Introduction
Breast cancer is the most frequently diagnosed type
of cancer in women and is the leading cause of cancer-associated
mortality in women worldwide (1).
In total, ≥70% of breast cancer cases are classified as estrogen
receptor (ER)α-positive breast cancer (2). Tamoxifen (TAM), a partial ER agonist,
is the gold standard first-line endocrine therapy for premenopausal
patients with ERα-positive breast cancer. Furthermore, the adjuvant
therapy of breast cancer with TAM for ~5 years can reduce the rate
of disease recurrence by one-half and annual breast cancer
mortality by one-third (3). Despite
these clear benefits, resistance to TAM therapy is a challenge, and
~40% of patients with ERα-positive breast cancer eventually develop
TAM resistance (4). It has been
revealed that the epigenetic regulation of gene expression is a
major cause of TAM resistance in the laboratory and clinic
(5). The methylation of CpG sites
in DNA promoter sequences is an important epigenetic mechanism that
may result in the transcriptional inactivation of genes and the
modulation of drug resistance in breast cancer (6). There is evidence to suggest that TAM
resistance is associated with the methylation and expression of the
N-acetyltransferase 1 (NAT1) gene (7,8).
However, the molecular mechanism involved in the regulation of NAT1
gene expression and methylation remains unknown.
Long non-coding RNA (lncRNA) H19 is an imprinted
oncofetal gene (9). The expression
level of lncRNA H19 is associated with the presence of ERs and
progesterone receptors in mammary and uterine cells (10,11).
The upregulation of H19 may be associated with the poor prognosis
of TAM-resistant breast cancer, and the downregulation of H19
expression has been shown to inhibit the expression of
transcription factors associated with the Wnt pathway and
epithelial-mesenchymal transition (12). In addition, Basak et al
(13) revealed that H19
upregulation was regulated by Notch and hepatocyte growth factor
signaling in endocrine therapy-resistant cells, and that the
inhibition of pathways regulating H19 expression significantly
overcame TAM and fulvestrant resistance. These aforementioned
findings have a certain significance for the present study.
While the mechanism by which H19 regulates
transcription remains largely unknown, Tsang and Kwok (14) reported that H19 regulated multidrug
resistance protein (MDR1) gene promoter methylation and induced
MDR1-associated chemotherapy resistance in hepatocellular carcinoma
cells. Thus, the alteration of target gene methylation may be a
molecular mechanism underlying the regulation of gene transcription
by H19.
Materials and methods
Cell culture and 4-hydroxytamoxifen
treatment
MCF-7 breast cancer cells were obtained from the
American Type Culture Collection and cultured as previously
described (10). Briefly, MCF-7
cells were cultured in RPMI-1640 supplemented with 10% FBS
(Biological Industries), 100 U/ml penicillin, 100 µg/ml
streptomycin and 20 mM L-glutamine (Invitrogen; Thermo Fisher
Scientific, Inc.). TAM-resistant MCF-7 cells were developed by the
treatment of wild-type MCF-7 cells with 1x10-6 M
4-hydroxytamoxifen (Sigma-Aldrich; Merck KGaA) for 21 days and then
1x10-7 M 4-hydroxytamoxifen for 6 months, as described
previously (15). All cells were
maintained in a humidified incubator with 5% CO2 at
37˚C. In the present study, 4-hydroxytamoxifen was used as it is
the main active metabolite of TAM. These 4-hydroxytamoxifen-treated
cells were named as MCF-7R and the corresponding parental cells as
MCF-7.
Cell viability assay
To determine viable cell numbers, cells were plated
in 96-well plates (5,000 cells/well) and treated with either 0.1%
dimethyl sulfoxide or 4-hydroxytamoxifen (1x10-10,
1x10-9, 1x10-8, 1x10-7 and
1x10-6 M) for 24, 48 and 72 h. Cell viability was
determined using an MTS assay and quantified by measuring the
absorbance at 490 nm. Cell viability is expressed as the fold of
the corresponding control. The half-maximal inhibitory
concentration (IC50) of 4-hydroxytamoxifen for cell
growth was determined by a dose-response experiment and calculated
relative to the corresponding control.
Cell transfection and knockdown of
gene expression by RNA interference
A synthetic RNA oligonucleotide targeting H19 was
obtained from Guangzhou RiboBio Co., Ltd. H19 lncRNA was knocked
down using a specific H19-targeting small interfering RNA (siRNA)
and a short hairpin RNA (shRNA) vector. The H19 hairpin siRNA
sequences were 5'-CATCAAAGACACCATCGGA-3'. The siRNA was subcloned
into a pSilencer 2.1-U6 neovector (shRNAH19; Guangzhou RiboBio Co.,
Ltd.). A negative control siRNA and negative control shRNA were
also purchased from Guangzhou RiboBio Co., Ltd.
Briefly, cells were plated in phenol red-free medium
containing 5% stripped FBS (Biological Industries) in 12-well
plates at a density of ~70%. Stripped FBS has been incubated with
activated carbon that removes non-polar, lipophilic material, such
as viruses, growth factors, hormones and cytokines regardless of
molecular weight. This method has little effect on salts, glucose
or amino acids. For the knockdown of H19, the cells were
transfected with 200 nM siRNA H19 for 24 h at 37˚C. Following
knockdown, the cells were treated with 4-hydroxytamoxifen for 48 h
at 37˚C at the concentrations indicated in the respective assay. To
generate a stable lncRNA H19 knockdown cell line, 1 µg H19 shRNA
was transfected into MCF-7R cells using Lipofectamine®
RNAiMAX reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
transfected cells were selected using G418 (Promega Corporation)
and a stable clone was obtained and verified for H19 expression.
All cells were maintained in a humidified incubator with 5%
CO2 at 37˚C. Following incubation for 48 h, the
transfected cells were harvested and utilized for subsequent
experiments.
Breast cancer tissue sample
collection
The current study included 30 primary premenopausal
patients (age, 41.22±3.67 years) with ER-positive breast cancer.
The patients underwent surgical resection at Xiangya Hospital
(Changsha, China) and were randomly enrolled from January 2012 to
December 2013. The resected tumor specimens were immediately frozen
in liquid nitrogen and kept at -80˚C until RNA extraction. The
collection and preservation of tumor and paired tumor adjacent
tissues samples and the obtaining of written informed consent were
approved by the Ethics Committee of Xiangya Hospital (project no.
CTXY-140001-5). Patients continued to take 20-40 mg tamoxifen daily
following surgery for ~1 year. Regular follow-up, including
clinical examination, chest X-ray and mammography, was performed
for ≥5 years.
Reverse transcription-quantitative-PCR
(RT-qPCR)
Total RNAs from harvested cells or patient tissues
were prepared using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol,
and the RNA concentration was determined using a NanoDrop™ 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). The quality of
the RNA samples was confirmed based on a ratio of absorbance values
at 260 and 230 nm of >1.7 and a ratio of absorbance values at
260 and 280 nm of between 1.8 and 2.0. Total RNA (500 ng) was
reversely transcribed to cDNA in a 20-µl reaction mixture
comprising 200 units reverse transcriptase, 50 pmol random hexamer,
1X PCR buffer [10 mM Tris/HCl (pH 9.0), 50 mM KCl and 1.5 mM
MgCl2] and 1 mM deoxynucleotide triphosphates (System
Biosciences, LLC). The reaction products were diluted to a volume
of 100 µl with distilled water prior to qPCR. The qPCR mixture
comprised 2 µl diluted reverse transcription product, 1X SYBR-Green
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
50 nM forward and reverse primers. qPCR was carried out using a
LightCycler® 480 Sequence Detection System (Roche
Diagnostics). Following an initial 10-min incubation at 95˚C,
thermocycling was carried out for 40 cycles of 95˚C for 15 sec and
60˚C for 1 min. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
was used as the reference gene. The primers used were as follows:
H19 forward, 5'-GTCCGGCCTTCCTGAACACCTT-3' and reverse,
5'-GCTTCACCTTCCAGAGCCGAT-3' (10);
NAT1 forward, 5'-AGCACTGGCATGATTCACCTTCT-3' and reverse,
5'-GAGGCTGCCACATCTGGTAT-3' and GAPDH forward,
5'-TTGATTTTGGAGGGATCTCGCTC-3' and reverse,
5'-GAGTCAACGGATTTGGTCGTATTG-3'. The level of RNA was expressed as a
fold of the control calculated using the 2-IICq method
(16).
Western blot analysis
Western blotting was conducted as previously
described with minor modifications (10,17).
Briefly, cells were collected 48 h after the transfection with H19
siRNA, shRNA or the respective negative control to create lncRNA
H19 knockdown cell lines. Total cellular proteins were extracted
from the harvested cells using a lysis buffer (62.5 mM Tris-HCl pH
6.8, 100 mM dithiothreitol, 2% SDS and 10% glycerol). The protein
concentrations were determined using the Bradford method with
Bio-Rad Protein Assay reagent following the manufacturer's
instructions (Bio-Rad Laboratories, Inc.). Then, 20 µg/lane cell
lysate was resolved using 12% sodium dodecyl sulfate polyacrylamide
electrophoresis and transferred to nitrocellulose membranes. The
blots were incubated in blocking buffer comprising 5% non-fat dry
milk in Tris-buffered saline with 0.5% Tween (TBS-T) at room
temperature for 2 h. After washing with TBS-T, the nitrocellulose
membranes were incubated with a specific antibody against NAT1
(anti-rabbit; 1:250, cat. no. ab109114; Abcam) or β-actin
(anti-mouse; 1:2,000; cat. no. AC-15; Sigma-Aldrich; Merck KGaA)
overnight at 4˚C. Following the incubation with a horseradish
peroxidase-conjugated secondary antibody (mouse anti-rabbit IgG;
1:10,000; cat. no. ab6728; Abcam) or rabbit anti-mouse IgG
(1:10,000; cat. no. ab106762; Abcam) at room temperature for 1 h,
the signal was detected using an ECL Western Blotting system
(Promega Corporation) and visualized using the ChemiDoc MP Imaging
System (cat. no. 12003154; Bio-Rad Laboratories, Inc.). β-actin was
used as the loading control.
Bisulfite genomic sequencing PCR (BSP)
assay
Genomic DNA was isolated from the peripheral blood
leucocytes with phenol-chloroform followed by ethanol
precipitation. Chromosomal DNA from the MCF-7 and MCF-7R cells was
treated with bisulfite using an EpiTect Bisulfite kit (Qiagen,
Inc.). The methylation status of six CpG sites in the promoter
region of the NAT1 gene was investigated (-790 to -1 of GenBank
accession no. AY338489 in which the major transcriptional site is
numbered +1) (8). A single ‘C’ at
the corresponding CpG site was considered as complete methylation,
a single ‘T’ as no methylation, and overlapping ‘C’ and ‘T’ as
partial methylation. The gene-specific promoter regions were
amplified via nested PCR from the bisulfite-treated DNA. The
analysis of DNA methylation, including the primer sequences and PCR
conditions used, was performed as previously described (8). The PCR products were purified by gel
electrophoresis using 1% agarose gel, ligated into a pGEM-T plasmid
(Promega Corporation) and transformed into DH5α cells (Invitrogen;
Thermo Fisher Scientific, Inc.) using standard heat-shock
procedures (18). Blue/white
screening was conducted and ≥10 positive bacterial clones were
sequenced as previously described (19). Quality control, sequence analysis
and data illustration were performed using BiQ Analyzer 3.0
software (20). The data have been
deposited in the CNGB Sequence Archive of the China National
GeneBank DataBase with accession number CNP0001707.
Analysis of public breast cancer
datasets
The importance of NAT1 in the outcome of patients
with breast cancer treated with TAM was assessed by analyzing the
relevance of NAT1 expression levels in breast cancer tissues to
patient outcomes in the public datasets GSE2990(21) and GSE20685(22). R script-generated data with
P<0.05 from the GEO2R analysis of the GEO database (https://www.ncbi.nlm.nih.gov/geo/geo2r/)
were obtained (23).
An online survival analysis tool was used to
evaluate the effect of the NAT1 gene on breast cancer prognosis.
Kaplan-Meier plots were generated using an online tool (http://kmplot.com/analysis/index.php?p=service&cancer=breast)
(24).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 21.0; IBM Corp.). Each experiment was performed
at least three times, and the data are presented as the mean ± SEM.
For parametric data, the Student's t-test was used to determine the
statistical significance between two groups, and one-way ANOVA
followed by a post hoc Tukey's test was used to compare differences
among multiple groups. Kaplan-Meier plots for disease-free survival
(DFS) were plotted and analyzed using log-rank tests. A total of 30
primary premenopausal patients were allocated to low and high H19
and NAT1 groups according to whether expression in the tumor tissue
was ≥1.5-fold lower or higher compared with paracancerous tissue.
The fold difference of expression between tumor and adjacent normal
tissue was ≥1.5 in every case. Pearson correlation was used to
measure the correlation between the expression levels of H19 and
NAT1. P<0.05 was considered to indicate a statistically
significant difference.
Results
Establishment of the MCF-7R cell
line
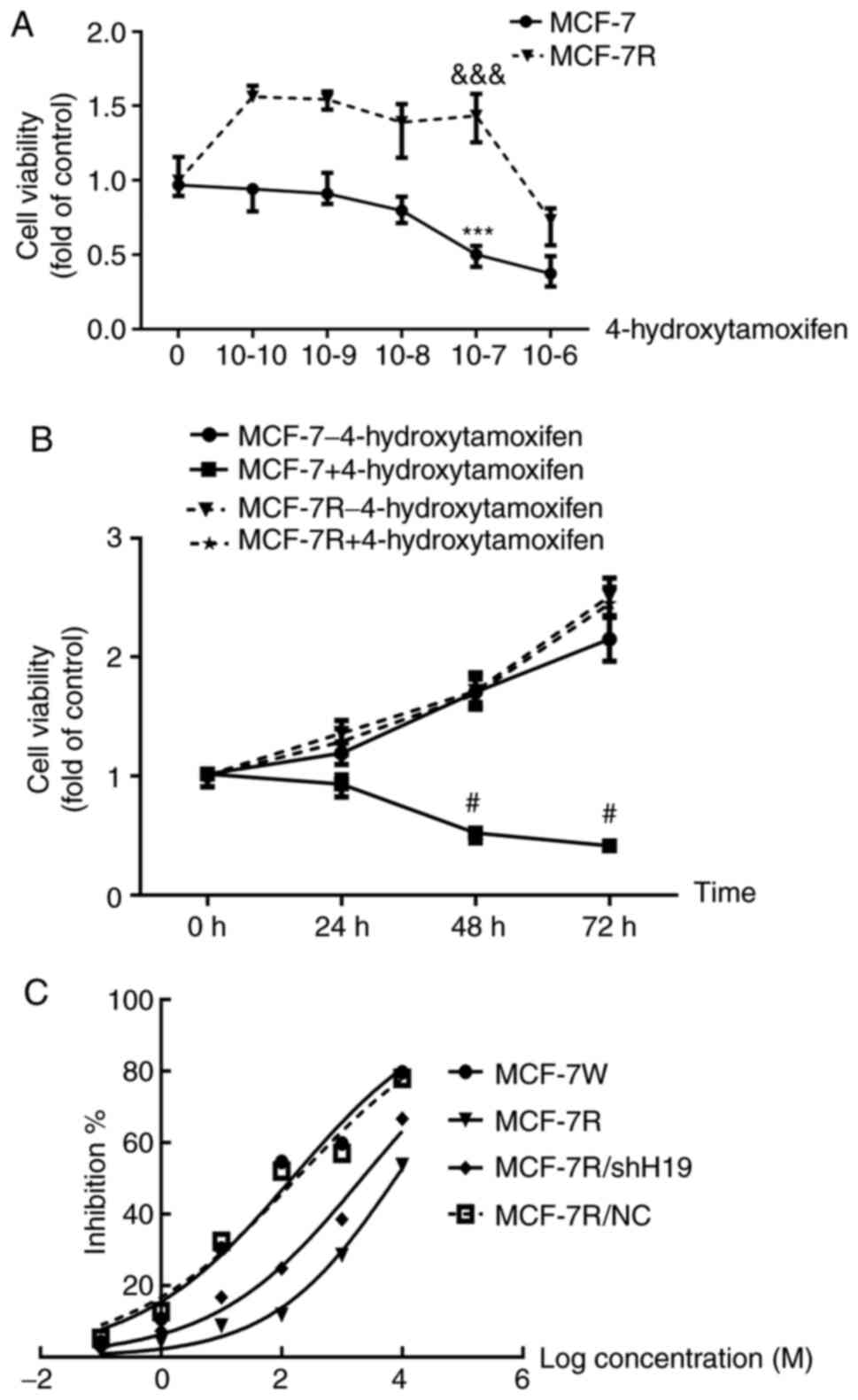
To analyze the sensitivity of the MCF-7 and MCF-7R
cell lines, the two cell lines were treated with different
concentrations of 4-hydroxytamoxifen for 24-72 h. The viability of
the MCF-7 cells was significantly inhibited by 4-hydroxytamoxifen
at a concentration of 1x10-7 M, while the viability of
MCF-7R cells was significantly promoted (Fig. 1A). Furthermore, comparison of the
MCF-7R and MCF-7 cells treated with 1x10-7 M
4-hydroxytamoxifen reveals that the proliferation of the MCF-7R
cells, as indicated by the viable cell number, was significantly
increased at 48 and 72 h (Fig. 1B).
These results suggest that MCF-7R cells have a significant
resistance to 4-hydroxytamoxifen. Furthermore, 4-hydroxytamoxifen
exhibited a dose- (Fig. 1A) and
time-dependent (Fig. 1B) inhibitory
effect on MCF-7 cells. The IC50 values were also
calculated and were 148.1 nM for the MCF-7 cells and 7.753 µM for
the MCF-7R cells (Fig. 1C).
Knockdown of lncRNA H19 restores TAM
sensitivity in MCF-7R cells
As lncRNA H19 is an estrogen-inducible gene
(10) and TAM is a partial ER
agonist, it was hypothesized that lncRNA H19 expression may be
elevated in MCF-7R cells. Therefore, the contribution of lncRNA H19
to the TAM resistance of MCF-7R breast cancer cells was
investigated. The knockdown of lncRNA H19 in MCF-7R cells was
conducted using H19-specific siRNA, which knocked down H19 lncRNA
expression by ~63% compared with that in the cells transfected with
non-specific (NS) siRNA (Fig. 2A).
Transfection with H19 siRNA significantly increased the sensitivity
of the MCF-7R cells to 4-hydroxytamoxifen compared with that of the
cells transfected with NS)siRNA (Fig.
2C). By contrast, transfection with NS siRNA, which did not
affect H19 expression, had no effect on the sensitivity of the
MCF-7R cells to 4-hydroxytamoxifen treatment.
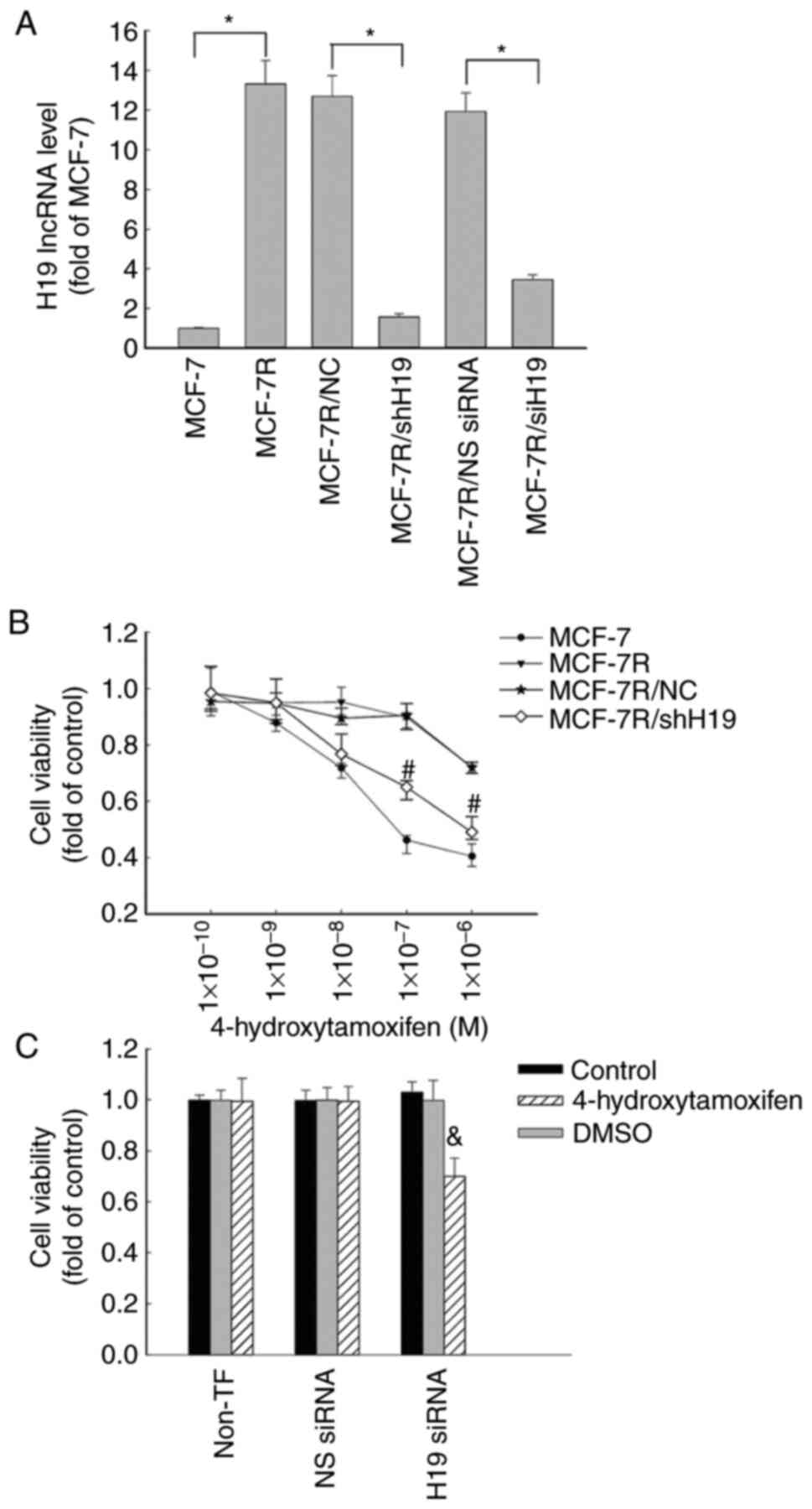 | Figure 2Increased expression of lncRNA H19 in
MCF-7R cells is associated with tamoxifen resistance. (A) MCF-7R
cells were transfected with siH19, NS siRNA, shH19 or NC shRNA. The
levels of lncRNA H19 were quantified using reverse
transcription-quantitative PCR in MCF-7R, H19-knockdown
MCF-7R/siH19, MCF-7R/shH19 and corresponding control cells and are
expressed as fold of that in the parental MCF-7 control.
*P<0.05 as indicated. (B) MCF-7, MCF-7R, MCF-7/NC and
H19-knockdown MCF-7R/shH19 cells were plated in 96-well plates and
treated with various concentrations of 4-hydroxytamoxifen for 48 h
for the determination of cell proliferation using an MTS assay.
#P<0.05 vs. the MCF-7R/NC group. (C) Transfected
MCF-7R/siH19 cells were treated with or without 1x10-7 M
4-hydroxytamoxifen for 48 h for the determination of cell
proliferation using an MTS assay. All data are presented as the
mean ± SEM of three independent experiments.
&P<0.05 vs. the NS siRNA/4-hydroxytamoxifen
group. MCF-7R, tamoxifen-resistant MCF-7 cells; lncRNA, long
non-coding RNA; siH19, siRNA targeting H19; NS, non-specific;
siRNA, small interfering RNA; shH19, shRNA targeting H19; NC,
negative control shRNA; shRNA, short hairpin RNA; non-TF,
non-transfected. |
To further investigate the function of lncRNA H19 in
TAM-resistant cells, MCF-7R/shH19 cells were established by the
stable transfection of MCF-7R cells using a specific H19 shRNA
expression vector to knock down H19 expression. The transfection
reduced the expression of H19 lncRNA by >90% in the MCF-7R/shH19
cells compared with the MCF-7R/NC cells, as revealed using RT-qPCR
analysis. However, H19 expression was not affected in the control
vector transfected MCF-7R/NC cells (Fig. 2A). When lncRNA H19 was knocked down
using shRNA, the MCF-7R cells were more sensitive to
4-hydroxytamoxifen therapy, with a significant decrease in cell
viability (Fig. 2B) and a reduction
in IC50 value from 7.753 µM in the MCF-7R cells to 2.155
µM in the MCF-7R/shH19 cells (Fig.
1C). These data suggest that lncRNA H19 may play a key role in
the 4-hydroxytamoxifen resistance of MCF-7R cells.
NAT1 expression is decreased in MCF-7R
cells and restored by lncRNA H19 knockdown
The mRNA and protein expression levels of NAT1 in
MCF-7R cells were significantly lower compared with those in MCF-7
cells (Fig. 3). Following the
knockdown of H19 in MCF-7R cells using H19 shRNA and siRNA, the
mRNA and protein expression levels of NAT1 were significantly
increased compared with those in the MCF-7R/NC and NS siRNA groups,
respectively (Fig. 3). These
results suggest that H19 serves an important role in TAM-resistant
breast cancer cells via the regulation of NAT1 gene expression.
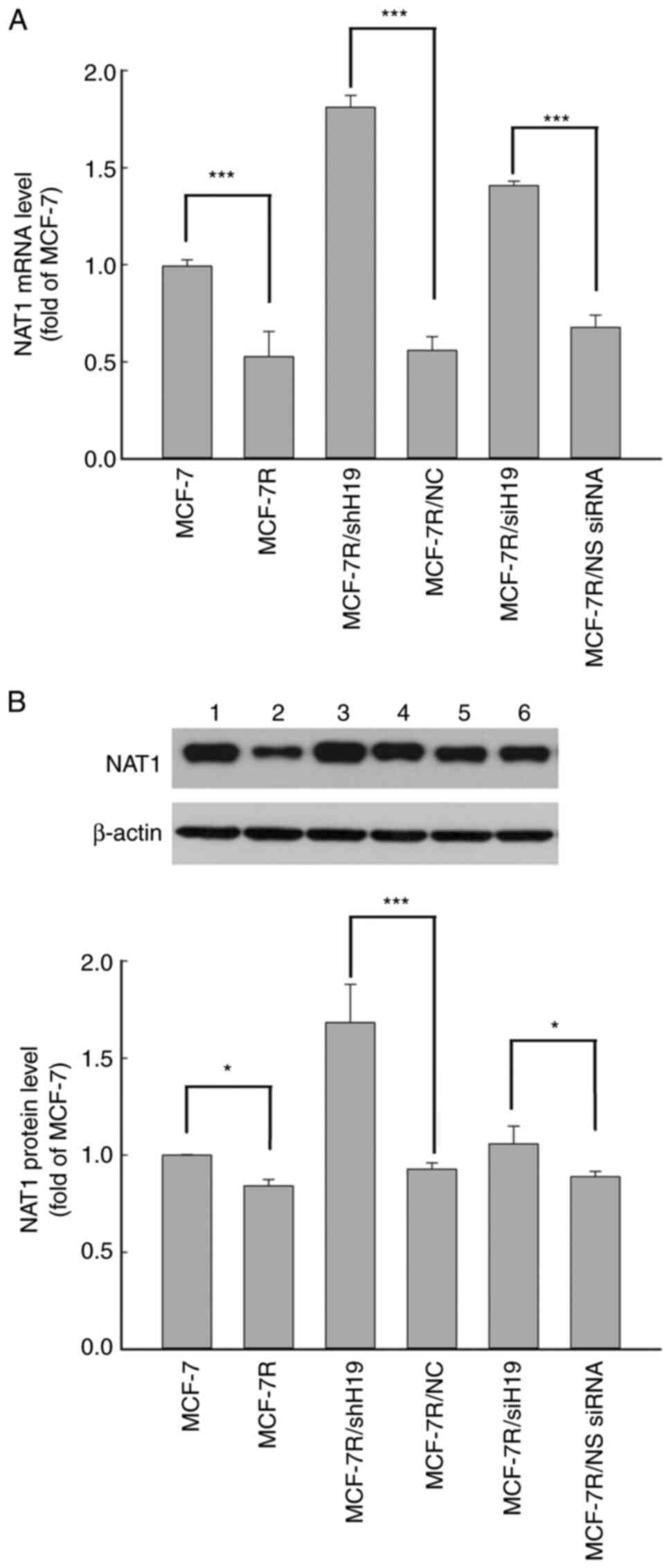 | Figure 3NAT1 expression is decreased in
MCF-7R cells and rescued by long non-coding RNA H19 knockdown. NAT1
(A) mRNA and (B) protein expression levels were detected using
reverse transcription-quantitative PCR and western blot analysis,
respectively, in various cell lines. Representative blots are shown
with lanes as follows: lane 1, MCF-7; lane 2, MCF-7R, lane 3,
MCF-7R/shH19; lane 4, MCF-7/NC; lane 5, MCF-7R/siH19; lane 6,
MCF-7R/NS siRNA. All data are presented as the mean ± SEM of three
independent experiments. *P<0.05 and
***P<0.05 as indicated. NAT1, N-acetyltransferase 1;
MCF-7R, tamoxifen-resistant MCF-7 cells; siH19, siRNA targeting
H19; NS, non-specific; siRNA, small interfering RNA; shH19, shRNA
targeting H19; NC, negative control shRNA; shRNA, short hairpin
RNA. |
lncRNA H19 regulation of NAT1 gene
promoter methylation in MCF-7R cells
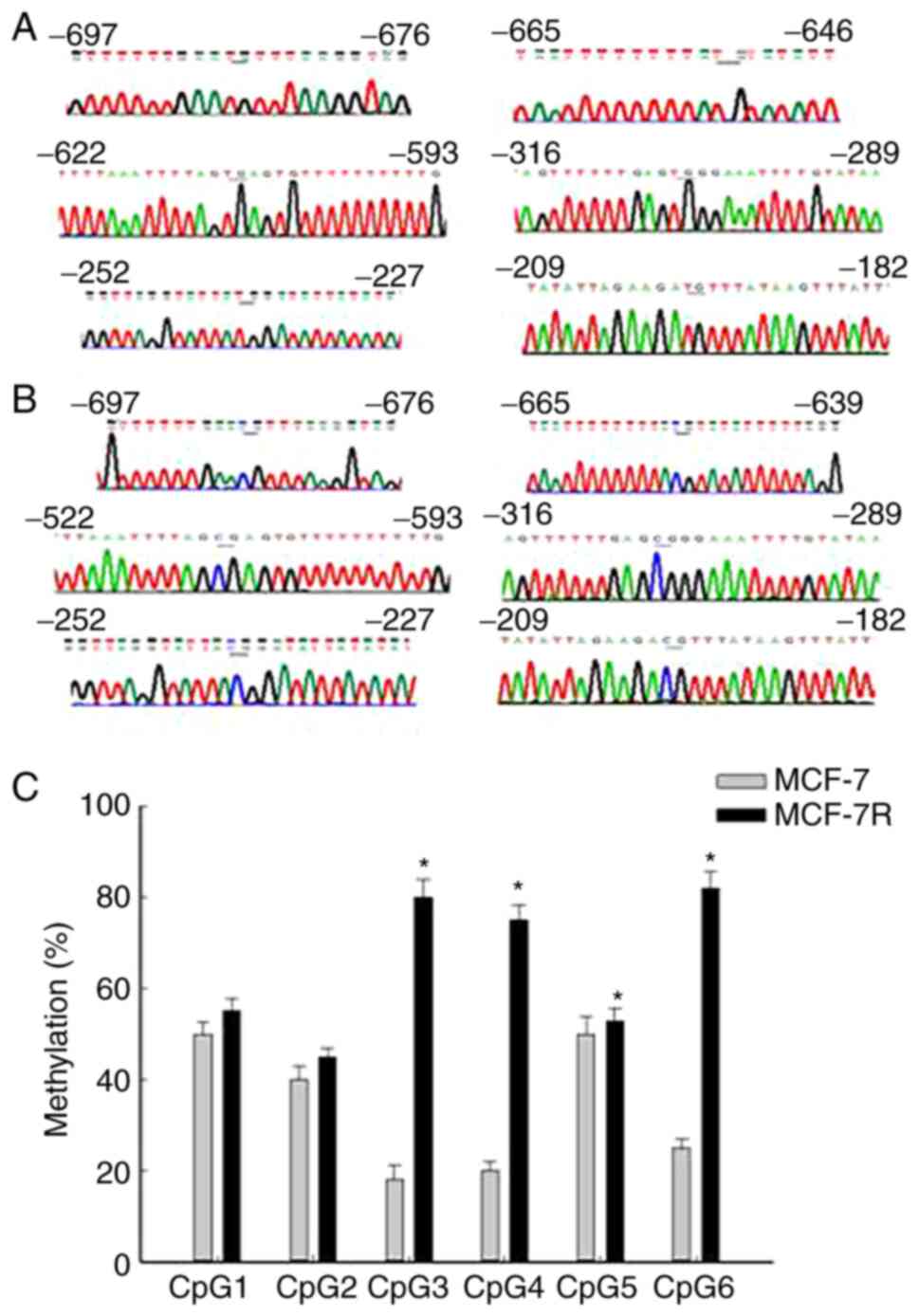
The methylation status of the NAT1 gene promoter in
MCF-7 and MCF-7R cells was examined using a BSP assay. Completely
unmethylated (Fig. 4A) and
completely methylated samples of the NAT1 gene were used (Fig. 4B). The methylation profiles of six
CpG sites were investigated in the NAT1 upstream promoter region
(8). The mean methylation levels in
the MCF-7R group ranged from 45.18% at CpG2 to 82.17% at CpG6
(Fig. 4C). Comparison of NAT1
methylation profiles between the MCF-7 and MCF-7R groups indicated
significant differences at CpG3, CpG4, CpG5 and CpG6 (Fig. 4C). The mean methylation rate of NAT1
in MCF-7R cells was significantly higher compared with that in
MCF-7 cells (Table I). Moreover,
H19 knockdown by siRNA transfection significantly decreased the
mean methylation rate of the NAT1 promoter in MCF-7R cells compared
with that in the untransfected cells (Table I). A higher methylation rate of NAT1
and decreased expression of NAT1 were observed in the MCF-7R group
compared with the MCF-7 group, and H19 knockdown reversed these
resistance-associated changes (Fig.
3 and Table I). This suggests
that H19 may regulate NAT1 and drug resistance by altering the
degree of NAT1 gene promoter methylation in MCF-7R cells.
 | Table IMethylation density of the NAT1 gene
after knockdown of H19 in MCF-7 and MCF-7R cells. |
Table I
Methylation density of the NAT1 gene
after knockdown of H19 in MCF-7 and MCF-7R cells.
| | Methylation density
(%) |
|---|
| Cell line | Non-TF | NS siRNA | siRNA H19 |
|---|
| MCF-7 | 30.78±2.2 | 31.61±0.35 | 36.5±2.92 |
| MCF-7R |
66.11±0.75a | 63.84±2.02 |
26.39±0.75b |
Tumor NAT1 expression is associated
with clinical outcome in patients with breast cancer treated with
TAM
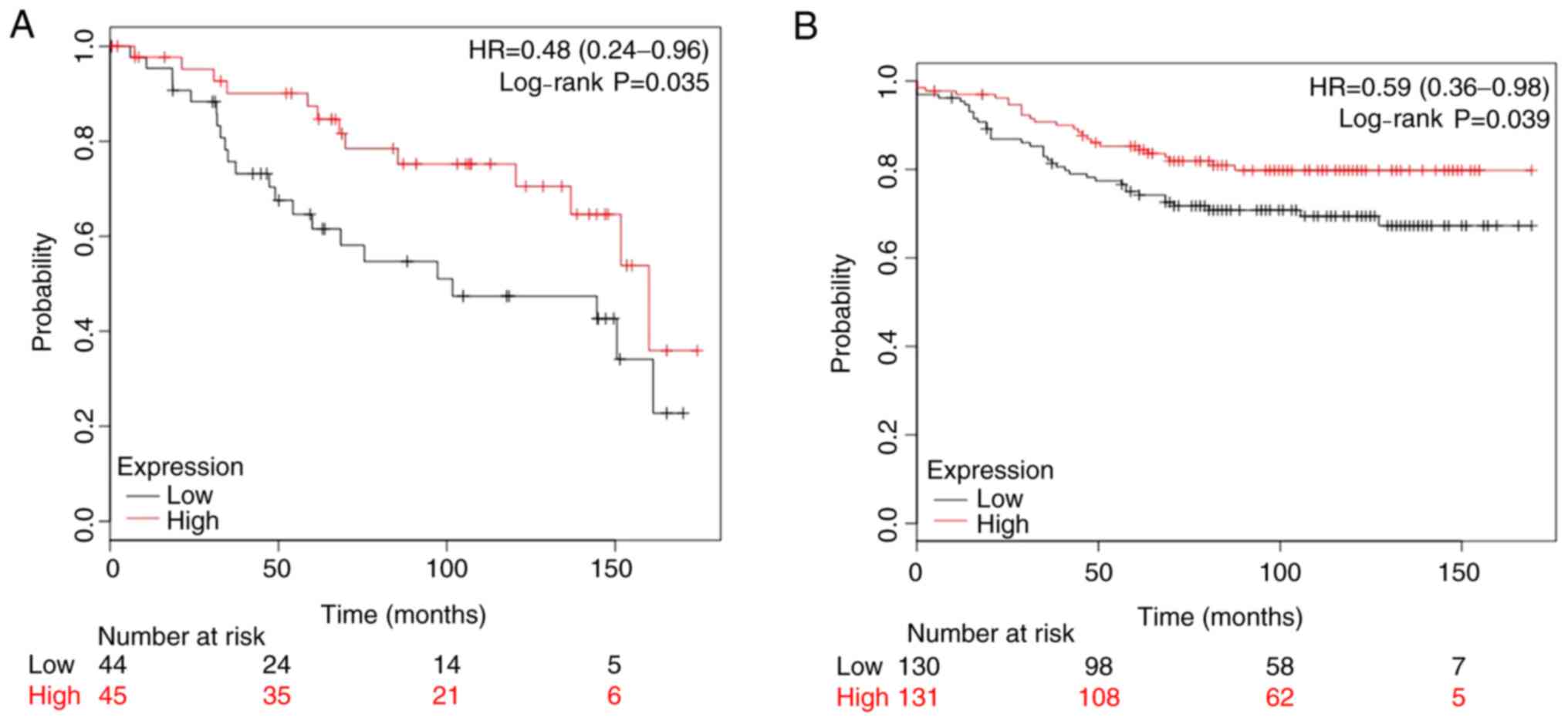
The present study analyzed two publicly available
datasets, GSE2990 (n=89) and GSE20685 (n=261), where patients with
ER-positive breast cancer had been treated with adjuvant TAM
monotherapy and ≥10-year follow-up data were available. The
Kaplan-Meier analysis demonstrated that low NAT1 expression was
associated with significantly shorter overall survival compared
with that of patients with high NAT1 expression (Fig. 5). With regard to the two datasets,
they indicated that a higher mRNA expression level of NAT1 was
favorable to the outcome of patients with breast cancer treated
with TAM therapy.
Patient characteristics and
responses
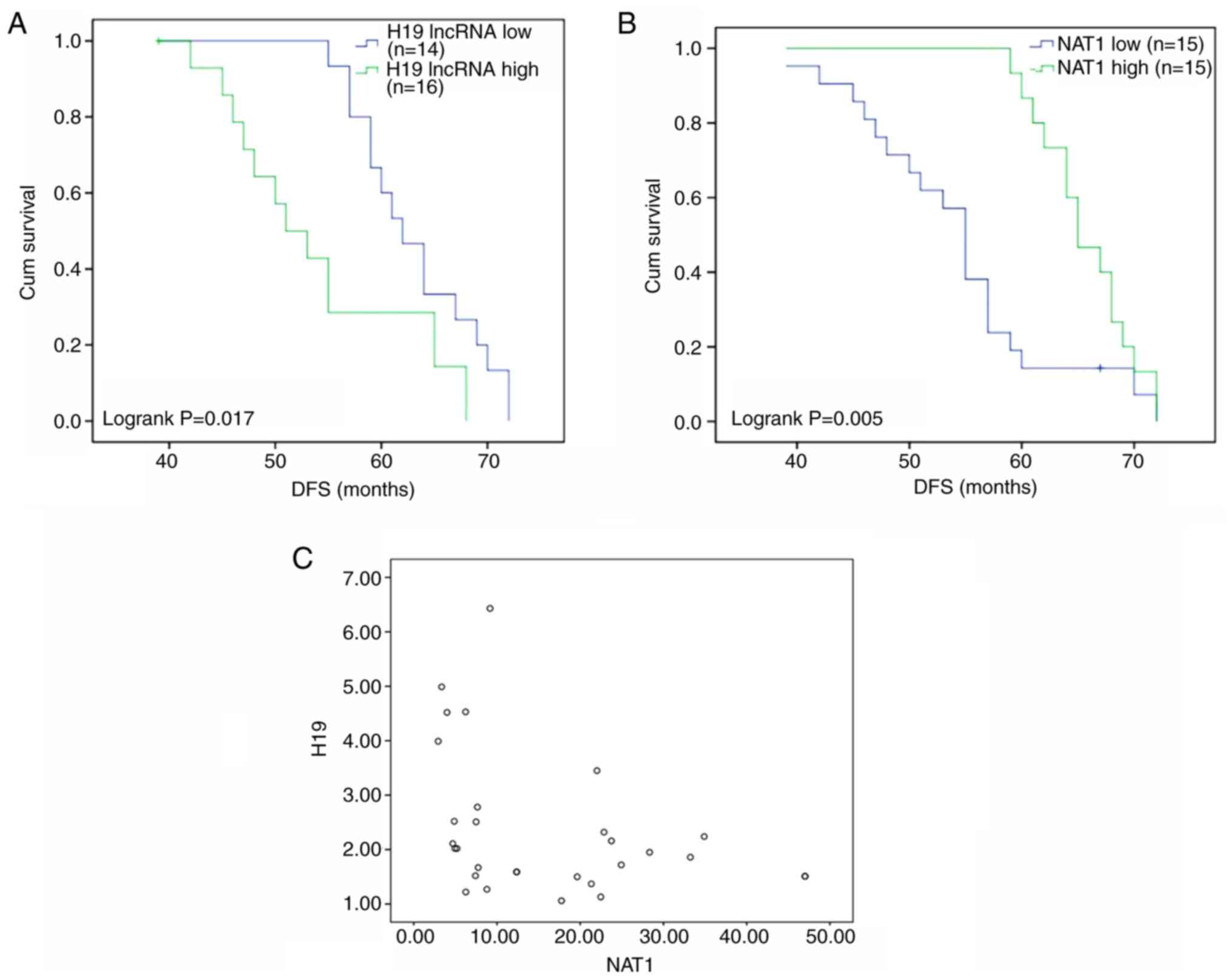
A total of 30 premenopausal patients with breast
cancer who took TAM for at least 1 year after surgery were followed
up for ≥5 years. These patients were allocated to low and high H19
and NAT1 groups according to whether expression in the tumor tissue
was ≥1.5-fold lower or higher compared with paracancerous tissue.
DFS was found to differ significantly according to the expression
levels of H19 and NAT1. The DFS times of patients with high and low
expression levels of H19 were 54.1 months (95% CI, 49.5-58.8) and
63.2 months (95% CI, 60.3-66.1), respectively (Fig. 6A), while the DFS times of patients
with high and low expression levels of NAT1 were 65.7 months (95%
CI, 63.6-67.8) and 54.5 months (95% CI, 50.7-58.3), respectively
(Fig. 6B). Furthermore, a
significant very weak negative correlation was identified between
the expression levels of H19 and NAT1 in the individual samples
(Pearson correlation coefficient, -0.0377; Fig. 6C). Collectively, these results
suggest that higher expression of NAT1 and lower expression of H19
are associated with improved outcomes in patients with breast
cancer treated with TAM.
Discussion
Although there have been advances in cancer therapy,
TAM remains the mostly widely used endocrine therapy for patients
with ER-positive breast cancer. However, >40% of breast cancer
cases develop resistance to endocrine therapy (4). Therefore, an improved understanding of
the cellular and molecular pathways of TAM resistance may
facilitate the development of strategies to overcome
resistance.
Previous studies have reported that lncRNAs are a
key component of gene regulatory networks and may serve a vital
role in tumorigenesis and TAM resistance (12,25,26).
The results of the present study suggest that H19 is associated
with TAM resistance in breast cancer. The modulating effect of H19
on NAT1 gene promoter methylation was further examined.
lncRNA H19 has been revealed to be an
estrogen-regulated gene (10). Zhou
et al (27) reported that
H19 regulates epithelial-mesenchymal and mesenchymal-epithelial
transition in breast cancer. The present study found that H19
expression was significantly upregulated in TAM-resistant (MCF-7R)
cells, which is consistent with previous reports (12,13).
Moreover, the knockdown of lncRNA H19 in the MCF-7R cells
ameliorated their resistance to 4-hydroxytamoxifen and promoted the
inhibitory effect of 4-hydroxytamoxifen on cell proliferation; the
knockdown of H19 enhanced the sensitivity to 4-hydroxytamoxifen
in vitro. In a study by Wang et al (28), it was also reported that the
knockdown of H19 significantly enhanced the sensitivity of
TAM-resistant MCF7 cells to TAM in vitro and in vivo,
and inhibited autophagy in these cells. It has also been observed
that H19 expression is increased in doxorubicin (Dox)-resistant
breast cancer (29). Furthermore,
H19 upregulation has been observed in Dox-resistant liver cancer
cells (14), TAM-resistant breast
cancer cells, fulvestrant-resistant breast cancer cells (13,28)
and cisplatin-resistant lung cancer cells (30). Based on these findings, lncRNA H19
has potential as a useful biomarker and drug resistance target.
The present study demonstrated that NAT1 was
significantly downregulated at the mRNA and protein levels in
TAM-resistant cells. These results are consistent with previous
reports that indicated the involvement of NAT1 in the TAM
resistance of breast cancer cells (8,9). The
significance of NAT1 in TAM resistance was further demonstrated in
the present study by the analysis of public breast cancer datasets,
which indicated that low NAT1 expression was associated with poor
survival in patients with breast cancer treated with TAM therapy.
The analysis of primary patients also found that the high and low
expression levels of NAT1 were associated with different DFS times
and TAM response rates. In addition, NAT1 was upregulated at the
mRNA and protein levels in TAM-resistant cells when H19 was knocked
down, indicating that H19 controls NAT1 expression.
The present study investigated the molecular
mechanism by which H19 modulates NAT1 gene expression; the
methylation status of the NAT1 promoter in MCF-7 and MCF-7R cells
was examined using the BSP method. The DNA methylation of NAT1 has
been identified to serve a critical role in cancer, including
breast cancer and colon adenocarcinoma (9,31).
Hypermethylation of the NAT1 gene may affect the initiation of TAM
resistance (9). The results of the
present study suggest that H19 may regulate NAT1 and thereby affect
drug resistance by altering the degree of NAT1 promoter methylation
in MCF-7R cells. The results also suggest that high H19 expression
is associated with poor prognosis in patients with breast cancer
treated with TAM therapy. Therefore, the present study indicates
that H19 serves a crucial role in TAM resistance, which may
facilitate the development of therapeutic strategies to ameliorate
the resistance of cancer to endocrine therapy. However, it must be
noted the sample size is the present study is small, which is a
limitation, and a larger sample size is required in future studies
to confirm the results.
Therefore, the present study suggests that the H19
gene regulates NAT1 expression in TAM-resistant cells via the
mediation of NAT1 promoter methylation. Further elucidation of the
role of H19 in the modulation of NAT1 gene expression and
methylation should improve our understanding of endocrine therapy
resistance, and may provide a theoretical basis for further studies
evaluating targeted drugs.
Acknowledgements
Not applicable.
Funding
This work was supported in part by grants from the National
Natural Science Foundation of China (grant nos. 81673516 and
81403021), Natural Science Foundation of Fujian (grant no.
2019J01177), Fujian Science and Technology Innovation Joint Fund
Project (grant no. 2017Y9067), High-level hospital foster grants
from Fujian Provincial Hospital (grant no. 2019HSJJ06), Fujian
Province Young and Middle-aged Talent Training Program (grant no.
2019-ZQN-35) and the Medical Science Research Project (grant no.
BJBQEKYJJ-B19001CS).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The BSP data have been deposited into the CNGB Sequence
Archive of the China National GeneBank DataBase with accession
number CNP0001707.
Authors' contributions
HS, GW and JZ participated in research design. HS,
YZ and YP conducted experiments. HS, JC, XW performed data
analysis. HS, JC, GW wrote or contributed to the writing of the
manuscript. HS and GW confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Xiangya Hospital (project no. CTXY-140001-5). Written informed
consent was obtained from all patients.
Patient consent for publication
The patients who participated in this study provided
signed consent for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand-binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Drăgănescu M and Carmocan C: Hormone
therapy in breast cancer. Chirurgia (Bucur). 112:413–417.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ring A and Dowsett M: Mechanisms of
tamoxifen resistance. Endocr Relat Cancer. 11:643–658.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Abdel-Hafiz HA: Epigenetic mechanisms of
tamoxifen resistance in luminal breast cancer. Diseases.
5(16)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lo PK and Sukumar S: Epigenomics and
breast cancer. Pharmacogenomics. 9:1789–1902. 2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kim SJ, Kang HS, Chang HL, Jung YC, Sim
HB, Lee KS, Ro J and Lee ES: Promoter hypomethylation of the
N-acetyltransferase 1 gene in breast cancer. Oncol Rep. 19:663–668.
2008.PubMed/NCBI
|
|
8
|
Kim SJ, Kang HS, Jung SY, Min SY, Lee S,
Kim SW, Kwon Y, Lee KS, Shin KH and Ro J: Methylation patterns of
genes coding for drug-metabolizing enzymes in tamoxifen-resistant
breast cancer tissues. J Mol Med (Berl). 88:1123–1131.
2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sun H, Wang G, Peng Y, Zeng Y, Zhu QN, Li
TL, Cai JQ, Zhou HH and Zhu YS: H19 lncRNA mediates
17β-estradiol-induced cell proliferation in MCF-7 breast cancer
cells. Oncol Rep. 33:3045–3052. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Adriaenssens E, Lottin S, Dugimont T,
Fauquette W, Coll J, Dupouy JP, Boilly B and Curgy JJ: Steroid
hormones modulate H19 gene expression in both mammary gland and
uterus. Oncogene. 18:4460–4473. 1999.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gao H, Hao G, Sun Y, Li L and Wang Y: Long
noncoding RNA H19 mediated the chemosensitivity of breast cancer
cells via Wnt pathway and EMT process. Onco Targets Ther.
11:8001–8012. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Basak P, Chatterjee S, Bhat V, Su A, Jin
H, Lee-Wing V, Liu Q, Hu P, Murphy LC and Raouf A: Long non-coding
RNA H19 acts as an estrogen receptor modulator that is required for
endocrine therapy resistance in ER+ breast cancer cells. Cell
Physiol Biochem. 51:1518–1532. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tsang WP and Kwok TT: Riboregulator H19
induction of MDR1-associated drug resistance in human
hepatocellular carcinoma cells. Oncogene. 26:4877–4881.
2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Coser KR, Wittner BS, Rosenthal NF,
Collins SC, Melas A, Smith SL, Mahoney CJ, Shioda K, Isselbacher
KJ, Ramaswamy S and Shioda T: Antiestrogen-resistant subclones of
MCF-7 human breast cancer cells are derived from a common
monoclonal drug-resistant progenitor. Proc Natl Acad Sci USA.
106:14536–14541. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Butcher NJ, Ilett KF and Minchin RF:
Substrate-dependent regulation of human Arylamine
N-Acetyltransferase-1 in cultured cells. Mol Pharmacol. 57:468–473.
2000.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Froger A and Hall JE: Transformation of
plasmid DNA into E. coli using the heat shock method. J Vis Exp:
Aug 1, 2007 (Epub ahead of print). doi: 10.3791/253.
|
|
19
|
Reed K, Hembruff SL, Laberge ML,
Villeneuve DJ, Côté GB and Parissenti AM: Hypermethylation of the
ABCB1 downstream gene promoter accompanies ABCB1 gene amplification
and increased expression in docetaxel-resistant MCF-7 breast tumor
cells. Epigenetics. 3:270–280. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bock C, Reither S, Mikeska T, Paulsen M,
Walter J and Lengauer T: BiQ Analyzer: Visualization and quality
control for DNA methylation data from bisulfite sequencing.
Bioinformatics. 21:4067–4068. 2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jiang X, Menon A, Wang S, Kim J and
Ohno-Machado L: Doubly optimized calibrated support vector machine
(DOC-SVM): An algorithm for joint optimization of discrimination
and calibration. PLoS One. 7(e48823)2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Meng L, Xu Y, Xu C and Zhang W: Biomarker
discovery to improve prediction of breast cancer survival: Using
gene expression profiling, meta-analysis, and tissue validation.
Onco Targets Ther. 9:6177–6185. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41 (Database
Issue):D991–D995. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Pénzváltó Z, Lánczky A, Lénárt J,
Meggyesházi N, Krenács T, Szoboszlai N, Denkert C, Pete I and
Győrffy B: MEK1 is associated with carboplatin resistance and is a
prognostic biomarker in epithelial ovarian cancer. BMC Cancer.
14(837)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Xue X, Yang YA, Zhang A, Fong KW, Kim J,
Song B, Li S, Zhao JC and Yu J: lncRNA HOTAIR enhances ER signaling
and confers tamoxifen resistance in breast cancer. Oncogene.
35:2746–2755. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Teunissen SF, Jager NG, Rosing H, Schinkel
AH, Schellens JH and Beijnen JH: Development and validation of a
quantitative assay for the determination of tamoxifen and its five
main phase I metabolites in human serum using liquid chromatography
coupled with tandem mass spectrometry. J Chromatogr B Analyt
Technol Biomed Life Sci. 879:1677–1685. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li
LY, Guan GH, Liu Q, Qian YH and Xie D: The lncRNA H19 mediates
breast cancer cell plasticity during EMT and MET plasticity by
differentially sponging miR-200b/c and let-7b. Sci Signal.
10(eaak9557)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang J, Xie S, Yang J, Xiong H, Jia Y,
Zhou Y, Chen Y, Ying X, Chen C, Ye C, et al: The long noncoding RNA
H19 promotes tamoxifen resistance in breast cancer via autophagy. J
Hematol Oncol. 12(81)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhu QN, Wang G, Guo Y, Peng Y, Zhang R,
Deng JL, Li ZX and Zhu YS: lncRNA H19 is a major mediator of
doxorubicin chemoresistance in breast cancer cells through a
cullin4A-MDR1 pathway. Oncotarget. 8:91990–92003. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang Q, Cheng N, Li X, Pan H, Li C, Ren S,
Su C, Cai W, Zhao C, Zhang L and Zhou C: Correlation of long
non-coding RNA H19 expression with cisplatin-resistance and
clinical outcome in lung adenocarcinoma. Oncotarget. 8:2558–2567.
2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shi C, Xie LY, Tang YP, Long L, Li JL, Hu
BL and Li KZ: Hypermethylation of N-Acetyltransferase 1 is a
prognostic biomarker in colon adenocarcinoma. Front Genet.
10(1097)2019.PubMed/NCBI View Article : Google Scholar
|