Introduction
White adipose tissue is a specialized organ that can
store fat and release lipids in response to various types of
signals (1). It has been shown that
adipose tissue regulates metabolic homeostasis by secreting
adipokines (2). However, the
excessive gathering of adipose tissues can lead to metabolic
disorders and promote obesity (3).
As of 2016, 39% of adults (≥18 years) were characterized as
overweight worldwide (4). Moreover,
obesity is associated with various other comorbidities, such as
hyperglycemia, diabetes, cardiovascular disease, osteoarthritis and
cancer (5). Hypertrophy and
hyperplasia of adipocytes are the main causes of obesity and can
also result in the development of metabolic disorders (6). Therefore, it is important to
understand the detailed mechanism of adipogenesis, which may
contribute to the prevention and treatment of obesity and its
associated metabolic diseases.
Adipogenesis is a pivotal process required for lipid
homeostasis, energy balance and obesity (7). A highly organized signaling cascade
involving numerous transcription factors, such as peroxisome
proliferator-activated receptor (PPAR)γ, CCAAT-enhancer-binding
protein (C/EBP)α and C/EBPβ, regulate adipocyte differentiation
(8). Increased expression levels of
PPARγ and C/EBPα are associated with lipid droplet formation and
induction of sterol regulatory element-binding protein 1 (SREBP1)
expression, as well as of markers of adipocyte maturity, including
adiponectin, fatty acid binding protein 4 (FABP4/aP2), perilipin A
and fatty acid synthase (9).
Previously, the Wnt/β-catenin signaling pathway has been discovered
to act as a molecular switch during adipogenesis (10); adipogenesis can be inhibited by
activating this pathway (11). A
recent study found that the adipogenesis regulated by the Wnt
pathway could be mediated by its effector TCF7L2(12). However, to the best of our
knowledge, the molecular mechanism underlying the association
between adipogenesis and the Wnt/β-catenin signaling pathway in
obesity still needs to be further clarified. Emerging evidence has
revealed that Wnt signaling is modulated by P2X7R (13,14).
SREBP1 is a transcription factor that is required
for adipogenesis, lipid homeostasis and cellular lipogenesis
(15,16). In our previous study, it was shown
that SREBP1 could bind to the promoter sites of the purinergic
receptor P2X ligand-gated ion channel 7 (P2RX7) based on analysis
of data from a database. P2RX7 is a ligand-gated cation channel
encoded by the P2X7 gene, which belongs to the P2X receptor family
(17). P2X7R is primarily expressed
on cells of hematopoietic, epithelial, mesenchymal and neural
lineage, and plays major roles in immunity, neoplasia, inflammation
and neurological functions (18).
In addition, P2X7R is associated with lipid metabolism (19). P2X7R was also discovered to play an
extensive role in controlling lipid storage and metabolism in
vivo (20). Suppression of CD36
attenuates adipogenesis via the P2X7R pathway in 3T3-L1 cells
(21), and Wnt3a has been shown to
mitigate acute lung injury by reducing P2X7 receptor-mediated
alveolar epithelial type I cell death (13). Therefore, it was hypothesized that
there may be an association between P2X7R, Wnt signaling and
adipogenesis. In the present study, the effects of P2X7R on 3T3-L1
preadipocyte proliferation and differentiation were examined in
vitro and in vivo, and the associated underlying
mechanism of action was also assessed.
Materials and methods
Experimental animals
Male 8-week-old C57BL/6J mice (n=10, weight: 19-20
g) were obtained from the Animal Core Facility and housed at 22±2˚C
with a relative humidity of 50±10% on a 12-h light/dark cycle, with
free access to food and water. Following 1 week of acclimatization,
the mice were randomly assigned into weight-matched groups and fed
either a normal chow diet (control; 15% fat) or a high-fat diet
(HFD; 60% fat; TROPHIC Animal Feed High-Tech Co., Ltd.). The mice
were fed a HFD for 12 weeks and blood samples (0.6 ml) were
collected from their eyeballs under anesthesia with 2%
pentobarbital sodium (50 mg/kg). The mice were then sacrificed by
cervical dislocation. The fat depots from visceral adipose tissues
were immediately removed, frozen in liquid nitrogen and stored at
-80˚C until required for further analysis. The present study was
approved by the Ethics Committee of Yancheng Third People's
Hospital (Yancheng, China).
Cell culture and adipocyte
differentiation
The 3T3-L1 cell line was purchased from the American
Type Culture Collection and cultured in DMEM (Sigma-Aldrich; Merck
KGaA) supplemented with 10% calf serum (Sigma-Aldrich; Merck KGaA).
The cells were cultured for 2 days to reach confluence (day 0).
Differentiation was induced using DMEM containing 10% fetal bovine
serum (BMG Labtech GmbH), 5 µg/ml insulin (Sigma-Aldrich; Merck
KGaA), 1 µM dexamethasone and 0.5 µM 3-isobutyl-1-methylxanthine
(Sigma-Aldrich; Merck KGaA) for 2 days. Subsequently, the medium
was changed to growth medium (DMEM containing 10% FBS and 5 µg/ml
insulin), which was used for an additional 2 days (21). The medium was replaced with growth
medium and replaced every other day until day 8.
Cell transfection
3T3-L1 cells (5x104 cells/well) were
seeded into 6-well plates 24 h prior to transfection and cultured
at 37˚C in a humidified incubator with 5% CO2.
Subsequently, the cells were transfected with small interfering RNA
(siRNA) targeting SREBP1 (siRNA-SREBP1-1,
5'-GCAAGUCAUUGUUACUAUAAG-3' and 5'-UAUAGUAACAAUGACUUGCUG-3';
siRNA-SREBP1-2, 5'-GACUGUGUGUCUACAACUACA-3' and
5'-UAGUUGUAGACACACAGUCAU-3'; siRNA-SREBP1-NC,
5'-GCAAGUAGCUCUAUUUAUAAG-3' and 5'-UAUAAAAUAGAGCUCUUGCUG-3'), P2X7R
(siRNA-P2X7R-1, 5'-GGAACGAUGUCUUGCAGUAUG-3' and
5'-UACUGCAAGACAUCGUUCCAG-3'; siRNA-P2X7R-2,
5'-GCAGUAUGAGACAAACAAAGU-3' and 5'-UUUGUUUGUCUCAUACUGCAA-3';
siRNA-P2X7-NC, 5'-GGUGUAACGAGUAUCAGUCUG-3' and
5'-GACUGAUACUCGUUACACCAG-3'), or Wnt3a (siRNA-Wnt3a-1,
5'-GGUCAGCACAUGACACUAAUG-3' and 5'-UUAGUGUCAUGUGCUGACCCG-3' and
siRNA-Wnt3a-2, 5'-GCUUUAAUCUAGCUGACAAGA-3' and
5'-UUGUCAGCUAGAUUAAAGCUG-3'; siRNA-Wnt3a-NC,
5'-CGGUCAGCCAACAUACUAGAUG-3' and 5'-UCUAGUAUGUUGUGCUGACCCG-3')
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Per
well, 50 pmol siRNAs were used. The siRNAs were designed and
purchased from Shanghai GenePharma Co., Ltd. The cells in the blank
control group were untransfected. Following incubation for 48 h at
37˚C, cells were used for subsequent experiments. Transfection
efficiency was assessed using reverse transcription-quantitative
PCR (RT-qPCR).
RT-qPCR
Total RNA from cells was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Total RNA was
reverse transcribed into cDNA using a PrimeScript RT Reagent kit
according to the manufacturer's protocol (Invitrogen; Thermo Fisher
Scientific, Inc.). qPCR was subsequently performed using SYBR-Green
Premix Ex Taq II (Takara Bio, Inc.), following the manufacturer's
instructions. GAPDH was used as the internal control. The primer
sequences used were as follows: SREBP1 forward,
5'-ACAGTGACTTCCCTGGCCTAT-3' and reverse,
5'-GCATGGACGGGTACATCTTCAA-3'; P2X7R forward,
5'-CACCGTGCTTACAGGTGCTA-3' and reverse, 5'-CGGTCTTGGGGAACTCCTTC-3';
PPARγ forward, 5'-GGGATCAGCTCCGTGGATCT-3' and reverse,
5'-TGCACTTTGGTACTCTTGAAGTT-3'; C/EBPα forward,
5'-GGCCAATGGCATCCAAAATA-3' and reverse, 5'-CCTTGGCGAATTCTGTGAGC-3';
fatty acid binding protein 4 (FABP4) forward,
5'-GCAGTGTTTCTGTGGCTGACAC-3' and reverse, 5'-GCCATGCACAGGGTCCA-3';
Wnt3a forward, 5'-AGCTACCCGATCTGGTGGTC-3' and reverse,
5'-CAAACTCGATGTCCTCGCTAC-3'; and GAPDH forward,
5'-TGCACCACCAACTGCTTAGC-3' and reverse,
5'-GGCATGGACTGTGGTCATGAG-3'. qPCR was performed on an ABI Prism
7500 Real-Time PCR Detection system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. The thermocycling conditions were as follows: Initial
denaturation for 5 min at 94˚C, 35 cycles of 94˚C denaturation for
30 sec, 55˚C annealing for 30 sec and 72˚C extension for 30 sec.
The 2-ΔΔCq method (22)
was used in each sample for relative quantification.
Western blot analysis
The cells were collected and lysed 48 h after the
aforementioned treatments using RIPA lysis buffer (Beyotime
Institute of Biotechnology). The protein concentration was
calculated using the BCA method. Equal amounts of proteins (30
µg/lane) were resolved using 10-15% SDS-gels by SDS-PAGE and
subsequently transferred onto PVDF membranes (MilliporeSigma). The
membranes were incubated with specific antibodies against the
following proteins: SREBP1 (1:1,000; cat. no. ab28481), PPARγ
(1:1,000; cat. no. ab272718), C/EBPα (1:1,000; cat. no. ab40764),
FABP4 (1:2,000; cat. no. ab92501), adipose triglyceride lipase
(ATGL; 1:1,000; cat. no. ab207799), phosphorylated
hormone-sensitive lipase (p-HSL; 1:100,000; cat. no. ab109400),
monoacylglycerol lipase (MGL; 1:2,000; cat. no. ab77398) and Wnt3a
(1:1,000; cat. no. ab219412; all purchased from Abcam), and
β-catenin (1:500; cat. no. 8480) and cyclin D1 (1:1,000; cat. no.
55506; both from Cell Signaling Technology, Inc.) at 4˚C overnight.
The membranes were then washed and incubated with HRP-conjugated
secondary antibodies (1:10,000; cat. no. 7074; Cell Signaling
Technology, Inc.) the following day at room temperature for 2 h.
Signals were visualized using chemiluminescence reagent
(MilliporeSigma), and densitometry analysis (ImageJ 1.46r; National
Institutes of Health) was performed to measure protein
expression.
Measurement of triglyceride (TG),
total cholesterol (TC) and glycerin levels
The collected blood was centrifuged at 3,000 x g at
4˚C for 15 min to obtain serum. In serum and/or mature 3T3-L1
cells, TG and TC contents were measured with an AdipoRed assay
reagent kit (Lonza Group, Ltd.) or Cholesterol assay kit (Abcam),
respectively, in accordance with the manufacturers' instructions.
Fluorescence was measured following 10 min of incubation on a
Victor3 plate reader (PerkinElmer, Inc.) with the following
settings: Excitation (Ex) 485/Emission (Em) 572 nm (for TG) and
Ex538/Em587 (for TC).
Differentiated 3T3-L1 adipocytes were serum-starved
for 2 h and treated with AZD1208 (cat. no. HY-15604;
MedChemExpress) or isoproterenol, a known lipolysis inducer, for 3
h. Culture medium was obtained and used to measure glycerol
contents with a free glycerol reagent (Sigma-Aldrich; Merck KGaA)
according to the manufacturer's instructions. The absorbance was
determined at 540 nm using a microplate reader.
Dual luciferase reporter assay
JASPAR software (jaspar.genereg.net/) was used to predict the promoter
region of P2X7R, which was a potential target of SREBP1. To confirm
this prediction, dual luciferase reporter assays were performed to
investigate the regulatory association between SREBP1 and P2X7R.
The promoter sites of P2X7R containing the SREBP1 binding site was
cloned into a pmirGLO dual luciferase reporter vectors (Promega
Corporation). The reporter vectors containing the P2X7R promoter
sites and siRNA-SREBP-1 or siRNA-NC were co-transfected into mature
3T3-L1 cells (8 days old) using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). Following 48 h of
incubation, the cells were lysed and the relative luciferase
activities were detected using a Dual Luciferase Reporter assay
system (Promega Corporation). Renilla luciferase was used as an
internal control for the normalization of luciferase reporter gene
in the experiment.
Oil Red O staining
The mature adipocytes were fixed in 10% (v/v)
formaldehyde solution for 30 min at room temperature and the cells
were stained with 0.5% Oil Red O for 30 min at room temperature.
The samples were washed three times with PBS and the cells were
visualized using a light microscope (Olympus Corporation;
magnification, x400). Oil red O was extracted with 100% isopropanol
and the concentration was determined by measuring the absorbance at
510 nm.
Statistical analysis
The data are expressed as the mean ± standard
deviation. GraphPad Prism 8.0 software (GraphPad Software, Inc.)
was used for all statistical analyses. Data were compared using a
one-way ANOVA, followed by a Tukey's post hoc test. Each experiment
was repeated at least three times. A unpaired Student's t-test was
used to compare differences between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
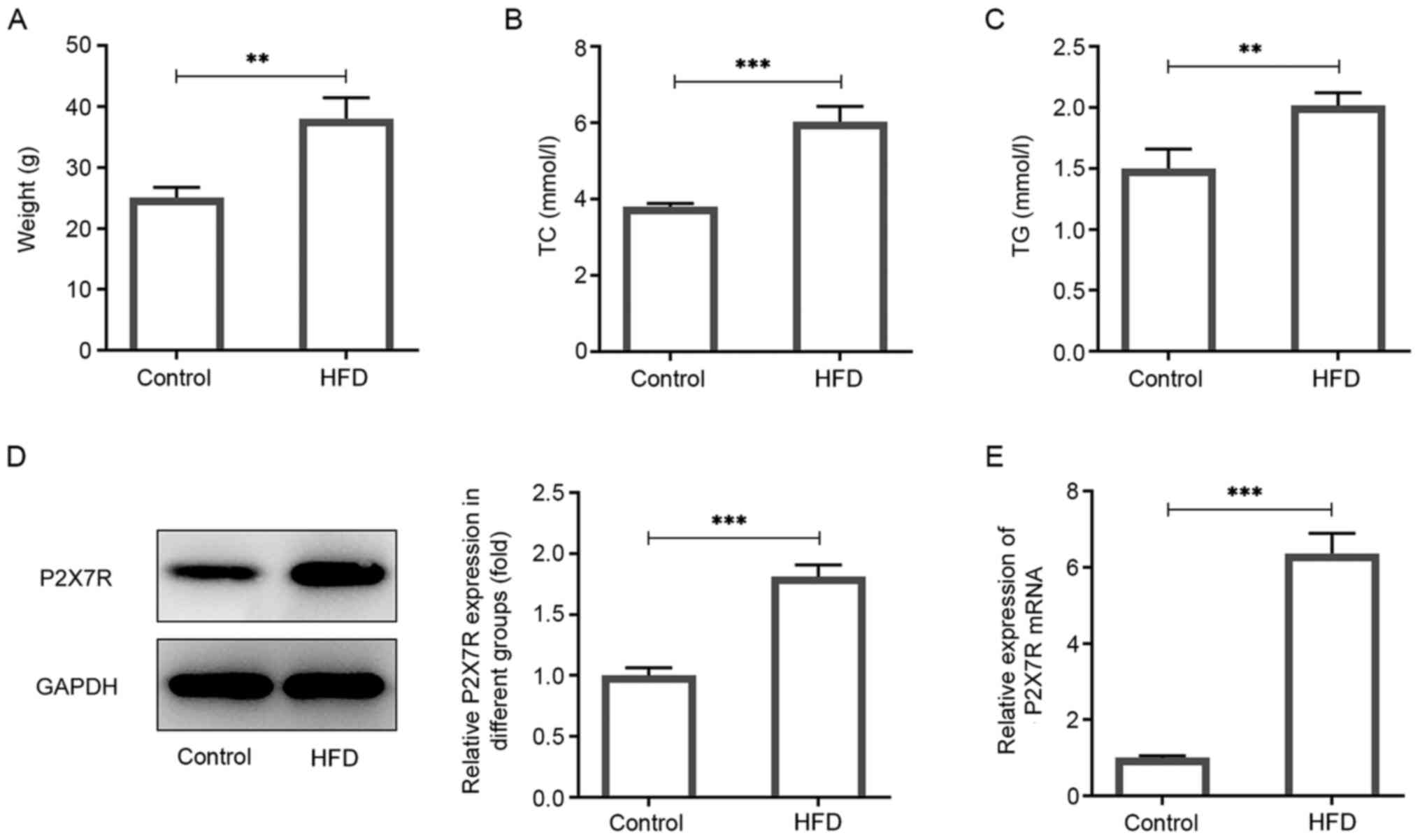
P2X7R is highly expressed in obese
mice
The expression levels of P2X7R were determined in
visceral adipose tissues of obese mice. C57BL/6J mice were randomly
separated into two groups according to their weight. The HFD group
was fed a HFD and the control group was fed a normal chow diet for
12 weeks. Mice in the HFD group had significantly increased body
weights (52%) compared with the control group (Fig. 1A). The TG and TC levels in serum
were also increased compared with those of the control group
(Fig. 1B and C). Western blotting and RT-qPCR analyses
indicated that the protein and mRNA levels of P2X7R was increased
in the HFD group (Fig. 1D and
E). Therefore, the data indicated
that P2X7R was highly expressed in obese mice.
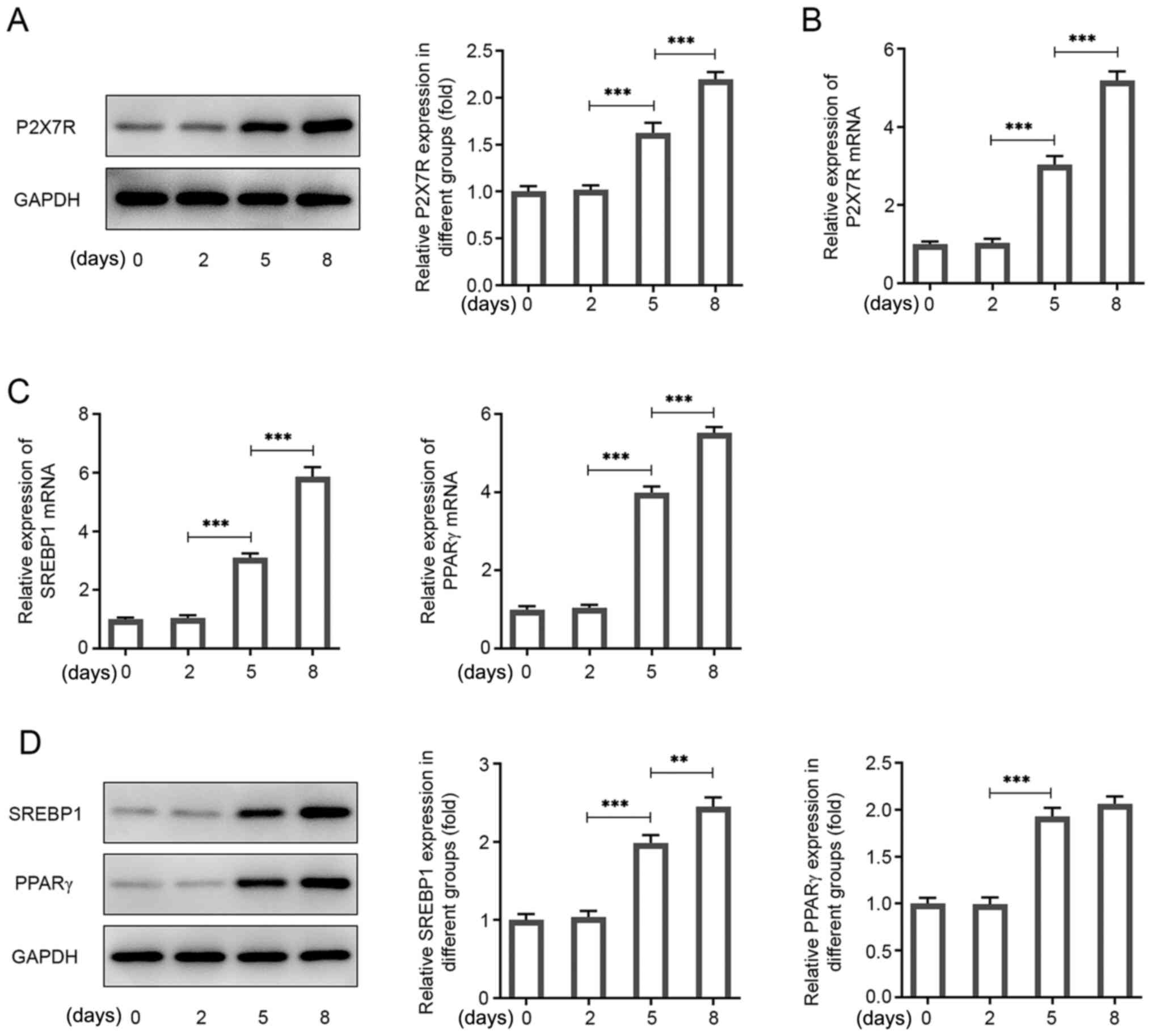
P2X7R is highly expressed during
adipogenic differentiation
Subsequently, the specific function of P2X7R during
adipogenesis was investigated by assessing P2X7R expression at
different time points during the differentiation of 3T3-L1 cells
into mature adipocytes. The P2X7R protein and mRNA levels were
increased following adipogenic induction and maintained at a high
level of expression during adipocyte differentiation (Fig. 2A and B). In addition, the expression levels of
the key transcription factor PPARγ, and of the SREBP1 gene, which
have been used as adipogenic markers (23-25)
are involved in lipogenesis, were assessed. Their expression levels
were significantly increased during adipogenic differentiation
(Fig. 2C and D). Mature 3T3-L1 cells were cultured for 8
days and used to perform further analysis.
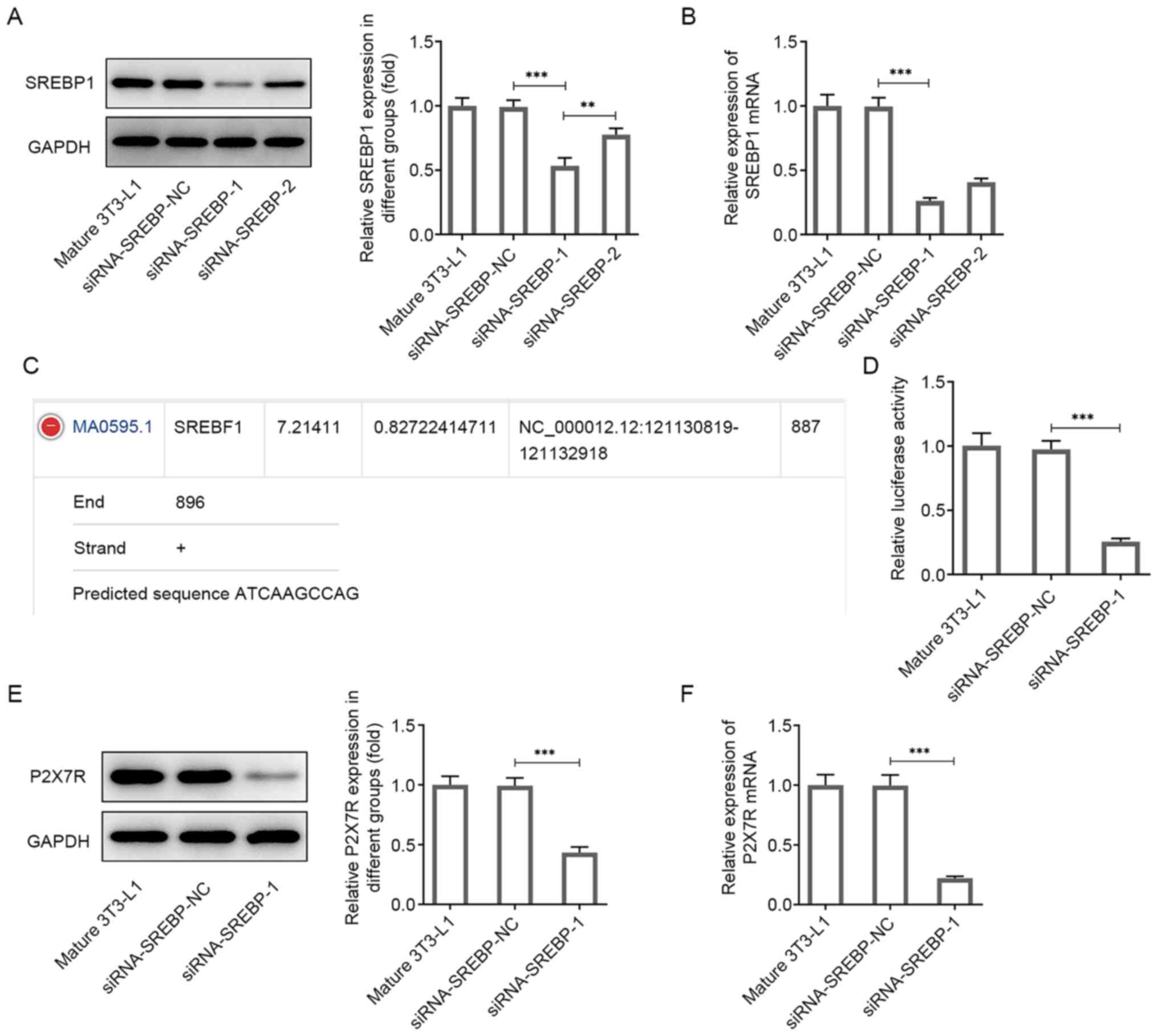
P2X7R expression levels are reduced
following knockdown of SREBP1 expression
To investigate the potential molecular mechanism of
P2X7R, its putative target genes were screened using bioinformatics
analysis. The data indicated that SREBP1, which is a lipid droplet
regulator, could bind to the promoter sites of P2X7R. To identify
whether SREBP1 participates in the adipogenic process, its
expression was knocked down by transfecting siRNA-SREBP1 into
mature 3T3-L1 cells following the induction of differentiation.
SREBP1 expression was significantly decreased following the
transfection of the cells with siRNA-SREBP1-1 and siRNA-SREBP1-2
compared with that of mature 3T3-L1 cells, as determined by western
blotting and RT-qPCR analyses (Fig.
3A and B). The siRNA-SREBP1-1
group had the lowest expression and was used in subsequent
experiments (Fig. 3A and B).
The JASPAR database predicted the binding of SREBP-1
to the P2X7R promoter (Fig. 3C).
Subsequently, dual luciferase reporter assays were performed to
confirm this prediction. The results indicated that siRNA-SREBP1
significantly inhibited the transcriptional activity of the P2X7R
promoter, whereas the negative control of SREBP1 (siRNA-NC)
abolished the inhibitory effect of P2X7R (Fig. 3D). Moreover, P2X7R expression was
significantly decreased following transfection of siRNA-SREBP1 into
3T3-L1 cells compared with the corresponding expression noted in
the control group (Fig. 3E and
F). Overall, these data
demonstrated an interaction between P2X7R and SREBP1 and indicated
that P2X7R expression was reduced following knockdown of
SREBP1.
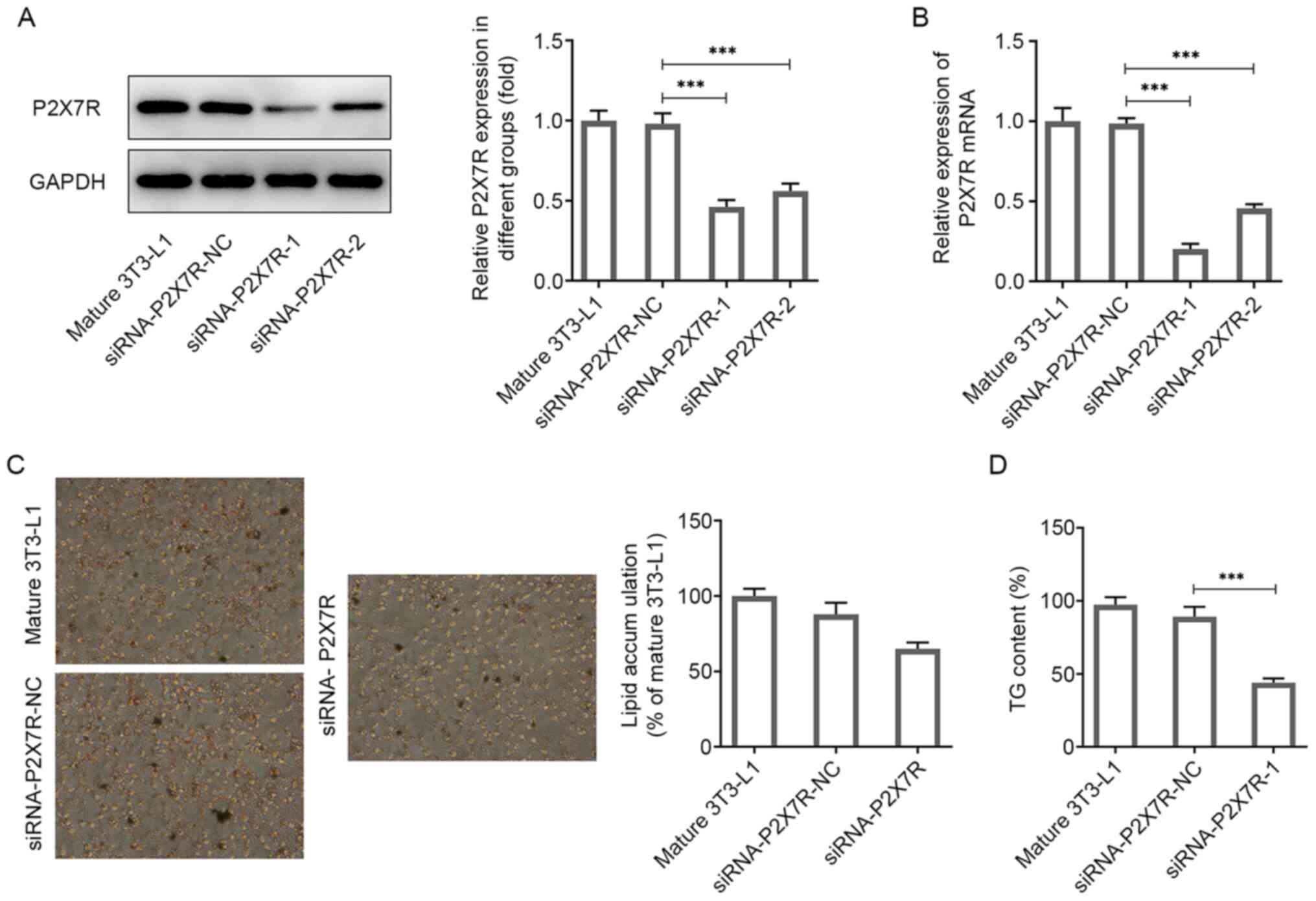
Suppression of P2X7R reduces
adipogenic differentiation and lipid accumulation
It has been shown that lipid deposition is
associated with the proliferation of preadipocytes and their
differentiation into adipose tissue cells (26). To demonstrate the function of P2X7R
in adipogenesis, its ability to affect the differentiation of
3T3-L1 preadipocytes was assessed following transfection of the
cells with siRNA-P2X7R or siRNA-NC. P2X7R expression was
significantly decreased following transfection of 3T3-L1 cells with
siRNA-P2X7R-1 and siRNA-P2X7R-2 compared with that observed in the
control group as determined by western blotting and RT-qPCR
analyses (Fig. 4A and B). The siRNA-P2X7R-1 group exhibited the
lowest expression and was used for subsequent experiments. In
addition, it was confirmed by Oil Red O staining that knockdown of
P2X7R expression reduced the adipogenic capacity of preadipocytes
(Fig. 4C). The AdipoRed assay
indicated a reduction in TG accumulation in the siRNA-P2X7R-1 group
(Fig. 4D). Collectively, the data
revealed that P2X7R promoted 3T3-L1 preadipocyte
differentiation.
To determine the effects of P2X7R on lipid
accumulation, RT-qPCR and western blot analyses were performed. The
adipogenic process is tightly controlled by PPARγ, C/EBPα and FABP4
(27,28). In the present study, the expression
levels of PPARγ, C/EBPα and FABP4 were significantly decreased
following transfection of the cells with siRNA-P2X7R-1 compared
with the mature 3T3-L1 group (Fig.
5A and B). In addition,
lipolysis involves the sequential activity of lipolytic enzymes
(ATGL, p-HSL and MGL) (29). The
expression levels of ATGL, p-HSL and MGL were significantly
increased following transfection of the cells with siRNA-P2X7R-1
compared with that observed in the mature 3T3-L1 cells (Fig. 5C). Moreover, glycerol levels were
increased 3-fold in mature 3T3-L1 cells following transfection with
siRNA-P2X7R-1 compared with the control cell group (Fig. 5D). Therefore, the data indicated
that P2X7R enhanced adipogenesis and reduced lipolysis in 3T3-L1
cells.
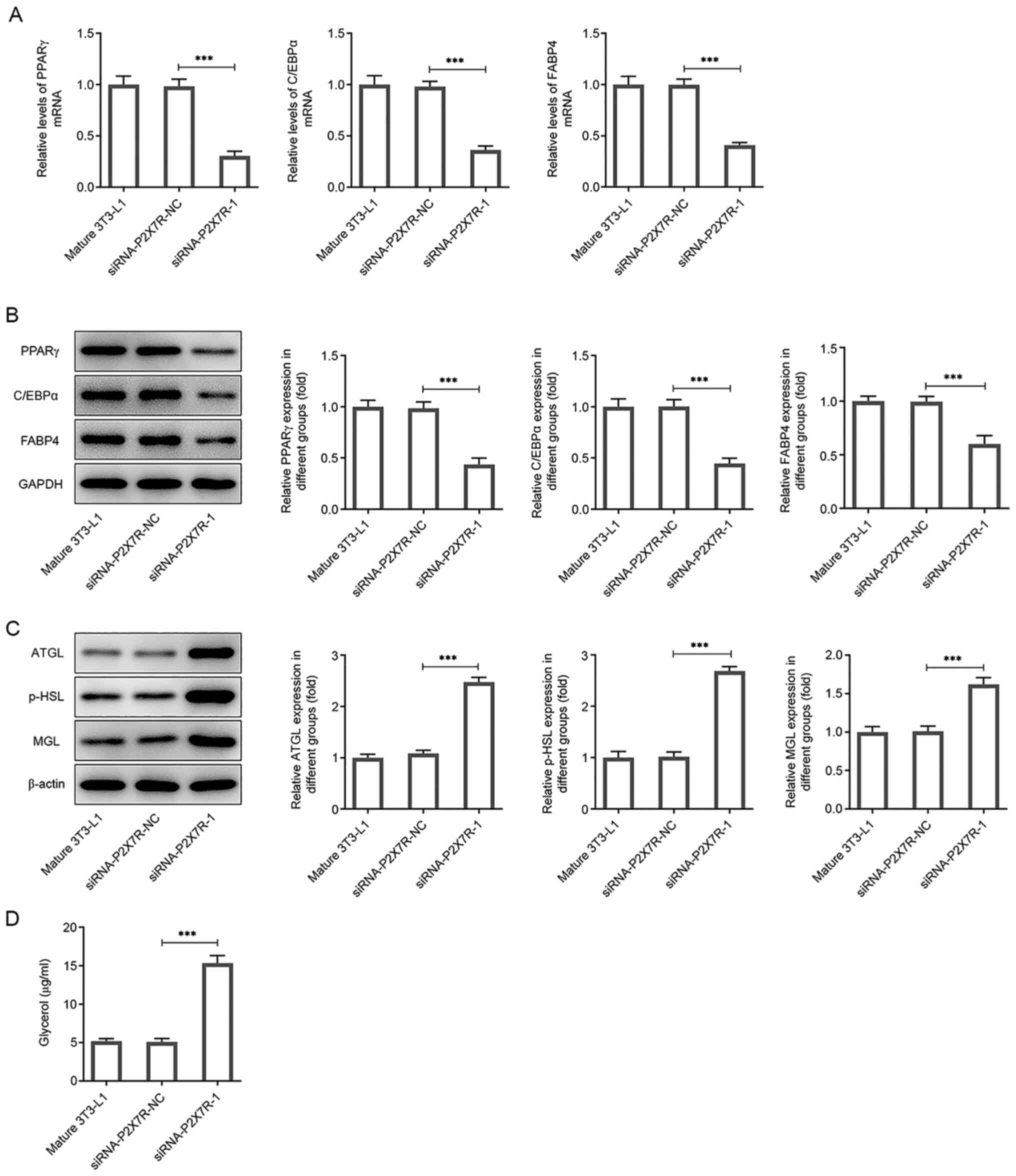 | Figure 5P2X7R knockdown reduces the
expression of lipogenesis-related proteins and increases the
expression levels of lipolysis-associated enzymes. (A) mRNA and (B)
protein expression levels of PPARγ, C/EBPα and FABP4 following
siRNA-mediated knockdown. (C) Protein expression levels of ATGL,
p-HSL and MGL following siRNA-mediated knockdown. (D) Glycerol
levels following siRNA-mediated knockdown of P2X7R. Data are
presented as the mean ± standard deviation.
***P<0.001. P2X7R, purinergic receptor P2X
ligand-gated ion channel 7; siRNA small interfering RNA; NC,
negative control; PPARγ, peroxisome proliferator-activated receptor
γ; C/EBPα, CCAAT-enhancer-binding protein α; FABP4, fatty acid
binding protein 4; ATGL, adipose triglyceride lipase; p-HSL,
phosphorylated hormone-sensitive lipase; MGL, monoacylglycerol
lipase. |
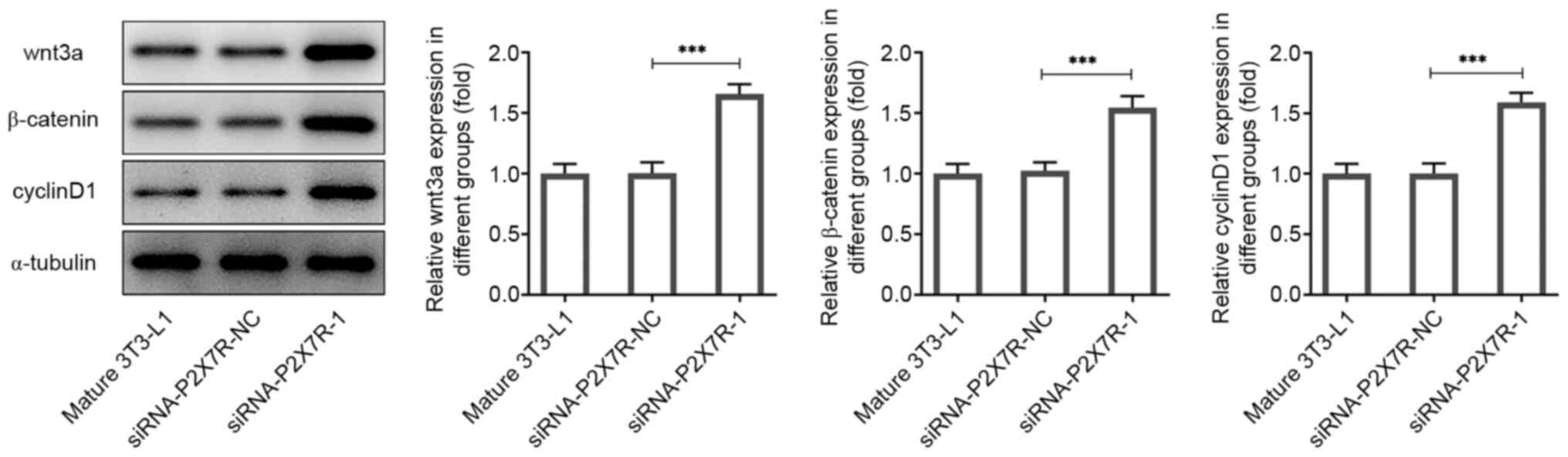
Suppression of P2X7R reduces
adipogenesis
Amongst the factors involved in adipocyte
differentiation, Wnt/β-catenin has been shown to suppress
adipogenesis by inhibiting the expression of PPARγ. In order to
further explore the mechanism of P2X7R in the regulation of
adipogenesis, the expression levels of the Wnt/β-catenin signaling
pathway-associated proteins were examined by western blotting
following P2X7R interference. Silencing of P2X7R expression
significantly promoted the expression levels of Wnt3a, β-catenin
and cyclin D1 (Fig. 6). Next, to
investigate the role of wnt3a in the effects of P2X7R on
adipogenesis-related markers and lipolysis-related markers, the
downregulation of Wnt3a was induced by the transfection of
siRNA-Wnt3a-1 or -2 into mature 3T3-L1 cells. As shown in Fig. 7A and B, the protein and mRNA levels of Wnt3a
were significantly reduced by siRNA-Wnt3a-1. Moreover, the
expression levels of Wnt3a could be reversed by transfection of
siRNA-Wnt3a-1 or siRNA-Wnt3a-2 compared with that observed in the
P2X7R interference group (Mature 3T3-L1 + siRNA-P2X7R-1; Fig. 7C and D). The mature 3T3-L1 + siRNA-P2X7R-1 +
siRNA-Wnt3a-1 group exhibited the lowest P2X7R expression levels
and was thus used for subsequent experiments.
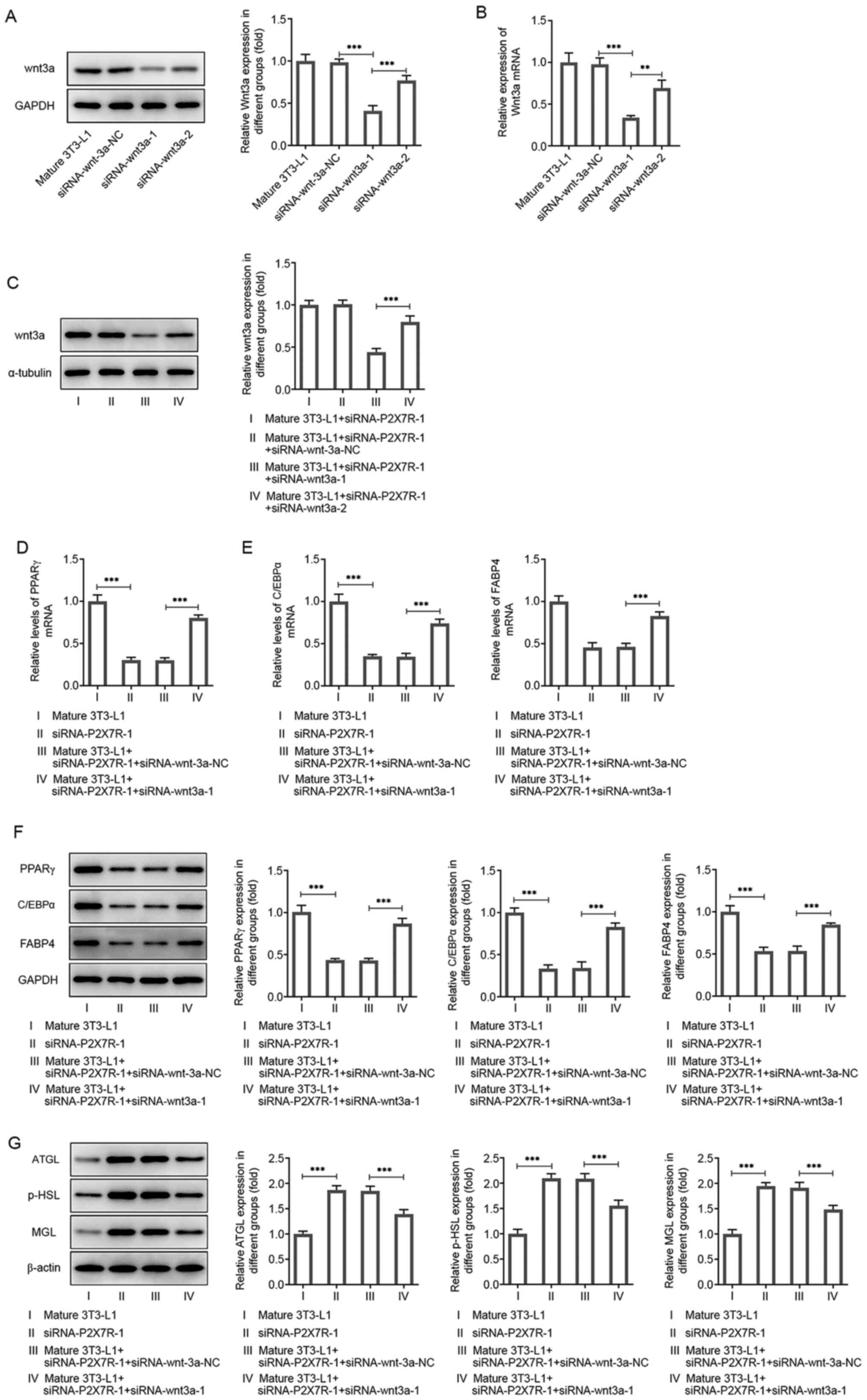 | Figure 7P2X7R regulates the Wnt/β-catenin
pathway. (A) Protein and (B) mRNA levels of Wnt3a in 3T3-L1 cells
following siRNA-wnt3a-1 or siRNA-wnt3a-2 transfection. (C) Protein
and (D) mRNA levels of Wnt3a in 3T3-L1 cells following
co-transfection of siRNA-P2XR-1 with siRNA-wnt3a-1 or
siRNA-wnt3a-2. (E) mRNA and (F) protein expression levels of PPARγ,
C/EBPα and FABP4. (G) Protein expression levels of ATGL, p-HSL and
MGL. Data are presented as the mean ± standard deviation.
**P<0.01, ***P<0.001. P2X7R, purinergic
receptor P2X ligand-gated ion channel 7; siRNA small interfering
RNA; NC, negative control; C/EBPα, CCAAT-enhancer-binding protein
α; ATGL, adipose triglyceride lipase; p-HSL, phosphorylated
hormone-sensitive lipase; MGL, monoacylglycerol lipase; PPARγ,
peroxisome proliferator-activated receptor γ; FABP4, fatty acid
binding protein 4. |
During adipogenesis, the expression levels of PPARγ,
C/EBPα and FABP4 were significantly decreased following
transfection of the cells with siRNA-P2X7R-1 compared with those of
the mature 3T3-L1 group (Fig.
7D-F). These effects could be reversed by addition of
siRNA-Wnt3a-1 (Fig. 7D-F). During
lipolysis, the protein expression levels of ATGL, p-HSL and MGL
were significantly increased following transfection of the cells
with siRNA-P2X7R-1 compared with those observed in the mature
3T3-L1 group. These effects could be reversed by transfection of
siRNA-Wnt3a-1 (Fig. 7G). In
summary, the results indicated that P2X7R enhanced adipogenesis and
reduced lipolysis in 3T3-L1 cells.
Discussion
Obesity is a serious public health problem and is
considered a risk factor for the development of numerous diseases,
such as hypertension, type 2 diabetes, coronary heart disease and
cancer (6). In the present study,
HFD mice were established and 3T3-L1 cells were induced in order to
assess adipocyte differentiation. P2X7R was highly expressed in
vivo and in vitro. SREBP1 was confirmed to affect the
transcription activities of P2X7R and then regulate its expression.
Inhibition of P2X7R significantly reduced the adipogenic capacity
of preadipocytes, decreased the expression of
adipogenesis-associated transcription factors (PPARγ, C/EBPα and
FABP4), increased the expression levels of lipolytic enzymes (ATGL,
p-HSL and MGL) and regulated the contents of TG, TC and glycerin in
mature 3T3-L1 cells. These effects could be reversed by application
of siRNA-Wnt3a. The results suggested that P2X7R modulation by
SREBP1, regulated adipogenesis and lipid degradation.
P2X7 receptors play an important role in multiple
biological functions and are present in various tissues (30). Previous in vitro studies have
shown that P2X7 suppressed adipocyte differentiation marker
expression by regulating phospholipase and sphingomyelinase
activity levels and consequently the production of bioactive lipid
signaling molecules (31,32). Metabolic syndrome, obesity and type
2 diabetes are relevant risk factors for the increased incidence of
chronic kidney disease. The latter is simulated in the laboratory
by persistent exposure to HFD (29). In the present study, the HFD mouse
model was established and 3T3-L1 cells were induced to
differentiate into adipocytes. P2X7R was highly expressed in the
HFD mouse model and in mature 3T3-L1 cells. The transcription
factors PPARγ and C/EBPα are critical for adipogenesis. SREBP1 is
another transcription factor, which is required for adipogenesis,
lipid homeostasis and cellular lipogenesis (21,33).
In adipocytes, the hydrolysis of TGs, which is considered as
lipolysis in intracellular lipid droplets, provides stock for fatty
acid oxidation. This catabolic process is activated by protein
kinase A and involves various lipolytic enzymes (ATGL, HSL and MGL)
(34). A previous study
demonstrated that P2X7R participates in the activation of the NLR
family pyrin domain containing 3 inflammasome in podocytes, which
may trigger cell damage due to obesity-associated glomerulopathy
(17). Loss of P2X7 nucleotide
receptor function leads to abnormal fat distribution in mice
(20). However, the effects of the
elevated expression of P2X7R have not been fully explored. Late
adipogenic factors, such as PPARγ and C/EBPα, are critical
transcriptional regulators of lipogenesis (35). A previous study showed that
downregulation of PPARγ, C/EBPα and FABP4 was involved in the
decrease in lipid accumulation (36). Lipolysis is primarily achieved
through the gradual hydrolysis of triglycerides via HSL, ATG and
MGL to generate free fatty acids (37). Inhibition of P2X7R significantly
reduced the adipogenic capacity of preadipocytes, decreased the
expression levels of adipogenesis-associated transcription factors
(PPARγ, C/EBPα and FABP4) and enhanced the expression levels of
lipolytic enzymes (ATGL, p-HSL and MGL) (35-37).
Therefore, it was concluded that P2X7R was involved in adipogenesis
and lipid degradation.
The Wnt family members are evolutionarily conserved
secretory lipoproteins, which play an important role in cell
proliferation, differentiation and polarity during embryogenesis
(38). Previously, the Wnt
signaling pathway has been confirmed to play major roles in a
series of additional developmental and physiological processes,
including adipocyte biology (11,39).
However, to the best of our knowledge, the definite function and
the underlying mechanism of the Wnt/β-catenin signaling pathway in
adipogenesis remains unclear. In the present study, silencing of
P2X7R expression promoted the expression of Wnt3a, β-catenin and
cyclin D1. The expression levels of Wnt3a were reversed to their
initial levels by the addition of siRNA-Wnt3a. It has been
previously shown that Wnt signaling prevents the induction of
adipogenic regulators, including C/EBPα and PPARγ during
preadipocyte differentiation. Previously, transient activation of
the Wnt/β-catenin signaling pathway was shown to inhibit adipogenic
regulators in bipotential ST2 cells, which preceded the Wnt-induced
increase in osteoblastogenic transcription factors (40). Knockdown of C/EBPα and PPARγ is a
primary mechanism through which Wnt signaling controls mesenchymal
cell fate (41,42). The present study indicated that the
expression levels of PPARγ, C/EBPα and FABP4 were significantly
decreased following transfection with siRNA-P2X7R compared with
those noted in the mature 3T3-L1 group. These effects could be
reversed by addition of siRNA-Wnt3a. Moreover, the protein
expression levels of ATGL, p-HSL and MGL were significantly
increased during lipolysis following transfection of 3T3-L1 cells
with siRNA-P2X7R compared with those of the mature 3T3-L1 group.
These effects were reversed by addition of siRNA-Wnt3a. Therefore,
the data indicated that P2X7R enhanced adipogenesis and reduced
lipolysis in 3T3-L1 cells. However, whether this role is related to
Wnt/β-catenin signaling pathway still requires further studies, and
this may be considered a limitation of the present study.
In summary, the present study confirmed that P2X7R
is a novel regulator of 3T3-L1 preadipocyte development. P2X7R
promoted 3T3-L1 mature cell adipogenesis and lipid degradation.
These findings may enhance the current understanding of
P2X7R-regulated adipogenesis and provide a novel model to assess
the pathogenesis of obesity.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL and LG conceived and designed the study, and
acquired, analyzed and interpreted the data, as well as drafting
the manuscript and revising it for important intellectual content.
JL, LG and QX performed the experiments and interpreted the data.
All authors have read and approved the final manuscript. JL and LG
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Yancheng Third People's Hospital (Yancheng, China;
approval no. 20200030).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bou M, Todorcevic M, Rodriguez J, Capilla
E, Gutierrez J and Navarro I: Interplay of adiponectin, TNFα and
insulin on gene expression, glucose uptake and PPARү, AKT and TOR
pathways in rainbow trout cultured adipocytes. Gen Comp Endocrinol.
205:218–225. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Saxton SN, Clark BJ, Withers SB, Eringa EC
and Heagerty AM: Mechanistic links between obesity, diabetes, and
blood pressure: Role of perivascular adipose tissue. Physiol Rev.
99:1701–1763. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kahn CR, Wang G and Lee KY: Altered
adipose tissue and adipocyte function in the pathogenesis of
metabolic syndrome. J Clin Invest. 129:3990–4000. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bleich SN, Vercammen KA, Zatz LY, Frelier
JM, Ebbeling CB and Peeters A: Interventions to prevent global
childhood overweight and obesity: A systematic review. Lancet
Diabetes Endocrinol. 6:332–346. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bhupathiraju SN and Hu FB: Epidemiology of
obesity and diabetes and their cardiovascular complications. Circ
Res. 118:1723–1735. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gallagher EJ and LeRoith D: Obesity and
diabetes: The increased risk of cancer and cancer-related
mortality. Physiol Rev. 95:727–748. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tang QQ and Lane MD: Adipogenesis: From
stem cell to adipocyte. Annu Rev Biochem. 81:715–736.
2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Haczeyni F, Bell-Anderson KS and Farrell
GC: Causes and mechanisms of adipocyte enlargement and adipose
expansion. Obes Rev. 19:406–420. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sawamoto A, Nakanishi M, Okuyama S,
Furukawa Y and Nakajima M: Heptamethoxyflavone inhibits
adipogenesis via enhancing PKA signaling. Eur J Pharmacol.
865(172758)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Im DU, Kim SC, Chau GC and Um SH:
Carbamazepine enhances adipogenesis by inhibiting Wnt/β-catenin
expression. Cells. 8(1460)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Prestwich TC and Macdougald OA:
Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr
Opin Cell Biol. 19:612–617. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen X, Ayala I, Shannon C, Fourcaudot M,
Acharya NK, Jenkinson CP, Heikkinen S and Norton L: The diabetes
gene and Wnt pathway effector TCF7L2 regulates adipocyte
development and function. Diabetes. 67:554–568. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Guo Y, Mishra A, Weng T, Chintagari NR,
Wang Y, Zhao C, Huang C and Liu L: Wnt3a mitigates acute lung
injury by reducing P2X7 receptor-mediated alveolar epithelial type
I cell death. Cell Death Dis. 5(e1286)2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sindhavajiva PR, Sastravaha P,
Arksornnukit M and Pavasant P: Purinergic 2X7 receptor activation
regulates WNT signaling in human mandibular-derived osteoblasts.
Arch Oral Biol. 81:167–174. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gnoni A, Siculella L, Paglialonga G,
Damiano F and Giudetti AM: 3,5-diiodo-L-thyronine increases de novo
lipogenesis in liver from hypothyroid rats by SREBP-1 and
ChREBP-mediated transcriptional mechanisms. IUBMB Life. 71:863–872.
2019.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Vadrot N, Duband-Goulet I, Cabet E,
Attanda W, Barateau A, Vicart P, Gerbal F, Briand N, Vigouroux C,
Oldenburg AR, et al: The p. R482W substitution in A-type lamins
deregulates SREBP1 activity in Dunnigan-type familial partial
lipodystrophy. Hum Mol Genet. 24:2096–2109. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hou XX, Dong HR, Sun LJ, Yang M, Cheng H
and Chen YP: Purinergic 2X7 receptor is involved in the podocyte
damage of obesity-related glomerulopathy via activating
nucleotide-binding and oligomerization domain-like receptor protein
3 inflammasome. Chin Med J (Engl). 131:2713–2725. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
North RA: Molecular physiology of P2X
receptors. Physiol Rev. 82:1013–1067. 2002.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Su QQ, Tian YY, Liu ZN, Ci LL and Lv XW:
Purinergic P2X7 receptor blockade mitigates alcohol-induced
steatohepatitis and intestinal injury by regulating MEK1/2-ERK1/2
signaling and egr-1 activity. Int Immunopharmacol. 66:52–61.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Beaucage KL, Xiao A, Pollmann SI, Grol MW,
Beach RJ, Holdsworth DW, Sims SM, Darling MR and Dixon SJ: Loss of
P2X7 nucleotide receptor function leads to abnormal fat
distribution in mice. Purinergic Signal. 10:291–304.
2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gao H, Li D, Yang P, Zhao L, Wei L, Chen Y
and Ruan XZ: Suppression of CD36 attenuates adipogenesis with a
reduction of P2X7 expression in 3T3-L1 cells. Biochem Biophys Res
Commun. 491:204–208. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Han Y, Lee SH, Lee IS and Lee KY:
Regulatory effects of 4-methoxychalcone on adipocyte
differentiation through PPARγ activation and reverse effect on
TNF-α in 3T3-L1 cells. Food Chem Toxicol. 106:17–24.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lee CW, Seo JY, Lee J, Choi JW, Cho S, Bae
JY, Sohng JK, Kim SO, Kim J and Park YI: 3-O-Glucosylation of
quercetin enhances inhibitory effects on the adipocyte
differentiation and lipogenesis. Biomed Pharmacother. 95:589–598.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ma J, Liao H, Lin Y, Huang K, Zhu J and
Wang Y: Molecular characterization, expression analysis of Chemerin
gene and its potential role in intramuscular adipocyte
differentiation of goat. Anim Biotechnol. 31:382–390.
2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Rosen ED and MacDougald OA: Adipocyte
differentiation from the inside out. Nat Rev Mol Cell Biol.
7:885–896. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Moseti D, Regassa A and Kim WK: Molecular
regulation of adipogenesis and potential anti-adipogenic bioactive
molecules. Int J Mol Sci. 17(124)2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang L, Zhang L, Wang X and Si H:
Anti-adipogenic effects and mechanisms of ginsenoside Rg3 in
pre-adipocytes and obese mice. Front Pharmacol.
8(113)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Jin H, Lee K, Chei S, Oh HJ, Lee KP and
Lee BY: Ecklonia stolonifera extract suppresses lipid accumulation
by promoting lipolysis and adipose browning in high-fat
diet-induced obese male mice. Cells. 9(871)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Di Virgilio F, Dal Ben D, Sarti AC,
Giuliani AL and Falzoni S: The P2X7 receptor in infection and
inflammation. Immunity. 47:15–31. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Coccurello R and Volonte C: P2X7 Receptor
in the management of energy homeostasis: Implications for obesity,
dyslipidemia, and insulin resistance. Front Endocrinol (Lausanne).
11(199)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Calderon-Dominguez M, Mir JF, Fucho R,
Weber M, Serra D and Herrero L: Fatty acid metabolism and the basis
of brown adipose tissue function. Adipocyte. 5:98–118.
2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Moon MH, Jeong JK, Lee YJ, Seol JW, Ahn
DC, Kim IS and Park SY: 18β-Glycyrrhetinic acid inhibits adipogenic
differentiation and stimulates lipolysis. Biochem Biophys Res
Commun. 420:805–810. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Seo YJ, Jin H, Lee K, Song JH, Chei S, Oh
HJ, Oh JH and Lee BY: Cardamonin suppresses lipogenesis by
activating protein kinase A-mediated browning of 3T3-L1 cells.
Phytomedicine. 65(153064)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lee JE, Schmidt H, Lai B and Ge K:
Transcriptional and epigenomic regulation of adipogenesis. Mol Cell
Biol. 39:e00601–e00618. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Walther TC and Farese RV Jr: Lipid
droplets and cellular lipid metabolism. Annu Rev Biochem.
81:687–714. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Suh HJ, Cho SY, Kim EY and Choi HS:
Blockade of lipid accumulation by silibinin in adipocytes and
zebrafish. Chem Biol Interact. 227:53–62. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xi FX, Wei CS, Xu YT, Ma L, He YL, Shi XE,
Yang GS and Yu TY: MicroRNA-214-3p targeting Ctnnb1 promotes 3T3-L1
preadipocyte differentiation by interfering with the Wnt/β-catenin
signaling pathway. Int J Mol Sci. 20(1816)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kang S, Bennett CN, Gerin I, Rapp LA,
Hankenson KD and Macdougald OA: Wnt signaling stimulates
osteoblastogenesis of mesenchymal precursors by suppressing
CCAAT/enhancer-binding protein alpha and peroxisome
proliferator-activated receptor gamma. J Biol Chem.
282:14515–14524. 2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liu J and Farmer SR: Regulating the
balance between peroxisome proliferator-activated receptor gamma
and beta-catenin signaling during adipogenesis. A glycogen synthase
kinase 3beta phosphorylation-defective mutant of beta-catenin
inhibits expression of a subset of adipogenic genes. J Biol Chem.
279:45020–45027. 2004.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu J, Wang H, Zuo Y and Farmer SR:
Functional interaction between peroxisome proliferator-activated
receptor gamma and beta-catenin. Mol Cell Biol. 26:5827–5837.
2006.PubMed/NCBI View Article : Google Scholar
|