Introduction
Osteoporosis (OP) is characterized by reduced bone
formation, increased bone resorption, and destruction of the bone
microstructure, resulting in reduced bone strength, increased
brittleness and metabolic osteopathic syndrome, which predisposes
patients to fractures (1,2). Currently, the effects of OP treatments
are not ideal, and additionally, more detailed basic research
studies are necessary to identify a new target for treatment.
Bone marrow mesenchymal stem cells (BMSCs) are a
subgroup of adult stem cells with multiple differentiation
potential found in the bone marrow matrix of mammals. BMSCs exhibit
the potential to differentiate into a variety of cell types, high
plasticity, the easy introduction and expression of exogenous genes
and other advantageous biological characteristics and can
differentiate into osteoblasts in an osteoblast-induced
microenvironment (3,4). Due to this characteristic, BMSCs play
an important role in the basic research of OP.
MicroRNAs (miRNAs or miRs), a type of endogenous
single-stranded noncoding small RNA that has been revealed in
previous studies to be widespread in animals and plants, are
important posttranscriptional regulators (5,6). In
previous studies, miRNAs have been revealed to be involved in the
regulation of a variety of physiological and pathological processes
in the body and have gradually become the focus of life science
research (7,8). miRNAs play an important regulatory
role in the proliferation and differentiation of osteoblasts,
osteoclasts and chondrocytes by regulating the expression of target
genes through relevant signaling pathways, ultimately affecting
bone production and metabolism (9,10).
miRNAs are expected to become potential gene therapy targets for
the clinical treatment of bone metabolic diseases and bone injury.
Previous studies have found stable miRNAs in circulating blood and
identified significantly different expression levels of various
miRNAs in the peripheral blood of patients with OP compared with
those in healthy subjects (11,12).
Our previous study demonstrated that the expression of miR-144-3p
in the peripheral blood and bone tissues of OP patients was
significantly decreased (13).
Members of the GATA family of transcription factors
are highly evolutionarily conserved, zinc finger domain-specific
transcription factors that bind a specific DNA sequence,
(A/T)GATA(A/G), and regulate a variety of biological processes.
GATA binding protein 4 (GATA 4) is one of six transcription factors
in the family (14). Most previous
studies of GATA4 have focused on its role in the heart (15,16).
Another previous study also revealed that GATA4 plays an important
role in the regulation of bone metabolism, especially in the
differentiation and formation of osteoblasts (17). After consulting the literature and
miRNA Base (13), it was
hypothesized that GATA4 is an upstream targeted regulator of
miR-144-3p.
In the present study, the role of miR-144-3p in
regulating the osteogenic differentiation of BMSCs and the effect
of GATA4 on osteogenic differentiation by targeting miR-144-3p was
demonstrated.
Materials and methods
Animals
A total of six male Sprague-Dawley rats aged 2-3
weeks and weighing 40-60 g, were obtained from the Experimental
Animal Centre of Guizhou Medical University, (Guizhou, China) and
were used in the present study. The rats were kept in a
temperature-controlled room, with a humidity of 40-70%, in a 12 h
light-dark cycle with free access to standard chow and tap water.
All animals were reared in a specific pathogen-free environment at
a comfortable temperature and humidity (18). The BMSC extraction process was
approved by the Experimental Animal Ethics Committee of Guizhou
Medical University (approval no. 1702032).
Identification rat BMSCs by using flow
cytometry
To identify the target cell, fluorescein
isothiocyanate (FITC)-conjugated CD90 (1:100; cat. no. 561973; BD
Biociences), CD45 (1:100; cat. no. 561867; BD Biosciences), and
phycoerythrin (PE)-conjugated CD 44 (1:100; cat. no. MA5-16908;
Thermo Fisher Scientific Inc.) were used to label the BMSC
membranes. A flow cytometer (FC 500; BD Biosciences) and FlowJo
software v.10.5.2 (BD Biosciences) were used for analysis.
Mesenchymal stem cell culture and
osteogenic differentiation
The isolation and culture of BMSCs were conducted as
previously described (13).
Briefly, the rats were weighed and 150 mg/kg of pentobarbital
sodium were administered intraperitoneally for euthanasia. After
euthanasia, BMSCs were isolated from bilateral femurs and tibia of
rats and cultured in a cell incubator containing 5% CO2
at 37˚C. Medium consisting of Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc.), 10 ml of fetal
bovine serum (FBS; Biological Industries), and 1 ml of secondary
antibody (cat. no. 30-002-CI; Beijing Solarbio Science &
Technology Co., Ltd.), at a ratio of 100:10:1 was prepared and
replaced every 2-3 days. When the cells reached 80% confluence,
they were digested with a trypsin-EDTA solution (Beijing Solarbio
Science & Technology Co., Ltd.) and passaged at a ratio of
1:2.
Osteogenic induction medium was prepared by adding
10% FBS, 100 mmol/l dexamethasone, 0.05 mmol/l vitamin C, and 10
mmol/l glycerophosphate (all purchased from Beijing Solarbio
Science & Technology Co., Ltd.) to 90 ml of high-glucose DMEM
and stored at 4˚C. During in vitro osteogenic
differentiation, cells were seeded in 6-well plates and induced by
osteogenic induction medium, which was replaced every 2-3 days.
Alkaline phosphatase (ALP) and
Alizarin Red S staining
BMSCs from generations 4 and 5 were inoculated in
6-well plates, and osteogenic induction medium was added when the
cells reached 70-80% confluence for induction. Alizarin Red S
(Beijing Solarbio Science & Technology Co., Ltd.) staining was
then carried out on days 0, 7, 14 and 21 strictly according to the
manufacturer's protocol. The medium in the plate was first
discarded, and the cells were washed 3 times with PBS. After
fixation with 4% paraformaldehyde for 10-15 min at room
temperature, the fixation solution was discarded and the cells were
washed 3 times with ddH2O. After dd (double distilled)
H2O was completely absorbed, an 4% Alizarin Red S
staining solution was slowly added and incubated with the cells at
37˚C in a humidified atmosphere of 5% CO2 for 20-30 min.
The dye was discarded and the cells were washed 3-5 times with
ddH2O. The appropriate amount of ddH2O was
added to each well to prevent the cells in the well from drying.
The cells were observed, and images were captured under a light
microscope (Leica DMi8 M/C/A; Leica Microsystems, GmbH)
(magnification, x100).
An ALP staining kit (Beyotime Institute of
Biotechnology) was used for staining according to the
manufacturer's instructions. The medium in 6-well plates was
discarded, and cells were washed 3 times with PBS. The reagent (75%
fixative solution) was used to fix the cells for 2-5 min at room
temperature, and the cells dried naturally. The fixative solution
was discarded, and the cells were rinsed with distilled water for
30 sec. After the distilled water had been discarded, the matrix
solution was added, and the cells were incubated at 37˚C in the
dark for 15 min. After the excess dye had been absorbed, 200 µl
color solution A was immediately added, followed by incubation for
5 min at room temperature and washing for 30 sec. The excess water
was discarded, and 10 µl color solution B was added, followed by
incubation for 5 min at room temperature and washing for 30 sec.
After the excess water had been discarded, dye was added for 30
sec, followed by a washing step for 30 sec, after which the excess
water was discarded. The cells were then observed under a light
microscope (Nikon TE-2000; Nikon Corporation) (magnification, x40)
and images were captured.
Transfection assays
All transfection reagents [the miR-144-3p
oligonucleotides (miR-144-3p mimic 5'-UACAGUAUAGAUGAUGUACU-3',
negative mimic control 5'-UUCUCCGAACGUGUCACGUTT-3, miR-144-3p
inhibitor 5'-AGUACAUCAUCUAUACUGUA3', inhibitor negative control
5'-CAGUACUUUUGUGUAGUACAA) and GP-siRNA-Mate Plus] were purchased
from Shanghai GenePharma Co., Ltd. BMSCs at generations 4-5 were
digested, seeded into 6-well plates at a density of
3x105 cells/ml and cultured to a confluence of 60-80%
for transfection. Prior to transfection, the transfection reagent
(RNA oligo) was prepared at the desired concentration (4 µl)
according to the manufacturer's instructions, mixed directly with
the GP-siRNA-Mate Plus transfection reagent at room temperature,
and left to rest for 10-15 min. The compound was added to 300 µl of
complete medium, mixed and added to a 6-well plate. After 6-8 h,
this solution was added to complete medium or osteogenic induction
medium. mRNA and protein expression was detected at 36-72 h after
transfection. For each group of tests, 3 wells containing the
compound were prepared, and the mean value was obtained. The
experiment was repeated three times.
Adenovirus infection
The cells were inoculated into a 48-well plate at a
density of 1x105 cells/well. A total of 250 µl of DMEM
containing 10% FBS, 100 µl adenovirus (Shanghai GenePharma Co.,
Ltd) and 100 µl Lipofectamine 2000® reagent (Invitrogen;
Thermo Fisher Scientific, Inc) was added to each well. After
mixing, the cells were cultured for 24 h in an incubator containing
5% CO2 at 37˚C. The adenovirus with AdGATA4 (Shanghai
GenePharma Co., Ltd.) in the 48-well plate was absorbed and 500 µl
of DMEM containing 10% FBS was added to each well, followed by
continued culture at 37˚C in an incubator containing 5%
CO2 for 48 h. After 48 h, RT-qPCR for detection of GATA
binding protein 4 and miR-144-3p expression was performed. The
adenovirus infection efficiency was observed under a confocal
microscope (Nikon eclipse 80i; Nikon Corporation) (magnification,
x40). B was the blank control group (untransfected cells).
Dual-luciferase reporter assay
For luciferase reporter assays, three plasmids were
designed: Plasmids containing one cis-GATA4 motif from the
miR-144 promoter (P1) or two cis-GATA4 motifs from the
miR-144 promoter (P2) and the pGL3-basic vector. The plasmids were
purchased from Shanghai GenePharma Co., Ltd. and the sequences are
listed in Tables I and II. After 293T cells (Shanghai Furi
Technology Co., Ltd.) had been cultured in a 10-cm culture dish
until reaching 80-90% confluence, the cells were diluted to
1x106 cells/ml. The cells were used to inoculate 12-well
plates at a density of 5x105 cells/well, mixed and
cultured for 24 h at 37˚C in an atmosphere containing 5%
CO2. The culture medium in the 12-well plate was
aspirated and the transfection mixture GP-transfect-Mate (Shanghai
GenePharma Co., Ltd.) was added dropwise into the 12-well plate at
room temperature for 20 min. After mixing, the mixture was
incubated for 5 h at 37˚C under 5% CO2 for 24 and 48 h,
and samples were collected. The cells were used for dual-luciferase
assays in which luciferase activity was assessed using the Dual-Glo
Luciferase Assay System (Promega Corporation). Three replicates
were performed for each group, the mean values were obtained, and
the detection time-point was 48 h. The experiment was repeated
three times.
 | Table IGene sequences of plasmid P1. |
Table I
Gene sequences of plasmid P1.
| pGL3-miR-144-p1
(568 bp) |
|---|
|
CTAGCATCTTCAGCCTTCAGTTTCATTCCCAGCACAGGAAACTAAGCAGA |
|
AGATAAACAAAAAGGGAGCCAAGCCTCAGCTTGTCTCAGGAAGCCAGC |
|
AGGCAAAGAGTTAAGAAGCAGGGACTTCTAGAACCCGGGAAAACGTGC |
|
CCCACCCAGGGGAGGGGCCAGAGGGTTAAAAGCCAAGCTGCTTGAGTG |
|
AGAAGAGACAAGGCAGGCTCTCCCTGTGCAGAGGATTCCCTGGACGAG |
|
GCTCCAGCTCCACTCCAGCTCCAGGTAAGCAGTCCTTGGAGTGGCTGTC |
|
AGCCTGCTTATAGGTCTGCCCAGAGGGAAGCTCCTGCCTCACAACTTCGT |
|
TTCTGCCTGTAACTCTGGATCCCTAAGAGACCCGAGTAGACCTTAGCTTC |
|
CTTCTCTAAGCCACCTGGGGTTATCCTGGACCACAGGATCAGGGAGATGC |
|
TGCTCTGGGAGGGAAGTGGAGGAGCAGAGGTAGGGACTTAGGTGTCCC |
|
TGACTGACCCTGAGCCAATCCCCTGGCTCACTCCAGGCCTGCTGCTCAC |
|
CTCCTCCTCCAGGACCTTGGCTGGGATATC |
 | Table IIGene sequences of plasmid P2. |
Table II
Gene sequences of plasmid P2.
| pGL3-miR-144-p2
(969 bp) |
|---|
|
TCAGCTTCCCAGCAGAGGCCCACTTGTCCACGGACCTTGCCAGAGGTGG |
|
CTTGCAAGCTTCAGCTCTGCCCACCCAGCTCAAACAGAGTCAAAGCCTA |
|
GGGATGGAGTCAGGCTGAGGGTACATGGAGCCTGCTCCCAGATAGATTC |
|
CATCTAGGTCCAGTTGCCAGGACCTCCCTGTCCTATTCAGATTCAACTAA |
|
CATTCCCATCATCACCCACAACAAACTGGGACCTTTCAGCGAGGCCCGA |
|
ACAACTAAGCCCTGAAGGACGTGGTGAGGGTCTTTCTCTTTCCTAGCCC |
|
AGTGGGGTAGGCAGGTAACCTTCTTTGTGGGGTTGGGGGTGAATGGTCA |
|
CCCACTTAGAAGACGGGAGGCAGGGTGGCGGTGTAGTGTCAGTGTTGG |
|
GGTTCTTGGCTAGCATCTTCAGCCTTCAGTTTCATTCCCAGCACAGGAAA |
|
CTAAGCAGAAGATAAACAAAAAGGGAGCCAAGCCTCAGCTTGTCTCAG |
|
GAAGCCAGCAGGCAAAGAGTTAAGAAGCAGGGACTTCTAGAACCCGGG |
|
AAAACGTGCCCCACCCAGGGGAGGGGCCAGAGGGTTAAAAGCCAAGCT |
|
GCTTGAGTGAGAAGAGACAAGGCAGGCTCTCCCTGTGCAGAGGATTCCC |
|
TGGACGAGGCTCCAGCTCCACTCCAGCTCCAGGTAAGCAGTCCTTGGAG |
|
TGGCTGTCAGCCTGCTTATAGGTCTGCCCAGAGGGAAGCTCCTGCCTCAC |
|
AACTTCGTTTCTGCCTGTAACTCTGGATCCCTAAGAGACCCGAGTAGACC |
|
TTAGCTTCCTTCTCTAAGCCACCTGGGGTTATCCTGGACCACAGGATCAG |
|
GGAGATGCTGCTCTGGGAGGGAAGTGGAGGAGCAGAGGTAGGGACTTA |
|
GGTGTCCCTGACTGACCCTGAGCCAATCCCCTGGCTCACTCCAGGCCTG |
|
CTGCTCACCTCCTCCTCCAGGACCTTGGCTGGGATATC |
Reverse transcription-quantitative
(RT-q)PCR for mRNA and miRNA
Cell samples to be collected were obtained, and the
culture medium was discarded. Then, cells were washed with PBS, and
TRIzol reagent (Thermo Fisher Scientific, Inc.) was used to extract
total RNA from the cells. A total of 2 µg of RNA was used for
reverse transcription. The total RNA was converted to cDNA with a
Custom Gene RT-qPCR Quantitation Kit according to the
manufacturer's instructions (Shanghai GenePharma Co., Ltd).
Quantitative PCR was performed using a Bio-Rad CFX 96 Touch
real-time PCR system (Bio-Rad Laboratories, Inc.) and was executed
using the TB Green Premix Ex Taq II (Shanghai GenePharma Co., Ltd.)
with 2 µl of cDNA template in a 25-µl final reaction mixture (95˚C
for 30 sec; 95˚C for 5 sec, 60˚C for 30 sec, 40 cycles). The
average threshold cycle (Ct) for each gene was determined from
triplicate reactions; the relative expression level of mRNA or
miRNAs was normalized to that of the internal controls, β-actin or
U6 using the 2-ΔΔCq method (19).
The primers specific for mRNAs were purchased from
Shanghai GenePharma Co., Ltd. and the sequences are listed in
Table III.
 | Table IIIPrimers used in reverse
transcription-quantitative PCR. |
Table III
Primers used in reverse
transcription-quantitative PCR.
| Gene | Forward primer | Reverse primer |
|---|
| Rat-miR-144-3p |
GCAGAGTACAGTATAGATGATG |
GTGCAGGGTCCGAGGT |
| rat-GATA4 |
GGGCTGTCATCTCACTATGGG |
TGGATAGCCTTGTGGGGAGA |
| rat-U6 |
CAGCACATATACTAAAATTGGAACG |
GTGCAGGGTCCGAGGT |
| rat-ALP |
CGATGGCTTTGGTACGGAGT |
TGCGGGACATAAGCGAGTTT |
| rat-RUNX2 |
TGAGATTTGTAGGCCGGAGC |
CTGAGGCGGTCAGAGAACAA |
| rat-ACTB |
CGTAAAGACCTCTATGCCAACA |
GGAGGAGCAATGATCTTGATCT |
Western blot analysis
After 72 h of transfection, the cells were washed
with PBS and lysed with RIPA lysis buffer (cat. no. KGP702; Nanjing
KeyGen Biotech Co., Ltd.). Protein determination was performed
using the bicinchoninic acid (BCA) method. Samples (20 µg/well)
were separated by 8 and 15% SDS-PAGE. After electrophoresis, a PVDF
membrane was immersed in methanol for 10 sec, and the gel and PVDF
membrane soaked in methanol were immersed in rapid electrophoretic
buffer for 10 min. Non-specific binding was blocked with 5% non-fat
skimmed milk in Tris-buffered saline plus 0.1% TBST (cat. no.
T1081; Beijing Solarbio Science & Technology Co., Ltd.) at 25˚C
for 2 h. Membranes were probed with primary antibodies overnight at
4˚C, including Runt-related transcription factor 2 (Runx2)
polyclonal antibody (1:1,000; cat. no. ab23981; Abcam) and ALP
monoclonal antibody (1:2,000; cat. no. ab194297; Abcam). β-actin
(1:10,000; cat. no. A5441; Sigma-Aldrich; Merck KGaA) was used as a
loading control. After washing with TBS containing 0.1% Tween- 20,
the immobilized primary antibodies were incubated with a
horseradish peroxidase-conjugated secondary goat anti-rabbit IgG
antibody (1:2,000; cat. no. ab205718; Abcam) for 1 h at 25˚C. Then,
SuperSignal West Pico chemiluminescent substrate (P0018S; Beyotime
Institute of Biotechnology) and a full-function FR-1800
luminescence and fluorescence biological image analysis system
(Shanghai Furi Technology Co., Ltd.) were used for
chemiluminescence detection. Gel-Pro Analyzer software version 4.0
(Media Cybernetics, Inc.) was used for analysis and processing.
Statistical analysis
All data are expressed as the mean ± standard
deviation (SD). Statistical analyses were performed with SPSS
(version 17.0; SPSS, Inc.). Comparisons between only two groups
were performed by the Student's t-test. One-way or two-way ANOVAs,
with Bonferroni's post hoc tests were performed for comparisons
among multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-144-3p is
downregulated during osteogenic differentiation
Our previous study revealed that the expression of
miR-144-3p in the serum and bone tissue of OP patients was
significantly increased compared with that of healthy individuals
(13). BMSCs in rats were induced
to undergo osteogenic differentiation to investigate whether the
expression of miR-144-3p would change during the osteogenic
differentiation of BMSCs in rats. The result of flow cytometry
demonstrated that BMSCs were positive for CD44 (95.48%), CD29
(99.31%) and negative for CD45 (0.20%) (Fig. S1). During osteogenic
differentiation of BMSCs, ALP staining and Alizarin Red staining
gradually deepened with time prolonging (Fig. 1A and B). As induction duration increased, the
mRNA expression of miR-144-3p was decreased, and the mRNA
expression of the osteogenic marker genes ALP and Runx2 was
significantly increased (Fig. 1C).
These results indicated that miR-144-3p was negatively associated
with the expression of osteogenic marker genes.
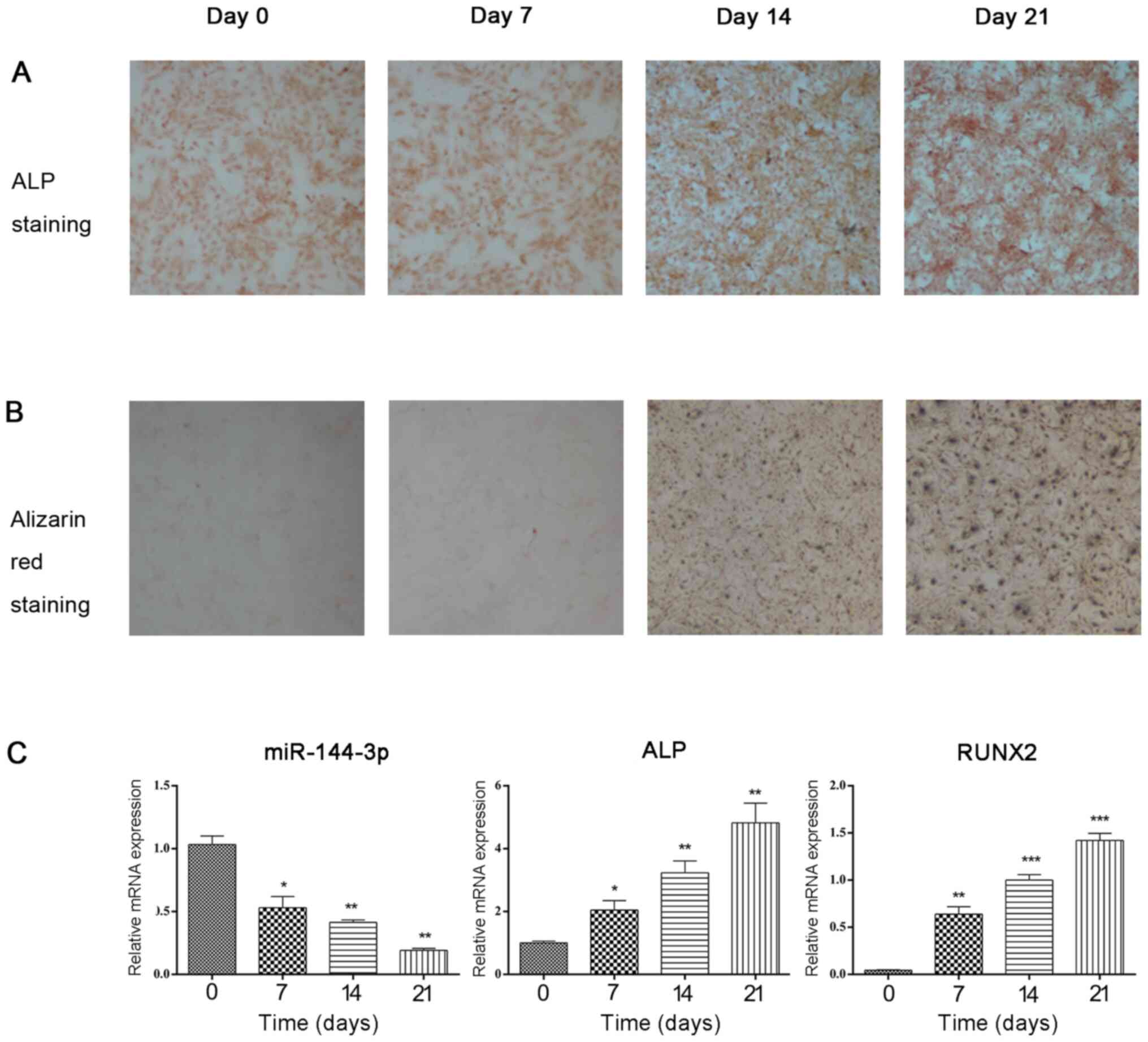 | Figure 1Expression of miR-144-3p is
downregulated during osteogenic differentiation. (A and B) During
osteogenic differentiation of BMSCs, ALP staining and Alizarin Red
staining gradually deepened with time prolonging. The results were
performed to ensure efficient osteogenic differentiation of BMSCs.
(C) miR-144-3p, ALP, and Runx2 expression levels in BMSCs were
quantitatively assessed by reverse transcription-quantitative PCR
at the indicated time-points (0, 7, 14 and 21 days) during
osteogenic differentiation. The data, normalized to β-actin (for
mRNA) or U6 (for miRNA), are the mean ± SD of three experiments.
miR, microRNA. *P<0.05, **P<0.01 and
***P<0.001. ALP, alkaline phosphatase; BMSCs, bone
marrow mesenchymal stem cells; RUNX2, Runt-related transcription
factor 2. |
miR-144-3p inhibits osteogenic
differentiation
As previously demonstrated, miR-144-3p was involved
in the osteogenic differentiation of BMSCs, but the specific
regulatory role was not further examined. In this part of the
study, the expression of miR-144-3p was regulated by cell
transfection, and the relationship between the expression of
miR-144-3p and osteogenic marker genes was analyzed to determine
whether miR-144-3p negatively regulates the differentiation of
osteoblasts.
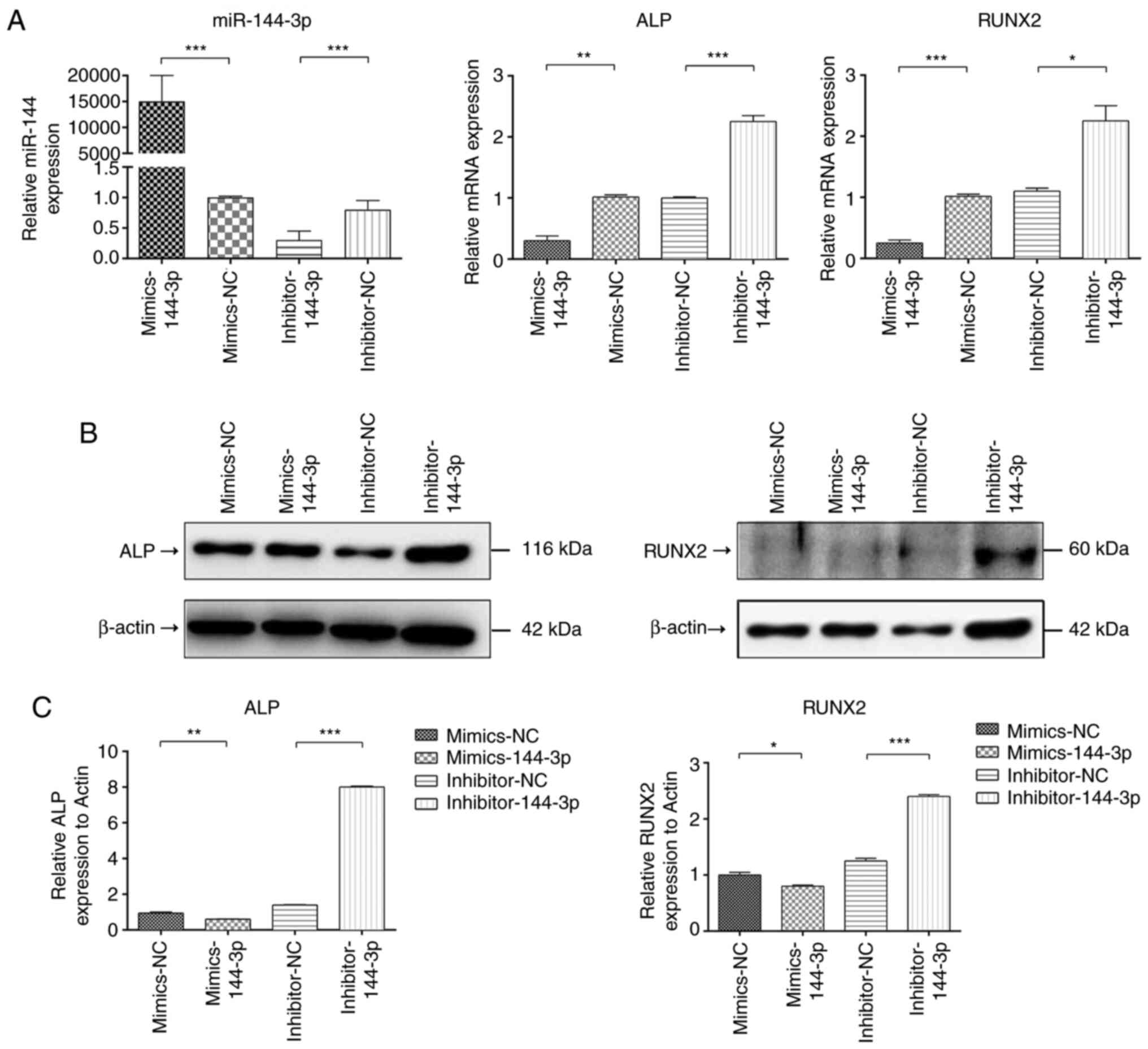
The mRNA expression of miR-144-3p was assessed at 48
h after transfection in BMSCs transfected with miR-144-3p mimic,
inhibitor and the corresponding controls. The expression of
miR-144-3p mRNA in the transfected miR-144-3p mimic group was
significantly increased compared with that in the corresponding
negative control (NC) treatment group, while the expression of
miR-144-3p mRNA in the miR-144-3p inhibitor group was significantly
decreased compared with that in the inhibitor NC group (Fig. 2A). After BMSCs were transfected with
the miR-144-3p mimic, the mRNA and protein expression of the
osteogenic marker genes ALP and Runx2 were significantly decreased
compared with that in the corresponding NC treatment group, while
the mRNA and protein expression of the osteogenic marker genes ALP
and Runx2 were significantly increased in the transfected
miR-144-3p inhibitor group compared with the inhibitor NC group
(Fig. 2A-C). These results
indicated that miR-144-3p was a negative regulatory factor in the
osteogenic differentiation of BMSCs and inhibited the expression of
osteogenic marker genes.
GATA4 directly targets miR-144-3p
In our previous study (13), it was revealed that miR-144-3p was
significantly downregulated during the osteogenic differentiation
of BMSCs in rats and plays a negative regulatory role in this
process. Huang et al demonstrated that miR-144-3p could
affect the osteogenic differentiation process in rat BMSCs by
regulating Smad4(20), which
demonstrated that Smad4 is a regulatory gene targeted by
miR-144-3p; however, the regulatory factors upstream of miR-144-3p
in the osteogenic differentiation process have not yet been
studied, to the best of our knowledge. As mentioned above, GATA4
may be an upstream regulator of miR-144-3p. To further demonstrate
the direct targeting relationship between GATA4 and miR-144-3p, an
adenovirus infection experiment was conducted and dual-luciferase
reporter gene detection was used.
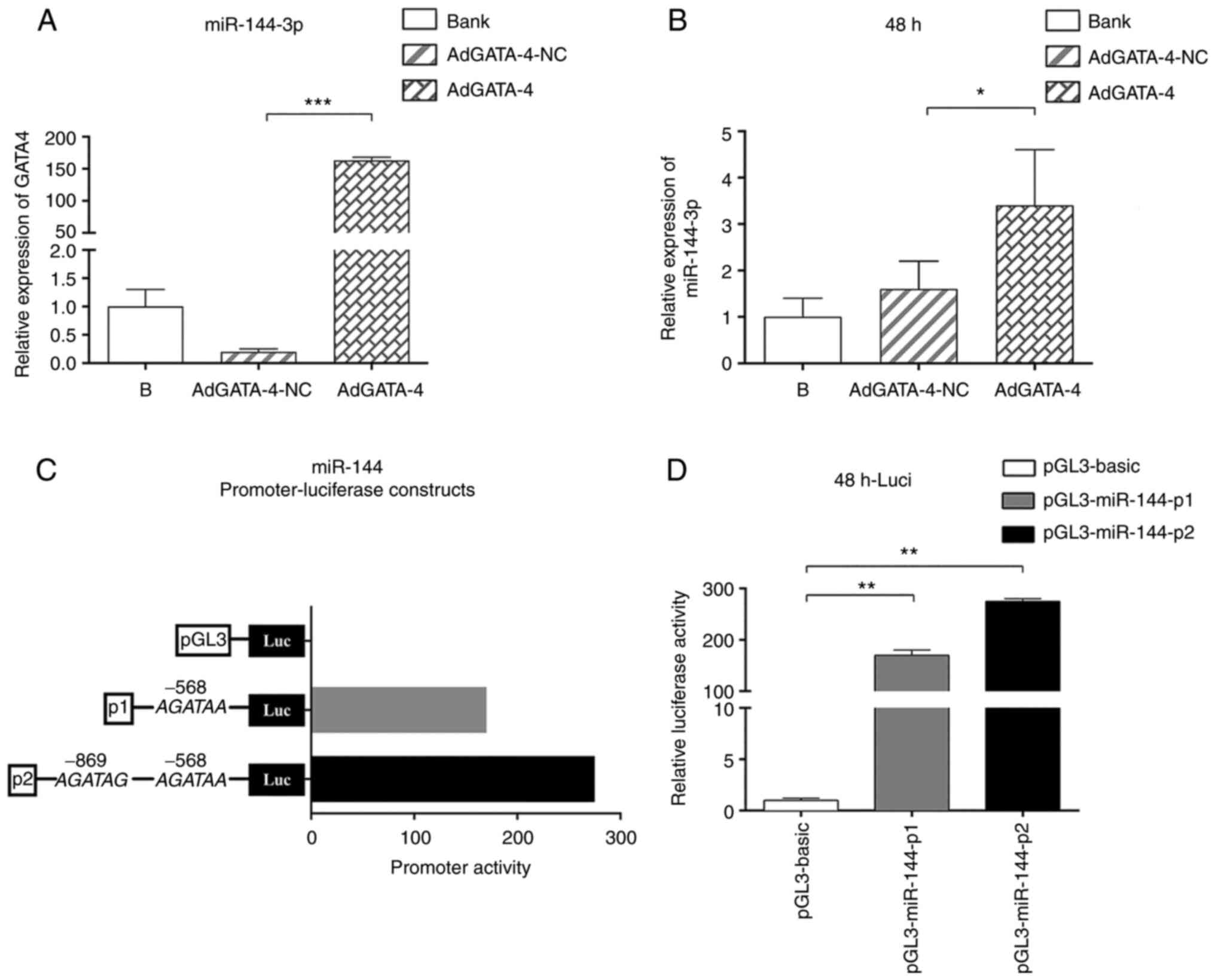
BMSCs were infected by adenovirus carrying AdGATA4,
and cell samples were collected after 48 h of infection for RT-qPCR
detection. The mRNA levels of GATA4 and miR-144-3p were increased
compared with those of the NC group (Fig. 3A and B). These differences were statistically
significant, indicating that GATA4 had a positive regulatory effect
on the expression of miR-144-3p. Next, whether GATA4 directly
regulates miR-144 promoter activity was investigated. Two miR-144
promoter-luciferase plasmids containing one (P1) or two (P2)
putative GATA-binding sites at positions -568 and -969 of the
miR-144 promoter region were generated (Fig. 3C). These luciferase reporter
constructs and the pGL3-basic vector were cotransfected into 293T
cells with AdGATA4. After 48 h of transfection, the luciferase
activity of plasmids P1 and P2 was significantly increased compared
with that of the control transfection group. Moreover, plasmid P2,
which contained two binding sites, exhibited stronger luciferase
activity compared with plasmid P1, which contained one binding site
(Fig. 3C and D). Therefore, the experimental results
indicated that the GATA4 regulatory site played a positive role in
regulating the regions of the miR-144 promoter at -568 and
-969.
Discussion
miRNAs are important posttranscriptional regulators
confirmed by numerous studies to be involved in the regulation of
bone metabolism (21,22). In normal human serum, the type and
amount of miRNAs are essentially stable. When tissues are
destroyed, miRNAs in tissues will be released into the blood,
causing an aberrant increase in the miRNA content in the serum
(23,24). In addition, when lesions occur in
the body, such as those due to tumors and OP, the miRNA content in
serum will also become aberrant (25,26).
Therefore, miRNAs can serve as biomarkers for the early diagnosis
of OP.
Previous basic studies on OP have revealed that the
selection of target miRNAs in BMSC osteogenic differentiation can
be achieved by miRNA microarray detection and analysis of miRNA
expression in BMSCs undergoing osteogenic differentiation (27) or by collecting blood samples from OP
patients and healthy controls for the screening of target miRNAs by
RT-qPCR (28). After a target gene
was identified, cellular experiments in which the target gene was
overexpressed or inhibited were conducted to further verify its
role in BMSC osteogenic differentiation (29). In our previous study (13), miR-144-3p was selected as a target
gene by assessing the first group of selected targets; determining
the 10 most likely candidate target genes, again through the
collection of blood samples from OP patients and healthy controls
and RT-qPCR detection of mRNA levels of candidate target genes; and
selecting the target genes with the greatest reliability, which
revealed miR-144-3p as a target gene. In the present study, during
osteogenic induction and differentiation, the expression of
miR-144-3p was decreased, while the expression of osteogenic marker
genes (ALP and Runx2) was increased. After further cell
transfection, the expression of endogenous miR-144-3p was
inhibited, osteogenic induction and differentiation continued, and
the expression of osteogenic marker genes was increased.
Overexpression of miR-144-3p inhibited the expression of osteogenic
markers. These results indicated that miR-144-3p negatively
regulated the osteogenic differentiation of BMSCs. Our previous
study revealed that miR-144-3p expression was significantly reduced
in the peripheral blood and bone tissue in patients with OP and
that through targeting RANK, miR-144-3p regulated osteoclast
formation, further affecting bone metabolism and the bone formation
process, leading to the occurrence of OP (13). Research on miR-144-3p is limited to
its targeting of RANK to regulate the osteoclast formation process,
and the role of miR-144-3p in the differentiation of BMSCs has not
yet been studied, to the best of our knowledge. Nevertheless, the
regulatory factors upstream of miR-144-3p in the osteogenic
differentiation process have not yet been studied, to the best of
our knowledge.
In previous years, most studies on GATA4 have
examined its role in regulating the differentiation of myocytes
from BMSCs. GATA4 has been revealed to also play an important role
in the regulation of bone metabolism, primarily by regulating the
differentiation of BMSCs into osteoblasts (30,31).
Song et al demonstrated that GATA4 could negatively regulate
osteogenic differentiation in BMSCs (17). Other previous studies have reported
that GATA4 could not only inhibit the expression of RANKL in the
process by which BMSCs differentiate into osteoblasts but also
regulate the differentiation of osteoclasts (32-35).
In addition, GATA4 has been revealed to participate in the
regulation of tooth development by affecting the expression of
osteogenesis regulatory factors and promoting the proliferation of
tooth mesenchymal cells (36).
Zhang et al reported that GATA4 could directly regulate the
miR-144/451 gene cluster in the mechanism of myocardial ischemia
and reperfusion and played a protective role in myocardial cells
(37). To further study the roles
of GATA4 and miR-144-3p in osteogenic differentiation, an
adenovirus infection experiment was designed. Adenovirus carrying
AdGATA4 was used to infect BMSCs, and RT-qPCR detection was
conducted at 48 h after infection. The mRNA expression levels of
AdGATA4 and miR-144-3p were increased compared with those in the NC
group. Then, by dual-luciferase assay, it was revealed that the
GATA4 regulatory site played a positive role in regulating the
regions of the miR-144 promoter at -568 and -969.
In conclusion, our results demonstrated that
miR-144-3p played a negative regulatory role in the osteogenic
differentiation of BMSCs and that GATA4 inhibited osteogenic
differentiation by targeting the expression of miR-144-3p. However,
the downstream mechanism of GATA4-mediated miR-144-3p regulation in
osteogenic differentiation requires further investigation. OP is
caused by the imbalance between bone formation and bone resorption
(38). In the present study, only
osteogenic differentiation was examined, and no relevant studies
were conducted on osteoclast differentiation. Future studies will
include in vivo experiments to investigate the effects of
miR-144-3p on bone formation and bone resorption. Thus, further
in vivo experiments are necessary to verify the clinical
application of miR-144-3p in the treatment of OP.
Supplementary Material
Identification of rat BSMCs by flow
cytometry. As shown, BMSCs were positive for CD44 (95.48%), CD29
(99.31%) and negative for CD45 (0.2%). BMSCs, bone marrow
mesenchymal stem cells.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 82060271),
the Guiyang Science and Technology Bureau of Guizhou Province
(grant no. 2016-100146) and the Science Foundation of Guizhou
Medical University (grant no. 19NSP046).
Availability of data and materials
All data generated and/or analyzed during the
present study are included in the published article.
Authors' contributions
TQ and CW conceived and designed the experiments. TQ
performed the experiments. TQ, HL, TL, LS and CC wrote the
manuscript. HL, TL and CC made substantial contributions to
conception and design, acquisition of data, and analysis and
interpretation of data. LS was involved in drafting the manuscript
and revising it critically for important intellectual content. LS
designed the experiments. CW reviewed and revised the manuscript
for important intellectual content. TQ and CW confirmed the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hendrickx G, Boudin E and Van Hul W: A
look behind the scenes: The risk and pathogenesis of primary
osteoporosis. Nat Rev Rheumatol. 11:462–474. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pouresmaeili F, Kamalidehghan B, Kamarehei
M and Goh YM: A comprehensive overview on osteoporosis and its risk
factors. Ther Clin Risk Manag. 14:2029–2049. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Paspaliaris V and Kolios G: Stem cells in
osteoporosis: From biology to new therapeutic approaches. Stem
Cells Int. 2019(1730978)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Liu J, Wu M, Feng G, Li R, Wang Y and Jiao
J: Downregulation of LINC00707 promotes osteogenic differentiation
of human bone marrow-derived mesenchymal stem cells by regulating
DKK1 via targeting miR-103a-3p. Int J Mol Med. 46:1029–1038.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shenoy A and Blelloch RH: Regulation of
microRNA function in somatic stem cell proliferation and
differentiation. Nat Rev Mol Cell Biol. 15:565–576. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
van Wijnen AJ, van de Peppel J, van
Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, Oursler MJ, Im HJ,
Taipaleenmäki H, Hesse E, et al: MicroRNA functions in osteogenesis
and dysfunctions in osteoporosis. Curr Osteoporos Rep. 11:72–82.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Nugent M: MicroRNA function and
dysregulation in bone tumors: The evidence to date. Cancer Manag
Res. 6:15–25. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Clark EA, Kalomoiris S, Nolta JA and
Fierro FA: Concise review: MicroRNA function in multipotent
mesenchymal stromal cells. Stem Cells. 32:1074–1082.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen H, Ji X, She F, Gao Y and Tang P:
miR-628-3p regulates osteoblast differentiation by targeting RUNX2:
Possible role in atrophic non-union. Int J Mol Med. 39:279–286.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cui X, Wang S, Cai H, Lin Y, Zheng X,
Zhang B and Xia C: Overexpression of microRNA-634 suppresses
survival and matrix synthesis of human osteoarthritis chondrocytes
by targeting PIK3R1. Sci Rep. 6(23117)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen J, Li K, Pang Q, Yang C, Zhang H, Wu
F, Cao H, Liu H, Wan Y, Xia W, et al: Identification of suitable
reference gene and biomarkers of serum miRNAs for osteoporosis. Sci
Rep. 6(36347)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Panach L, Mifsut D, Tarín JJ, Cano A and
García-Pérez M: Serum circulating microRNAs as biomarkers of
osteoporotic fracture. Calcif Tissue Int. 97:495–505.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang C, He H, Wang L, Jiang Y and Xu Y:
Reduced miR-144-3p expression in serum and bone mediates
osteoporosis pathogenesis by targeting RANK. Biochem Cell Biol.
96:627–635. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Laforest B and Nemer M: GATA5 interacts
with GATA4 and GATA6 in outflow tract development. Dev Biol.
358:368–378. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhou P, He A and Pu WT: Regulation of
GATA4 transcriptional activity in cardiovascular development and
disease. Curr Top Dev Biol. 100:143–169. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li Q, Shen P, Zeng S and Liu P: TIEG1
inhibits angiotensin II-induced cardiomyocyte hypertrophy by
inhibiting transcription factor GATA4. J Cardiovasc Pharmacol.
66:196–203. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Song I, Kim K, Kim JH, Lee YK, Jung HJ,
Byun HO, Yoon G and Kim N: GATA4 negatively regulates osteoblast
differentiation by downregulation of Runx2. BMB Rep. 47:463–468.
2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Snykers S, Vanhaecke T and Rogiers V:
Isolation of rat bone marrow stem cells. Methods Mol Biol.
320:265–372. 2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Huang C, Geng J, Wei X, Zhang R and Jiang
S: miR-144-3p regulates osteogenic differentiation and
proliferation of murine mesenchymal stem cells by specifically
targeting Smad4. FEBS Lett. 590:795–807. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yuan Y, Zhang L, Tong X, Zhang M, Zhao Y,
Guo J, Lei L, Chen X, Tickner J, Xu J, et al: Mechanical stress
regulates bone metabolism through microRNAs. J Cell Physiology.
232:1239–1245. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Xie Y, Zhang L, Gao Y, Ge W and Tang P:
The multiple roles of microrna-223 in regulating bone metabolism.
Molecules. 20:19433–19448. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sugimachi K, Matsumura T, Hirata H, Uchi
R, Ueda M, Ueo H, Shinden Y, Iguchi T, Eguchi H, Shirabe K, et al:
Identification of a bona fide microRNA biomarker in serum exosomes
that predicts hepatocellular carcinoma recurrence after liver
transplantation. Br J Cancer. 112:532–538. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Witwer KW: Circulating microRNA biomarker
studies: Pitfalls and potential solutions. Clin Chem. 61:56–63.
2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang X, Guo B, Li Q, Peng J, Yang Z, Wang
A, Li D, Hou Z, Lv K, Kan G, et al: miR-214 targets ATF4 to inhibit
bone formation. Nat Med. 19:93–100. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Matsumura T, Sugimachi K, Iinuma H,
Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano
Y, et al: Exosomal microRNA in serum is a novel biomarker of
recurrence in human colorectal cancer. Br J Cancer. 113:275–281.
2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Huang K, Fu J, Zhou W, Li W, Dong S, Yu S,
Hu Z, Wang H and Xie Z: MicroRNA-125b regulates osteogenic
differentiation of mesenchymal stem cells by targeting Cbfβ in
vitro. Biochimie. 102:47–55. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li Z, Zhang W and Huang Y: miRNA-133a is
involved in the regulation of postmenopausal osteoporosis through
promoting osteoclast differentiation. Acta Biochim Biophys Sin
(Shanghai). 50:273–280. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen T, Che X, Han P, Lu J, Wang C, Liang
B, Hou Z, Wei X, Wei L and Li P: MicroRNA-1 promotes cartilage
matrix synthesis and regulates chondrocyte differentiation via
post-transcriptional suppression of Ihh expression. Mol Med Rep.
22:2404–2414. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhong L, Chiusa M, Cadar AG, Lin A,
Samaras S, Davidson JM and Lim CC: Targeted inhibition of ANKRD1
disrupts sarcomeric ERK-GATA4 signal transduction and abrogates
phenylephrine-induced cardiomyocyte hypertrophy. Cardiovasc Res.
106:261–271. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Song I, Jeong BC, Choi YJ, Chung YS and
Kim N: GATA4 negatively regulates bone sialoprotein expression in
osteoblasts. BMB Rep. 49:343–348. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Khalid AB, Slayden AV, Kumpati J, Perry
CD, Berryhill SB, Crawford JA, Fatima I, Morselli M, Pellegrini M,
Miranda-Carboni GA and Krum SA: GATA4 represses RANKL in
osteoblasts via multiple long-range enhancers to regulate
osteoclast differentiation. Bone. 116:78–86. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Khalid AB, Slayden AV, Kumpati J, Perry
CD, Osuna MAL, Arroyo SR, Miranda-Carboni GA and Krum SA: GATA4
directly regulates runx2 expression and osteoblast differentiation.
JBMR Plus. 2:81–91. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Miranda-Carboni GA, Guemes M, Bailey S,
Anaya E, Corselli M, Peault B and Krum SA: GATA4 regulates estrogen
receptor-alpha-mediated osteoblast transcription. Mol Endocrino.
25:1126–1136. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Güemes M, Garcia AJ, Rigueur D, Runke S,
Wang W, Zhao G, Mayorga VH, Atti E, Tetradis S, Péault B, et al:
GATA4 is essential for bone mineralization via ERα and TGFβ/BMP
pathways. J Bone Miner Res. 29:2676–2687. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Guo S, Zhang Y, Zhou T, Wang D, Weng Y,
Wang L and Ma J: Role of GATA binding protein 4 (GATA4) in the
regulation of tooth development via GNAI3. Sci Rep.
7(1534)2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu
WT, Jegga AG and Fan GC: Synergistic effects of the GATA-4-mediated
miR-144/451 cluster in protection against simulated
ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell
Cardiol. 49:841–850. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kureel J, Dixit M, Tyagi AM, Mansoori MN,
Srivastava K, Raghuvanshi A, Maurya R, Trivedi R, Goel A and Singh
D: miR-542-3p suppresses osteoblast cell proliferation and
differentiation, targets BMP-7 signaling and inhibits bone
formation. Cell Death Dis. 5(e1050)2014.PubMed/NCBI View Article : Google Scholar
|