Introduction
Liver cancer ranks sixth among the most common
cancer types globally and 905,677 new cases were diagnosed in
2020(1). It is also one of the
three major causes of cancer-related death, seriously threatening
the health and safety of individuals worldwide, particularly in
China and East Asia (2,3). Research indicated that liver cancer
is mainly caused by long-term alcohol consumption, obesity,
exposure to poisons, hepatitis C virus and, importantly, hepatitis
B virus (HBV) infection (4). HBV
is a hepatotropic DNA virus with carcinogenic effects. Patients
infected with HBV may develop serious diseases such as hepatitis,
cirrhosis and even liver cancer. According to reports, at least
50-60% of liver cancer cases worldwide are caused by persistent HBV
infection (5). Furthermore, due to
the high replication level and mutation rate of HBV, the
post-operative effects in HBV-positive patients with liver cancer
are not ideal (6). Therefore, it
is necessary to further analyze the occurrence and development
mechanisms in HBV-positive liver cancer to identify novel
therapeutic targets.
Circular RNAs (circRNAs) are newly discovered
endogenous non-coding RNAs (ncRNAs) with a covalent closed-loop
structure that are able to regulate gene transcription and are
widely found in the cytoplasm of eukaryotic animals. As it contains
neither 5' caps nor 3' polyadenylated tails, is not sensitive to
RNase R digestion and is more stable than linear RNA (7). In recent years, a large body of
experimental evidence has pointed out that the abnormal expression
of circRNAs is closely related to various diseases, including
cancer. CircRNAs have a dual regulatory role in cancer development,
both as tumor suppressors (8) and
as proto-oncogenes (9). In
addition, certain circRNAs have been proven to actively participate
in the pathogenesis of HBV-positive liver cancer, such as
circ_0009582(10) and
circRNA_101764(11). However,
studies on circRNAs in HBV-positive liver cancer remain limited and
further exploration is required.
In terms of biological functions, circRNAs regulate
gene expression and protein translation by binding to RNA-binding
proteins (12). For instance, in
glioblastoma multiforme, circSMARCA5 interacts with SRSF1,
negatively controls its expression and ultimately affects
angiogenesis and VEGFA mRNA splicing (13). Furthermore, circRNAs may function
as important competitive endogenous RNAs (ceRNAs) through microRNA
(miRNA) response elements (MREs) to competitively adsorb miRNA and
affect mRNA expression. For instance, the level of circRNA-UBE2G1
is significantly increased in osteoarthritis tissues and combines
with miR-37 as a ceRNA to increase the expression of HIF-1α and
promote the development of osteoarthritis (14). CircHDAC9 participates in the
occurrence and development of Alzheimer's disease by targeting the
miR-138/Sirt1 pathway (15);
circFAT1(e2) is able to act as a ‘sponge’ for miR-181b, inhibiting
its expression, leading to HK2 imbalance and promoting osteosarcoma
metastasis (16). However, the
expression profile of dysregulated circRNAs in HBV-positive liver
cancer and its underlying mechanisms remain to be fully
elucidated.
In the present study, 1,493 differentially expressed
circRNAs in HBV-positive liver cancer cells were screened through
microarray analysis and the specific functions of differentially
expressed circRNAs were analyzed through bioinformatics. Among the
differentially expressed circRNAs, hsa_circ_0066966 was
significantly increased in HBV-positive liver cancer cells.
Finally, it was demonstrated that abnormal expression of
hsa_circ_0066966 not only promoted the proliferation of
HBV-positive liver cancer cells but also positively regulated their
migration, which provided a novel perspective for the study of the
mechanisms involved in HBV-positive liver cancer.
Materials and methods
Cell lines and culture
The four liver cancer cell lines used for the
experimental part of the present study were all obtained from the
Chinese Academy of Sciences, including two HBV-positive liver
cancer cell lines (HepG2.2.15 and Hep3B) and two HBV-negative liver
cancer cell lines (HepG2 and Huh7). All of the 4 cell lines had
been authenticated by short tandem repeat profiling. According to
the supplier's instructions, the cells were cultured in RPM1-1640
medium with 10% fetal bovine serum (FBS), 1% penicillin and 1%
streptomycin (all from Invitrogen; Thermo Fisher Scientific, Inc.)
at 37˚C in an incubator with 5% CO2.
Microarray analysis
The HBV-positive liver cancer cell lines HepG2.2.15
and Hep3B and the two HBV-negative liver cancer cell lines HepG2
and Huh7 were selected for the experiments performed in parallel.
One experimental replicate was used for each cell line. In short,
total RNA was isolated using the RNeasy Total RNA Isolation Kit
(Qiagen GmbH) according to the manufacturer's protocol and purified
by using an RNeasy Mini Kit (Qiagen GmbH). RNA samples were then
used to generate biotinylated cRNA targets for the human ceRNA
array v3.0 (Sinotech Genomics). The biotinylated cRNA targets were
then hybridized with the slides. After hybridization, slides were
scanned on the Agilent Microarray Scanner (Agilent Technologies,
Inc.). Data were extracted with Feature Extraction software 10.7
(Agilent Technologies, Inc.). Raw data were normalized by a
Quantile algorithm with the R package ‘limma’. The circRNA data
were then extracted from all of the normalized data. The
dysregulated circRNAs in HBV-positive liver cancer cells and
HBV-negative liver cancer cells were screened using a threshold of
absolute fold-change [FC (abs)] ≥2 and the resulting differentially
expressed circRNAs were used for subsequent analysis. The original
data of the microarray analysis were deposited as a Gene Expression
Omnibus (GEO) dataset under the accession no. GSE181988 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181988).
Annotation of circRNA/miRNA
interactions
Arraystar's homemade miRNA target prediction
software based on TargetScan (17)
and miRanda (18) was employed to
predict miRNAs that may interact with circRNAs. The output from the
miRNA target prediction software also detailed annotations
regarding circRNA/miRNA interactions.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses
In order to further explore potential molecular
mechanisms involving abnormal expression of circRNAs, GO and KEGG
enrichment analyses of host genes of dysregulated circRNAs were
performed using the R package clusterProfiler. P<0.05 was
considered to indicate statistically significant enrichment of
these genes.
Reverse transcription-quantitative PCR
(RT-qPCR)
The levels of circRNAs in liver cancer cells were
detected by RT-qPCR, with GAPDH used as an internal reference gene
transcript. TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to extract total RNA from the four liver cancer cell
lines as per the manufacturer's instructions, followed by RT of RNA
into cDNA using the PrimeScript RT kit (Takara Bio, Inc.).
Subsequently, according to the manufacturer's protocol, SYBR Premix
Ex Taq™ (Takara Bio, Inc.) was used to perform qPCR on an Applied
Biosystems 7500 Real-Time PCR platform (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using the following thermocycling
conditions: Initial denaturation at 95˚C for 5 min, followed by 40
cycles of 95˚C for 5 sec and 60˚C for 1 min. Finally, the
expression levels of circRNAs in liver cancer cells were identified
using the 2-ΔΔCq method (19). The PCR primer sequences for the 10
circRNAs are listed in Table
SI.
Cell transfection
Small interfering RNA (siRNA) against
hsa_circ_0066966 (si-hsa_circ_0066966) and negative control siRNA
(si-NC) were acquired from GenePharma. hsa_circ_0066966 expression
plasmid (named as hsa_circ_0066966) and its corresponding negative
control (named NC) were obtained from Guangzhou RiboBio Co., Ltd.
The liver cancer cells were first seeded in a 6-well plate and then
cultured until 60-70% confluent. Next, the plasmids and
oligonucleotides were transfected into HBV-negative liver cancer
cells and HBV-positive liver cancer cells using Lipofectamine
2000® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) following the manufacturer's protocols. The cells were
harvested 48 h later and overexpression and interference efficiency
were tested by RT-qPCR.
MTT assay
Cell proliferation ability after transfection was
detected using the MTT assay (Invitrogen; Thermo Fisher Scientific,
Inc.). In brief, transfected cells were seeded into a 96-well
flat-bottom plate at a density of 4x103 cells/well and
incubated for 24, 48, 72 or 96 h, followed by treatment with MTT at
37˚C under 5% CO2 for 4 h. Finally, the medium in each
well was removed and dimethyl sulfoxide was added. The optical
absorbance at 492 nm was recorded using an ELX-800 University
Microplate Reader (BioTek, Inc.).
Cell migration assay
Cell migration was detected using
Transwell® assays. After transfection, 5x104
cells were resuspended in 200 µl RPMI 1640 medium without FBS and
then cultured in the upper Transwell chambers (8 µm pore size; EMD
Millipore). At the same time, RPMI 1640 medium with 10% FBS (as a
chemoattractant) was placed into the lower Transwell chambers.
After incubating the chamber at 37˚C and 5% CO2 for 24
h, liver cancer cells in the upper chamber were removed and cells
that had migrated to the bottom of the membrane were stained with
0.1% crystal violet (20% methanol) for 15 min at room temperature.
Finally, the cells were imaged using a microscope (Olympus
Corporation) and five fields were randomly selected for cell
counting.
Statistical analysis
SPSS v.21.0 (IBM Corporation) was used for data
analysis. Values are expressed as the mean ± standard deviation.
Differences between the two groups were analyzed using Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Differentially expressed circRNAs
between HBV-positive liver cancer cells and HBV-negative liver
cancer cells
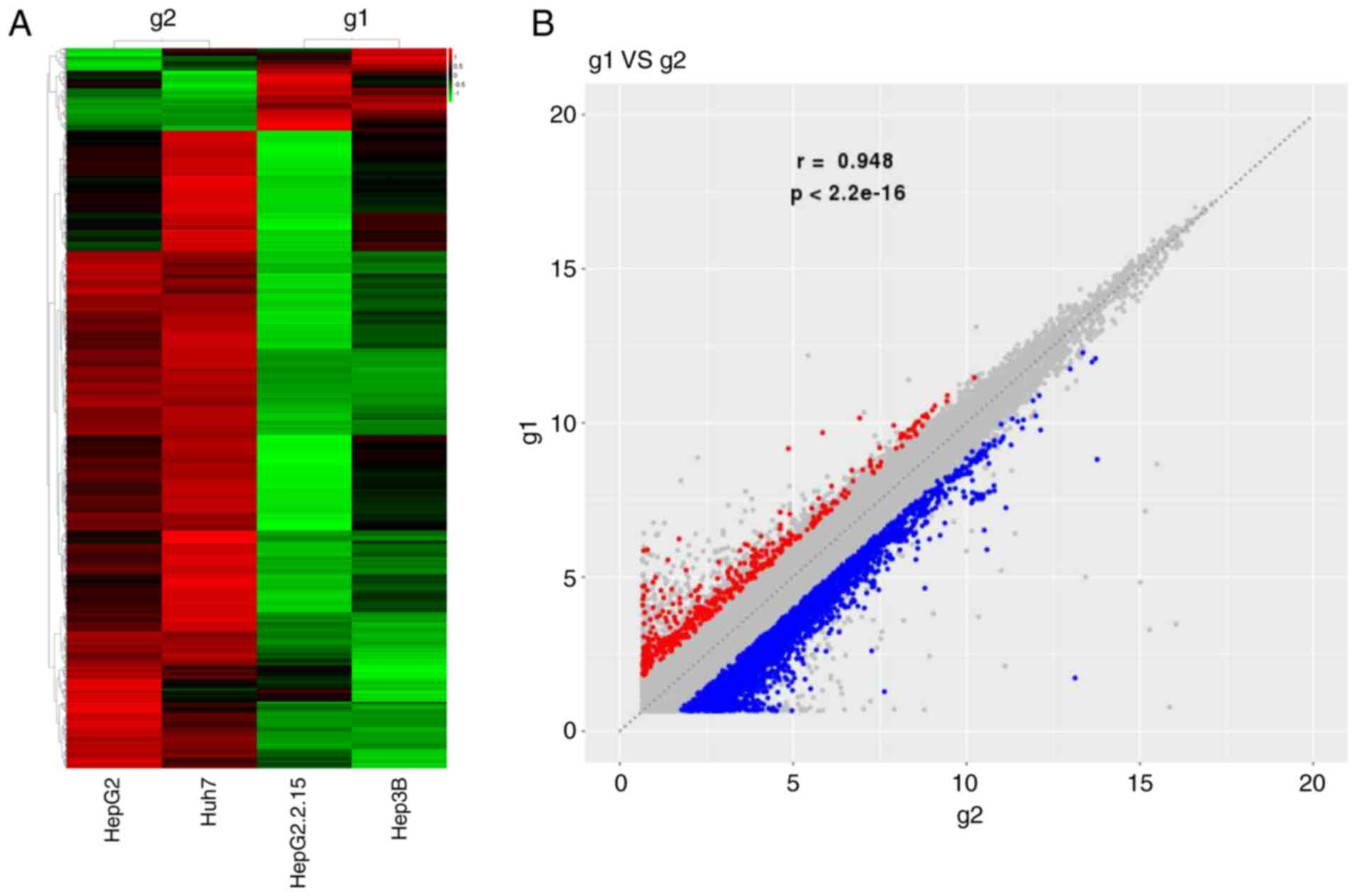
Heat-map analysis indicated that, compared with
HBV-negative liver cancer cells, a total of 1,493 abnormally
expressed circRNAs [FC (abs) ≥2] were present in HBV-positive liver
cancer cells, of which 171 were upregulated and 1,322 were
significantly decreased (Fig. 1).
In the present experiments, two types of liver cancer cell lines
were used in the HBV-positive/negative liver cancer cell groups,
hence it was not possible to obtain P-values involving changes in
circRNA expression. Table I lists
detailed information concerning the top-10 dysregulated circRNAs in
HBV-positive liver cancer cells and their cognate genes. Among
these, hsa_circ_0079954 exhibited the greatest level of
upregulation [FC (abs)=34.087], while hsa_circ_0030525 was the most
downregulated circRNA [FC (abs)=81.771]. Their host genes were HECW
and MYCBP2, respectively. In addition, the cognate gene of
hsa_circ_0066966, golgin B1 (GOLGB1), and the host gene of
hsa_circ_0088524, GOLGA1, are both Golgi-related proteins.
 | Table ITop 10 dysregulated circRNAs in
hepatitis B virus-positive liver cancer cells. |
Table I
Top 10 dysregulated circRNAs in
hepatitis B virus-positive liver cancer cells.
| CircRNA |
Log2FC | FC | FC abs | Direction of
dysregulation | Host gene |
|---|
|
hsa_circ_0030525 | -6.354 | 0.012 | 81.771 | Down | MYCBP2 |
|
hsa_circ_0079954 | 5.091 | 34.087 | 34.087 | Up | HECW1 |
|
hsa_circ_0090095 | -4.953 | 0.032 | 30.970 | Down | ACOT9 |
|
hsa_circ_0060534 | 4.306 | 19.787 | 19.787 | Up | SLPI |
|
hsa_circ_0032138 | -4.163 | 0.056 | 17.911 | Down | HIF1A |
|
hsa_circ_0066966 | 3.950 | 15.460 | 15.460 | Up | GOLGB1 |
|
hsa_circ_0085289 | -3.889 | 0.068 | 14.812 | Down | CTHRC1 |
|
hsa_circ_0088524 | 3.876 | 14.682 | 14.682 | Up | GOLGA1 |
|
hsa_circ_0062852 | -3.858 | 0.069 | 14.498 | Down | MORC2 |
|
hsa_circ_0091095 | -3.666 | 0.079 | 12.696 | Down | ATRX |
Potential circRNA/miRNA
interactions
To investigate the possible mechanisms involved in
differentially expressed circRNAs in HBV-positive liver cancer
cells, the MREs of the top-10 dysregulated circRNAs in HBV-positive
liver cancer cells were predicted using the miRNA target prediction
software from Arraystar Inc. based on TargetScan and miRanda and
the top-10 dysregulated circRNAs were annotated in detail using the
circRNA/miRNA interaction information (Table II). According to the results, the
most upregulated circRNA, hsa_circ_0079954, targeted the MREs of
the following miRNAs: hsa-miR-383-3p, hsa-miR-7161-3p,
hsa-miR-3065-3p, hsa-miR-127-5p and hsa-miR-4291. Similarly, the
most downregulated circRNA, hsa_circ_0030525, targeted the MREs of
the following miRNAs: hsa-miR-391, hsa-miR-487a-5p, hsa-miR-27b-3p,
hsa-miR-1273g-5p and hsa-miR-199a-5p. It is also worth noting that
both hsa_circ_0066966 and hsa_circ_0088524 interacted with
hsa-miR-3619-5p.
 | Table IIMREs of top 10 dysregulated circRNAs
in hepatitis B virus-positive liver cancer cells. |
Table II
MREs of top 10 dysregulated circRNAs
in hepatitis B virus-positive liver cancer cells.
| CircRNA | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|
|
hsa_circ_0030525 | hsa-miR-3918 |
hsa-miR-487a-5p | hsa-miR-27b-3p |
hsa-miR-1273g-5p |
hsa-miR-199a-5p |
|
hsa_circ_0079954 | hsa-miR-383-3p |
hsa-miR-7161-3p |
hsa-miR-3065-3p | hsa-miR-127-5p | hsa-miR-4291 |
|
hsa_circ_0090095 |
hsa-miR-7153-5p | hsa-miR-8070 |
hsa-miR-3925-5p |
hsa-miR-455-3p.1 | hsa-miR-212-3p |
|
hsa_circ_0060534 | hsa-miR-4480 |
hsa-miR-6791-3p |
hsa-miR-4776-5p | hsa-miR-6165 |
hsa-miR-664a-3p |
|
hsa_circ_0032138 | hsa-miR-5708 | hsa-miR-22-5p |
hsa-miR-6504-5p |
hsa-miR-664a-5p | hsa-miR-149-5p |
|
hsa_circ_0066966 | hsa-miR-214-3p | hsa-miR-922 | hsa-miR-646 |
hsa-miR-3619-5p |
hsa-miR-374a-3p |
|
hsa_circ_0085289 | hsa-miR-338-3p | hsa-miR-29a-3p |
hsa-miR-5586-5p |
hsa-miR-6507-3p | hsa-miR-29c-3p |
|
hsa_circ_0088524 | hsa-miR-3689e |
hsa-miR-6740-3p |
hsa-miR-6809-3p |
hsa-miR-3619-5p |
hsa-miR-3689b-5p |
|
hsa_circ_0062852 | hsa-miR-4277 |
hsa-miR-514b-3p | hsa-miR-584-3p |
hsa-miR-514a-3p | hsa-miR-409-3p |
|
hsa_circ_0091095 | hsa-miR-377-3p | hsa-miR-3133 | hsa-miR-4511 |
hsa-miR-5195-3p | hsa-miR-145-5p |
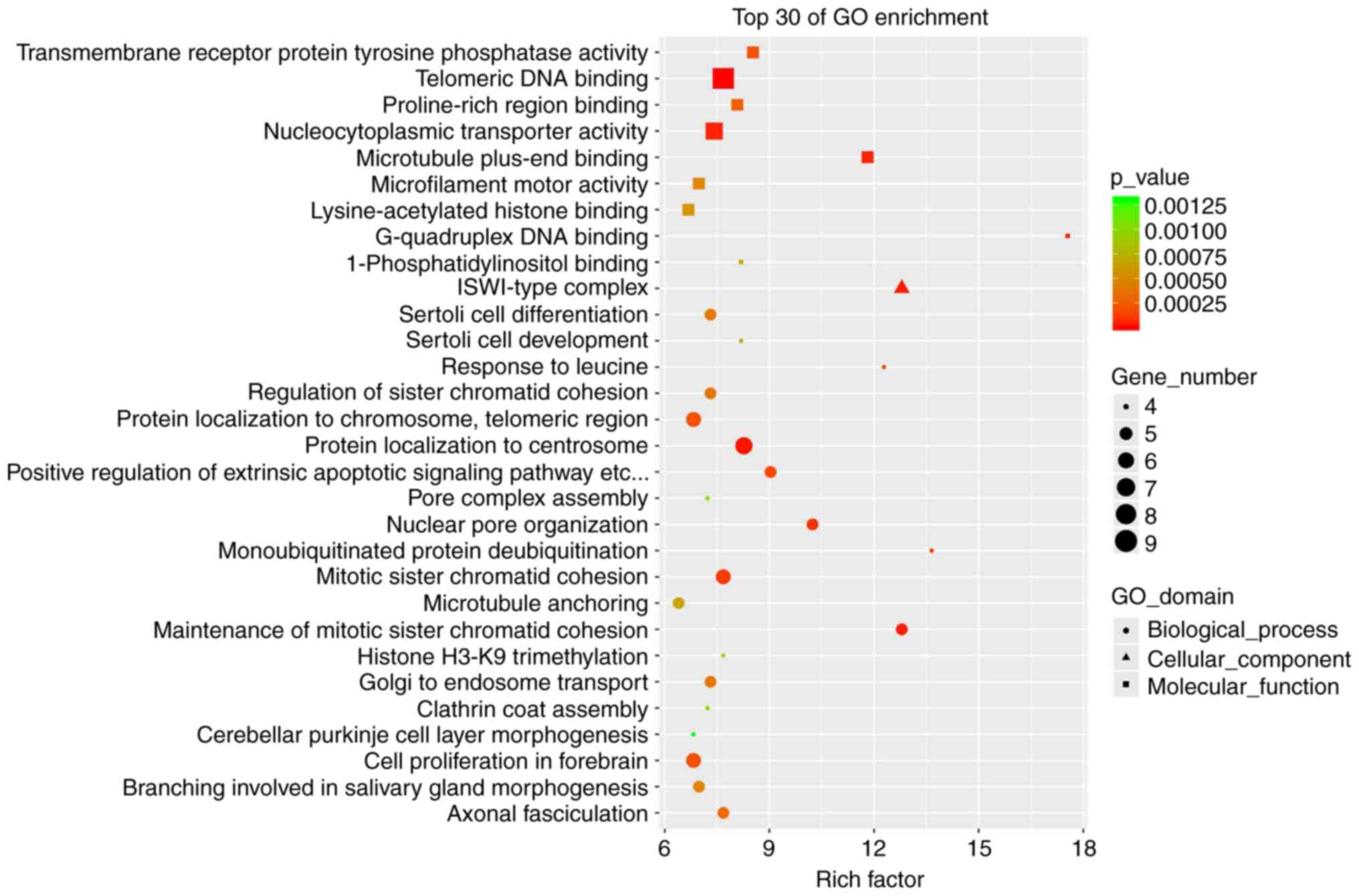
GO enrichment analysis of genes
encoding dysregulated circRNAs
In order to annotate and classify circRNAs and to
further understand the important biological roles of abnormally
expressed circRNAs, GO enrichment analysis was performed on the
cognate genes of differentially expressed circRNAs. Fig. 2 lists the top-30 enriched GO terms.
GO analysis suggested that in the category molecular function,
transmembrane receptor protein tyrosine phosphatase activity,
telomeric DNA binding, proline-rich region binding,
nucleocytoplasmic transporter activity and microtubule plus-end
binding were the most enriched terms. In the category biological
process, genes were mainly enriched in Sertoli cell differentiation
and development, responses to leucine, regulation of sister
chromatid cohesion and protein localization to chromosomes and
telomeric regions. In the category cellular component, genes were
mainly enriched in the ISWI-type complex (Fig. 2).
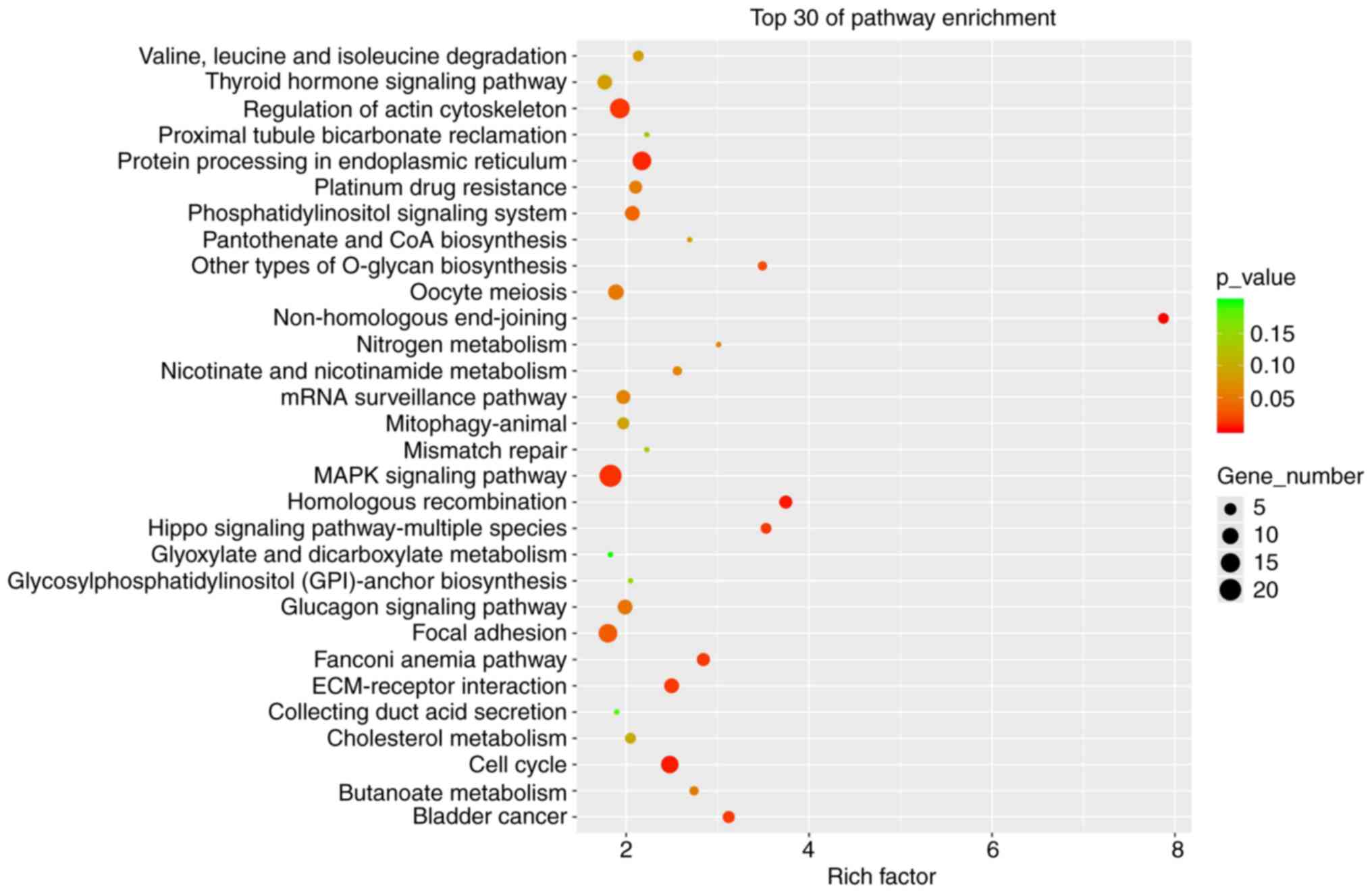
KEGG pathway enrichment analysis of
dysregulated circRNA cognate genes
KEGG pathway enrichment analysis was performed on
the cognate genes of dysregulated circRNAs to further explore the
signaling pathways that differentially expressed circRNAs may be
involved in. Fig. 3 lists the
top-30 pathways of the KEGG enrichment. The results indicated that
pathways such as non-homologous end-joining, homologous
recombination, Hippo signaling pathway in multiple species, cell
cycle and MAPK signaling were significantly enriched. Among these,
the cell cycle, MAPK and Hippo signaling pathways are related to a
variety of cancers. In addition, pathways directly related to
bladder cancer were also enriched (Fig. 3), which implies that the
differentially expressed circRNAs identified have a high functional
correlation with cancer.
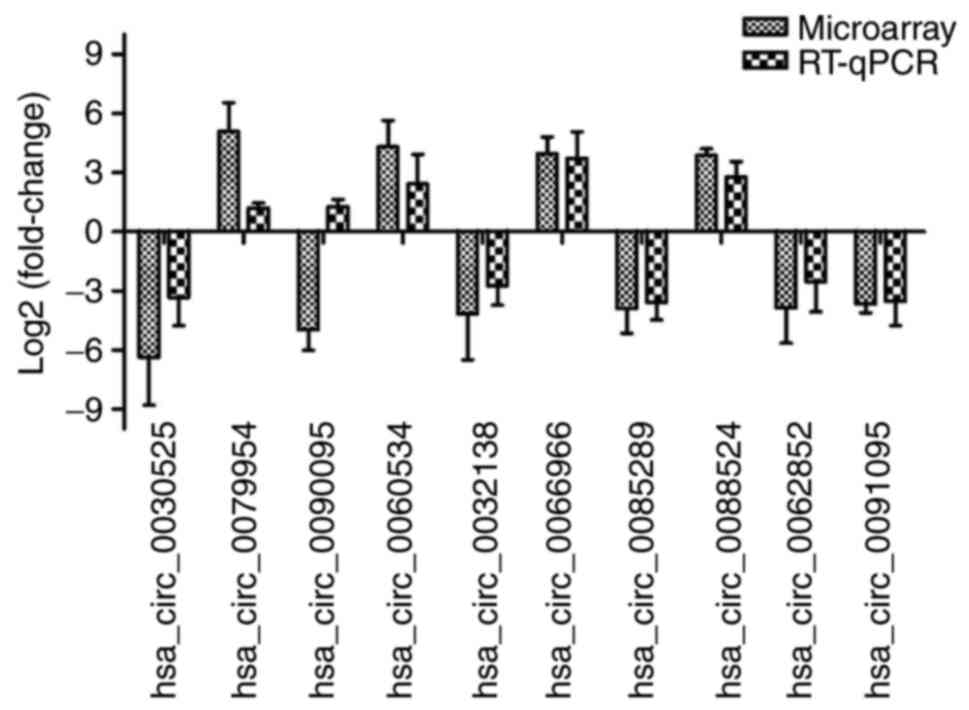
RT-qPCR validation of the top-10
dysregulated circRNAs in HBV-positive liver cancer cells
To further support the reliability of the microarray
results, the expression of the top-10 dysregulated circRNAs in
HBV-positive liver cancer cells was verified by RT-qPCR. The
results indicated that only the expression trend of
hsa_circ_0090095 was opposite to the microarray results. The
expression trends of the remaining 9 circRNAs (hsa_circ_0030525,
hsa_circ_0079954, hsa_circ_0060534, hsa_circ_0032138,
hsa_circ_0066966, hsa_circ_0085289, hsa_circ_0088524,
hsa_circ_0062852 and hsa_circ_0091095) were similar to their
microarray results, and the values of Log2FC of the
circRNAs were not significantly different between the RT-qPCR and
microarray data (P>0.05), indicating that the microarray
analysis results were reliable. In addition, the absolute value of
Log2FC of hsa_circ_0066966 was the highest (Fig. 4), and thus, this circRNA was used
for further research.
Knockdown of hsa_circ_0066966 in
HBV-positive liver cancer cells inhibits cellular proliferation and
migration
hsa_circ_0066966 (chr3:121441108-121448187) was
derived from the GOLGB1 gene. The function of hsa_circ_0066966 in
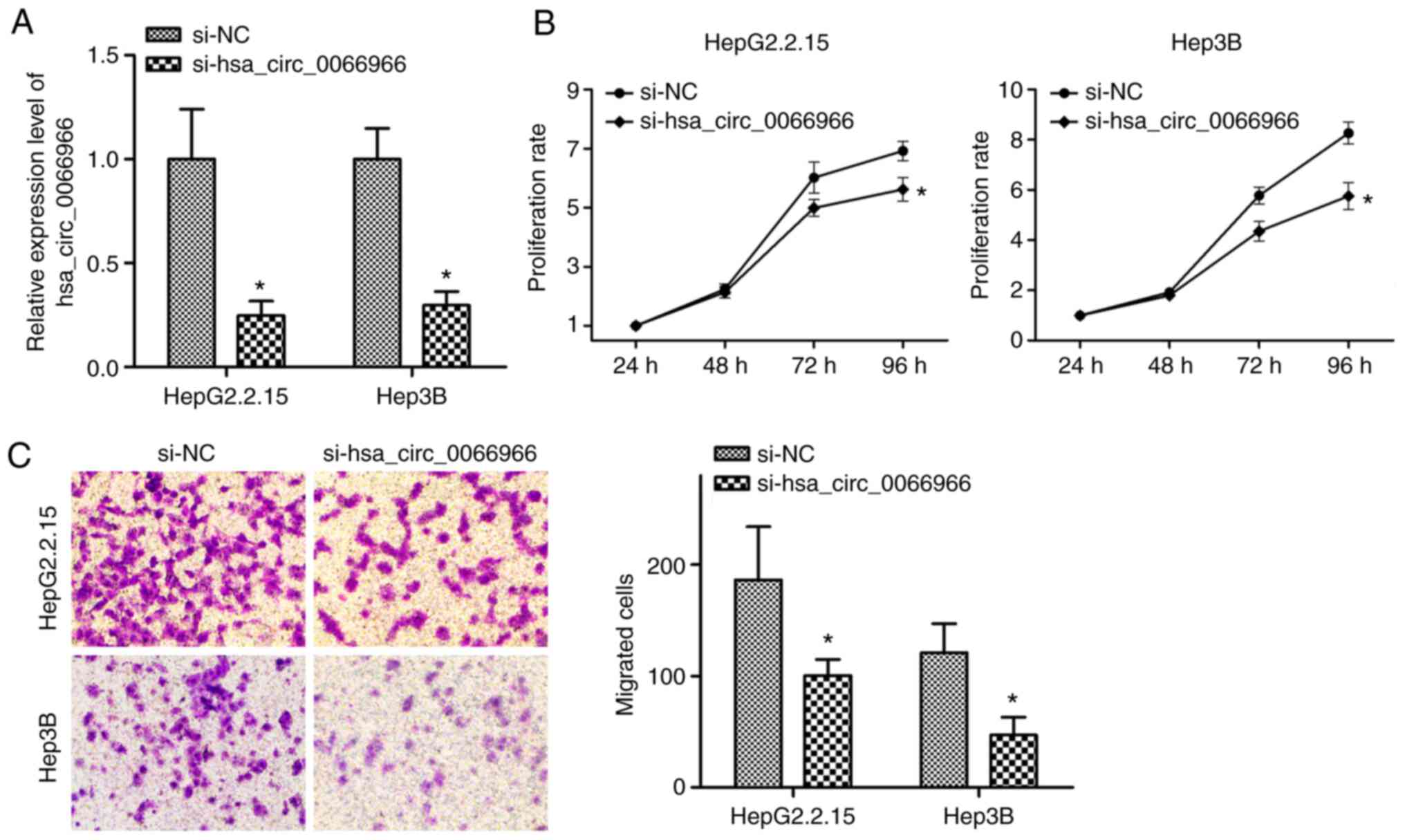
HBV-positive liver cancer cells was explored. Si-hsa_circ_0066966
was transfected into the corresponding liver cancer cells and
RT-qPCR was used to verify the knockdown efficiency. The results
indicated that si-hsa_circ_0066966 transfection significantly
reduced hsa_circ_0066966 levels in HepG2.2.15 and Hep3B cells
(P<0.05; Fig. 5A). Next, MTT
assays were performed and it was revealed that in the two
HBV-positive liver cancer cell lines, hsa_circ_0066966 silencing
significantly inhibited cell proliferation at 96 h (P<0.05;
Fig. 5B). The experiment was
followed by Transwell assays, which indicated that, compared with
the si-NC group, hsa_circ_0066966 silencing significantly inhibited
the migration ability of HepG2.2.15 and Hep3B cells (P<0.05;
Fig. 5C). The above results
demonstrated that knockdown of hsa_circ_0066966 not only inhibited
the proliferation of HBV-positive liver cancer cells but also
affected their migration.
Overexpression of hsa_circ_0066966 in
HBV-negative liver cancer cells promotes cellular proliferation and
migration
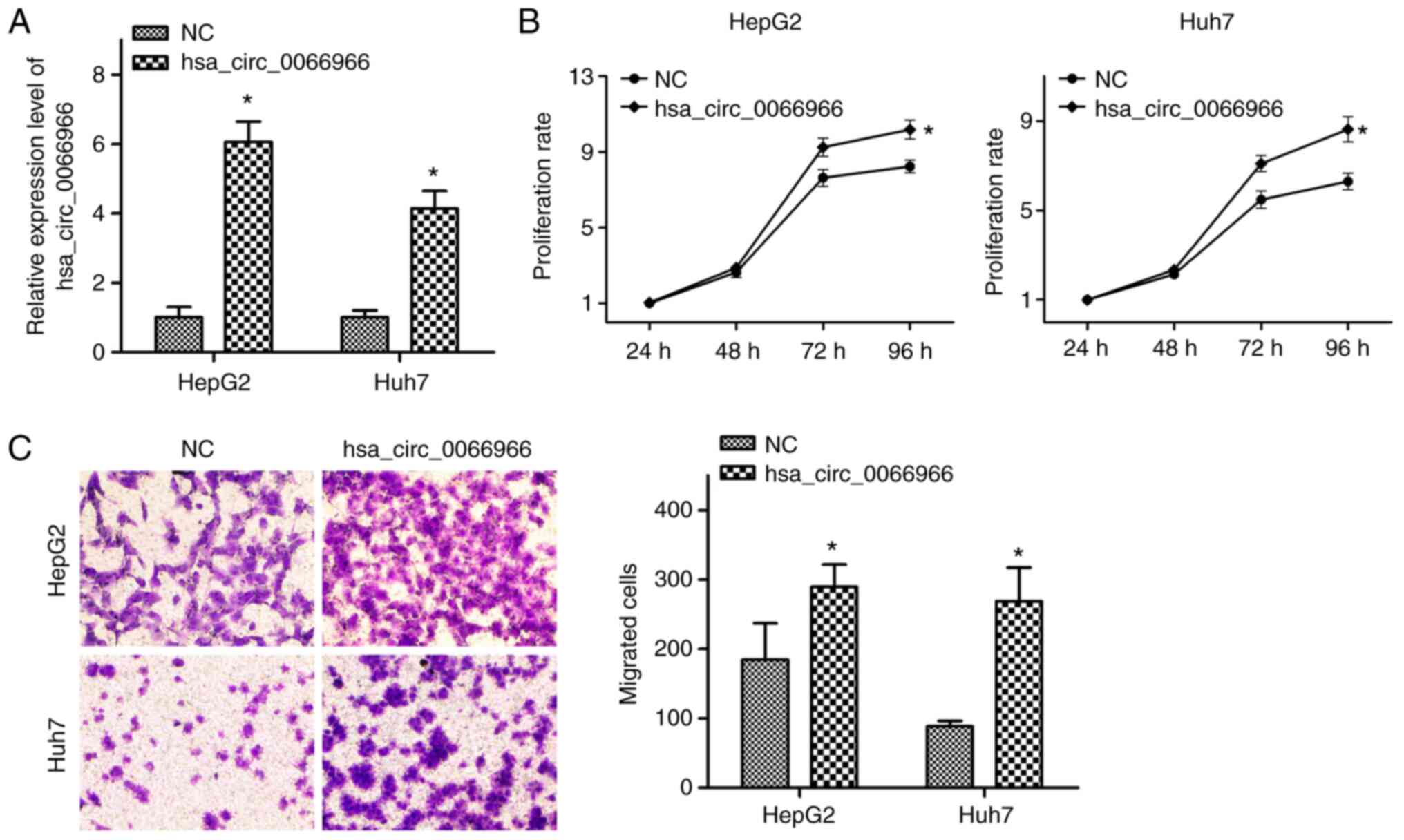
Finally, in order to explore whether
hsa_circ_0066966 has a role in HBV-negative liver cancer cells, an
hsa_circ_0066966 overexpression plasmid was constructed. The
results confirmed that after cells were transfected with
hsa_circ_0066966 plasmid, the levels of hsa_circ_0066966 increased
significantly (P<0.05; Fig.
6A). According to the results of the MTT assays,
hsa_circ_0066966 overexpression in HepG2 and Huh7 cells
significantly promoted the proliferative capacity at 96 h
(P<0.05; Fig. 6B). It was then
explored whether hsa_circ_0066966 is able to regulate the migration
of HBV-negative liver cancer cells. The Transwell assays indicated
that, compared with the NC group, the migration ability of
HBV-negative liver cancer cells in the hsa_circ_0066966 group was
significantly enhanced (P<0.05; Fig. 6C). In conclusion, overexpression of
hsa_circ_0066966 had a significant stimulatory effect on the
proliferation and migration of HBV-negative liver cancer cells.
Discussion
CircRNAs are a type of stable ncRNA. Continuous
research has pointed out that circRNAs have vital roles in a
considerable number of diseases, particularly cancer. For instance,
hsa_circRNA_000166 has a significant promoting effect on migration
and invasion by colon cancer cells (20) and hsa_circ_0131242 positively
regulates the occurrence of triple-negative breast cancer through
sponging hsa-miR-2682(21). In
addition, other studies further demonstrated that circCYFIP2
affects the level of E2F1 by regulating miR-1205 and ultimately
mediates metastasis in gastric cancer (22). Due to the gradual increase in
morbidity and mortality of HBV-positive liver cancer cases, there
is an urgent requirement to identify key targets in the
pathogenesis of HBV-positive liver cancer. Therefore, exploring the
expression of circRNAs in HBV-positive liver cancer may further
augment therapeutic approaches. In the present experiments, by
determining the expression of dysregulated circRNAs in HBV-positive
liver cancer cells through microarray analysis, a total of 1,493
differentially expressed circRNAs [FC (abs) ≥2] were identified, of
which 171 were significantly increased; the remaining 1,322 were
abnormally decreased, indicating that abnormal expression of
circRNAs was significantly associated with the occurrence of
HBV-positive liver cancer.
In the microarray analysis, only two samples were
included in each group, unlike the usually used 3 samples or 3
repeats. In fact, it was intended to compare 3 types of
HBV-positive liver cancer cells with 3 HBV-negative liver cancer
cells, but the third HBV-positive liver cancer cell line was not
possible to obtain. In addition, compared to using three replicate
samples, using two different samples in each group improves the
screening of those circRNAs exerting important oncogenic roles.
Therefore, two different cell lines were used for microarray in
each group.
As is widely known, circRNAs with MREs are able to
bind and adsorb the corresponding miRNA molecules through
competitive binding, thereby exerting a ‘sponging’ effect to reduce
the negative regulation of miRNA on gene expression and thus
regulating the expression of target genes (12). For instance, circRNA 000554 has an
inhibitory effect on epithelial-mesenchymal transition in breast
cancer and regulates the level of ZFP36 via its sponging effect on
miR-182(23). Furthermore,
circPACRGL, derived from exosomes, regulates the expression of
TGF-β1 through competitive binding with miR-142-3p and miR-506-3p,
and promotes colorectal cancer progression (24). In the present study, the five MREs
of the top-10 differentially expressed circRNAs were predicted by
bioinformatics analysis. Of note, hsa_circ_0066966 and
hsa_circ_0088524 shared the same MRE, hsa-miR-3619-5p, which has
been proven to have tumor-suppressive effects in
cisplatin-resistant cutaneous squamous cell carcinoma and bladder
cancer (25,26). Furthermore, Tan et al
(27) predicted that circZFR may
be used as a ‘sponge’ for hsa-miR-3619-5p and promote the
development of liver cancer. Hence, it was speculated that
hsa_circ_0066966 and hsa_circ_0088524 simultaneously competitively
bind hsa-miR-3619-5p to inhibit its expression and accelerate the
processes of HBV-positive liver cancer. In addition, two other
MREs, hsa-miR-214-3p and hsa-miR-646, of hsa_circ_0066966 were also
reported to be closely involved in liver cancer progression. For
instance, hsa-miR-214-3p has been indicated to be sponged by lncRNA
HOXA11-AS (28) and
hsa_circ_0008450(29), which has
an oncogenic role in liver cancer tumorigenesis, and ectopic
expression of hsa-miR-214-3p suppressed liver cancer cell
proliferation and invasion (28).
Another study indicated that hsa-miR-646 is an MRE of circ_0000267
and co-transfection with circ_0000267 and miR-646 significantly
reversed the oncogenic biological behavior of liver cancer cells
induced by circ_0000267(30).
Therefore, the two MREs may be a potential means for
hsa_circ_0066966 to exert its oncogenic role in HBV-positive liver
cancer.
To further clarify the role of differentially
expressed circRNAs in HBV-positive liver cancer, GO and KEGG
pathway enrichment analysis was performed. GO enrichment analysis
revealed that the most significantly enriched items were Sertoli
cell differentiation and development and telomeric DNA binding,
indicating that the differentially expressed circRNAs may
participate in these biological functions. In addition, KEGG
pathway enrichment analysis revealed that pathways related to
cancer were significantly enriched, such as the MAPK and Hippo
signaling pathways. The triggering of liver cancer involves
activation of the MAPK signaling pathway (31). Furthermore, the Hippo signaling
pathway was reported to have a certain relationship with the
occurrence and development of liver cancer (32). Another study indicated that the
circular RNA LPAR3 affected the migration of esophageal squamous
cell carcinoma cells by activating the MAPK signaling pathway,
while regulating cellular invasion (33). At the same time, hsa_circ_0128846
regulates the MST1, LATS1 and YAP proteins in the Hippo signaling
pathway, ultimately promoting the progression of colorectal cancer
(34). In the present study, the
MAPK and Hippo signaling pathways significantly enriched cognate
circRNA genes with dysregulated expression, indicating that circRNA
may have a role in the progression and prognosis of HBV-positive
liver cancer by regulating the MAPK and Hippo pathways.
Next, the expression of the top-10 dysregulated
circRNAs in HBV-positive liver cancer cells was verified. The
RT-qPCR results indicated that among the top-10 dysregulated
circRNAs identified by microarray analysis, only the expression
trend of hsa_circ_0090095 was opposite to the microarray results,
suggesting that the microarray results were reliable. It also
determined that hsa_circ_0066966 had the highest absolute value of
Log2FC, with significantly increased levels in
HBV-positive liver cancer cells. Although there is no relevant
literature regarding the function of hsa_circ_0066966, GOLGB1, the
gene encoding hsa_circ_0066966, has been proven to positively
influence the occurrence and development of liver cancer (35). Therefore, hsa_circ_0066966 was
speculated to regulate the progression of liver cancer. CircRNAs
have been demonstrated to regulate the proliferation and migration
of cancer cells to varying degrees. For instance,
hsa-circRNA-103809 promoted the proliferation of liver cancer cells
by miR-1270(36) and circ_0091579
may also actively regulate proliferation, migration and invasion of
liver cancer cells (37). In the
present study, the function of hsa_circ_0066966 in HBV-positive and
-negative liver cancer cells was investigated. The results
suggested that if the expression of hsa_circ_0066966 was
diminished, a decrease in the proliferation and migration ability
of HBV-positive liver cancer cells was observed. By contrast,
increased expression of hsa_circ_0066966 not only enhanced the
proliferative capacity of HBV-negative liver cancer cells but also
significantly improved their migratory ability. These results are
consistent with those of a previous study, suggesting that
hsa_circ_0066966's cognate gene positively regulates the
proliferation and migration of HBV-positive liver cancer cells and
has an inhibitory effect on apoptosis (35), which may be a potential mechanism
involved in HBV-positive liver cancer.
To date, it remains elusive how HBV influences
circRNA expression, but the important roles of the major protein of
HBV, HBx, in cellular transcriptional regulation may provide
certain clues to this. First, a previous genome-wide analysis of
HBx chromatin recruitment in HBV-replicating cells (38) revealed that there are specific loci
for HBx to bind to a large number of target sequences, including
protein-coding genes and ncRNAs, which are enriched in pathways of
cell metabolism, chromatin dynamics and cancer (38). Furthermore, HBx is able to interact
with multiple transcription factors, including ATF/CREB, ATF3,
c/EBP, NF-IL-6, ETS, EGR, SMAD4, OCT1, RXR receptor, p53,
chromatin-modifying enzymes (CBP, p300 and PCAF) and components of
the basal transcriptional machinery (RPB5, TFIIB, TBP and TFIIH)
(39). The extensive functions of
HBx indicate that alterations to the expression of circRNAs in
HBV-positive cells may be attributed to the direct regulation of
HBx on certain circRNAs or interactions with transcription factors,
directly regulating the transcription of circRNAs, which awaits
further investigation.
However, it should be acknowledged that the present
study had certain limitations. First, the sample number used for
microarray was <3 in each group and one additional HBV-negative
and HBV-positive liver cancer cell line may be used for future
studies. In addition, the specific mechanisms involving
hsa_circ_0066966 still require to be investigated. In the meantime,
it may be useful to perform related animal experiments and further
demonstrate the clinical significance.
In summary, the present study identified
dysregulated circRNAs in HBV-positive liver cancer cells through
microarray analysis and screened 171 circRNAs with significantly
increased expression and 1,322 circRNAs with significantly
decreased expression. In addition, it was demonstrated that
hsa_circ_0066966 was abnormally expressed in HBV-positive liver
cancer cells and was closely related to liver cancer cell
proliferation and migration. Therefore, hsa_circ_0066966 may
provide novel research directions for investigating mechanisms
involving HBV-positive liver cancer in the future.
Supplementary Material
Primer sequences for the top 10
dysregulated circRNAs (5'-3').
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Natural Science Research
Project of the Shanghai Minhang Science and Technology Committee
(grant nos. 2018MHZ074 and 2019MHZ069).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The original data of the microarray analysis were
deposited as a GEO dataset under accession no. GSE181988
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181988).
Authors' contributions
XY designed the study. YY performed the experiments
and wrote the manuscript. XY revised the manuscript. YY and DX
contributed to analyzing the data. XY and DX confirm the
authenticity of all the raw data. All authors reviewed the results.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
de Mattos AZ, Debes JD, Boonstra A, Yang
JD, Balderramo DC, Sartori GDP and de Mattos AA: Current impact of
viral hepatitis on liver cancer development: The challenge remains.
World J Gastroenterol. 27:3556–3567. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bravi F, Bosetti C, Tavani A, Gallus S and
La Vecchia C: Coffee reduces risk for hepatocellular carcinoma: An
updated meta-analysis. Clin Gastroenterol Hepatol. 11:1413–1421.e1.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tangkijvanich P, Mahachai V, Komolmit P,
Fongsarun J, Theamboonlers A and Poovorawan Y: Hepatitis B virus
genotypes and hepatocellular carcinoma in Thailand. World J
Gastroenterol. 11:2238–2243. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Du Y, Su T, Ding Y and Cao G: Effects of
antiviral therapy on the recurrence of hepatocellular carcinoma
after curative resection or liver transplantation. Hepat Mon.
12(e6031)2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16(94)2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
He YX, Ju H, Li N, Jiang YF, Zhao WJ, Song
TT and Ren WH: Association between hsa_circ_0006156 expression and
incidence of gastric cancer. Eur Rev Med Pharmacol Sci.
24:3030–3036. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen KH, Pan JF, Chen ZX, Pan D, Gao T,
Huang M and Huang JN: Effects of hsa_circ_0000711 expression level
on proliferation and apoptosis of hepatoma cells. Eur Rev Med
Pharmacol Sci. 24:4161–4171. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wu C, Deng L, Zhuo H, Chen X, Tan Z, Han
S, Tang J, Qian X and Yao A: Circulating circRNA predicting the
occurrence of hepatocellular carcinoma in patients with HBV
infection. J Cell Mol Med. 24:10216–10222. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang S, Cui S, Zhao W, Qian Z, Liu H, Chen
Y, Lv F and Ding HG: Screening and bioinformatics analysis of
circular RNA expression profiles in hepatitis B-related
hepatocellular carcinoma. Cancer Biomark. 22:631–640.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cortes-Lopez M and Miura P: Emerging
functions of circular RNAs. Yale J Biol Med. 89:527–537.
2016.PubMed/NCBI
|
|
13
|
Barbagallo D, Caponnetto A, Brex D,
Mirabella F, Barbagallo C, Lauretta G, Morrone A, Certo F, Broggi
G, Caltabiano R, et al: CircSMARCA5 regulates VEGFA mRNA splicing
and angiogenesis in glioblastoma multiforme through the binding of
SRSF1. Cancers (Basel). 11(194)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen G, Liu T, Yu B, Wang B and Peng Q:
CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through
miR-373/HIF-1a axis. Cell Cycle. 19:1696–1705. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lu Y, Tan L and Wang X: Circular
HDAC9/microRNA-138/Sirtuin-1 pathway mediates synaptic and amyloid
precursor protein processing deficits in Alzheimer's disease.
Neurosci Bull. 35:877–888. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gu H, Cheng X, Xu J, Zhou K, Bian C, Chen
G and Yin X: Circular RNA circFAT1(e2) promotes osteosarcoma
progression and metastasis by sponging miR-181b and Regulating HK2
Expression. Biomed Res Int. 2020(3589871)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5(R1)2003.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao G and Dai GJ: Hsa_circRNA_000166
Promotes cell proliferation, migration and invasion by regulating
miR-330-5p/ELK1 in colon cancer. Onco Targets Ther. 13:5529–5539.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li Y, Shi P, Zheng T, Ying Z and Jiang D:
Circular RNA hsa_circ_0131242 promotes triple-negative breast
cancer progression by sponging hsa-miR-2682. Onco Targets Ther.
13:4791–4798. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lin J, Liao S, Li E, Liu Z, Zheng R, Wu X
and Zeng W: circCYFIP2 acts as a sponge of miR-1205 and affects the
expression of its target gene E2F1 to regulate gastric cancer
metastasis. Mol Ther Nucleic Acids. 21:121–132. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mao Y, Lv M, Cao W, Liu X, Cui J, Wang Y,
Wang Y, Nie G, Liu X and Wang H: Circular RNA 000554 represses
epithelial-mesenchymal transition in breast cancer by regulating
microRNA-182/ZFP36 axis. FASEB J:. 34:11405–11420. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng
B, Chen C, Chang W, Ping Y, Ji P, et al: Exosomal circPACRGL
promotes progression of colorectal cancer via the
miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer.
19(117)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang M, Luo H and Hui L: MiR-3619-5p
hampers proliferation and cisplatin resistance in cutaneous
squamous-cell carcinoma via KPNA4. Biochem Biophys Res Commun.
513:419–425. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang Q, Miao S, Han X, Li C, Zhang M, Cui
K, Xiong T, Chen Z, Wang C and Xu H: MicroRNA-3619-5p suppresses
bladder carcinoma progression by directly targeting beta-catenin
and CDK2 and activating p21. Cell Death Dis. 9(960)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tan A, Li Q and Chen L: CircZFR promotes
hepatocellular carcinoma progression through regulating
miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch
Biochem Biophys. 661:196–202. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhan M, He K, Xiao J, Liu F, Wang H, Xia
Z, Duan X, Huang R, Li Y, He X, et al: LncRNA HOXA11-AS promotes
hepatocellular carcinoma progression by repressing miR-214-3p. J
Cell Mol Med. 22:3758–3767. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lin T, Dai Y, Guo X, Chen W, Zhao J, Cao L
and Wu Z: Silencing of hsa_circ_0008450 represses hepatocellular
carcinoma progression through regulation of microRNA-214-3p/EZH2
Axis. Cancer Manag Res. 11:9133–9143. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Pan H, Tang L, Jiang H, Li X, Wang R, Gao
J and Li Q: Enhanced expression of circ_0000267 in hepatocellular
carcinoma indicates poor prognosis and facilitates cell progression
by sponging miR-646. J Cell Biochem: Feb 5, 2019 (Epub ahead of
print).
|
|
31
|
Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM,
Hu YW, Lin L, Chen J, Zheng L and Wang Q: HBx-related long
non-coding RNA DBH-AS1 promotes cell proliferation and survival by
activating MAPK signaling in hepatocellular carcinoma. Oncotarget.
6:33791–33804. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhao X, Qin W, Jiang Y, Yang Z, Yuan B,
Dai R, Shen H, Chen Y, Fu J and Wang H: ACADL plays a
tumor-suppressor role by targeting Hippo/YAP signaling in
hepatocellular carcinoma. NPJ Precis Oncol. 4(7)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Shi Y, Fang N, Li Y, Guo Z, Jiang W, He Y,
Ma Z and Chen Y: Circular RNA LPAR3 sponges microRNA-198 to
facilitate esophageal cancer migration, invasion, and metastasis.
Cancer Sci. 111:2824–2836. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang X, Chen Y, Liu W, Liu T and Sun D:
Hsa_circ_0128846 promotes tumorigenesis of colorectal cancer by
sponging hsa-miR-1184 and releasing AJUBA and inactivating
Hippo/YAP signalling. J Cell Mol Med. 24:9908–9924. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Choi JH, Kim MJ, Park YK, Im JY, Kwon SM,
Kim HC, Woo HG and Wang HJ: Mutations acquired by hepatocellular
carcinoma recurrence give rise to an aggressive phenotype.
Oncotarget. 8:22903–22916. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cao Y, Tao Q, Kao X and Zhu X:
Hsa-circRNA-103809 promotes hepatocellular carcinoma development
via MicroRNA-1270/PLAG1 like zinc finger 2 axis. Dig Dis Sci.
66:1524–1532. 2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu W, Yin C and Liu Y: Circular RNA
circ_0091579 promotes hepatocellular carcinoma proliferation,
migration, invasion, and glycolysis through miR-490-5p/CASC3 Axis.
Cancer Biother Radiopharm: Jul 14, 2020 (Epub ahead of print).
|
|
38
|
Guerrieri F, Belloni L, D'Andrea D,
Pediconi N, Le Pera L, Testoni B, Scisciani C, Floriot O, Zoulim F,
Tramontano A and Levrero M: Genome-wide identification of direct
HBx genomic targets. BMC Genomics. 18(184)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Levrero M and Zucman-Rossi J: Mechanisms
of HBV-induced hepatocellular carcinoma. J Hepatol. 64 (Suppl
1):S84–S101. 2016.PubMed/NCBI View Article : Google Scholar
|