 3'-UTR. Furthermore, results showed that miR-361 could
specifically inhibit staphylococcal nuclease domain containing-1
gene expression and growth of colorectal tumor bodies.
Collectively, these results indicated that miR-361 serves a role in
suppressing the growth of multiple tumor types.
The present study also identified an inhibitory role
of miR-361 in PC. MTT assay results demonstrated that after miR-361
overexpression using transfection with mimics, the viability of
AsPC-1 and PANC-1 cells significantly decreased. Flow cytometry
also identified that miR-361 could induce PC cell apoptosis.
Similar effects were also found with regard to the migratory
abilities of PC cells, which were inhibited by miR-361, suggesting
that miR-361 served an inhibitory role in PC. However, a previous
report revealed that miR-361 overexpression promoted PC cell
migration and invasion in vitro (PANC-1, SW1990, BxPC-3 and
CFPAC-1 cells) (30), while
conversely, the present study identified an inhibitory effect of
miR-361 on PC development in vivo. In addition, miR-361
overexpression was found to reduce PC cell proliferation and
metastasis. Therefore, it was hypothesized that the discrepancies
between these two studies may be attributed to the differences in
cell lines and patient cohorts used.
The generic MAPK/JNK signaling pathway is involved
in four different cascades, named according to their MAPK/JNK tier
constituents: ERK1/2, ERK5, p38 MAPK and c-Jun aminoterminal
kinases (34). Furthermore, ERK and
MAPK/JNK pathways are associated with cell apoptosis, senescence,
migration, differentiation and proliferation (34). It has been reported that the natural
compound Oblongifolin C effectively suppresses PC cell
proliferation by inducing apoptosis and G0/G1
arrest via amelioration of the Src proto-oncogene/MAPK/ERK
signaling pathways (35).
Sulforaphane is an active component found in cruciferous vegetables
and can repress cell colony formation and proliferation, as well as
induce apoptosis via caspase-3 activation in PC cells (36). Previous studies have also shown that
sulforaphane suppresses AKT and MAPK/JNK phosphorylation and
stimulates forkhead transcription factors, causing apoptosis and
cell cycle arrest (36). The
proto-oncogene Kirsten rat sarcoma viral oncogene (KRAS) serves an
important role in PC formation and development and the ERK/MAPK/JNK
signaling pathway participates in modulating KRAS expression in PC
(37). Thus, these studies support
the conclusion that MAPK/JNK exerts a protective role in PC growth
and metastasis. In the current study, MAPK/JNK expression was
repressed via transfection with the miR-361 mimic. Overexpression
of MAPK/JNK restored the proliferative and migratory capabilities
in PC cells suppressed by miR-361, suggesting that MAPK/JNK is
involved in mediating the activity of miR-361 during PC
development.
In conclusion, the present results indicated that
miR-361 may possess tumor-suppressing effects that result in
impaired major malignant capabilities in PC cell lines and in PC
cell proliferation in vivo. Furthermore, the results
suggested that decreased miR-361 expression in PC tumors and cells
was associated with cancer cell proliferation impairment. Thus,
miR-361 may be considered as a prognostic and diagnostic biomarker
for PC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW conceived the current study and designed the
experiments. ZX, YL and WZ contributed to the data collection,
performed data analysis and interpreted the results. JW wrote the
manuscript. TJ contributed to the data collection and analysis
during the revision of the article. JW and YL confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Board of
Ethics Review of the No. 215 Hospital of the Shaanxi Nuclear
Industry (Xianyang, China). All patients provided written informed
consent prior to enrollment in the current study. The experimental
protocol involving animals was approved by the Animal Ethics
Committee of the No. 215 Hospital of Shaanxi Nuclear Industry.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Guo X and Cui Z: Current diagnosis and
treatment of pancreatic cancer in China. Pancreas. 31:13–22.
2005.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Long J, Luo GP, Xiao ZW, Liu ZQ, Guo M,
Liu L, Liu C, Xu J, Gao YT, Zheng Y, et al: Cancer statistics:
Current diagnosis and treatment of pancreatic cancer in Shanghai,
China. Cancer Lett. 346:273–277. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bosetti C, Bertuccio P, Negri E, La
Vecchia C, Zeegers MP and Boffetta P: Pancreatic cancer: overview
of descriptive epidemiology. Mol Carcinog. 51:3–13. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yang S, Zhang Y, Zhao X, Wang J and Shang
J: microRNA-361 targets Wilms' tumor 1 to inhibit the growth,
migration and invasion of non-small-cell lung cancer cells. Mol Med
Rep. 14:5415–5421. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lin P, Pang Q, Wang P, Lv X, Liu L and Li
A: The targeted regulation of Gli1 by miR-361 to inhibit
epithelia-mesenchymal transition and invasion of esophageal
carcinoma cells. Cancer Biomark. 21:489–498. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Isaji S, Kawarada Y and Uemoto S:
Classification of pancreatic cancer: Comparison of Japanese and
UICC classifications. Pancreas. 28:231–234. 2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Peng DY, Song H and Liu LB:
Resveratrol-downregulated phosphorylated liver kinase B1 is
involved in senescence of acute myeloid leukemia stem cells. J
Huazhong Univ Sci Technolog Med Sci. 35:485–489. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4(e05005)2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kang KA, Piao MJ, Madduma Hewage SR, Ryu
YS, Oh MC, Kwon TK, Chae S and Hyun JW: Fisetin induces apoptosis
and endoplasmic reticulum stress in human non-small cell lung
cancer through inhibition of the MAPK signaling pathway. Tumour
Biol. 37:9615–9624. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mondal A, Biswas R, Rhee YH, Kim J and Ahn
JC: Sulforaphene promotes Bax/Bcl2, MAPK-dependent human gastric
cancer AGS cells apoptosis and inhibits migration via EGFR,
p-ERK1/2 down-regulation. Gen Physiol Biophys. 35:25–34.
2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yeo TP, Hruban RH, Leach SD, Wilentz RE,
Sohn TA, Kern SE, Iacobuzio-Donahue CA, Maitra A, Goggins M, Canto
MI, et al: Pancreatic cancer. Curr Probl Cancer. 26:176–275.
2002.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yonemori K, Kurahara H, Maemura K and
Natsugoe S: MicroRNA in pancreatic cancer. J Hum Genet. 62:33–40.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Furtek SL, Backos DS, Matheson CJ and
Reigan P: Strategies and approaches of targeting STAT3 for cancer
treatment. ACS Chem Biol. 11:308–318. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kıvanç G, Kalliope ND, Ezgi KA, Katrin JC,
Jiaoyu A, Marina L and Hana A: The role of autophagy in pancreatic
cancer: From bench to the dark bedside. Cell.
9(1063)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xu Z, Li C, Qu H, Li H, Gu Q and Xu J:
MicroRNA-195 inhibits the proliferation and invasion of pancreatic
cancer cells by targeting the fatty acid synthase/Wnt signaling
pathway. Tumour Biol. 39(1010428317711324)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang G, Liu D, Long G, Shi L, Qiu H, Hu
G, Hu G and Liu S: Downregulation of microRNA-181d had suppressive
effect on pancreatic cancer development through inverse regulation
of KNAIN2. Tumour Biol. 39(1010428317698364)2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY,
Fang C, Huang Q and Tian L: microRNA-7 impairs autophagy-derived
pools of glucose to suppress pancreatic cancer progression. Cancer
Lett. 400:69–78. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kanitz A, Imig J, Dziunycz PJ, Primorac A,
Galgano A, Hofbauer GF, Gerber AP and Detmar M: The expression
levels of microRNA-361-5p and its target VEGFA are inversely
correlated in human cutaneous squamous cell carcinoma. PLoS One.
7(e49568)2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sun JJ, Chen GY and Xie ZT:
MicroRNA-361-5p inhibits cancer cell growth by targeting CXCR6 in
hepatocellular carcinoma. Cell Physiol Biochem. 38:777–785.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ma F, Song H, Guo B, Zhang Y, Zheng Y, Lin
C, Wu Y, Guan G, Sha R and Zhou Q: miR-361-5p inhibits colorectal
and gastric cancer growth and metastasis by targeting
staphylococcal nuclease domain containing-1. Oncotarget.
6(17404)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu D, Tao T, Xu B, Chen S, Liu C, Zhang
L, Lu K, Huang Y, Jiang L, Zhang X, et al: MiR-361-5p acts as a
tumor suppressor in prostate cancer by targeting signal transducer
and activator of transcription-6 (STAT6). Biochem Biophys Res
Commun. 445:151–156. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen W, Wang J, Liu S, Wang S, Cheng Y,
Zhou W, Duan C and Zhang C: MicroRNA-361-3p suppresses tumor cell
proliferation and metastasis by directly targeting SH2B1 in NSCLC.
J Exp Clin Cancer Res. 35(76)2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu J, Zhu J, Xiao Z, Wang X and Luo J:
BBOX1-AS1 contributes to colorectal cancer progression by sponging
hsa-miR-361-3p and targeting SH2B1. FEBS Open Bio: Jan 27, 2020
(Epub ahead of print).
|
|
30
|
Hu J, Li L, Chen H, Zhang G, Liu H, Kong
R, Chen H, Wang Y, Li Y, Tian F, et al: MiR-361-3p regulates
ERK1/2-induced EMT via DUSP2 mRNA degradation in pancreatic ductal
adenocarcinoma. Cell Death Dis. 9(807)2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhao D and Cui Z: MicroRNA-361-3p
regulates retinoblastoma cell proliferation and stemness by
targeting hedgehog signaling. Exp Ther Med. 17:1154–1162.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lago TS, Silva JA, Lago EL, Carvalho EM,
Zanette DL and Castellucci LC: The miRNA 361-3p, a regulator of
GZMB and TNF is associated with therapeutic failure and longer time
healing of cutaneous Leishmaniasis Caused by L. (viannia)
braziliensis. Front Immunol. 9(2621)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ji Y, Wang D, Zhang B and Lu H: MiR-361-3p
inhibits β-amyloid accumulation and attenuates cognitive deficits
through targeting BACE1 in Alzheimer's disease. J Integr Neurosci.
18:285–291. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sun Y, Liu WZ, Liu T, Feng X, Yang N and
Zhou HF: Signaling pathway of MAPK/ERK in cell proliferation,
differentiation, migration, senescence and apoptosis. J Recept
Signal Transduct Res. 35:600–604. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li Y, Xi Z, Chen X, Cai S, Liang C, Wang
Z, Li Y, Tan H, Lao Y and Xu H: Natural compound Oblongifolin C
confers gemcitabine resistance in pancreatic cancer by
downregulating Src/MAPK/ERK pathways. Cell Death Dis.
9(538)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Roy SK, Srivastava RK and Shankar S:
Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of
FOXO transcription factor, leading to cell cycle arrest and
apoptosis in pancreatic cancer. J Mol Signal. 5(10)2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yang K, Li Y, Lian G, Lin H, Shang C, Zeng
L, Chen S, Li J, Huang C, Huang K and Chen Y: KRAS promotes tumor
metastasis and chemoresistance by repressing RKIP via the MAPK-ERK
pathway in pancreatic cancer. Int J Cancer. 142:2323–2334.
2018.PubMed/NCBI View Article : Google Scholar
|