Introduction
Sepsis refers to the life-threatening organ
dysfunction caused by the uncontrolled immune response of the host
to infection, which is one of the main causes of death worldwide
(1,2). Using data from 1,553 reports from
high-income countries with cases between 1979 and 2015, the
researchers initially extrapolated that there are an estimated 31.5
million cases of sepsis and 19.4 million cases of severe sepsis
globally, with a potential 5.3 million deaths per year (3). Acute lung injury (ALI) is one of the
most common complications of sepsis. According to statistics, the
fatality rate of ALI may account for 34.9% of the total number of
individuals admitted to hospital; thus, ALI has attracted extensive
attention from healthcare researchers (4). ALI refers to the damage of alveolar
epithelial and capillary endothelial cells caused by direct and
indirect factors, resulting in acute hypoxic respiratory
insufficiency. The most important pathological feature of ALI is an
extensive inflammatory reaction (5). Although treatments of ALI have
improved in recent years, there are currently no drugs approved by
the Food and Drug Administration or the China Food and Drug
Administration for ALI treatment (6). Therefore, discovering therapeutic
targets and drugs for the treatment of ALI and reducing the
mortality rates associated with ALI have become the focus of
current research.
Stomatin (STOM) protein is a unidirectional,
low-polymerized lipid raft-related protein, also known as human
erythrocyte integrated membrane protein (7). Its main function is to regulate the
activity of ion channels, glucose transporters and cytoskeleton
protein recombination (8,9). Previous studies have identified that
STOM is abnormally expressed in a variety of malignant tumors, such
as lung cancer and oral squamous carcinoma, suggesting that it may
be involved in the occurrence and development of tumors (10,11).
However, to the best of our knowledge, there are few studies that
focus on the role of STOM in diseases other than tumors. Moreover,
results of a previous study demonstrated that STOM expression is
increased in children with sepsis, and that STOM may potentially be
used as a transcriptional marker for the diagnosis of sepsis
(12). In addition, the expression
level of STOM is significantly increased in lung alveolar
epithelial cells that have been damaged due to ischemia/reperfusion
injury (13). Hypoxia leads to an
increase in the levels of reactive oxygen species (ROS) and
oxidative stress, which then promotes the occurrence of cellular
inflammation (14). Therefore, it
was hypothesized that STOM may affect inflammation and oxidative
stress in sepsis-treated alveolar epithelial cells.
CD36 is a pattern recognition receptor that is
expressed in various types of cells, including microglial, immune
and tumor cells (15). CD36
mediates lipid uptake, immune recognition, inflammation, molecular
adhesion and apoptosis (15).
Results of a previous study revealed that CD36 regulates the innate
immune response during pneumonia, tuberculosis, malaria, human
immunodeficiency virus and sepsis in a ligand-mediated manner
(16). A prior study indicated
that ETS domain-containing protein Elk-1 B domain works primarily
through CD36, and it reduces lung barrier dysfunction, neutrophil
migration into the lung and lung inflammation caused by
lipopolysaccharide (LPS) (17).
Moreover, the inhibition of CD36 significantly suppresses
LPS-induced inflammation and ALI (17). Thus, it is important to determine
whether STOM promotes the injury of alveolar epithelial cells
induced by sepsis by regulating CD36.
In the present study, the expression levels and
biological functions of STOM and CD36 in the LPS-treated alveolar
epithelial cell line, MLE-12, were analyzed. The results of the
present study may provide a novel theoretical basis for
STOM-knockdown in the treatment of sepsis-induced ALI.
Materials and methods
Cell culture and treatment
The alveolar epithelial cell line MLE-12 was
purchased from the BeNa Culture Collection; Beijing Beina Chunglian
Institute of Biotechnology. Cells were cultured in DMEM/F12 Coon's
medium (DMEM/F12; Thermo Fisher Scientific, Inc.) containing 10%
FBS (Thermo Fisher Scientific, Inc.) and maintained in a 5%
CO2 incubator at 37˚C. After the cells were passaged
three times, cells in logarithmic phase were selected for follow-up
assays.
The sepsis cell model was established using MLE-12
cells. Cells in the logarithmic phase were routinely cultured for
24 h at 37˚C. When the degree of cell fusion reached 90%, 100 ng/ml
LPS (Sigma-Aldrich; Merck KGaA) was added to the culture for 24 h
at 37˚C based on data from a previous study (18).
Bioinformatics analysis
Microarray data were obtained from the GEO database
(www.ncbi.nlm.nih.gov/geo). Gene
expression profiles of the neonatal sepsis samples public dataset
GSE145227 was downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi)
(19). The GSE145227 dataset
contained 10 samples from children with sepsis and 12 samples from
healthy controls. Affymetrix Human lncRNA Array (version 1.0;
Affymetrix; Thermo Fisher Scientific, Inc.) was used to detect the
expression levels of long non-coding (lnc)RNAs and mRNAs in both
septic and control groups. Subsequently, the data were analyzed
using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r) and GraphPad
Prism 8.0 software (GraphPad Software, Inc.). Search tool for
recurring instances of neighboring genes (STRING) database (version
11.0; https://version-11-0.string-db.org/) was used to
analyze the potential association between STOM and CD36.
Cell transfection
Small interfering si(RNA) against STOM (si-STOM;
si-STOM-1, 1 µg, 5'-GTGTTTCTAAAGATGGAATTTCA-3'; si-STOM-2, 1 µg,
5'-GAGTCATCTATTCTGATTATTTG-3') and the scrambled negative control
group (si-NC; 1 µg; 5'-ACTTGCGCTTGCGAAAATCTATATAGC-3') were
constructed by Guangzhou RiboBio Co., Ltd. The CD36-overexpression
vector (Ov-CD36; 50 nM) and empty control vector (Ov-NC; 50 nM)
were constructed by Shanghai GenePharma Co., Ltd. siRNA and
plasmids were transfected into MLE-12 cells were inoculated into
six-well plates (5x105 cells/well), and then
transfection was performed for 8 h at 37˚C using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
After transfection for 48 h, the transfection efficiency of cells
in each group was detected via reverse transcription-quantitative
(RT-q)PCR. Subsequent experiments were completed within 48 h.
Reverse transcription quantitative PCR
(RT-qPCR) analysis
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from cells.
Following which, 5 µg total RNA was used as the control and cDNA
was synthesized using AMV reverse transcription kit (Promega
Corporation) according to the manufacturer's protocol. qPCR was
performed on an ABI Prism 7900 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using a
SYBR® Green kit (Qiagen, Inc.). The relative expression
level of the target gene was calculated using the 2-ΔΔCq
method (20). GAPDH was used for
normalization. The following thermocycling conditions were used:
Initial denaturation at 95˚C for 10 min; followed by 37 cycles of
denaturation at 95˚C for 15 sec and annealing at 60˚C for 1 min;
extension for 10 min at 65˚C. The sequences of PCR primers were as
follows: CD36 Forward, 5'-AAGCCAGGTATTGCAGTTCTTT-3' and reverse,
5'-GCATTTGCTGATGTCTAGCACA-3'; STOM forward,
5'-GTGACTCTCGACAATGTAAC-3' and reverse, 5'-TGATCTCATAACGGAGGCAG-3';
and GAPDH forward, 5'-TGTCAAGCTCATTTCCTGGTAT-3' and reverse,
5'-CTCTCTTCCTCTTGTGCTCTTG-3'.
ELISA
Transfected cells were centrifuged at 1,000 x g for
5 min at 4˚C. The supernatant was collected and used for ELISA. The
levels of malondialdehyde (MDA), superoxide dismutase (SOD),
lactate dehydrogenase (LDH), TNF-α and ROS in the cell supernatant
were measured using the MDA assay kit (cat. no. A003-1-2; Nanjing
Jiancheng Bioengineering Institute), SOD Assay Kit (cat. no.
A001-3-2; Nanjing Jiancheng Bioengineering Institute), LDH
Cytotoxicity Assay Kit (cat. no. C0016; Beyotime Institute of
Biotechnology), TNF-α assay kit (cat. no. H052-1; Nanjing Jiancheng
Bioengineering Institute) and ROS assay kit (cat. no. S0033S;
Beyotime Institute of Biotechnology), respectively, according to
the manufacturer's protocol. These samples were measured using a
Hitachi spectrophotometer (F-7000 FL spectrophotometer; Hitachi,
Ltd.).
Cell Counting Kit-8 (CCK-8) assay
Cells were inoculated on 96-well plates at a density
of 4x104 cells/well. The cells were cultured in DMEM/F12
medium containing 10% FBS for 24, 48 and 72 h at 37˚C.
Subsequently, 10 µl CCK-8 solution (cat. no. C0038; Beyotime
Institute of Biotechnology) was added to each well to culture for 1
h. The absorbance of cells in each well was detected at 450 nm
using an automatic enzyme labeling instrument.
RNA immunoprecipitation (RIP)
assay
The RIP assay was performed using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (including RIP buffer
and beads; cat. no. 17-700; MilliporeSigma) according to the
manufacturer's protocol. Cells were incubated with RIP buffer
supplied with the kit in an ice bath for 20 min. A total of 20 µl
magnetic beads were applied for incubation of cell extracts and
coated with 1 µg Ago2 antibody (cat. no. 03-110; Merck KGaA) or 2
µg IgG antibody supplied with the kit for 12 h at 40˚C.
Immunoprecipitated complexes were collected by adding 500 µl lysate
per 200 µl RIP buffer, followed by centrifugation at 10,000 x g for
10 min at 4˚C. and RT-qPCR was subsequently performed.
Western blotting
Total protein was extracted from MLE-12 cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology). The
protein concentration was detected using a BCA protein quantitative
kit (Pierce; Thermo Fisher Scientific, Inc.). A total of 25 µg
protein per lane was separated via SDS-PAGE on a 10% gel, and
subsequently transferred to PVDF membranes. These membranes were
blocked with 5% non-fat milk for 2 h at room temperature. Membranes
were incubated with primary antibodies (1:1,000) against STOM (cat.
no. ab166623), CD36 (cat. no. ab133625), TNF-α (cat. no. ab215188),
IL-6 (cat. no. ab233706) and GAPDH (cat. no. ab9485) (all Abcam)
overnight at 4˚C. After the membranes were washed with TBST (0.1%
Tween-20) three times, membranes were incubated with HRP-conjugated
goat anti-rabbit (1:2,000; cat. no. 14708; Cell Signaling
Technology, Inc.) secondary antibody for 2 h at room temperature.
ECL reagent (Pierce; Thermo Fisher Scientific, Inc.) was used to
detect the protein bands. ImageJ software 1.4.3 (National
Institutes of Health) was used to detect the relative gray value of
the target proteins.
Statistical analysis
GraphPad Prism 8.0 software (GraphPad Software,
Inc.) was used for statistical analysis. The data are expressed as
the mean ± standard deviation (unless otherwise shown). One-way
ANOVA followed by Tukey's post hoc test was used to analyze the
comparison between multiple groups. An unpaired Student's t-test
was used to compare differences between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
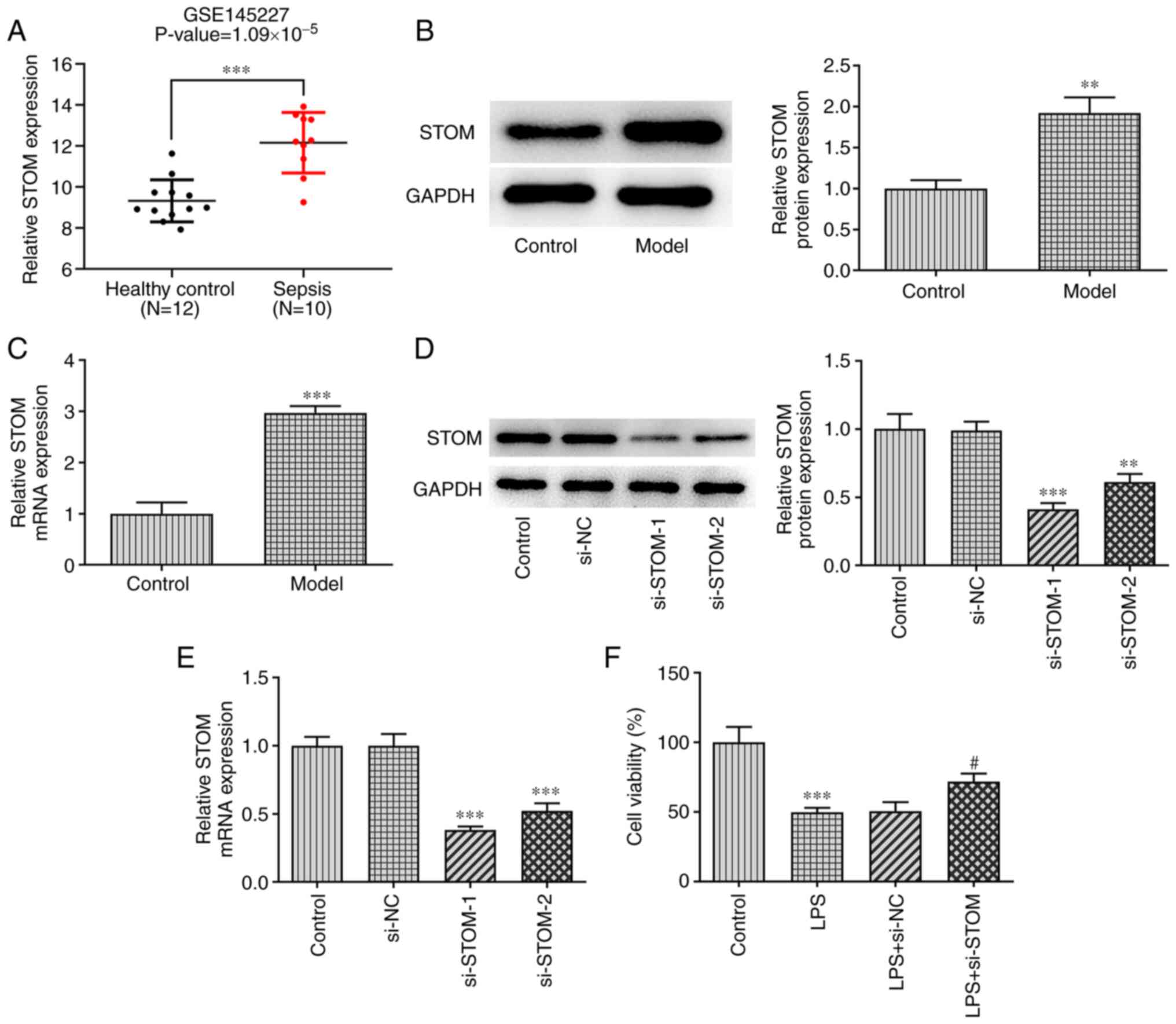
STOM is upregulated in LPS-treated
MLE-12 cells and STOM-knockdown promotes the viability of
LPS-treated MLE-12 cells
GEO2R analysis of the GeneChip GSE145227 was
employed to confirm that the expression level of STOM was
significantly increased in the peripheral blood of newborns with
sepsis compared with in the peripheral blood of healthy newborns
(Fig. 1A). Subsequently, the
results of RT-qPCR and western blotting demonstrated that the
expression level of STOM in the sepsis model group was
significantly higher compared with the control group (Fig. 1B and C). These results suggested that STOM may
be involved in LPS-induced damage of MLE-12 cells.
The expression levels of STOM in MLE-12 cells were
decreased following transfection with si-STOM. As demonstrated in
Fig. 1D and E, the expression levels of STOM in the
si-NC group were similar to the control group; whereas STOM
expression was decreased in both the si-STOM-1 and si-STOM-2 groups
compared with the control group, with si-STOM-1 exhibiting an
increased level of transfection efficiency compared with si-STOM-2.
Therefore, si-STOM-1 was selected for follow-up experiments. A
CCK-8 assay was applied to detect cell viability. As presented in
Fig. 1F, the results demonstrated
that treatment with LPS significantly inhibited cell viability
compared with the control group, while STOM-knockdown reduced the
inhibitory effect of LPS on cell viability compared with LPS+si-NC
group.
STOM-knockdown reduces oxidative
stress and inflammation in LPS-treated MLE-12 cells
Oxidative stress and inflammation levels were
detected using ELISA and western blotting assays. As demonstrated
in Fig. 2A-D, compared with the
LPS + si-NC group, the levels of LDH, MDA and ROS in the LPS +
si-STOM group were significantly decreased, whereas the level of
SOD was significantly increased. Moreover, treatment with LPS
induced high levels of LDH, MDA and ROS, and low levels of SOD
compared with the control group. The results of western blotting
and RT-qPCR analyses demonstrated that compared with the control
group, LPS significantly increased the expression levels of the
pro-inflammatory cytokines TNF-α and IL-6, whereas these changes
were partially reversed by STOM-knockdown (Fig. 2E). Similar results were observed
using ELISA. As presented in Fig.
2F, results of ELISA revealed that compared with the LPS +
si-NC group, STOM-knockdown significantly decreased the levels of
the pro-inflammatory cytokines TNF-α and IL-6. These results
indicated that STOM-knockdown reduced oxidative stress and
inflammation in MLE-12 cells treated with LPS.
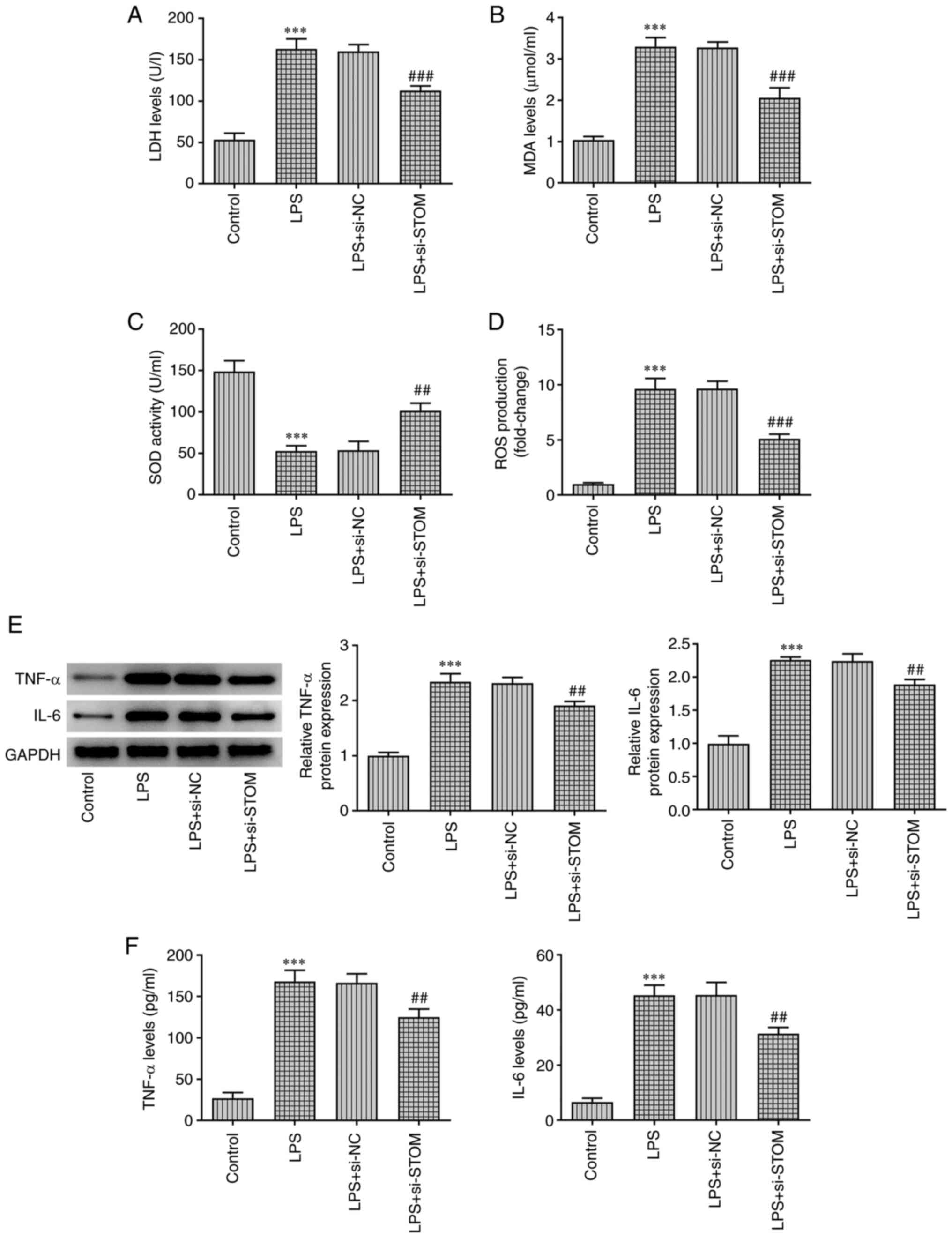 | Figure 2STOM-knockdown reduces oxidative
stress and inflammation in LPS-induced MLE-12 cells. MLE-12 cells
were treated with LPS, followed by transfection with si-NC and
si-STOM. Detection of (A) LDH, (B) MDA, (C) SOD and (D) ROS levels
in MLE-12 cells. Detection of TNF-α and IL-6 (E) protein expression
levels and (F) content levels in MLE-12 cells. The results are
representative of at least three independent experiments.
***P<0.001 vs. Control group; ##P<0.01,
###P<0.001 vs. LPS + si-NC group. LPS,
lipopolysaccharide; si-, small interfering RNA; NC, negative
control; LDH, lactate dehydrogenase; MDA, malondialdehyde; SOD,
superoxide dismutase; ROS, reactive oxygen species; STOM,
stomatin. |
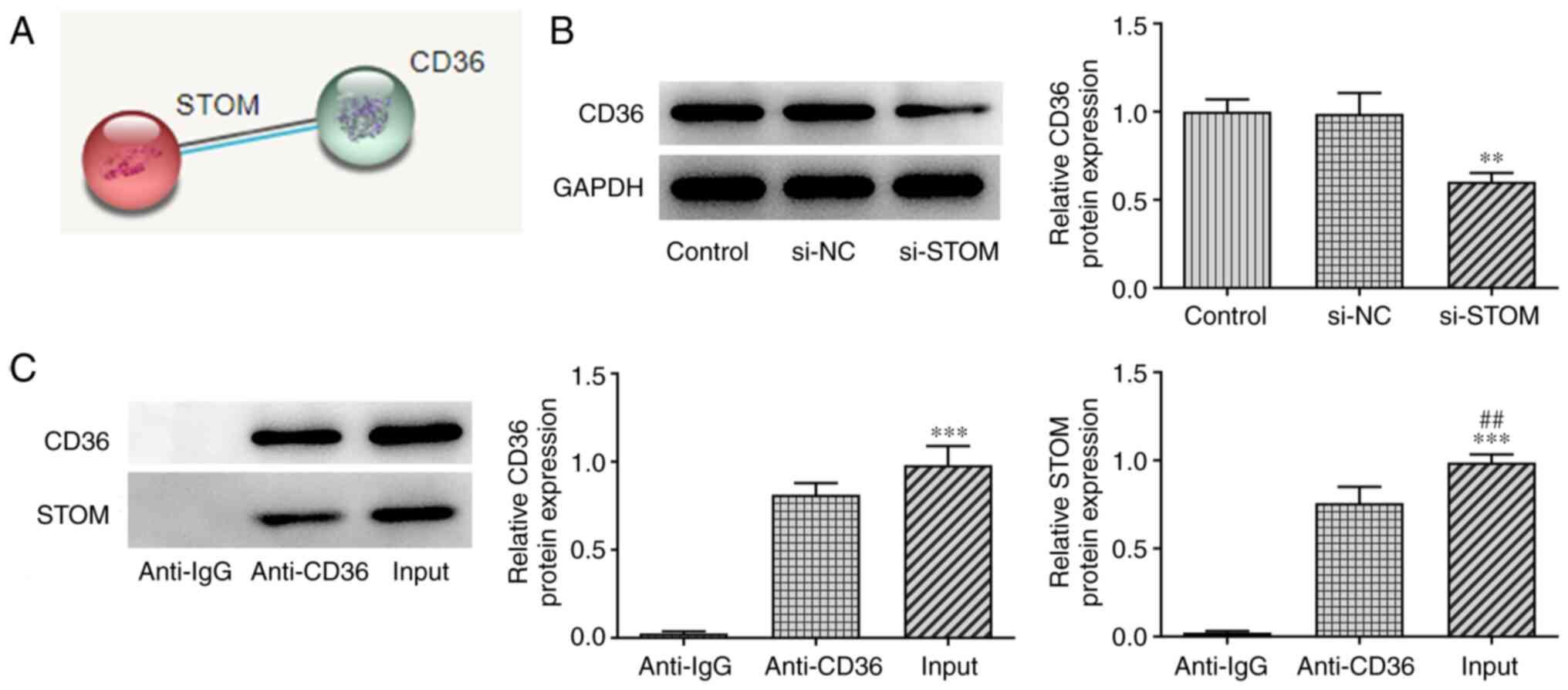
STOM positively regulates CD36
To further explore the effects of STOM on the damage
of MLE-12 cells treated with LPS, the downstream target gene of
STOM, CD36, was screened for using the STRING database (Fig. 3A). The interaction between STOM and
CD36 was verified using a RIP assay (Fig. 3C). The results revealed that CD36
and STOM bound strongly. Moreover, as demonstrated in Fig. 3B, STOM-knockdown significantly
decreased the expression of CD36 compared with the control. Thus,
results of the present study demonstrated that STOM positively
regulated CD36.
STOM promotes oxidative stress and
inflammation in LPS-treated MLE-12 cells by upregulating CD36
Lastly, a CD36-overexpressing plasmid (Ov-CD36) was
constructed. As displayed in Fig.
4A and B, compared with the
Ov-NC group, both the protein and mRNA expression levels of CD36
were significantly increased in the Ov-CD36 group. Detection of
oxidative stress levels using the kits revealed a significant
decrease in LDH levels after interfering with STOM compared with
the LPS + si-NC group. However, the inhibitory effect of
interfering with STOM on LDH was reversed after CD36 overexpression
(Fig. 4C). Additionally, the trend
of MDA and ROS among groups was similar to that of LDH (Fig. 4D and F). By contrast, following interference
with STOM, SOD levels were upregulated compared with the LPS +
si-NC group, but following CD36 overexpression, SOD levels were
decreased (LPS + si-STOM + Ov-CD36 vs. LPS + si-STOM + Ov-NC;
Fig. 4E). As demonstrated in
Fig. 4C-F, the overexpression of
CD36 reversed the inhibitory effect of STOM-knockdown on oxidative
stress. As demonstrated in Fig. 4G
and H, TNF-α and IL-6 expression
was suppressed after knockdown of STOM compared with the LPS +
si-NC group. However, overexpression of CD36 significantly
increased TNF-α and IL-6 expression compared with the knockdown of
STOM group. Therefore, it was clear that overexpression of CD36
alleviated the inhibitory effect of STOM-knockdown on inflammation.
Collectively, these results demonstrated that STOM aggravated
LPS-induced damage of MLE-12 cells by promoting the expression of
CD36.
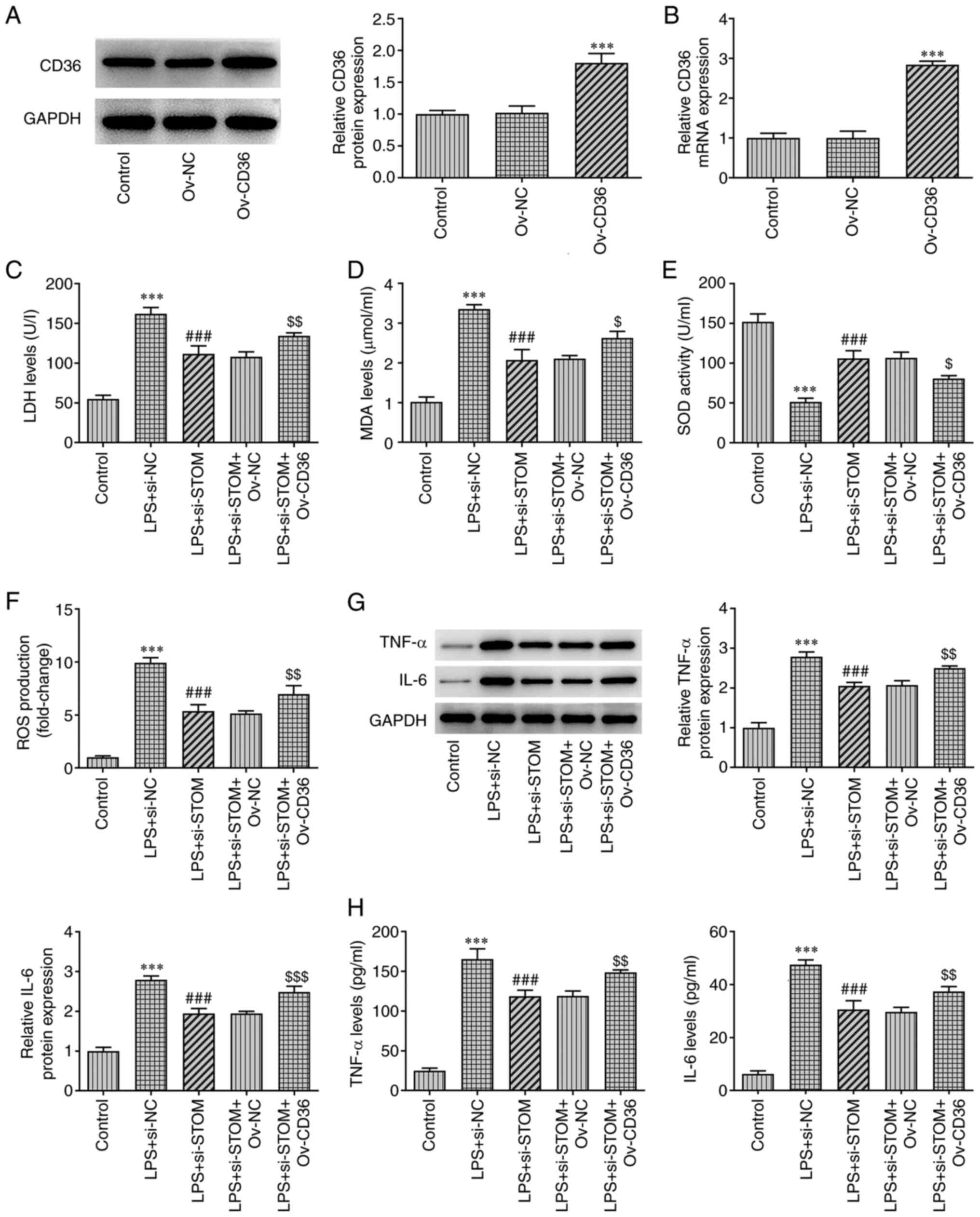 | Figure 4STOM promotes oxidative stress and
inflammation in LPS-induced MLE-12 cells. Detection of CD36 (A)
protein and (B) mRNA levels in MLE-12 cells transfected with Ov-NC
and Ov-CD36. Detection of (C) LDH, (D) MDA, (E) SOD and (F) ROS
levels in LPS-induced MLE-12 cells transfected with Ov-NC and
Ov-CD36. Detection of TNF-α and IL-6 (G) protein and (H) mRNA
levels in MLE-12 cells. The results are representative of at least
three independent experiments. ***P<0.001 vs. Control
group; ###P<0.001 vs. LPS + si-NC group;
$P<0.05, $$P<0.01,
$$$P<0.001 vs. LPS + si-STOM + Ov-NC group. LPS,
lipopolysaccharide; Ov-, overexpression; NC, negative control; LDH,
lactate dehydrogenase; MDA, malondialdehyde; SOD, superoxide
dismutase; ROS, reactive oxygen species; si-, small interfering
RNA; STOM, stomatin. |
Discussion
Alveolar epithelial cells act as important barriers
against external pathogens and are closely associated with the
occurrence and development of lung diseases, such as lung injury
and lung fibrosis (21). High
levels of oxidative stress and inflammation in alveolar epithelial
cells destroy the integrity of lung alveolar epithelial cells and
induce ALI (22). Bacterial
endotoxin LPS is the main component of the outer wall of
Gram-negative bacteria, which promotes apoptosis and induces ALI
(23). In the present study,
STOM-knockdown significantly suppressed LPS-induced oxidative
stress and inflammation in MLE-12 cells by downregulating the
expression level of CD36. These results suggested that STOM may
play a notable role in LPS-induced MLE-12 cell injury.
STOM is a unidirectional lipid raft-related protein
isolated from the plasma membrane of normal human erythrocytes
(24). To date, there is
relatively limited direct research on the function of STOM. The
analysis of GeneChip GSE145227 of neonatal patients with sepsis
demonstrated that, compared with healthy newborns, the expression
levels of STOM in the peripheral blood of newborns with sepsis were
significantly increased (25). In
addition, STOM is highly expressed in alveolar epithelial cells
treated under hypoxic conditions (10), and oxidative stress and
inflammation are caused by hypoxia (14). STOM-like protein 2 (SLP-2) is one
of the most widely studied homologs of STOM. Studies have reported
that SLP-2 is upregulated in a variety of cancer types, including
esophageal squamous cell carcinoma (26), epithelial ovarian cancer (27) and non-small cell lung cancer,
amongst others (28), and that
SLP-2-silencing significantly reduces cancer progression (29). Notably, SLP-2 acts as a
pro-inflammatory factor in patients with colitis and liver cancer
(30,31). Thus, STOM may affect the oxidative
stress and inflammation of alveolar epithelial cells induced by
sepsis. In the present study, STOM was highly expressed in
LPS-treated MLE-12 cells compared with the control group.
STOM-knockdown significantly enhanced the viability of LPS-treated
MLE-12 cells, and suppressed oxidative stress and inflammation,
compared with the LPS-treated group. These results suggested that
STOM promoted oxidative stress and inflammation in LPS-induced
MLE-12 cells.
The effects of STOM on sepsis-induced MLE-12 cell
injury were further explored, and results obtained from the STRING
database demonstrated that CD36 interacts with STOM. CD36 is a
single-stranded transmembrane cell surface protein widely found in
various cells, including monocytes, macrophages and microvascular
endothelial cells (15,32). CD36 plays a key role in the
transport of fatty acids in the liver by participating in the
transmembrane transport of long-chain fatty acids (33). A previous study reported that CD36
promotes the inflammation of macrophages and microglia by binding
to toll-like receptors (34).
Notably, CD36-silencing effectively reduces LPS-induced
inflammation and ALI (25). The
results of the present study demonstrated that STOM positively
regulated the expression level of CD36. Furthermore, overexpression
of CD36 partially reversed the promoting effects of STOM-knockdown
on oxidative stress and inflammation in LPS-treated MLE-12 cells.
There are some limitations to the present study, such as the lack
of multiple cell lines used to validate the findings, and likewise,
the lack of in vivo experiments.
In conclusion, the expression levels of STOM in
mouse alveolar epithelial cells treated with LPS were significantly
increased compared with those in the control group, and
STOM-knockdown reduced the levels of oxidative stress and
inflammation by binding to CD36 in ALI. These results preliminarily
confirmed the role of STOM to be a valuable biomarker in the
progression of ALI. Thus, results obtained during the present study
may provide a theoretical basis for understanding the regulatory
mechanisms underlying ALI, and provide support for the development
of novel treatment options for ALI in the future. Moreover, further
in vitro experiments will be carried out using alternative
cell lines to MLE-12, and these will be supplemented with in
vivo experiments, to further confirm the role of STOM in ALI in
future investigations.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Both KW and LW were involved in the protocol design
and experiments used in the present study. All authors were
responsible for analysis and interpretation of data. KW was
responsible for writing and editing the manuscript. KW and LW
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Singer M, Deutschman CS, Seymour CW,
Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche
JD, Coopersmith CM, et al: The third international consensus
definitions for sepsis and septic shock (Sepsis-3). JAMA.
315:801–810. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Salomao R, Ferreira BL, Salomão MC, Santos
SS, Azevedo LCP and Brunialti MKC: Sepsis: Evolving concepts and
challenges. Braz J Med Biol Res. 52(e8595)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fleischmann C, Scherag A, Adhikari NK,
Hartog CS, Tsaganos T, Schlattmann P, Angus DC and Reinhart K:
International Forum of Acute Care Trialists. Assessment of global
incidence and mortality of hospital-treated sepsis. Current
estimates and limitations. Am J Respir Crit Care Med. 193:259–272.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Thompson BT, Chambers RC and Liu KD: Acute
respiratory distress syndrome. N Engl J Med. 377:562–572.
2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sawa T: The molecular mechanism of acute
lung injury caused by pseudomonas aeruginosa: From bacterial
pathogenesis to host response. J Intensive Care.
2(10)2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bellani G, Laffey JG, Pham T, Fan E,
Brochard L, Esteban A, Gattinoni L, van Haren F, Larsson A, McAuley
DF, et al: Epidemiology, patterns of care, and mortality for
patients with acute respiratory distress syndrome in intensive care
units in 50 countries. JAMA. 315:788–800. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Stewart GW: Stomatin. Int J Biochem Cell
Biol. 9:271–274. 1997.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Genetet S, Desrames A, Chouali Y, Ripoche
P, Lopez C and Mouro-Chanteloup I: Stomatin modulates the activity
of the anion exchanger 1 (AE1, SLC4A1). Sci Rep.
7(46170)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nagarajan A, Dogra SK, Sun L, Gandotra N,
Ho T, Cai G, Cline G, Kumar P, Cowles RA and Wajapeyee N:
Paraoxonase 2 facilitates pancreatic cancer growth and metastasis
by stimulating GLUT1-mediated glucose transport. Mol Cell.
67:685–701.e686. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen JC, Cai HY, Wang Y, Ma YY, Song LN,
Yin LJ, Cao DM, Diao F, Li YD and Lu J: Up-regulation of stomatin
expression by hypoxia and glucocorticoid stabilizes
membrane-associated actin in alveolar epithelial cells. J Cell Mol
Med. 17:863–872. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang D, Qi H, Li A, Deng F, Xu Y, Hu Z and
Liu Q: Coexisting overexpression of STOML1 and STOML2 proteins may
be associated with pathology of oral squamous cell carcinoma. Oral
Surg Oral Med Oral Pathol Oral Radiol. 129:591–599.e593.
2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Long G and Yang C: A sixgene support
vector machine classifier contributes to the diagnosis of pediatric
septic shock. Mol Med Rep. 21:1561–1571. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang M, Li C and Shi W: Stomatin-like
protein-2 confers neuroprotection effect in oxygen-glucose
deprivation/reoxygenation-injured neurons by regulating AMPK/Nrf2
signalling. J Drug Target. 28:600–608. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
McGarry T, Biniecka M, Veale DJ and Fearon
U: Hypoxia, oxidative stress and inflammation. Free Radic Biol Med.
125:15–24. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang J and Li Y: CD36 tango in cancer:
Signaling pathways and functions. Theranostics. 9:4893–4908.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Banesh S and Trivedi V: Therapeutic
potentials of scavenger receptor cd36 mediated innate immune
responses against infectious and non-infectious diseases. Curr Drug
Discov Technol. 17:299–317. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bocharov AV, Wu T, Baranova IN, Birukova
AA, Sviridov D, Vishnyakova TG, Remaley AT, Eggerman TL, Patterson
AP and Birukov KG: Synthetic amphipathic helical peptides targeting
CD36 attenuate lipopolysaccharide-induced inflammation and acute
lung injury. J Immunol. 197:611–619. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li J and Liu S: LncRNA GAS5 suppresses
inflammatory responses and apoptosis of alveolar epithelial cells
by targeting miR-429/DUSP1. Exp Mol Pathol.
113(104357)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bai Z, Li Y, Li Y, Pan J, Wang J and Fang
F: Long noncoding RNA and messenger RNA abnormalities in pediatric
sepsis: A preliminary study. BMC Med Genomics.
13(36)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shao L, Meng D, Yang F, Song H and Tang D:
Irisin-mediated protective effect on LPS-induced acute lung injury
via suppressing inflammation and apoptosis of alveolar epithelial
cells. Biochem Biophys Res Commun. 487:194–200. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hu Q, Wang Q, Han C and Yang Y: Sufentanil
attenuates inflammation and oxidative stress in sepsis-induced
acute lung injury by downregulating KNG1 expression. Mol Med Rep.
22:4298–4306. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zeng M, Huang C, Zheng H, Chen Q, He W and
Deng Y: Effects of ghrelin on iNOS-derived NO promoted LPS-induced
pulmonary alveolar epithelial A549 cells apoptosis. Cell Physiol
Biochem. 49:1840–1855. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Basu A, Harper S, Pesciotta EN, Speicher
KD, Chakrabarti A and Speicher DW: Proteome analysis of the
triton-insoluble erythrocyte membrane skeleton. J Proteomics.
128:298–305. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li Y, Li Y, Bai Z, Pan J, Wang J and Fang
F: Identification of potential transcriptomic markers in developing
pediatric sepsis: A weighted gene co-expression network analysis
and a case-control validation study. J Transl Med.
15(254)2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cao W, Zhang B, Ding F, Zhang W, Sun B and
Liu Z: Expression of SLP-2 was associated with invasion of
esophageal squamous cell carcinoma. PLoS One.
8(e63890)2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Guo XY, Guo HF and Guo HM: Clinical
significance of SLP-2 in epithelial ovarian cancer and its
regulatory effect on the notch signaling pathway. Eur Rev Med
Pharmacol Sci. 24:1666–1671. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yang CT, Li JM, Li LF, Ko YS and Chen JT:
Stomatin-like protein 2 regulates survivin expression in non-small
cell lung cancer cells through β-catenin signaling pathway. Cell
Death Dis. 9(425)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhou C, Li Y, Wang G, Niu W, Zhang J, Wang
G, Zhao Q and Fan L: Enhanced SLP-2 promotes invasion and
metastasis by regulating Wnt/β-catenin signal pathway in colorectal
cancer and predicts poor prognosis. Pathol Res Pract. 215:57–67.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kucuk I, Tanoglu A, Öncü K, Yılmaz I, Kara
M, Beyazıt Y, Akyol T, Kaplan M, Özarı HO and Yazgan Y:
Immunohistochemical activity of prohibitin-2 and stomatin-like
protein-2 in patients with ulcerative colitis. Turk J
Gastroenterol. 27:233–238. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Pu X, Dong C, Zhu W, Li W and Jiang H:
Silencing stomatin-like protein 2 attenuates tumor progression and
inflammatory response through repressing CD14 in liver cancer. Onco
Targets Ther. 12:7361–7373. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Park YM: CD36, a scavenger receptor
implicated in atherosclerosis. Exp Mol Med. 46(e99)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Pepino MY, Kuda O, Samovski D and Abumrad
NA: Structure-function of CD36 and importance of fatty acid signal
transduction in fat metabolism. Annu Rev Nutr. 34:281–303.
2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li X, Zhang X, Pang L, Yao L, ShangGuan Z
and Pan Y: Agaricus bisporus-derived β-glucan enter macrophages and
adipocytes by CD36 receptor. Nat Prod Res. 34:3253–3256.
2020.PubMed/NCBI View Article : Google Scholar
|