Introduction
Exosomes are membranous extracellular vesicles
50-100 nm in size, which are involved in cellular communication via
the delivery of proteins, lipids and RNA. Tumor derived exosomes
could inhibit immune cell proliferation and affect antigen
presentation and activation of immune cells (1). In the field of cancer, exosomes from
cancer cells may present both immunosuppressive and
immunostimulatory properties (2,3).
Increasing evidence has shown that exosomes are closely associated
with cancer (4) and a previous
study demonstrated that exosomes can serve as possible drug
delivery vehicles in cancer therapy (5). However, the role of exosomes derived
from human umbilical serum in cancer development has not been
investigated.
Since pregnancy involves extensive communication
among cells and tissues, secreted molecules are thought to
participate in cellular signaling, which is important to
placentation (6). Extracellular
vesicles mediate cell communication. In the process of human
pregnancy, elevated levels of circulating maternal exosomes are
observed (6). A previous study has
reported that exosomes isolated from the human umbilical cord may
enhance the angiogenic activities of endothelial cells and
endothelial cell proliferation, mostly through microRNA (miRNA)
(7). Due to their different
cellular origins, exosomes may carry distinct RNAs and protein
cargos (8).
A previous study has suggested that exosomes carry
bioactive molecules, such as miRNAs, which may participate in the
crosstalk between the placenta and maternal tissue (9). It has been demonstrated that
differentially expressed maternal serum exosomal miRNA molecules
can affect cell growth and organ development pathways, and that
umbilical serum exosomal miRNA is involved in cellular and
embryonic development (10). Thus,
different proteins found in exosomes may also be involved in
physiological or pathological states (11). Comparing umbilical serum exosomes
(UEs) with maternal serum exosomes (MEs) may lead to improved
understanding of the differences between the umbilical cord blood
and the peripheral blood environments.
A previous study has revealed that human serum may
induce hepatoma cell cycle arrest (12). Moreover, to the best of our
knowledge, the protein profile of exosomes derived from umbilical
cord serum (UCS) using mass spectrometry (MS)/MS has never been
described. Thus, the aim of the present study was to explore the
potential differences in the composition and function of the
proteins found in UEs and MEs, and to investigate the biological
effects of UEs on liver cancer cells.
Materials and methods
Umbilical cord blood extraction
Serum samples were collected at Xijing Hospital
(Xi'an, China) from March 2019 to May 2019. The present study was
approved by the ethics committee of Xijing Hospital (approval no.
20190105-1) and performed in accordance with their guidelines, with
written informed consent obtained for the use of patient tissue and
specimens. All experimental protocols were performed in accordance
with the relevant Good Laboratory Practice guidelines and
regulations of our laboratory. A total of eight healthy pregnant
women from Xijing Hospital who were pregnant for the first time and
had a natural vaginal delivery were enrolled. The average age of
these patients was 29 years old. None of these patients had
complications or a history of smoking or alcohol abuse. None of
them had taken medication within three months of the study or
experienced any infectious diseases. After delivery of the fetus,
umbilical cord blood was rapidly collected. The needle was inserted
into the umbilical vein and was connected to a negative pressure
bag. After insertion of the needle, the umbilical cord blood flowed
into the bag through negative pressure. Maternal blood samples were
collected from ulnar vein to extract MEs, which were extracted from
the same patients as the UEs were extracted from. The samples were
centrifuged 1 h later at 1,000 x g for 30 min at room temperature,
after which the upper layer of clear serum was collected. The
samples were then stored at -130˚C.
Exosome isolation
UEs and MEs were isolated using the ExoQuick™
Exosome Precipitation Solution (cat. no. EXOQ5A-1; System
Biosciences). The procedure of this kit is simple to conduct, and
it is efficient at isolating exosomes. Briefly, serum samples were
centrifuged at 3,000 x g for 15 min at 4˚C to remove clotted
materials and cell debris. A volume of 250 µl ExoQuick Exosome
Precipitation Solution was then added to 1-ml serum supernatants
(1:4 ratio of the ExoQuick solution to serum supernatants), and the
mixture was refrigerated for 30 min. The mixture was then
centrifuged at 1,500 x g for 30 min at 4˚C, and the supernatants
were discarded. The residual solution was centrifuged at 1,500 x g
for 5 min at 4˚C, and the supernatant was removed. The exosome
pellet was resuspended in 500 µl PBS and stored at -80˚C.
Trypsin digestion
Dithiothreitol was added to the protein samples to
make the final concentration 5 mM, and then reduced for 30 min at
56˚C. Iodoacetamide was then added to the samples to adjust the
concentration to 11 mM, and the samples were incubated for 15 min
in the dark at room temperature. The protein sample was then
diluted by adding 200 mM TEAB (tetraethyl ammonium bromide) to urea
concentration <2 M. Finally, trypsin was added at a mass ratio
(trypsin:protein) of 1:50 for digestion at 37˚C overnight. The
following day, trypsin was added at a mass ratio of 1:100 for
further digestion for 4 h.
TMT (tandem mass tags) labeling
The trypsin-digested peptides were desalted using
solid phase extraction cartridges (Strata X C18; Phenomenex) and
freeze-dried in a vacuum. The samples were then dissolved with 0.5
M TEAB and the peptides were labeled according to the
manufacturer's instructions (TMTsixplex™ Isobaric Label Reagent;
cat. no. 90068; Thermo Fisher Scientific, Inc.).
High-performance liquid chromatography
(HPLC) fractionation
Agilent 1260 Infinity Liquid Chromatography System
was used at 35˚C. High-pH reverse-phase HPLC was performed to
separate the peptides using an Agilent 300 Extend C18 column
(Agilent Technologies, Inc.). The column size was ZORBAX Extend-C18
Agilent Analytical 4.6x250-mm 5-Micron 80A. The composition of
mobile phase (two solvents) were: Buffer A, 98% water and 2%
acetonitrile; buffer B, 98% acetonitrile and 2% water. The flow
rate was used at 1 ml/min. Briefly, the peptides were fractionated
into 60 fractions with acetonitrile (8-32%) at pH 9 for 60 min. The
sample quantity was 1,000 µl. Pre-loading: Dried sample dissolved
in buffer A, and then centrifuged at 18,000 x g for 5 min. Sample
combination method: The samples were collected from the 11th min to
the 64th min when they flowed out of the liquid phase, and then the
first component were combined with the 11th, 29th, 47th and 64th
min samples.
LC-MS/MS analysis
The samples were dissolved with 0.1% aqueous formic
acid as the mobile phase using liquid chromatography. The peptides
were separated with EASY-nLC 1000 UPLC (Ultra Performance Liquid
Chromatography; Thermo Fisher Scientific, Inc.). Mobile phase A was
an aqueous solution containing 0.1% formic acid and 2%
acetonitrile, and mobile phase B was an aqueous solution containing
0.1% formic acid and 90% acetonitrile. The peptides separated by
UPLC were then subjected to an Nanospray Ion source. The ion source
voltage was set at 2.0 kV. An Orbitrap (Thermo Fisher Scientific,
Inc.) was used to detect and analyze the peptide parent ions and
their secondary fragments. The data acquisition mode used a
dependent scanning Data Dependent Acquisition program and the
primary scanning range is 350-1,800 m/z. Finally, the peptides were
analyzed by tandem MS/MS in a Q Exactive™ Plus instrument (Thermo
Fisher Scientific, Inc.).
Database search
The MaxQuant (v1.5.2.8,; http://www.maxquant.org/) search engine was used to
process the mass spectrometry data. The cleavage enzyme was set as
trypsin/P, and up to two missing cleavages were allowed. The
minimum length of the peptide segment was set to seven amino acids,
and the maximum modification number of the peptide segment was set
to five.
Bioinformatics
The quantitative value of the peptide in each sample
was calculated used the ratio of the labeling reporter ion
intensities in MS/MS spectra from raw data sets. The median of
unique peptides for a given protein was considered as the protein
quantitation in each sample. Ratio of protein quantitative value
between two samples was considered as the protein expression ratio.
In order to calculate the significant P-value of differentially
expressed proteins, firstly, perform log2 transform on the unique
peptide quantitative values of the protein in the two samples
(making the data conform to the normal distribution), and then the
two-sample two-tailed T test method was used to calculate P-value.
Proteins with P<0.05 and expression ratio >1.5 were regarded
as upregulated, while proteins with P<0.05 and expression ratio
<1.5 were regarded as downregulated.
Gene Ontology (GO) annotation
GO analysis is a kind of bioinformatics tool for
characterizing molecular function (MF), cellular component (CC) and
biological processes (BP) of genes (13). The protein IDs were determined by
MaxQuant (v1.5.2.8; http://www.maxquant.org/) database. Then the protein
ID was matched to the UniProt ID based on the UniProt-GOA database
and the UniProt ID was used to match the GO ID. Finally, the
corresponding information from the UniProt-GOA database was
retrieved based on the GO ID. UniProt-GOA database (http://www.ebi.ac.uk/GOA/) and InterProScan v5.14
software (http://www.ebi.ac.uk/interpro/) were used in this
section. The enriched proteins based on GO analysis can be divided
into three categories: Biological process, cellular component and
molecular function. Two-tailed Fisher's exact test was used to
determine the GO terms associated with the differentially expressed
proteins and P<0.05 was considered significant.
Domain annotation
The InterProScan v5.14 software (http://www.ebi.ac.uk/interpro/) and the InterPro
domain database (http://www.ebi.ac.uk/interpro/) were used to analyze
the enrichment of the functional domains of the differentially
expressed proteins. Two-tailed Fisher's exact test was used to test
the enrichment of the differentially expressed proteins and
P<0.05 was considered significant.
Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway annotation
The identified proteins were annotated using KAAS
(https://www.genome.jp/tools/kaas/)
and the annotated proteins were matched to the corresponding
pathways using KEGG mapper. Pathway enrichment analysis was based
on the KEGG database (https://www.kegg.jp/). Two-tailed Fisher's exact test
was used to determine the enriched KEGG pathways associated with
the differentially expressed proteins and P<0.05 was considered
significant.
Subcellular localization
The cells of eukaryotic organisms are elaborately
subdivided into functionally distinct membrane bound compartments
(14). Some major constituents of
eukaryotic cells are: extracellular space, cytoplasm, nucleus,
mitochondria, Golgi apparatus, endoplasmic reticulum (ER),
peroxisome, vacuoles, cytoskeleton, nucleoplasm, nucleolus, nuclear
matrix and ribosomes. Wolfpsort v0.2 software (https://wolfpsort.hgc.jp/) was used to predict
subcellular localization of the identified proteins.
Enrichment-based hierarchical
clustering
To identify the functional correlation of the
differentially expressed proteins in the comparison groups,
hierarchical clustering based on the functional enrichment of the
differentially expressed proteins in different groups was used to
investigate the potential associations and differences in specific
functions (GO terms, KEGG pathways and protein domains) of the
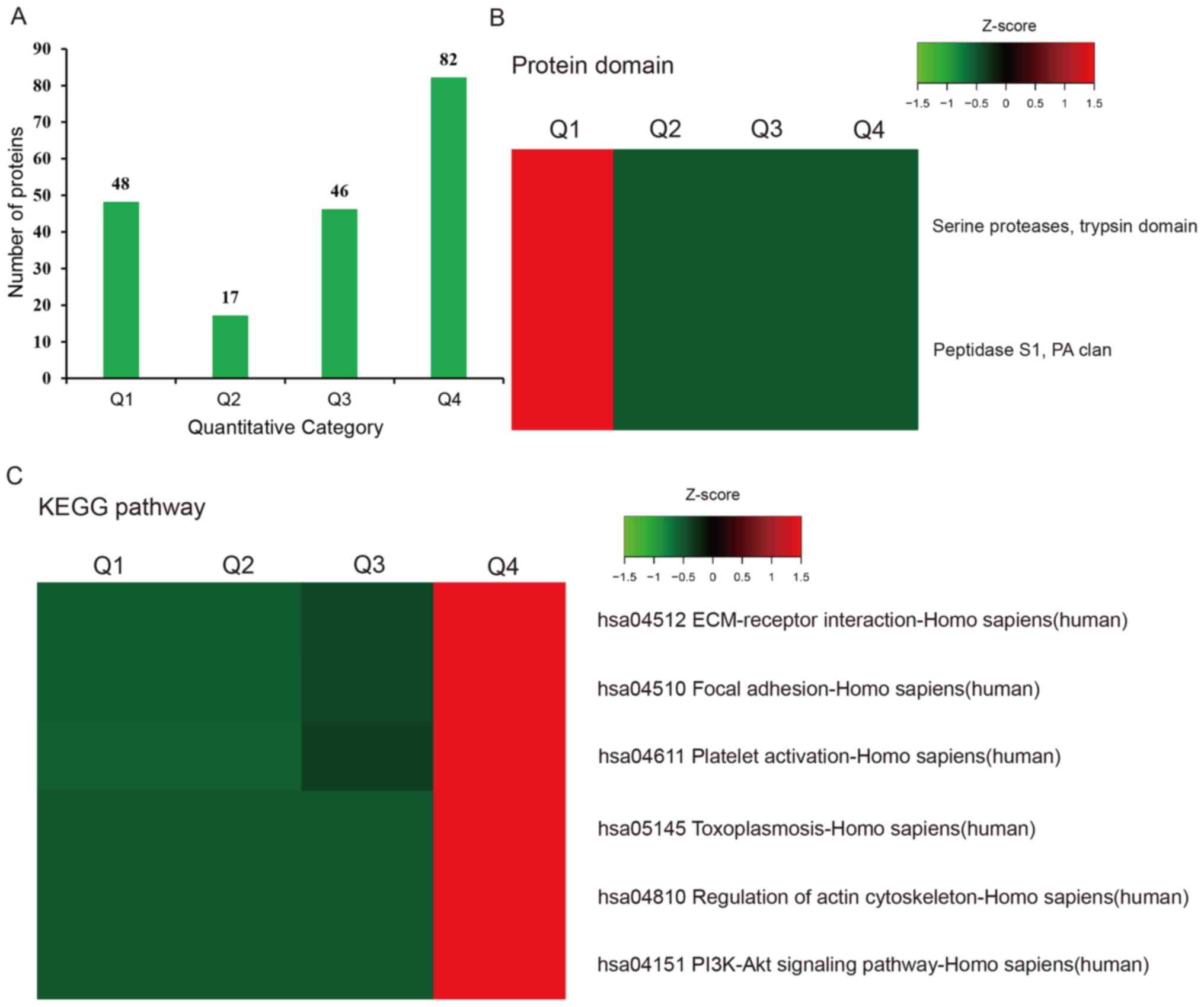
proteins. Differentially expressed proteins were categorized into
four groups according to their fold change: Q1 (0-0.5), Q2
(0.5-0.67), Q3 (1.5-2) and Q4 (>2), with P<0.05 in all cases.
The functional annotation following enrichment, together with the
corresponding enrichment P-value, were first collected. The
functional classifications that were enriched in at least one of
the clusters (with P<0.05) were then screened. The filtered
P-value data matrix was -log10 transformed. The
transformed data matrix was classified by Z transformation for each
functional category. Lastly, the dataset obtained following Z
transformation was analyzed using one-way hierarchical
clustering.
Transmission electron microscopy
The exosomes were fixed with 2.5% glutaraldehyde in
PBS for 1 h at 4˚C. The samples were then centrifuged for 10 min at
7,000 x g to remove glutaraldehyde at 4˚C. And the samples were
embedded by epoxy resin 618 at 35˚C for 3 h. Subsequently, the
samples were stained with 1% phosphotungstic acid for 5 min at room
temperature and fixed on copper mesh formvar grids.
Cell culture and Cell Counting Kit-8
(CCK-8) assay
The Huh-7 and MHCC97H hepatocellular carcinoma (HCC)
cell lines were purchased from iCell Bioscience, Inc. The liver
cancer cell line HepG2 and the HCC cell line Hep3B were purchased
from Shanghai GeneChem Co., Ltd. The HepG2 cell line was
authenticated by STR profiling. HCC cells and HepG2 liver cancer
cell line were cultured in Dulbecco's modified Eagle's medium
(HyClone; Cytiva) supplemented with 10% fetal bovine serum
(HyClone; Cytiva) at 37˚C in 5% CO2.
Cell viability was measured using the CCK-8 kit
(Sigma-Aldrich; Merck KGaA). Cells were seeded in 96-well plates
(3x103 cells per well) and treated with UEs or MEs at
different dilution ratios (1:5, 1:10, 1:20, 1:40, 1:80) for 24 h at
37˚C. As a negative control, cells were incubated with medium only.
Subsequently, 10 µl CCK-8 reagent was added to the cells for 2 h at
37˚C. A microplate reader was used to measure the optical density
at 450 nm.
Western blot analysis
Cells and exosomes were lysed using RIPA buffer
(Thermo Fisher Scientific, Inc.) with proteinase and phosphatase
inhibitors (Roche Diagnostics). Total protein was quantified using
a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.).
The mass of protein loaded per lane was 50 µg. Total protein was
separated by SDS-PAGE on 10% gels, then transferred to
polyvinylidene fluoride membranes (EMD Millipore). The membranes
were blocked with 5% non-fat milk at room temperature for 1 h and
incubated with primary antibodies overnight at 4˚C. The following
antibodies were used: i) Anti-CD63 (cat. no. ab134045; Abcam); ii)
anti-CD9 (cat. no. ab236630; Abcam); iii) anti-caspase-3 (cat. no.
#9662; Cell Signaling Technology, Inc.); iv) anti-cleaved-caspase-3
(cat. no. #9661; Cell Signaling Technology, Inc.); and v) and
anti-GAPDH antibody (cat. no. #5174; Cell Signaling Technology,
Inc.). All primary antibodies were monoclonal and used at 1:1,000
dilution. The membranes were incubated with secondary antibodies
for 1 h on the second day at room temperature. The
peroxidase-conjugated anti-rabbit secondary antibodies (cat. no.
A6154; Sigma-Aldrich; Merck KGaA) was used in 1:5,000 dilution.
Membranes were finally treated with enhanced chemiluminescence
reagent (Thermo Fisher Scientific, Inc.) and exposed to
chemiluminescence imaging systems (version 5.1l Bio-Rad
Laboratories).
Flow cytometry
After treatment with UEs (dilution ratio, 1:5) or
MEs (dilution ratios, 1:5) for 24 h at 37˚C, cells were digested
and resuspended in PBS at 1x106/ml. The samples were
then incubated with 10 µl Annexin V-FITC (Sigma-Aldrich; Merck
KGaA) and 10 µl propidium iodide (Sigma-Aldrich; Merck KGaA) for
10-15 min at room temperature in the dark. Finally, flow cytometry
(BD FACSCalibur; BD Biosciences) was used to analyze the cells. And
the data was analyzed by DIVA version 11.0 software (BD
Biosciences).
Statistical analysis
Statistical analysis was performed using SPSS 22.0
(IBM Corp.). A paired Student's t-test was used to compare the
differences between two groups. Comparisons among three groups were
analyzed using one-way ANOVA followed by Bonferroni correction.
P<0.05 was considered to indicate a statistically significant
difference.
Results
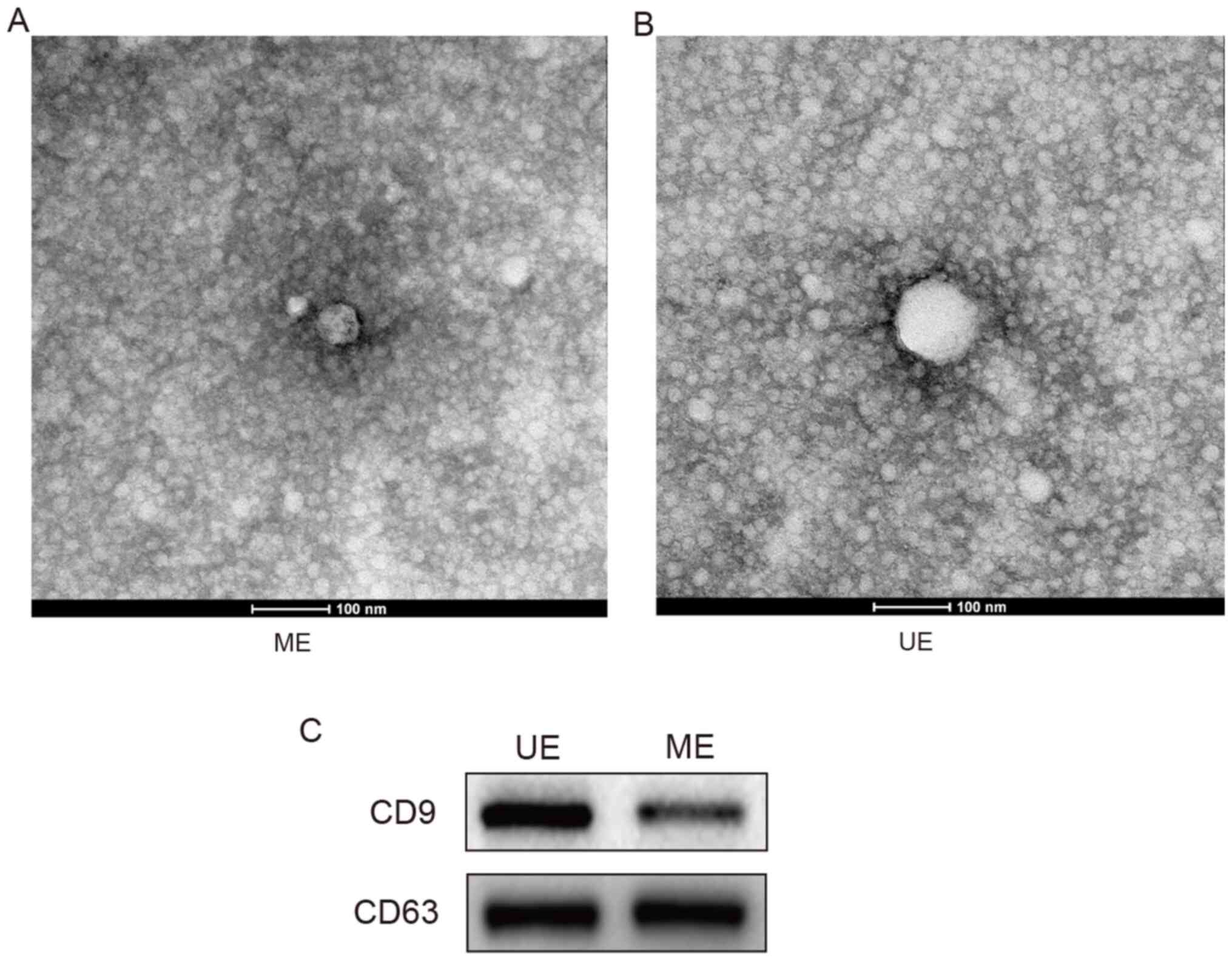
Exosome isolation from serum
samples
Exosomes were isolated from serum samples and
identified using transmission electron microscopy and western blot
analysis (15). The isolated
extracellular vesicles were found to have a diameter of 50-100 nm
(Fig. 1A and B). CD9 and CD63 were detected in the
protein samples extracted from the exosomes (Fig. 1C). The protein concentration in the
exosome samples was 1.5±0.13 µg/µl in the UE group and 1.1±0.10
µg/µl in the ME group.
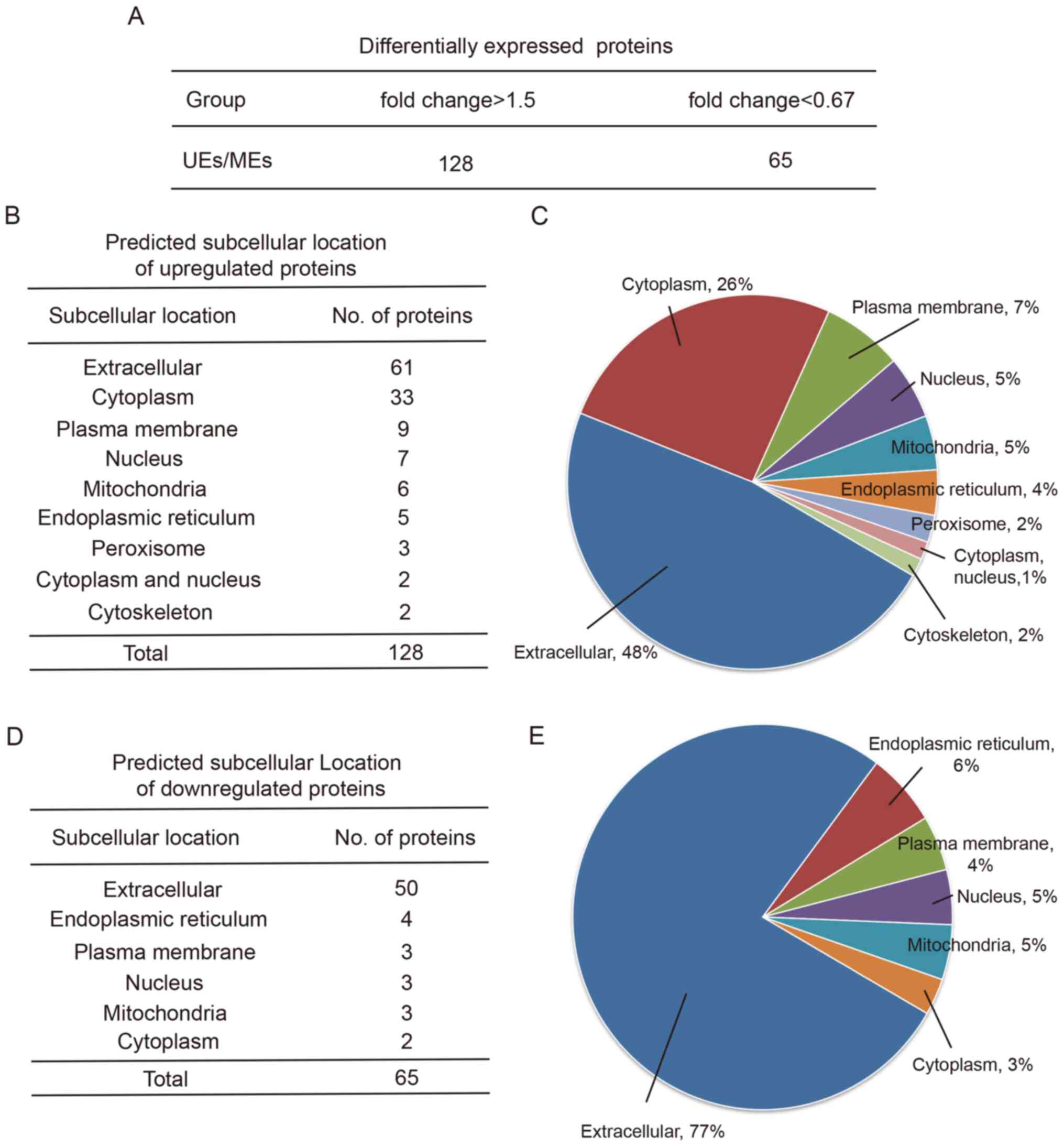
Comparison of the protein profiles
between UEs and MEs
The protein profiles of the exosomes from the serum
samples were then obtained, and 471 proteins were identified. Of
these, 193 proteins were differentially expressed between the two
groups, including 128 upregulated proteins and 65 downregulated
proteins in UEs relative to MEs (Fig.
2A). Representative mass spectra data of some of the
differentially expressed proteins are provided in Table SI. Representative mass spectra
images of the identified proteins are provided in Fig. S1. Wolfpsort was then used to
predict subcellular localization. The upregulated proteins were
predicted to be from the extracellular region (48%), followed by
the cytoplasm (26%), plasma membrane (7%), nucleus (5%),
mitochondria (5%), endoplasmic reticulum (4%), peroxisome (2%),
cytoplasm and nucleus (1%), and cytoskeleton (2%) (Fig. 2B and C). The downregulated proteins were from
the extracellular region (77%), followed by the endoplasmic
reticulum (6%), nucleus (5%), mitochondria (5%), plasma membrane
(4%) and cytoplasm (3%) (Fig. 2D
and E).
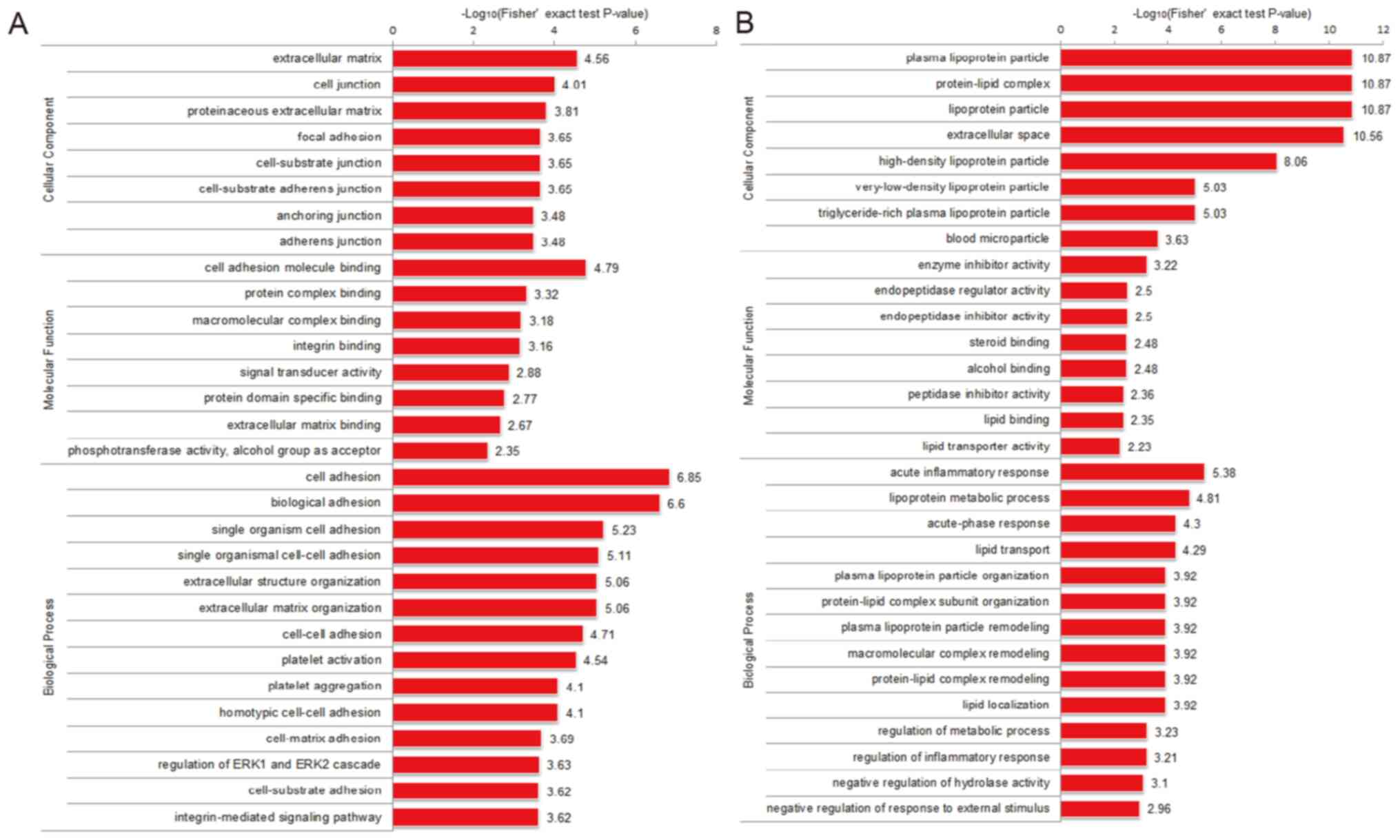
Functional enrichment analysis of
differentially expressed proteins
The differentially expressed proteins identified in
the present study were examined by GO analysis and categorized as
cellular component, molecular function and biological process
terms. The upregulated proteins derived from UEs were mostly
enriched in the ‘extracellular matrix’ cellular component, ‘cell
adhesion molecule binding’ molecular function, and ‘cell adhesion’
and ‘biological adhesion’ biological processes (Fig. 3A). The downregulated proteins were
mainly enriched in ‘plasma lipoprotein particles’, ‘protein-lipid
complex’ and ‘lipoprotein particle’ in the cellular component
category, as well as the ‘enzyme inhibitor activity’ molecular
function and the ‘acute inflammatory response’ biological process
(Fig. 3B). Thus, the differentially
expressed proteins in the UCS may participate in cell adhesion and
cell junctions.
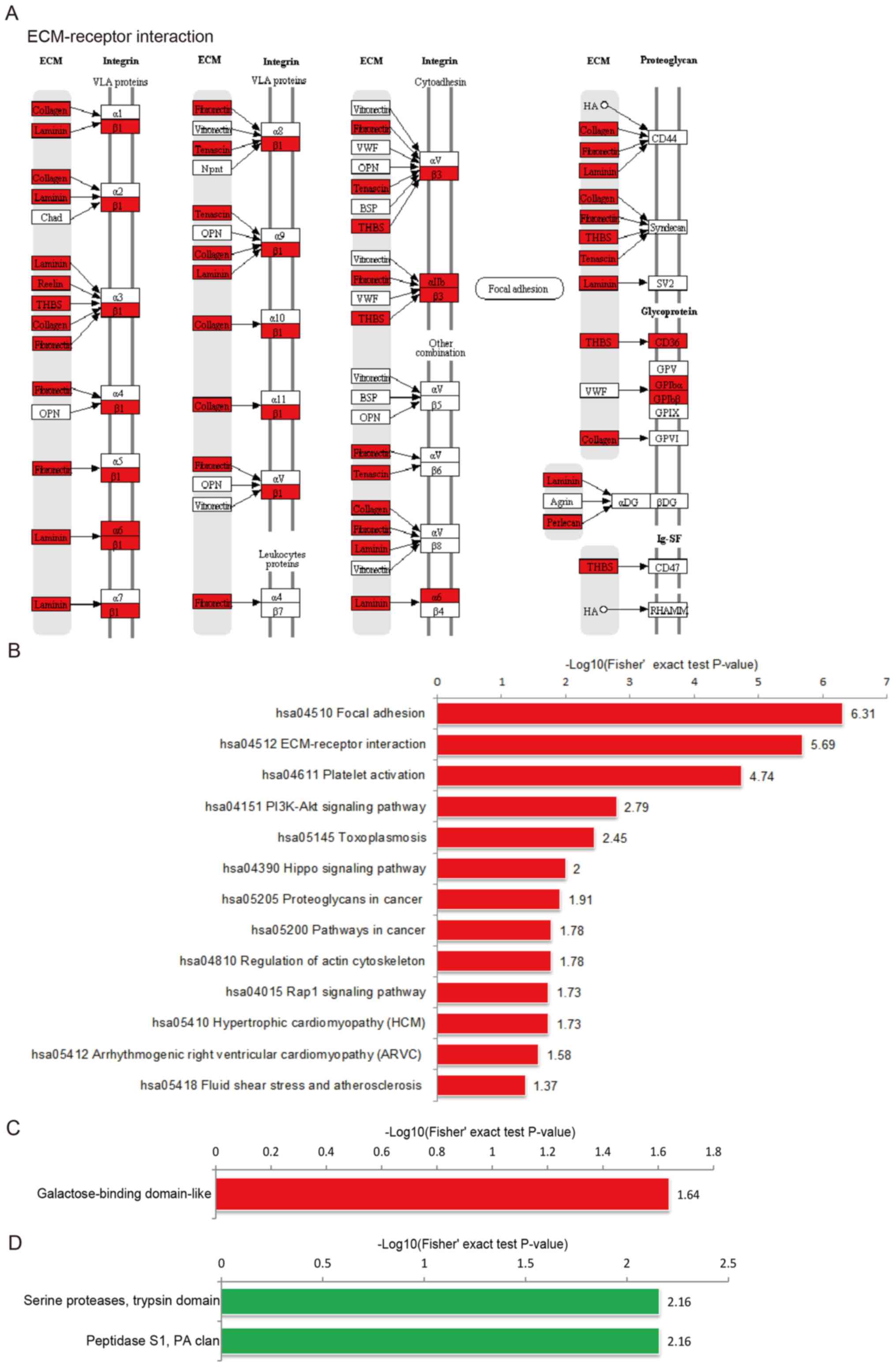
KEGG pathway enrichment of
differentially expressed proteins
KEGG pathway analysis predicted that the enriched
proteins derived from UCS were associated with the ‘ECM-receptor
interaction’ pathways (Fig. 4A),
including the ‘ECM-integrin’ and the ‘ECM-proteoglycan’ cascades.
According to the functional enrichment analysis based on the KEGG
pathway, ‘focal adhesion’ and ‘ECM-receptor interaction’ were the
most enriched pathways (Fig.
4B).
Protein domain enrichment of
differentially expressed proteins
Protein domains are elementary units of protein
structure and evolution, which mediate most (~75%) protein
interactions (16,17). The length of the domain is usually
between 25 amino acids and 500 amino acids (18-20).
The enriched domain of the upregulated proteins was mainly
‘galactose-binding domain-like’, whereas the downregulated proteins
were enriched in the ‘serine proteases, trypsin domain’ and
‘peptidase S1, PA clan’ protein domains (Fig. 4C and D).
Hierarchical clustering of the
functional annotation of differentially expressed proteins
Hierarchical clustering based on GO analysis in the
cellular component category showed that the differentially
expressed proteins of Q1 were mainly involved in, for example,
‘very-low-density lipoprotein particle’, ‘chylomicron’ and
‘high-density lipoprotein particle’ (Fig. S2). Q2 were mainly involved in
‘membrane attack complex’ and ‘pore complex’ in the cellular
component category. Q3 were mainly involved in ‘blood
microparticle’ in the cellular component category. While Q4 were
mainly involved in ‘proteinaceous extracellular matrix’,
‘extracellular matrix’ and ‘cell junction’ in the cellular
component category (Fig. S2).
Hierarchical clustering based on GO analysis in the biological
process category showed that the differentially expressed proteins
of Q1 were mainly involved in ‘acute inflammatory response’ and
‘acute-phase response’ (Fig. S3).
Q2 were mainly involved in ‘negative regulation of response to
external stimulus’ and ‘regulation of proteolysis’ in the
biological process category. Q3 were mainly involved in ‘positive
regulation of immune response’ and ‘protein activation cascade’ in
the biological process category. Q4 were mainly involved in
‘positive regulation of response to external stimulus’ in the
biological process category (Fig.
S3). Hierarchical clustering based on GO analysis in the
molecular function category showed that the differentially
expressed proteins of Q1 were mainly involved in ‘steroid binding’
(Fig. S4). Q2 were mainly involved
in ‘peptidase inhibitor activity’ and ‘endopeptidase regulator
activity’ in the molecular function category. Q3 were mainly
involved in ‘endopeptidase activity’ and ‘hydrolase activity’ in
the molecular function category. Q4 were mainly involved in
‘protein complex binding’ and ‘integrin binding’ in the molecular
function category (Fig. S4).
Hierarchical clustering based on protein domain
analysis revealed that UCS proteins of Q1 were mostly enriched with
‘serine proteases, trypsin domain’ and ‘peptidase S1, PA clan’,
while Q2-Q4 showed a lower degree of enrichment with ‘serine
proteases, trypsin domain’ and ‘peptidase S1, PA clan’ (Fig. 5B).
Hierarchical clustering based on KEGG pathway
analysis showed that UCS proteins of Q4 were mainly involved in
‘ECM-receptor interaction’, ‘focal adhesion’, ‘platelet
activation’, ‘toxoplasmosis’, ‘regulation of actin cytoskeleton’
and ‘PI3K-Akt signaling pathway’, while Q1-Q3 showed a lower degree
of enrichment in these terms (Fig.
5C).
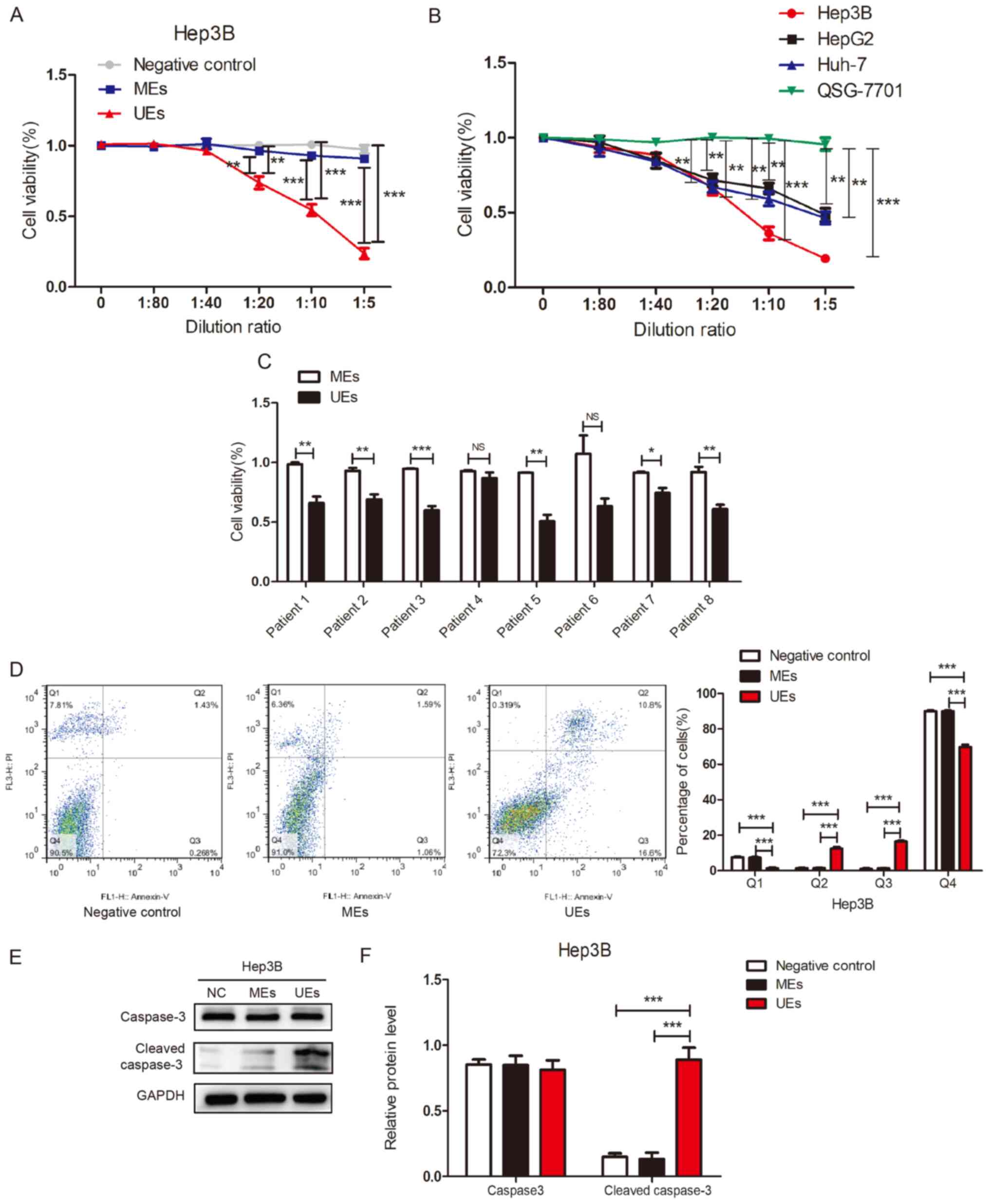
Effect of UEs on liver cancer
cells
To investigate the effects of exosomes derived from
human UCS on liver cancer cells, a CCK-8 assay was conducted to
detect cell viability following treatment with UE. As shown in
Fig. 6A, Hep3B cells were cultured
with exosomes at different dilution ratios. Cell viability was
significantly reduced in Hep3B cells treated with UEs compared with
cells treated with MEs or with the negative control group. The
inhibitory effect of UEs started at a dilution ratio of 1:20.
Subsequently, cell viability was detected in three liver cancer
cell lines following treatment with UEs at different dilution
ratios. The results showed that exosomes from human UCS could
reduce the viability of liver cancer cells compared with the normal
human liver cell line (QSG-7701) (Fig.
6B). To further validate this result, cell viability was
detected in Hep3B cells treated with MEs and UEs (n=8 in each
group). In 6 out 8 cases, UEs significantly suppressed cell
viability compared with MEs (Fig.
6C). These results indicated that exosomes derived from human
UCS could suppress liver cancer cell viability.
UEs may induce apoptosis of HCC
cells
To investigate the effect of UEs on cell apoptosis,
flow cytometry was used to determine apoptotic cell death in HCC
cells. UE treatment significantly increased the percentage of early
apoptosis (lower-right quadrant) and late apoptosis (upper-right
quadrant) (Figs. 6D and S5). To further demonstrate that UEs can
induce apoptosis of HCC cells, the protein expression levels of
caspase-3 and cleaved-caspase-3 were determined. The results
suggested that the expression of cleaved caspase-3 was
significantly higher in the UE group than in the ME and the
negative control groups (Fig. 6E
and F). This indicated that
exosomes isolated from UCS could induce apoptosis of HCC cells.
Discussion
Exosomes are subcellular vesicles consisting mainly
of phospholipids, proteins, cholesterol, ceramide and
sphingolipids. The morphological and biochemical properties of
exosomes vary according to their cellular origin (21,22).
Exosomes are extracted and isolated mainly by ultrafast
centrifugation, immunomagnetic bead separation, precipitation or
filtration, among which ultrafast centrifugation is considered the
most common method to separate exosomes at present, and is also the
gold standard for exosome extraction and identification (23,24).
Exosomes were extracted from UCS and maternal serum
samples. Transmission electron microscopy and western blotting were
then used to identify the exosomes. Nearly all of the isolated
extracellular vesicles had a diameter of 50-100 nm according to the
transmission electron microscopy results (25). In addition, CD9 and CD63 were
detected in the protein samples extracted from the exosomes. CD9
and CD63 are molecular markers of exosomes (26). These results confirmed that exosomes
were successfully isolated from UCS and maternal serum samples.
Exosomes can be found in several types of body
fluids, such as blood, saliva, urine, cerebrospinal fluid and milk
(27,28). Compared with the relatively simple
embryonic environment of umbilical cord blood, the physiological or
pathological environment of peripheral blood is more complex
(29). Therefore, comparing UEs
with MEs may help us better understand the differences between
umbilical cord blood and the peripheral blood environment. A recent
study revealed that UEs and MEs may enhance endothelial cell
proliferation and migration (30);
however, the role of exosomes isolated from the human umbilical
cord in cancer development has not been investigated. To explore
the potential differences in the composition and function of
proteins from UEs and MEs, a proteomic analysis of exosomes was
conducted by mass spectrometry and bioinformatics analysis. To
study the biological effects of UEs on liver cancer cells and to
explore their potential value as a new approach for liver cancer
biotherapy, MEs were used as a control.
According to the results of the proteomic analysis
of exosomes by mass spectrometry and bioinformatics analysis, UEs
were enriched with proteins that were involved in ‘ECM-receptor
interaction’. According to the functional enrichment analysis based
on KEGG pathways, these differentially expressed proteins were
associated with the integrin family. Integrin expression levels
have been revealed to be closely associated with tumor development.
The integrin family may be closely related to cell metastasis and
proliferation (31,32). Integrin β1 has been reported to be
mainly involved in adhesion between cells and the extracellular
matrix (33), and its activity may
affect the distribution and differentiation of stem cells (34). Moreover, integrin β1 has been shown
to be significantly upregulated in liver cancer and may serve as
bidirectional transducers of extracellular and intracellular
signals in the processes of cell adhesion (35). Therefore, a large number of
differentially expressed proteins in exosomes derived from UCS may
be related to the differentiation of a variety of stem cells, and
may also be involved in cell adhesion, migration, apoptosis and
other biological processes.
To investigate the effects of UEs on liver cancer
cells, CCK-8 assays were conducted to detect cell viability in
different groups. Exosomes derived from human UCS could suppress
liver cancer cell viability, especially Hep3B cells. However,
exosomes from UCS had no significant effect on the migration of
these cells (data not shown). Flow cytometry was used to determine
apoptotic cell death of HCC cells to investigate the effect of UEs
on cell apoptosis. The results indicated that UEs could induce
apoptosis of HCC cells. Exosomes often contain signaling molecules.
In the present bioinformatics analysis, KEGG pathways associated
with signaling were identified, such as ‘PI3K-AKT signaling’
pathway. However, whether they are related to apoptosis induced by
exosomes is unknown.
In conclusion, exosomes were isolated from UCS and
maternal serum samples, and to the best of our knowledge, the
present study was the first to conduct a proteomic analysis of
these samples using mass spectrometry. The present study
demonstrated that UCS was enriched with proteins involved in
ECM-receptor interactions. These differentially expressed proteins
may act on the integrin family, which could be related to cell
metastasis and proliferation. These findings indicated that
exosomes derived from human UCS can suppress the viability of liver
cancer cells and induce apoptosis of HCC cells. Further studies are
needed to elucidate the mechanism through which UEs suppress liver
cancer cell viability and induce apoptosis of HCC cells.
Supplementary Material
Representative mass spectra images of
the identified proteins. ADIPOQ, adiponectin, C1Q and collagen
domain containing; THBS4, thrombospondin 4; GP1BB, glycoprotein Ib
platelet subunit β; APOF, apolipoprotein F; PSG9, pregnancy
specific β-1-glycoprotein 9; APCS, amyloid P component, serum.
A heatmap was generated from the
hierarchical clustering of the Gene Ontology analysis results,
showing that umbilical cord serum proteins were enriched in
cellular component terms that were related to cell junction, cell
adhesion and extracellular matrix components. The differentially
expressed proteins were categorized into four groups according to
their fold change: Q1 (0-0.5), Q2 (0.5-0.67), Q3 (1.5-2) and Q4
(>2), with P<0.05 in all cases. The functional annotation
following enrichment, together with the corresponding enrichment
P-value, were first collected. The functional classifications that
were enriched in at least one of the clusters (with P<0.05) were
then screened. The filtered P-value data matrix was -log10
transformed. The transformed data matrix was classified by Z
transformation for each functional category. Lastly, the dataset
obtained following Z transformation was analyzed using one-way
hierarchical clustering.
A heatmap was generated from
hierarchical clustering of the Gene Ontology analysis results,
showing that umbilical cord serum proteins were enriched in
biological process terms that were related to gene regulation, as
well as cell growth, differentiation, motility and adhesion. The
differentially expressed proteins were categorized into four groups
according to their fold change: Q1 (0-0.5), Q2 (0.5-0.67), Q3
(1.5-2) and Q4 (>2), with P<0.05 in all cases.
A heatmap was generated from
hierarchical clustering of the Gene Ontology analysis results,
showing that umbilical cord serum proteins were enriched in
molecular function terms that were related to molecule binding,
including integrin binding and enzyme binding. The differentially
expressed proteins were categorized into four groups according to
their fold change: Q1 (0-0.5), Q2 (0.5-0.67), Q3 (1.5-2) and Q4
(>2), with P<0.05 in all cases.
Flow cytometry was used to determine
apoptotic cell death of MHCC97H cells treated with exosomes from
the negative control, ME and UE groups. The data are presented as
the mean ± SEM. *P<0.05, **P<0.01,
***P<0.001. ME, maternal serum exosome; UE, umbilical
serum exosome; PI, propidium iodide. The different quadrants Q1,
Q2, Q3 and Q4 represent necrotic cells, late apoptotic cells, early
apoptotic cells and viable cells, respectively.
Representative mass spectra of some
differentially expressed proteins.
Acknowledgements
The authors thank Dr Mingzuo Jiang (Department of
Gastroenterology and Hepatology, Jinling Hospital, Medical School
of Nanjing University) for their technical help.
Funding
Funding: This study was supported by grants from The National
Key R&D Program of China (grant no. 2017YFC1308600), The Key
Research and Development Program of Shaanxi Province (grant no.
2017ZDXM-SF-024) and The National Natural Science Foundation of
China (grant nos. 81670563 and 31900566).
Availability of data and materials
The data supporting the findings of the article is
available in the PRIDE database (https://www.ebi.ac.uk/pride/) under accession no.
PXD025079.
Authors' contributions
DZ designed the study. DZ, CF and RY performed the
experiments. WL and YC contributed to data analysis. MJ, YC and XL
performed data interpretation. YC contributed to the critical
revision of the manuscript. DZ wrote the manuscript. DZ and CF
confirm the authenticity of all the raw data. All authors approved
the final version of the manuscript.
Ethics approval and consent to
participate
This study was approved by The Ethics Committee of
Xijing Hospital. Written informed consent was obtained from the
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Olejarz W, Dominiak A, Żołnierzak A,
Kubiak-Tomaszewska G and Lorenc T: Tumor-Derived exosomes in
immunosuppression and immunotherapy. J Immunol Res.
2020(6272498)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen G, Huang AC, Zhang W, Zhang G, Wu M,
Xu W, Yu Z, Yang J, Wang B, Sun H, et al: Exosomal PD-L1
contributes to immunosuppression and is associated with anti-PD-1
response. Nature. 560:382–386. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Morishita M, Takahashi Y, Matsumoto A,
Nishikawa M and Takakura Y: Exosome-based tumor antigens-adjuvant
co-delivery utilizing genetically engineered tumor cell-derived
exosomes with immunostimulatory CpG DNA. Biomaterials. 111:55–65.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yu DD, Wu Y, Shen HY, Lv MM, Chen WX,
Zhang XH, Zhong SL, Tang JH and Zhao JH: Exosomes in development,
metastasis and drug resistance of breast cancer. Cancer Sci.
106:959–964. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kibria G, Ramos EK, Wan Y, Gius DR and Liu
H: Exosomes as a drug delivery system in cancer therapy: Potential
and challenges. Mol Pharm. 15:3625–3633. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mincheva-Nilsson L and Baranov V:
Placenta-derived exosomes and syncytiotrophoblast microparticles
and their role in human reproduction: Immune modulation for
pregnancy success. Am J Reprod Immunol. 72:440–457. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hu Y, Rao SS, Wang ZX, Cao J, Tan YJ, Luo
J, Li HM, Zhang WS, Chen CY and Xie H: Exosomes from human
umbilical cord blood accelerate cutaneous wound healing through
miR-21-3p-mediated promotion of angiogenesis and fibroblast
function. Theranostics. 8:169–184. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
La Marca V and Fierabracci A: Insights
into the diagnostic potential of extracellular vesicles and their
miRNA signature from liquid biopsy as early biomarkers of diabetic
micro/macrovascular complications. Int J Mol Sci.
18(1974)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Salomon C and Rice GE: Role of exosomes in
placental homeostasis and pregnancy disorders. Prog Mol Biol Transl
Sci. 145:163–179. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cleys ER, Halleran JL, McWhorter E,
Hergenreder J, Enriquez VA, da Silveira JC, Bruemmer JE, Winger QA
and Bouma GJ: Identification of microRNAs in exosomes isolated from
serum and umbilical cord blood, as well as placentomes of
gestational day 90 pregnant sheep. Mol Reprod Dev. 81:983–993.
2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Steenbergen RH, Joyce MA, Thomas BS, Jones
D, Law J, Russell R, Houghton M and Tyrrell DL: Human serum leads
to differentiation of human hepatoma cells, restoration of
very-low-density lipoprotein secretion, and a 1000-fold increase in
HCV Japanese fulminant hepatitis type 1 titers. Hepatology.
58:1907–1917. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Piao C, Zhang Q, Jin D, Wang L, Tang C,
Zhang N, Lian F and Tong X: A study on the mechanism of milkvetch
root in the treatment of diabetic nephropathy based on network
pharmacology. Evid Based Complement Alternat Med.
2020(6754761)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xie S, Du Y, Zhang Y, Wang Z, Zhang D, He
L, Qiu L, Jiang J and Tan W: Aptamer-based optical manipulation of
protein subcellular localization in cells. Nat Commun.
11(1347)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Andre Mdo R, Pedro A and Lyden D: Cancer
exosomes as mediators of drug resistance. Methods Mol Biol.
1395:229–239. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Przytycka T, Davis G, Song N and Durand D:
Graph theoretical insights into evolution of multidomain proteins.
J Comput Biol. 13:351–363. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Peterson TA, Nehrt NL, Park D and Kann MG:
Incorporating molecular and functional context into the analysis
and prioritization of human variants associated with cancer. J Am
Med Inform Assoc. 19:275–283. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Dawson N, Sillitoe I, Marsden RL and
Orengo CA: The classification of protein domains. Methods Mol Biol.
1525:137–164. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Mehrotra P, Ami VKG and Srinivasan N:
Clustering of multi-domain protein sequences. Proteins. 86:759–776.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sawyer N, Watkins AM and Arora PS: Protein
domain mimics as modulators of protein-protein interactions. Acc
Chem Res. 50:1313–1322. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Corrado C, Raimondo S, Chiesi A, Ciccia F,
De Leo G and Alessandro R: Exosomes as intercellular signaling
organelles involved in health and disease: Basic science and
clinical applications. Int J Mol Sci. 14:5338–5366. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Barile L and Vassalli G: Exosomes: Therapy
delivery tools and biomarkers of diseases. Pharmacol Ther.
174:63–78. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wu X, Showiheen SAA, Sun AR, Crawford R,
Xiao Y, Mao X and Prasadam I: Exosomes extraction and
identification. Methods Mol Biol. 2054:81–91. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tang YT, Huang YY, Zheng L, Qin SH, Xu XP,
An TX, Xu Y, Wu YS, Hu XM, Ping BH and Wang Q: Comparison of
isolation methods of exosomes and exosomal RNA from cell culture
medium and serum. Int J Mol Med. 40:834–844. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Cizmar P and Yuana Y: Detection and
characterization of extracellular vesicles by transmission and
cryo-transmission electron microscopy. Methods Mol Biol.
1660:221–232. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Khushman M, Bhardwaj A, Patel GK, Laurini
JA, Roveda K, Tan MC, Patton MC, Singh S, Taylor W and Singh AP:
Exosomal markers (CD63 and CD9) expression pattern using
immunohistochemistry in resected malignant and nonmalignant
pancreatic specimens. Pancreas. 46:782–788. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Flaumenhaft R: Formation and fate of
platelet microparticles. Blood Cells Mol Dis. 36:182–187.
2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zmigrodzka M, Guzera M, Miskiewicz A,
Jagielski D and Winnicka A: The biology of extracellular vesicles
with focus on platelet microparticles and their role in cancer
development and progression. Tumour Biol. 37:14391–14401.
2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Sadovsky Y, Mouillet JF, Ouyang Y, Bayer A
and Coyne CB: The function of TrophomiRs and other MicroRNAs in the
human placenta. Cold Spring Harb Perspect Med.
5(a023036)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jia L, Zhou X, Huang X, Xu X, Jia Y, Wu Y,
Yao J, Wu Y and Wang K: Maternal and umbilical cord serum-derived
exosomes enhance endothelial cell proliferation and migration.
FASEB J. 32:4534–4543. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ata R and Antonescu CN: Integrins and cell
metabolism: An intimate relationship impacting cancer. Int J Mol
Sci. 18(189)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yin HL, Wu CC, Lin CH, Chai CY, Hou MF,
Chang SJ, Tsai HP, Hung WC, Pan MR and Luo CW: β1 integrin as a
prognostic and predictive marker in triple-negative breast cancer.
Int J Mol Sci. 17(1432)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Begum A, Ewachiw T, Jung C, Huang A,
Norberg KJ, Marchionni L, McMillan R, Penchev V, Rajeshkumar NV,
Maitra A, et al: The extracellular matrix and focal adhesion kinase
signaling regulate cancer stem cell function in pancreatic ductal
adenocarcinoma. PLoS One. 12(e0180181)2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Quisenberry CR, Nazempour A, Van Wie BJ
and Abu-Lail NI: Evaluation of β1-integrin expression on
chondrogenically differentiating human adipose-derived stem cells
using atomic force microscopy. Biointerphases.
11(021005)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li Y, Ren Z, Wang Y, Dang YZ, Meng BX,
Wang GD, Zhang J, Wu J and Wen N: ADAM17 promotes cell migration
and invasion through the integrin β1 pathway in hepatocellular
carcinoma. Exp Cell Res. 370:373–382. 2018.PubMed/NCBI View Article : Google Scholar
|