Introduction
In recent years, the production of extended-spectrum
β-lactamases (ESBL) has become the main mechanism of resistance to
β-lactam and other antibiotics in Enterobacteriaceae and
Acinetobacter baumannii (A. baumannii) (1). ESBL enzymes confer resistance to
penicillins, cephalosporins, monobactams and other antibiotic
classes (2). Multidrug-resistant
(MDR) patterns due to ESBL production in pathogenic bacteria are
now becoming prevalent in hospitals worldwide, posing a public
health challenge, including treatment failure, prolonged hospital
stay and increased mortality rates (3-5).
Recently, >300 different ESBL types have been
described in Gram-negative bacteria (6,7). The
blaTEM and blaSHV types have
been recognized as the most prevalent ESBL genes conferring
antibiotic resistance in pathogenic bacteria worldwide (8-10).
Previous studies have revealed that the number of clinical isolates
harboring the blaCTX-M gene type has also
increased in the last few years (2,11).
The blaCTX-M family includes >130 β-lactamase
variants classified into five distinct groups:
blaCTX-M-1, blaCTX-M-2,
blaCTX-M-8, blaCTX-M-9 and
blaCTX-M-25 (12).
The genetic diversity of ESBL-producing
Enterobacteriaceae and A. baumannii has progressively
increased, posing challenges to hospital authorities due to their
ability to confer antibiotic susceptibility and limit therapeutic
options (13,14). The characterization of resistance
genes encoding ESBL-producing microorganisms is a powerful tool for
developing evidence-based guidelines for combating antibiotic
resistance in the clinical setting (11).
In Saudi Arabia, Gram-negative bacteria-harboring
ESBL resistance genes have been studied, with most studies emerging
from the central and eastern regions (5,15-19).
Despite the spread of antibiotic resistance among bacterial
pathogens in the southern region, data on the distribution of ESBL
resistance genes and their resistance profile among
Enterobacteriaceae and A. baumannii remain limited
(20,21). A previous study performed at the
Aseer Central Hospital, a regional hospital in the southern region,
identified MDR patterns among 98.1% of Acinetobacter species
recovered from patients at intensive care units (22). In addition, another study conducted
in the same hospital identified a high distribution of class D
carbapenemase-encoding genes in A. baumannii, mainly
ISAba1/OXA-23 and ISAba1/OXA-24 carbapenemases, which is alarming
and presents an emerging public health threat (23). The emergence of MDR A.
baumannii bacteremia in the southern region of Saudi Arabia has
been well documented as an important health problem (22,24,25).
A recent study reported the high frequency of MDR Gram-negative
bacteria and a rate of ESBL production of 27% in patients at the
King Abdullah Hospital, a referral hospital in Bisha, in the
southern region of the country (20).
Due to the lack of information on the genotyping of
ESBL producers and their MDR patterns in southern Saudi Arabia, the
present study aimed to determine the antibiotic susceptibility
patterns and distribution of ESBL genes in
Enterobacteriaceae and A. baumannii isolates
collected from clinical specimens of patients. The findings of the
present study facilitated the implementation of infection control
measures and provided epidemiological data to prevent spreading of
MDR bacteria. The data also provided guidelines for the use of
antibiotics in the clinical settings and improved the management of
patients suffering from infections caused by
Enterobacteriaceae and A. baumannii.
Materials and methods
Study design and setting
A cross-sectional study was conducted between
September 2017 and August 2018 at the King Abdullah Hospital
(Bisha, Saudi Arabia). A total of 274 Enterobacteriaceae and
A. baumannii were recovered from patients as a part of the
treatment and diagnosis procedure for infections. The clinical
samples were collected from the patients as a part of routine
investigations of infectious agents in the hospital microbiology
laboratory. Therefore, consent letters were not obtained from the
patients as per the study nature. Various clinical samples of urine
(n=96), sputum (n=84), wound swab (n=51), blood (n=27), high
vaginal swab (n=5), tracheal aspirate (n=5), umbilical discharge
(n=3), cerebrospinal fluid (n=2) and eye swab (n=1) collected from
274 patients suffering from different bacterial infections were
included in the study. Discharge from umbilical stump was collected
from neonatal and/or infant patients in the wards using sterile
swab soaked with normal saline and submitted to the laboratory.
High vaginal swab was taken from female patients by clinicians and
sent for microbiological examinations. Clinical specimens with
incomplete personal information of the patients were excluded from
the study. The average age of the patients was 46.0±25.5 years,
including 158 females and 116 males. In total, 32 patients were
aged 2-17 years, 97 were aged 18-40 years, 57 were aged 41-65 years
and 88 were aged >65years. Patients of <2 years old were
excluded from the study. The hospital is a referral hospital in the
southern region (365 beds) with different specialties serving the
Bisha province and the surrounding areas. The Research and Ethics
committee at the College of Medicine, University of Bisha (Bisha,
Saudi Arabia) reviewed and approved the present study protocol
(approval no. UBCOM/1438-05/04).
Isolation and identification of
bacteria
Enterobacteriaceae and A. baumannii
were collected from the microbiology laboratory of King Abdullah
Hospital during the routine processing of the clinical specimens of
patients. Preliminary isolation and identification of bacteria were
based on conventional microbiological methods. Briefly, Isolation
of bacterial pathogens from specimens of urine, stool, sputum and
other body fluids were performed by inoculating one loopful of each
sample onto MacConkey agar plates (Oxoid, Ltd.) using a sterile
disposable plastic loop (10-µl loop). Specimens of wounds, eye,
umbilical and vaginal swabs were inoculated directly onto MacConkey
agar plates by streaking them onto a small area of the plate. A
disposable sterile plastic loop (1-µl loop) was used for
cross-streaking to spread the inoculum over the surface of each
plate to obtain single colonies of the suspected bacterial
pathogen. Specimen of blood (5 ml) were extracted under aseptic
conditions, transferred immediately into sterile bottles containing
brain heart infusion broth (Oxoid, Ltd.), incubated at 37˚C with 5%
CO2 and examine daily for turbidity for ≤7 days. If
turbidity was observed, a 10-µl loopful of the blood sample was
subcultured onto MacConkey agar plates for isolation of the
suspected Gram-negative pathogen after aerobic incubation at 37˚C
for 24 h. The isolate was tentatively identified based on the
colony morphology, gram staining and oxidase test and the API 20 E
Gram-Negative Microbial Identification Kit (cat. no. 20160;
bioMerieux SA). Then, full identification of bacterial isolates was
confirmed using the Phoenix system identification assay (Becton,
Dickinson and Company). One single bacterial isolate from the
clinical sample of each patient was included in the present
study.
Screening of ESBL production
Phenotypic ESBL production among isolates was
examined using a double-disc synergy test (DDST) as previously
described (26) and the decreased
susceptibility to cefuroxime, ceftazidime and cefotaxime was
examined according to the Clinical and Laboratory Standard
Institute (CLSI) recommendations (27). Bacterial isolates yielded positive
results with DDST, when subjected to a multiplex-PCR amplification
assay to detect blaTEM, blaSHV,
blaCTX-M and blaOXA-1
resistance genes.
Antibiotic susceptibility testing of
ESBL-producing bacteria
Antibiotic susceptibility testing of the
Enterobacteriaceae and A. baumannii was performed
using the Kirby-Bauer disk diffusion method on Mueller-Hinton agar
medium (Oxoid) as per the CLSI guidelines (27). The following antibiotics with known
concentrations recommended by the CLSI were examined: Amikacin (30
µg), amoxicillin/clavulanate (20/10 µg), aztreonam (30 µg),
cefepime (30 µg), cefotaxime (30 µg), cefoxitin (30 µg),
ceftazidime (30 µg), cefuroxime (30 µg), ciprofloxacin (5 µg),
colistin (10 µg), gentamicin (10 µg), imipenem (10 µg), meropenem
(10 µg), nitrofurantoin (50 µg), piperacillin (100 µg),
piperacillin/tazobactam (100/10 µg), tobramycin (10 µg) and
trimethoprim/sulfamethoxazole (23.75 µg/1.25 µg; Oxoid). E.
coli American Type Culture Collection (ATCC) 25922 served as a
control strain for antibiotic susceptibility examination. The final
antibiotic susceptibility results of bacterial pathogens were
interpreted using the 2017 CLSI breakpoints to categorize the
isolates as susceptible or resistant. All intermediate results were
considered resistant strains. ‘A susceptible category indicates
that the isolates of the patient respond to the usually achievable
concentrations of that antibiotic when the dosage is recommended to
treat that type of infection and bacterial species. Conversely, the
resistant category indicates that the isolates of the patient are
not inhibited by the usually achievable concentrations of that
antibiotic with the dosages usually used with that drug’ (27). Isolates were defined as MDR when
they were resistant to at least three antibiotics from different
classes.
Multiplex-PCR for the detection of
ESBL genes
Multiplex-PCR runs were performed to identify the
blaTEM, blaSHV,
blaCTX-M and blaOXA-1 genes. A
pair of six specific oligonucleotide primers (Eurofins Scientific)
were used in the PCR reaction, as previously described (28). DNA was extracted from
ESBL-producing isolates using the boiling method as previously
described (29). The amplification
of ESBL genes was then carried out in a total reaction volume of 50
µl. Each reaction mixture contained 25 µl HotStarTaq Plus Master
Mix (cat. no. 203643; Qiagen GmbH), 4 µl DNA template, a variable
volume of a specific primer group (Table I) and 9 µl nuclease-free water. The
Eppendorf Master cycler Gradient instrument (Eppendorf) was used
for the amplification of target genes with the following optimal
cycling conditions: Initial heat activation at 95˚C for 5 min, 35
cycles of denaturation at 94˚C for 45 sec, annealing at 53˚C for 45
sec, extension at 72˚C for 1 min and a final extension at 72˚C for
10 min. The amplification products were visualized under
ultraviolet illumination at a wavelength of 312 nm, after running
at 85 volts for 60 min on 2% agarose containing ethidium bromide (1
µg/ml). A 100-bp DNA ladder (cat. no. 239045; Qiagen GmbH) was used
as a standard molecular weight to determine the size of PCR
products. DNA from reference blaCTX-M,
blaTEM, blaSHV and
blaOXA-like-positive strains was used as a
positive control.
 | Table IThe frequency of extended-spectrum
β-lactamase resistance genes among Gram-negative bacteria (n=72) as
detected by multiplex PCR using different primers. |
Table I
The frequency of extended-spectrum
β-lactamase resistance genes among Gram-negative bacteria (n=72) as
detected by multiplex PCR using different primers.
| | Oligonucleotide
primer (28) |
|---|
| Resistance
gene | N (%) | Sequence | Length | Band size (base
pairs) |
|---|
|
blaTEM | 61 (84.7) | F:
5'-CATTTCCGTGTCGCCCTTATTC-3' | 22 | 800 |
| | | R:
5'-CGTTCATCCATAGTTGCCTGAC-3' | 22 | |
|
blaCTX-M | 24 (33.3) | | | |
|
blaCTX-M | 20 (27.2) | F:
5'-TTAGGAARTGTGCCGCTGYA-3' | 20 | 688 |
|
group 1 | | R:
5'-CGATATCGTTGGTGGTRCCAT-3' | 21 | |
|
blaCTX-M | 3 (4.2) | F:
5'-CGTTAACGGCACGATGAC-3' | 18 | 404 |
|
group 2 | | R:
5'-CGATATCGTTGGTGGTRCCAT-3' | 21 | |
|
blaCTX-M | 3 (4.2) | F:
5'-TCAAGCCTGCCGATCTGGT-3' | 19 | 561 |
|
group 9 | | R:
5'-TGATTCTCGCCGCTGAAG-3' | 18 | |
|
blaCTX-M | 0.0 | F:
5'-AACRCRCAGACGCTCTAC-3' | 18 | 326 |
|
group
8/25 | | R:
5'-TCGAGCCGGAASGTGTYAT-3' | 19 | |
|
blaSHV | 2 (2.7) | F:
5'-AGCCGCTTGAGCAAATTAAAC-3' | 21 | 713 |
| | | R:
5'-ATCCCGCAGATAAATCACCAC-3' | 21 | |
|
blaOXA-1 | 1 (1.4) | F:
5'-GGCACCAGATTCAACTTTCAAG-3' | 22 | 564 |
| | | R:
5'-GACCCCAAGTTTCCTGTAAGTG-3' | 22 | |
DNA sequencing
Random PCR products of
blaTEM-, blaSHV-,
blaCTX-M- and blaOXA-1-positive
samples were selected for DNA sequencing to identify the specific
gene subtypes. A total of ~30 µl PCR products were sealed in
sterile Eppendorf tubes and sent to Macrogen, Inc. for sequencing.
PCR products were sequenced on an ABI PRISM® 3730XL
Analyzer (96 capillary types) using the same primer sets (Table I). The results were obtained from
the website of the company. Similarities in the nucleotide
sequences were compared on the GenBank database of the National
Center for Biotechnology Information website using the Basic Local
Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The
sequences obtained for blaTEM,
blaCTX-M and blaSHV genes have
been deposited in the GenBank database (http://getentry.ddbj.nig.ac.jp/) under accession
numbers LC636038 to LC636063.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
(IBM Corp.). Simple descriptive statistics were used to calculate
antibiotic-resistant patterns, and the prevalence and distribution
of ESBL resistance genes. Outcome data were stated as proportions
in frequencies and means ± standard deviation.
Results
Bacterial isolates
A total of 274 Enterobacteriaceae and A.
baumannii isolates obtained from the clinical samples of
patients were used in the present study. The isolates were
recovered from samples of urine, sputum, wound swab, blood, high
vaginal swab, tracheal aspirate, umbilical discharge, eye swab and
cerebral spinal fluid. This indicated that
Enterobacteriaceae and A. baumannii could cause
infections throughout body systems and sites, including
genito-urinary, respiratory, bloodstream, central nervous and soft
tissue infections, which was consistent with a recent study
(30).
Resistance patterns of
Enterobacteriaceae and A. baumannii
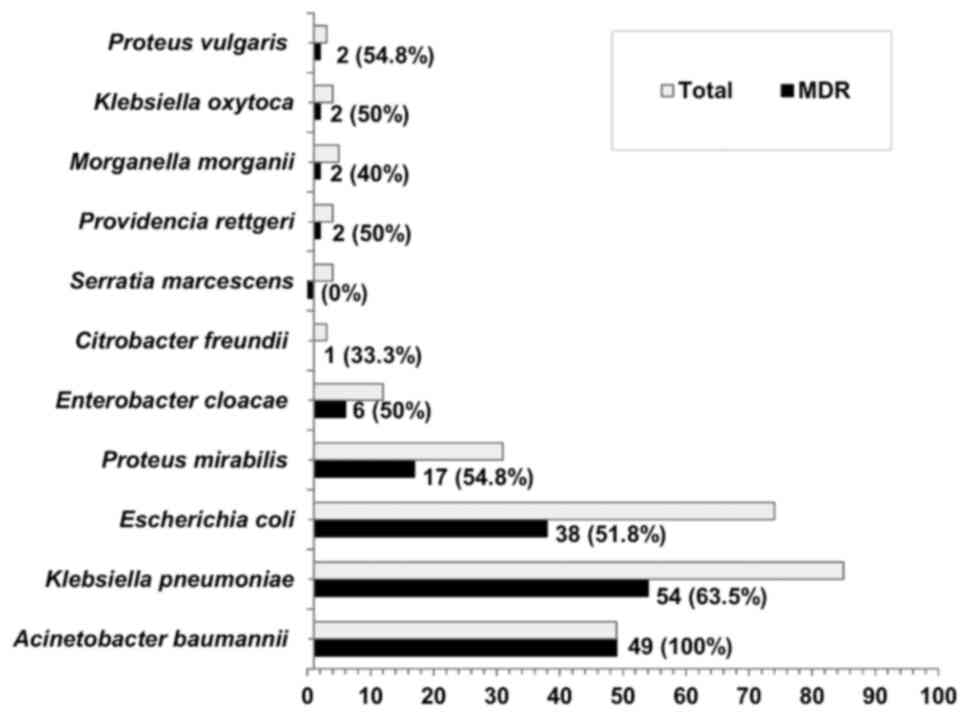
Out of the 274 isolates, 63.1% (n=173) exhibited MDR
patterns to different classes of antibiotics. Fig. 1 illustrated the MDR patterns among
different isolates. The MDR pattern was determined in 100% of A.
baumannii, 63.5% of Klebsiella pneumoniae (K.
pneumoniae), 54.8% of Proteus mirabilis (P. mirabilis)
and 51.8% of E. coli isolates.
Table II
summarized the antibiotic susceptibility of MDR isolates. A.
baumannii exhibited the highest resistance rates to the tested
antibiotics (86% for amikacin, 88% for gentamicin and 100% for
cefuroxime). K. pneumoniae exhibited the highest resistance
rates for cefuroxime (98%), aztreonam and
trimethoprim/sulfamethoxazole (87% each), and cefotaxime (83%).
E. coli exhibited high resistance rates for
trimethoprim/sulfamethoxazole (92%), cefuroxime (87%) and
ceftazidime (71%). P. mirabilis exhibited high resistance
rates for trimethoprim/sulfamethoxazole (100%),
amoxicillin/clavulanate, cefotaxime, cefuroxime (88% each),
cefepime, ciprofloxacin (82% each) and ofloxacin (77%).
 | Table IIPercentage of antimicrobial
resistance among multidrug-resistant Acinetobacter baumannii
and Enterobacteriaceae family members. |
Table II
Percentage of antimicrobial
resistance among multidrug-resistant Acinetobacter baumannii
and Enterobacteriaceae family members.
| | A.
baumannii | K.
pneumoniae | E. coli | Proteus
mirabilis | Enterobacter
cloacae | Citrobacter
freundii | Providencia
rettgeri | Proteus
vulgaris | Morganella
morganii | K.
oxytoca |
|---|
| Agent | (n=49) | (n=54) | (n=38) | (n=17) | (n=6) | (n=1) | (n=2) | (n=2) | (n=2) | (n=2) |
| Amikacin | 86(42) | 41(22) | 16(6) | 35(6) | 33(2) | 0.0 (0) | 50(1) | 50(1) | 50(1) | 50(1) |
|
Amoxicillin/clavulanate | 92(45) | 69(37) | 37(14) | 88(15) | 50(3) | 100(1) | 50(1) | 50(1) | 50(1) | 50(1) |
| Aztreonam | 96(47) | 87(47) | 68(26) | 65(11) | 83(5) | 100(1) | 50(1) | 50(1) | 100(2) | 100(2) |
| Cefepime | 94(46) | 82(44) | 66(25) | 82(14) | 50(3) | 0.0 (0) | 50(1) | 0.0 (0) | 50(1) | 0.0 (0) |
| Cefotaxime | 96(47) | 83(45) | 66(25) | 88(15) | 33(2) | 100(1) | 50(1) | 50(1) | 50(1) | 0.0 (0) |
| Ceftazidime | 94(46) | 80(43) | 71(27) | 59(10) | 50(3) | 0.0 (0) | 0.0 (0) | 50(1) | 50(1) | 0.0 (0) |
| Cefuroxime | 100(49) | 98(53) | 87(33) | 88(15) | 83(5) | 100(1) | 100(2) | 100(2) | 100(2) | 50(1) |
| Ciprofloxacin | 96(47) | 61(33) | 58(22) | 82(14) | 33(2) | 100(1) | 100(2) | 0.0 (0) | 50(1) | 100(2) |
| Colistin | 6(3) | 4.0(2) | 3.0(1) | 6.0(1) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 100(2) | 0.0 (0) |
| Foxitin | 92(45) | 46(25) | 18(7) | 59(10) | 50(3) | 100(1) | 0.0 (0) | 0.0 (0) | 100(2) | 50(1) |
| Gentamicin | 88(43) | 61(33) | 37(14) | 53(9) | 17(1) | 0.0 (0) | 50(1) | 50(1) | 100(2) | 0.0 (0) |
| Imipenem | 94(46) | 35(19) | 13(5) | 29(5) | 17(1) | 0.0 (0) | 50(1) | 50(1) | 100(2) | 0.0 (0) |
| Meropenem | 92(45) | 33(18) | 13(5) | 24(4) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 100(2) | 50(1) |
| Nitrofurantoin | 96(47) | 69(37) | 37(14) | 71(12) | 67(4) | 0.0 (0) | 100(2) | 0.0 (0) | 50(1) | 50(1) |
| Ofloxacin | 94(46) | 63(34) | 50(19) | 77(13) | 33(2) | 100(1) | 100(2) | 0.0 (0) | 50(1) | 100(2) |
| Piperacillin | 94(46) | 61(33) | 47(18) | 65(11) | 50(3) | 0.0 (0) | 50(1) | 50(1) | 100(2) | 50(1) |
|
Piperacillin/tazobactam | 94(46) | 54(29) | 40(15) | 35(6) | 50(3) | 0.0 (0) | 50(1) | 50(1) | 100(2) | 50(1) |
| Tobramicin | 90(44) | 59(32) | 37(14) | 53(9) | 33(2) | 0.0 (0) | 50(1) | 50(1) | 50(1) | 50(1) |
|
Trimethoprim/sulfamethoxazole | 96(47) | 87(47) | 92(35) | 100(17) | 100(6) | 100(1) | 100(2) | 50(1) | 50(1) | 100(2) |
Distribution of ESBL genes among MDR
isolates
Of the 173 MDR Enterobacteriaceae and A.
baumannii isolates, 78 (45.1%) exhibited ESBL production. Of
the 78 isolates, 72 (92.3%) carried ESBL genes as determined using
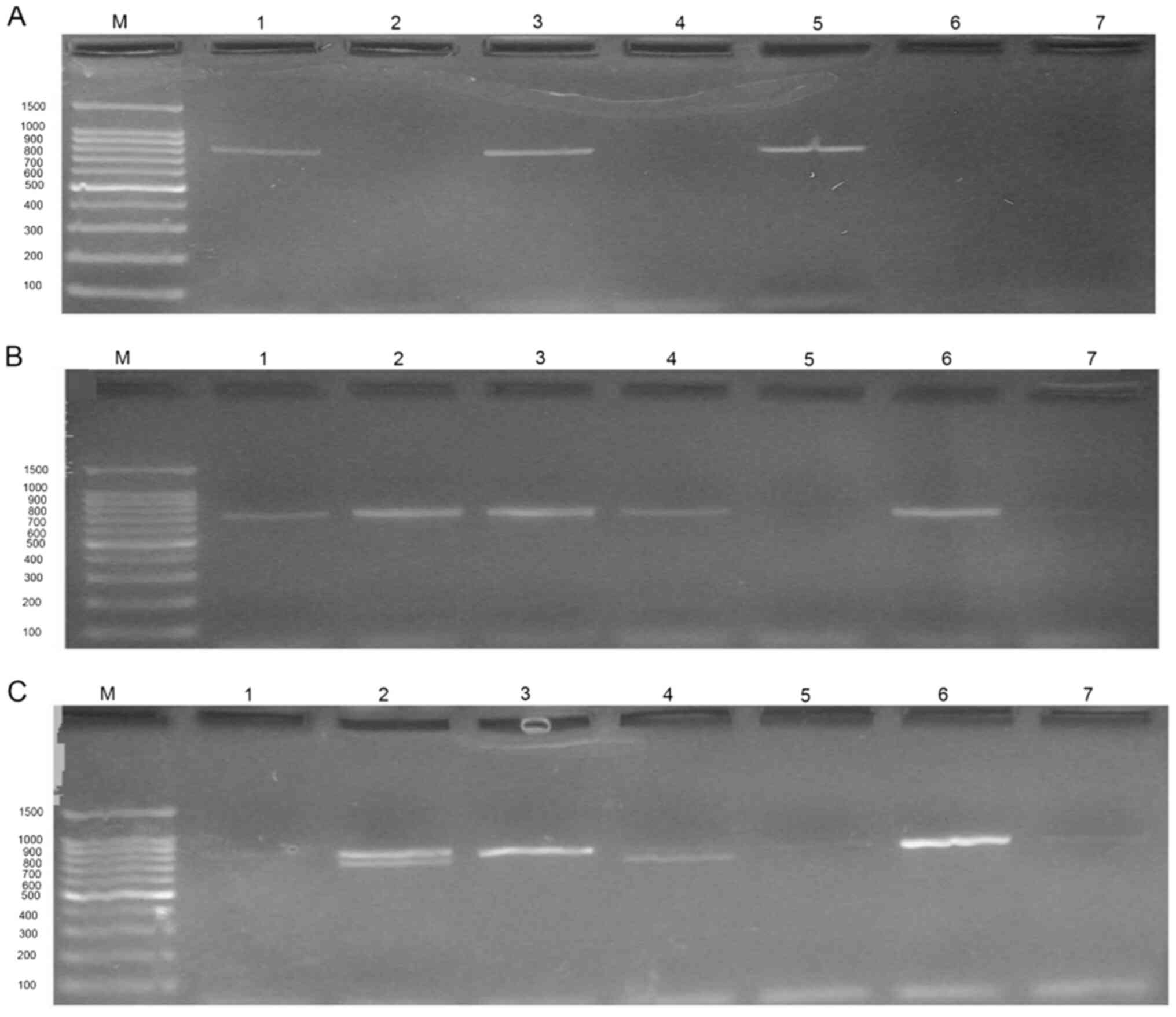
multiplex-PCR. Fig. 2 presents an
example of multiplex-PCR results revealed during the present study.
Of the 72 isolates, 53 were Enterobacteriaceae (K.
pneumoniae, 28; E. coli, 18; P. mirabilis, 5 and
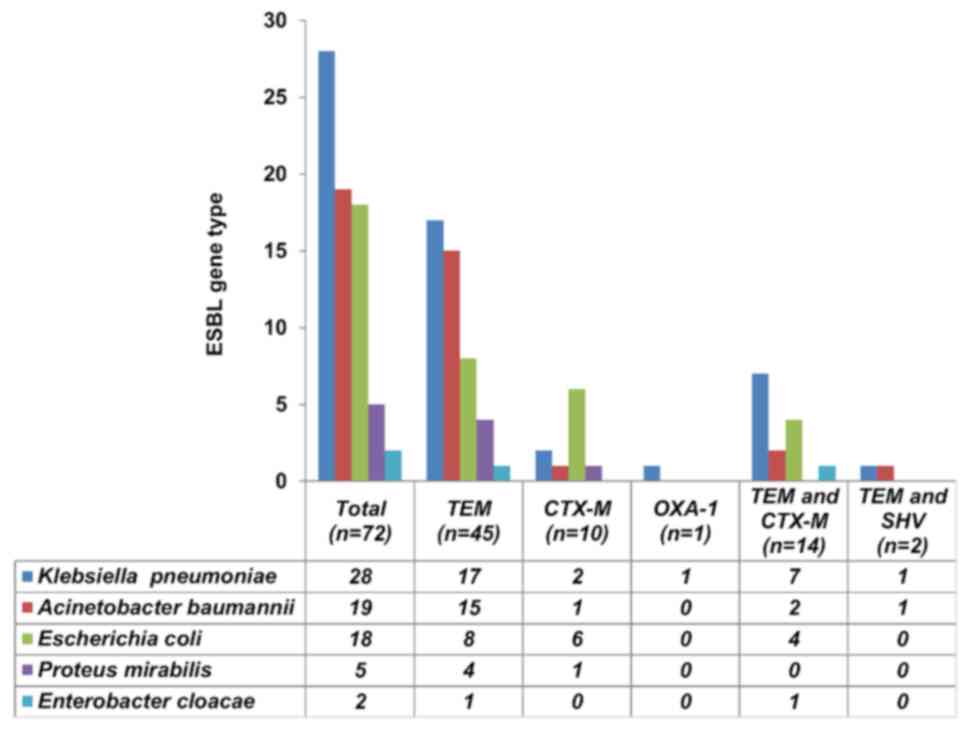
Enterobacter cloacae, 2) and 19 were A. baumannii
(Fig. 3). Collectively, the most
prevalent ESBL resistance genes in the isolates were
blaTEM (84.7%) followed by
blaCTX-M (33.3%), blaSHV (2.7%)
and blaOXA-1 (1.4%). The most frequent
blaCTX-M group was the
blaCTX-M-group 1 (27.2%), followed by the
blaCTX-M-group 2 and 9 (4.2%), whereas none of
the isolates carried the blaCTX-M-group 8/25
(Table I).
Frequency of ESBL genes in
Enterobacteriaceae and A. baumannii
Fig. 3 illustrates
the distribution of ESBL genes among Gram-negative bacterial
isolates. A single blaTEM gene was predominantly
produced by K. pneumoniae (60.7%), A. baumannii
(78.9%) and P. mirabilis (80%), while
blaCTX-M was commonly produced by E. coli
(33.3%).
Out of the total PCR-positive isolates (n=72), the
co-existence of two different genes in a single isolate was
revealed in 16 (22.2%) strains. This combination was mainly
observed between blaTEM and
blaCTX-M (19.4%; 14/72) and between
blaTEM and blaSHV (2.8%, 2/72)
genes. Among the isolates carrying a combination of
blaTEM and blaCTX-M genes
(n=14), the majority were K. pneumoniae (50%, n=7) and E.
coli (28.6%, n=4; Fig. 3).
Distribution of ESBL genes according
to isolate sources
The distribution of various ESBL resistance genes
among the clinical samples of patients is revealed in Table III. Bacterial isolates encoding
various ESBL resistance genes were commonly recovered from sputum
(n=30), followed by urine (n=18), wound (n=13) and blood (n=8)
specimens. A single blaTEM gene was frequently
detected among isolates from sputum (73.3%; 22/30), urine (61.1%;
11/18) and wound (53.8%; 7/13) specimens. The highest frequency of
a single blaCTX-M gene was detected among wound
isolates (23.1%; 3/13) compared with sputum (10%), blood (12.5%;
1/8) and urine (11.1%; 2/18) isolates. The co-existence of
blaTEM and blaCTX-M genes was
observed in 37.5% (3/8) of blood isolates, 23.1% (3/13) of wound
isolates and 22.2% (4/18) of urine isolates.
 | Table IIIDistribution of extended-spectrum
β-lactamase resistance genes in Gram-negative isolates recovered
from clinical samples of patients. |
Table III
Distribution of extended-spectrum
β-lactamase resistance genes in Gram-negative isolates recovered
from clinical samples of patients.
| | Single gene | Combined genes | |
|---|
| Source of
Gram-negative bacteria |
blaTEM (n=45) |
blaCTX-M (n=10) |
blaOXA-1 (n=1) |
blaTEM and
blaCTX-M (n=14) |
blaTEM and
blaSHV (n=2) | Total |
|---|
| Sputum (n=30) | 22 | 3 | 1 | 3 | 1 | 30 |
| Urine (n=18) | 11 | 2 | 0 | 4 | 0 | 18 |
| Wound (n=13) | 7 | 3 | 0 | 3 | 0 | 13 |
| Blood (n=8) | 4 | 1 | 0 | 3 | 0 | 8 |
| Tracheal aspirate
(n=2) | 0 | 1 | 0 | 1 | 0 | 2 |
| Eye swab (n=1) | 1 | 0 | 0 | 0 | 0 | 1 |
Sequencing analysis of resistance
genes encoding ESBL producers
The sequencing analysis of the
blaCTX-M, blaTEM and
blaSHV types is revealed in Table IV. Out of the 20
blaCTX-M family members, the majority (75%; n=15)
carried the blaCTX-M-15 subtype.
blaCTX-M-15 was identified among K.
pneumoniae (n=7) and E. coli (n=5) isolates. Out of the
18 blaTEM family members, TEM-1 was the most
prevalent variant (83.3%, n=15). This subtype was common among
A. baumannii (n=7) and K. pneumoniae (n=5)
isolates.
 | Table IVSequencing analysis results of
blaCTX-M, blaTEM and
blaSHV genes produced by
Enterobacteriaceae members and Acinetobacter
baumannii. |
Table IV
Sequencing analysis results of
blaCTX-M, blaTEM and
blaSHV genes produced by
Enterobacteriaceae members and Acinetobacter
baumannii.
| ESBL gene | Total (n=40) | K. pneumoniae
(n=13) | E. coli
(n=12) | P. mirabilis
(n=2) | E. cloacae
(n=3) | A. baumannii
(n=10) |
|---|
|
blaCTX-M subtype
(n=20) | | | | | | |
|
CTX-M-15 | 15 | 7 | 5 | 1 | 1 | 1 |
|
CTX-M-71 | 1 | 0 | 1 | 0 | 0 | 0 |
|
CTX-M-101 | 1 | 0 | 0 | 0 | 0 | 1 |
|
CTX-M-127 | 1 | 0 | 1 | 0 | 0 | 0 |
|
CTX-M-181 | 1 | 0 | 0 | 0 | 1 | 0 |
|
CTX-M-182 | 1 | 0 | 1 | 0 | 0 | 0 |
|
blaTEM subtype
(n=18) | | | | | | |
|
TEM-1 | 15 | 5 | 2 | 0 | 1 | 7 |
|
TEM-115 | 1 | 0 | 1 | 0 | 0 | 0 |
|
TEM-159 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TEM-169 | 1 | 0 | 1 | 0 | 0 | 0 |
|
blaSHV subtype
(n=2) | | | | | | |
|
SHV-28 | 1 | 0 | 0 | 0 | 0 | 1 |
|
SHV-226 | 1 | 1 | 0 | 0 | 0 | 0 |
Discussion
The emergence of MDR patterns due to ESBL-producing
Enterobacteriaceae and A. baumannii is becoming a
global concern (13,31). The present study determined
antibiotic susceptibility patterns and characterized ESBL genes
among clinical isolates of Enterobacteriaceae and A.
baumannii. Collectively, 63.1% of isolates exhibited MDR to
different antibiotics. In addition, MDR patterns were identified in
63.5% of K. pneumoniae and 51.8% of E. coli isolates.
In Riyadh, the capital of Saudi Arabia, MDR patterns were
identified in 67% of uropathogenic E. coli isolates at a
tertiary healthcare center (19).
These values were higher than those reported in Libya, where the
MDR phenotype was detected in 33.2% of E. coli and 42% of
K. pneumoniae isolates from patients with urinary tract
infections (32). The results were
also consistent with the observed high prevalence of MDR patterns
(100%)among clinical isolates of Enterobacteriaceae carrying
ESBL resistance genes collected in Ethiopia (33).
In the present study, the overall resistance rates
of MDR Enterobacteriaceae and A. baumannii were very
high for most examined antibiotics, except for colistin. Previous
studies have reported that co-resistance to several antibiotic
classes of penicillins, cephalosporins, aminoglycosides,
fluoroquinolones, trimethoprim/sulfamethoxazole and carbapenems was
common among ESBL-producing Gram-negative bacteria (7,19,33,34).
These findings indicated that the emergence of ESBL-producing
microorganisms could cause susceptibility to various
antibiotics.
A multiplex-PCR assay has been proposed to rapidly
detect several resistance genes encoding ESBL-producing
Gram-negative bacteria (28,35).
However, complete gene sequencing for the blaTEM,
blaSHV, blaCTX-M and
blaOXA-1 types is essential to differentiate
narrow-spectrum β-lactamases from ESBL (35). In the present study, among 81
phenotypically identified ESBL isolates, 88.9% carried one or more
of the following resistant genes: blaTEM,
blaCTX-M, blaSHV and
blaOXA. Previous studies have reported that
bacteria carrying ESBL genes confer resistance to extended-spectrum
cephalosporins, β-lactam agents and other antibiotic classes
(7,19,20).
This phenomenon may pose serious public health risks, as it would
result in substantial limitations in therapeutic options. Thus,
appropriate control measures, including establishing screening
strategies for identifying ESBL-producing bacteria, are required to
prevent such strains.
blaTEM was the most prevalent
gene detected in ESBL-producing Gram-negative bacteria in the
present study. This was inconsistent with a study from the Eastern
region of Saudi Arabia, where blaCTX-M (97.4%)
was more frequent than blaSHV (23.1%) and
blaTEM (0.0%) in Enterobacteriaceae
(16). Similarly, the predominance
of the blaCTX-M type in ESBL-producing
Gram-negative bacteria in the Eastern region have been documented
by other studies (4,36). Worldwide studies have reported
different ESBL resistance genes produced by Gram-negative bacteria.
For instance, blaCTX-M was the most prevalent
type in the Asian Pacific region, followed by
blaSHV and blaTEM (1). In Nigeria, the most frequent gene
types among isolates from patients with surgical site infections
were blaSHV, blaCTX-M and
blaOXA (37). In
Burkina Faso, the most prevalent ESBL resistance genes were
blaCTX-M (40.1%), blaTEM
(26.2%) and blaSHV (5.9%) in
Enterobacteriaceae (38).
These results, coupled with the present findings, revealed that the
prevalence of ESBL gene types can vary between locations and
geographical regions.
MDR ESBL-producing K. pneumoniae and A.
baumannii have become common causes of healthcare-related
infections (11,31). In the present study, the prevalence
of blaTEM in K. pneumoniae was revealed to
be 60.7%. Increasing rates of the blaTEM gene
have been reported among clinical isolates of K. pneumoniae
in Al-Qassim (70.9%) (8) and
Riyadh (54.05%), in the Central region of Saudi Arabia (17). On the other hand, the frequency of
blaTEM was high among A. baumannii
isolates, which was consistent with a previous study from the
Makkah city in the western region of the country (31). These high rates, which indicated
the dissemination of such ESBL-producing isolates, is alarming for
multiple hospitals. The high prevalence of the
blaTEM gene detected in K. pneumoniae and
A. baumannii isolates may increase the incidence rate of
infection caused by these ESBL producers across different regions.
This renders extensive surveillance studies in local and national
hospitals in Saudi Arabia necessary to understand the transmission
and epidemiology of resistance genes encoding ESBL-producing
bacteria. However, using molecular methods in local hospitals to
detect resistance genes may help develop effective new
antimicrobial treatments against ESBL producers and improve the
infection control system. In the present study, E. coli
commonly produced the blaCTX-M gene (33.3%).
Similarly, the predominance of E. coli carrying
blaCTX-M genes has been reported in the western
region of Saudi Arabia (39).
Previous studies revealed that blaCTX-M is the
most prevalent gene among uropathogenic E. coli isolates
from patients with hospital and community-acquired infections
(19,29). Furthermore, ESBL-producing E.
coli collected from fecal colonization was revealed to produce
the blaCTX-M gene as it has been reported in a
previous study (40). Several
factors and mechanisms contribute to the spread of bacterial clones
carrying the blaCTX-M gene in Saudi Arabia,
including plasmid dissemination and the clonal spread of bacterial
strains, the frequent use of cephalosporins and the large
population of migrant workers (13). According to Yasir et al
(39), the high diversity in the
E. coli clones may have arisen due to the fact that ~50% of
the population of Saudi Arabia are expatriates from developing
countries, including Pakistan, India, Bangladesh, the Philippines
and African countries where self-medication in patients is
evident.
The present study revealed that
blaCTX-M-15 was the most frequent subtype of the
blaCTX-M type. This was consistent with data from
Saudi Arabia (6,16,17)
and several other parts of the world (1,2).
These findings indicated that blaCTX-M-15 is a
public health concern, since it is the most widespread gene
worldwide. The emergence of the blaCTX-M-15
variant has been revealed to be attributed to the horizontal gene
transfer of genetic elements and the clonal expansion of
microorganisms (19,41). Furthermore, the widespread and
unnecessary use of ceftriaxone and cefotaxime have contributed to
the emergence and spread of blaCTX-M resistance
genes (2).
Multiple ESBL resistance genes in a single
bacterium render that strain more difficult to treat with several
antibiotic drugs (16). In the
present study, the co-existence of two different ESBL genes in the
same strain was detected in 22.2% of isolates. However, the most
common combination of ESBL resistance genes was between
blaTEM and blaCTX-M, which was
consistent with studies from Pakistan (42) and Algeria (12). However, the combined production of
blaTEM and blaCTX-M genes was
more frequently detected in K. pneumoniae (50%) and E.
coli (28.6%) isolates. These figures were lower than those
reported in Nepal, where two or more ESBL genes were present in
100% of Klebsiella spp. and 56.2% of E. coli clinical
isolates from a teaching hospital (7). The value reported in the present
study was considerably higher than the 3.4% reported in
uropathogenic E. coli from the Eastern region of Saudi
Arabia (29).
The present findings revealed that sputum was the
most frequent source of various ESBL resistance genes in
Gram-negative bacteria. This may be due to the several sputum
samples collected from patients at intensive care unit (ICU) wards.
A previous study indicated that the characterization of antibiotic
susceptibility of bacterial pathogens from the sputum of patients
in the ICU with ventilator-associated pneumonia can help control
this type of infection (43). In
addition, it is known that most patients admitted to the ICU are
immunocompromised and/or undergoing invasive procedures, which
would lead to prolonged antibiotic therapy (25). The extended stay, selective
pressure and frequent use of antibiotic treatment of patients in
the ICU contribute to the increase in ESBL producers (43).
The present study has several limitations that need
to be addressed in future studies. Firstly, the study was
laboratory-based; therefore, clinical data of the patients were not
obtained to analyze the risk factors for ESBL infection and
understand the epidemiological spread of ESBL genes. Secondly, the
study was conducted in a single center in southern Saudi Arabia.
The results can therefore not be representative of all parts of the
southern region. Multicenter studies are required to confirm these
findings. Thirdly, the AmpC β-lactamase class and other types of
ESBL enzymes, (such as blaVEB,
blaPER, blaGES and
blaBEL) which confer significant antibiotic
resistance among Gram-negative bacteria, were not examined.
In conclusion, the ESBL resistance genes were a
significant cause of MDR patterns and conferred susceptibility to
various antibiotic agents in Enterobacteriaceae and A.
baumannii. The present study reported high levels of various
resistance genes in ESBL-producing isolates, with
blaTEM being the most prevalent type. In
addition, the co-existence of two different ESBL genes has been
frequently detected in a single bacterial pathogen (12,42).
The blaCTX-M-15 gene is the predominant variant
among isolates carrying the blaCTX-M type. The
emergence of various ESBL-resistant and coexisting genes in
Enterobacteriaceae and A. baumannii is alarming and
may significantly limit the efficacy of therapeutic options in
hospital settings. However, extensive surveillance studies at both
the local and national levels are urgently required to obtain an
understanding of the transmission and epidemiology of resistance
genes in ESBL-producing bacteria. Using molecular methods at local
hospitals to detect resistance genes in ESBL-producing bacteria is
recommended to improve the infection control system and help set
effective antibiotic therapy plans.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Deanship of Scientific
Research at University of Bisha (Bisha, Saudi Arabia), as a part of
project number UB-12-1438.
Availability of data and materials
All data generated or analyzed during the study are
included in this published article. The datasets generated during
the present study are available in the Genbank repository,
(http://getentry.ddbj.nig.ac.jp/;
accession numbers LC636038- LC636063).
Authors' contributions
MEI, TBA, MA and BKE conceived the idea of the
study and developed the protocol. MEI and MA designed and conducted
the study. MEI and TBA analyzed and interpreted the data and wrote
the initial draft. MEI, TBA, MA, BKE reviewed the literature. BKE
revised the study for important intellectual contents. MEI and BKE
confirm the authenticity of all the raw data. All authors have read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The Research and Ethics Committee at the College of
Medicine, University of Bisha (Bisha, Saudi Arabia) reviewed and
approved the present study protocol.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sheng WH, Badal RE and Hsueh PR: SMART
Program. Distribution of extended-spectrum β-lactamases, AmpC
β-lactamases, and carbapenemases among Enterobacteriaceae
isolates causing intra-abdominal infections in the Asia-Pacific
region: Results of the study for Monitoring Antimicrobial
Resistance Trends (SMART). Antimicrob Agents Chemother.
57:2981–2988. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Manyahi J, Moyo SJ, Tellevik MG, Ndugulile
F, Urassa W, Blomberg B and Langeland N: Detection of CTX-M-15
beta-lactamases in Enterobacteriaceae causing hospital- and
community-acquired urinary tract infections as early as. 2004, in
Dar es Salaam, Tanzania. BMC Infect Dis. 17(282)2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jiang W, Yang W, Zhao X, Wang N and Ren H:
Klebsiella pneumoniae presents antimicrobial drug resistance
for β-lactam through the ESBL/PBP signaling pathway. Exp Ther Med.
19:2449–2456. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hassan MI, Alkharsah KR, Alzahrani AJ,
Obeid OE, Khamis AH and Diab A: Detection of extended spectrum
beta-lactamases-producing isolates and effect of AmpC overlapping.
J Infect Dev Ctries. 7:618–629. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Marie MA, John J, Krishnappa LG and
Gopalkrishnan S: Molecular characterization of the β-lactamases in
Escherichia coli and Klebsiella pneumoniae from a
tertiary care hospital in Riyadh, Saudi Arabia. Microbiol Immunol.
57:805–810. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yezli S, Shibl AM and Memish ZA: The
molecular basis of β-lactamase production in Gram-negative bacteria
from Saudi Arabia. J Med Microbiol. 64:127–136. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Parajuli NP, Maharjan P, Joshi G and
Khanal PR: Emerging Perils of Extended Spectrum β-Lactamase
Producing Enterobacteriaceae Clinical Isolates in a Teaching
Hospital of Nepal. BioMed Res Int. 2016:p1–7. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tawfik AF, Alswailem AM, Shibl AM and
Al-Agamy MHM: Prevalence and genetic characteristics of TEM, SHV,
and CTX-M in clinical Klebsiella pneumoniae isolates from
Saudi Arabia. Microb Drug Resist. 17:383–388. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Dirar MH, Bilal NE, Ibrahim ME and Hamid
ME: Prevalence of extended-spectrum β-lactamase (ESBL) and
molecular detection of blaTEM,
blaSHV and blaCTX-M genotypes
among Enterobacteriaceae isolates from patients in Khartoum,
Sudan. Pan Afr Med J. 37(213)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shahcheraghi F: Haleh Moezi MMF:
Distribution of TEM and SHV beta-lactamase genes among
Klebsiella pneumoniae strains isolated from patients in
Tehran. Med Sci Monit. 13:BR247–BR251. 2007.PubMed/NCBI
|
|
11
|
Somily AM, Habib HA, Absar MM, Arshad MZ,
Manneh K, Al Subaie SS, Al Hedaithy MA, Sayyed SB, Shakoor Z and
Murray TS: ESBL-producing Escherichia coli and Klebsiella
pneumoniae at a tertiary care hospital in Saudi Arabia. J
Infect Dev Ctries. 8:1129–1136. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nedjai S, Barguigua A, Djahmi N, Jamali L,
Zerouali K, Dekhil M and Timinouni M: Prevalence and
characterization of extended spectrum beta-lactamase-producing
Enterobacter cloacae strains in Algeria. J Infect Dev
Ctries. 7:804–811. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zowawi HM, Balkhy HH, Walsh TR and
Paterson DL: β-lactamase production in key gram-negative pathogen
isolates from the Arabian Peninsula. Clin Microbiol Rev.
26:361–380. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Alyamani EJ, Khiyami AM, Booq RY, Majrashi
MA, Bahwerth FS and Rechkina E: The occurrence of ESBL-producing
Escherichia coli carrying aminoglycoside resistance genes in
urinary tract infections in Saudi Arabia. Ann Clin Microbiol
Antimicrob. 16(1)2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Al-Agamy MHM, Shibl AM and Tawfik AF:
Prevalence and molecular characterization of extended-spectrum
beta-lactamase-producing Klebsiella pneumoniae in Riyadh,
Saudi Arabia. Ann Saudi Med. 29:253–257. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hassan H and Abdalhamid B: Molecular
characterization of extended-spectrum beta-lactamase producing
Enterobacteriaceae in a Saudi Arabian tertiary hospital. J
Infect Dev Ctries. 8:282–288. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Al-Qahtani AA, Al-Agamy MH, Ali MS,
Al-Ahdal MN, Aljohi MA and Shibl AM: Characterization of
extended-spectrum beta-lactamase-producing Klebsiella
pneumoniae from Riyadh, Saudi Arabia. J Chemother. 26:139–145.
2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Al-Agamy MH, Shibl AM, Hafez MM, Al-Ahdal
MN, Memish ZA and Khubnani H: Molecular characteristics of
extended-spectrum β-lactamase-producing Escherichia coli in
Riyadh: Emergence of CTX-M-15-producing E. coli ST131. Ann
Clin Microbiol Antimicrob. 13(4)2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Alqasim A, Abu Jaffal A and Alyousef AA:
Prevalence of Multidrug Resistance and Extended-Spectrum
β-lactamase Carriage of Clinical Uropathogenic Escherichia
coli Isolates in Riyadh, Saudi Arabia. Int J Microbiol.
2018(3026851)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ibrahim ME, Abbas M, Al-Shahrai AM and
Elamin BK: Phenotypic Characterization and Antibiotic Resistance
Patterns of Extended-Spectrum β-Lactamase- and AmpC
β-Lactamase-Producing Gram-Negative Bacteria in a Referral
Hospital, Saudi Arabia. Can J Infect Dis Med Microbiol.
2019(6054694)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Al-Garni SM, Ghonaim MM, Ahmed MMM,
Al-Ghamdi AS and Ganai FA: Risk factors and molecular features of
extended-spectrum beta-lactamase producing bacteria at southwest of
Saudi Arabia. Saudi Med J. 39:1186–1194. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Al Bshabshe A, Joseph MRP, Al Hussein A,
Haimour W and Hamid ME: Multidrug resistance Acinetobacter species
at the intensive care unit, Aseer Central Hospital, Saudi Arabia: A
one year analysis. Asian Pac J Trop Med. 9:903–908. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Elabd FM, Al-Ayed MSZ, Asaad AM, Alsareii
SA, Qureshi MA and Musa HA-A: Molecular characterization of
oxacillinases among carbapenem-resistant Acinetobacter
baumannii nosocomial isolates in a Saudi hospital. J Infect
Public Health. 8:242–247. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Almaghrabi MK, Joseph MRP, Assiry MM and
Hamid ME: Multidrug-Resistant Acinetobacter baumannii: An
Emerging Health Threat in Aseer Region, Kingdom of Saudi Arabia.
Can J Infect Dis Med Microbiol. 2018(9182747)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ibrahim ME: High antimicrobial resistant
rates among Gram-negative pathogens in intensive care units. A
retrospective study at a tertiary care hospital in Southwest Saudi
Arabia. Saudi Med J. 39:1035–1043. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jarlier V, Nicolas MH, Fournier G and
Philippon A: Extended broad-spectrum beta-lactamases conferring
transferable resistance to newer beta-lactam agents in
Enterobacteriaceae: Hospital prevalence and susceptibility
patterns. Rev Infect Dis. 10:867–878. 1988.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Clinical and Laboratory Standards
Institute (CLSI): Performance Standards for Antimicrobial
Susceptibility Testing; Twenty-Seventh Informational Supplement.
Document M100-S27. CLSI, Wayne, PA, 2017.
|
|
28
|
Dallenne C, Da Costa A, Decré D, Favier C
and Arlet G: Development of a set of multiplex PCR assays for the
detection of genes encoding important beta-lactamases in
Enterobacteriaceae. J Antimicrob Chemother. 65:490–495.
2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Mashwal FA, El Safi SH, George SK, Adam AA
and Jebakumar AZ: Incidence and molecular characterization of the
extended spectrum beta lactamase-producing Escherichia coli
isolated from urinary tract infections in Eastern Saudi Arabia.
Saudi Med J. 38:811–815. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gupta V, Ye G, Olesky M, Lawrence K,
Murray J and Yu K: Trends in resistant Enterobacteriaceae
and Acinetobacter species in hospitalized patients in the
United States: 2013-2017. BMC Infect Dis. 19(742)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Alyamani EJ, Khiyami MA, Booq RY, Alnafjan
BM, Altammami MA and Bahwerth FS: Molecular characterization of
extended-spectrum beta-lactamases (ESBLs) produced by clinical
isolates of Acinetobacter baumannii in Saudi Arabia. Ann
Clin Microbiol Antimicrob. 14(38)2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Abujnah AA, Zorgani A, Sabri MAM,
Mohammady HE, Khalek RA and Ghenghesh KS: Multidrug resistance and
extended-spectrum β-lactamases genes among Escherichia coli
from patients with urinary tract infections in Northwestern Libya.
Libyan J Med. 10(26412)2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zeynudin A, Pritsch M, Schubert S,
Messerer M, Liegl G, Hoelscher M, Belachew T and Wieser A:
Prevalence and antibiotic susceptibility pattern of CTX-M type
extended-spectrum β-lactamases among clinical isolates of
gram-negative bacilli in Jimma, Ethiopia. BMC Infect Dis.
18(524)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Dirar M, Bilal N, Ibrahim ME and Hamid M:
Resistance Patterns and Phenotypic Detection of β -lactamase
Enzymes among Enterobacteriaceae Isolates from Referral
Hospitals in Khartoum State, Sudan Bacterial isolates and
antimicrobial susceptibility testing. Cureus.
12(e7260)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Woodford N: Rapid characterization of
beta-lactamases by multiplex PCR. Methods Mol Biol. 642:181–192.
2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bindayna K, Khanfar HS, Senok AC and Botta
GA: Predominance of CTX-M genotype among extended spectrum beta
lactamase isolates in a tertiary hospital in Saudi Arabia. Saudi
Med J. 31:859–863. 2010.PubMed/NCBI
|
|
37
|
Olowo-Okere A, Ibrahim YKE and Olayinka
BO: Molecular characterisation of ESBL producing Gram-negative
bacterial isolates from surgical wounds of patients at a hospital
in north central Nigeria. J Glob Antimicrob Resist. 14:85–89.
2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kpoda DS, Ajayi A, Somda M, Traore O,
Guessennd N, Ouattara AS, Sangare L, Traore AS and Dosso M:
Distribution of resistance genes encoding ESBLs in
Enterobacteriaceae isolated from biological samples in
health centers in Ouagadougou, Burkina Faso. BMC Res Notes.
11(471)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yasir M, Ajlan AM, Shakil S, Jiman-Fatani
AA, Almasaudi SB, Farman M, Baazeem ZM, Baabdullah R, Alawi M,
Al-Abdullah N, et al: Molecular characterization, antimicrobial
resistance and clinico-bioinformatics approaches to address the
problem of extended-spectrum β-lactamase-producing Escherichia
coli in western Saudi Arabia. Sci Rep. 8(14847)2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Al-Agamy MH, El Mahdy TS and Shibl AM:
Fecal Colonization with Extended-Spectrum Beta-Lactamase and
AmpC-Producing Escherichia coli. BioMed Res Int.
2016(3704150)2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Barguigua A, El Otmani F, Talmi M,
Bourjilat F, Haouzane F, Zerouali K and Timinouni M:
Characterization of extended-spectrum β-lactamase-producing
Escherichia coli and Klebsiella pneumoniae isolates
from the community in Morocco. J Med Microbiol. 60:1344–1352.
2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Habeeb MA, Sarwar Y, Ali A, Salman M and
Haque A: Rapid emergence of ESBL producers in E. coli
causing urinary and wound infections in Pakistan. Pak J Med Sci.
29:540–544. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ning BT, Zhang CM, Liu T, Ye S, Yang ZH
and Chen ZJ: Pathogenic analysis of sputum from
ventilator-associated pneumonia in a pediatric intensive care unit.
Exp Ther Med. 5:367–371. 2013.PubMed/NCBI View Article : Google Scholar
|