Introduction
Coronary heart disease (CHD) poses a serious threat
to human life and health. The number of patients with CHD in China
has reached 11 million and is still increasing (1). Since the first successful
percutaneous transluminal coronary angioplasty was performed by
Gruentzig in 1977(2), coronary
artery intervention has undergone >40 years of development.
However, after coronary stent implantation, in-stent restenosis
(ISR) continues to be a serious obstacle faced by cardiologists
(3). Early identification of
patients at high risk of ISR and strengthening antiplatelet
management, coupled with effective risk factor (smoking, diabetes,
hypertension and hyperlipidemia) management for these patients, are
considered to be effective in preventing ISR (4). However, factors that can effectively
predict the risk of ISR have remained elusive.
Long non-coding RNAs (lncRNAs) belong to a family of
functional RNA molecules that are >200 nucleotides long and do
not encode proteins (5). Despite
being non-coding, lncRNAs have been reported to regulate a number
of cellular and tissue processes, including growth and development,
organ formation, bone marrow hematopoiesis, cell apoptosis and cell
proliferation (5,6). In addition, lncRNAs have been
reported to serve a key role in a number of pathophysiological
mechanisms, such as renal artery stenosis and renal failure
(7). In the cardiovascular field,
lncRNAs play a key regulatory role in the occurrence and
development of CHD, hypertension and cardiac insufficiency
(5). Ballantyne et al
(6) previously found that the
expression of lncRNA smooth muscle cell-enriched was increased in
plaques and plasma from patients with CHD, which could accelerate
the progression of atherosclerotic plaques by promoting the
proliferation and migration of vascular smooth muscle cells
(VSMCs). In addition, a previous study used gene chip technology to
study heart tissues from six patients with congenital heart defects
and indicated that the lncRNA forkhead box F1 adjacent non-coding
developmental regulatory RNA interacted with trithorax-group
proteins/mixed lineage leukemia complexes, leading to H3K27
trimethylation and reduction in the expression of target genes in
the heart and cardiac dysplasia (5). In addition, a previous study by Wang
et al (8) revealed that
plasma levels of the lncRNA antisense non-coding RNA at the INK4
locus (ANRIL) were significantly increased in patients with ISR
compared with patients without ISR, suggesting that ANRIL may be a
prognostic factor for ISR.
In the present study, plasma samples were collected
from three patients with ISR and another three patients without
ISR. RNA sequencing was used to identify lncRNAs that were
significantly differentially expressed between the two groups. From
these, ANRIL and homeobox A11-antisense (HOXA11-AS) were selected
for their dysregulation in patients with ISR compared with patients
without ISR for further investigation of their potential to predict
ISR in patients with single-vessel lesions who received
drug-eluting stents (DES).
Materials and methods
Patients
The inclusion criterium in the present study was
patients with single coronary artery vessel lesions who received
DES treatment. The exclusion criteria were: i) Patients with
tumors; ii) patients who died during the follow-up; and iii)
patients could not undergo another coronary angiography because of
other diseases; or iv) patients who refused to undergo a coronary
angiography review. The patients were followed up for 2-5
years.
A total of 410 patients with single-vessel lesions
who received DES at The Affiliated Hospital of Shaoxing University
and Shaoxing People's Hospital (Shaoxing, China) between September
2015 and September 2017 were included in the present study. The
following 114 patients were excluded: i) 20 patients died during
the follow-up period; ii) 6 patients were diagnosed with tumors;
iii) 24 patients could not undergo another coronary angiography
because of other diseases, such as leukemia and thrombocytopenia;
and iv) 64 patients refused to undergo a coronary angiography
review. Following angiography results, ISR was defined as stenosis
>50%. A total of 244 individuals were enrolled into the control
group and 52 patients were enrolled in the ISR group. Written
informed consent was obtained from all patients and the study
protocol was approved on February 1, 2015 by the Ethics Committee
of the Affiliated Hospital of Shaoxing University (approval no.
2015002).
Data collection
Health habits and medical history of the patients
were recorded by a Dr ZJ. The age, sex, weight (body mass index),
smoking history, systolic and diastolic blood pressure (SBP and
DBP), in addition to other clinical diseases, such as diabetes and
myocardial infarction, were also recorded and presented in Table I. Echocardiography was performed by
ZJ and LL. The levels of N-terminal pro B-type natriuretic peptide
(cat. no. JN19299; Shanghai Jining Shiye), high-sensitivity
C-reactive protein (hs-CRP; cat. no. JN18144; Shanghai Jining
Shiye) and creatinine (cat. no. JN17987; Shanghai Jining Shiye)
were tested by enzyme immunoassay in the central laboratory of The
Affiliated Hospital of Shaoxing University (Shaoxing, China).
 | Table IBaseline characteristics of patients
with drug-eluting stent implantation. |
Table I
Baseline characteristics of patients
with drug-eluting stent implantation.
| Parameter | Total (n=296) | Non-ISR (n=244) | ISR (n=52) | P-values |
|---|
| Male, n (%) | 155 (52.4) | 123 (50.8) | 32 (61.5) | 0.169 |
| Age, years | 66.1±6.3 | 65.4±7.2 | 69.2±5.1 | 0.231 |
| Body mass index,
kg/m2 | 21.4±2.3 | 21.9±2.9 | 20.8±2.5 | 0.301 |
| Systolic blood
pressure, mmHg | 142.5±18.2 | 132.5±16.5 | 148.2±17.0 | <0.001 |
| Diastolic blood
pressure, mmHg | 84.8±9.5 | 82.9±7.3 | 94.5±9.5 | 0.042 |
| Acute coronary
syndrome, n (%) | 82 (27.7) | 58 (23.8) | 24 (46.2) | 0.002 |
| Diabetes, n (%) | 124 (41.9) | 88 (36.1) | 36 (69.2) | <0.001 |
| Smoking, n (%) | 92 (31.1) | 79 (32.3) | 13 (25.0) | 0.327 |
| High-sensitivity
C-reactive protein (mg/l) | 4.5±0.7 | 5.4±0.5 | 4.2±0.5 | 0.311 |
| Low-density
lipoprotein (mmol/l) | 2.8±0.36 | 2.4±0.4 | 3.0±0.3 | 0.048 |
| Lnc ANRIL relative
expression | 1.68±0.05 | 1.55±0.08 | 2.44±0.1 | <0.001 |
| Lnc HOXA11-AS
relative expression | 9.04±0.36 | 9.28±0.3 | 7.53±0.4 | <0.001 |
Sample collection
A total of 6 h later after percutaneous coronary
intervention (PCI), 5 ml whole blood was collected from each
participant. The separation procedure was performed immediately
after sample collection. Blood samples were centrifuged at 1,000 x
g for 10 min at 4˚C to separate the blood cells. The supernatant
was then centrifuged at 13,000 x g for 10 min at 4˚C to completely
remove all cellular contaminants. The plasma was aliquoted into
microcentrifuge tubes and stored at -80˚C until further
analysis.
RNA sequencing
Equal volumes of serum were collected from three
patients with ISR and three patients without ISR in six separate
pools. Total RNA was extracted with TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions, and its quality and quantity were assessed using
NanoDrop™ 2000c (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). The following RNA sequencing protocol was performed by
Lianchuan BioTech Co., Ltd. Following DNase treatment (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions, cDNA libraries were constructed in a strand-specific
manner from 4 µg of DNase-treated RNA using TruSeq Stranded Total
RNA Library Prep kit (cat. no. 20020597; Illumina, Inc.) according
to the manufacturer's instructions. The samples were denatured to
single-stranded DNA with 0.1 M NaOH (final density, 8 pM;
concentrations measured by RT-qPCR) and amplified using TruSeqSR
Cluster Kit v3-cBot-HS (cat. no. GD-401-3001; Illumina, Inc.)
according to the manufacturer's instructions. The libraries were
pooled and sequenced on the Illumina NovaSeq 6000 system (Illumina,
Inc.) using 150 cycles of paired-end sequencing. High-quality reads
were obtained by trimming adapter sequences, invalid and
low-quality reads from the raw reads (quality control). The clean
reads were then mapped to the human genome by HISAT2 software
(v2.1.0; http://ccb.jhu.edu/software/hisat2) using default
parameters. Next, transcript assemblies were constructed using
StringTie software (v1.3.6; The Center for Computational Biology at
Johns Hopkins University) to merge transcripts, and DESeq2 software
(v2.11.40.2; Bioconductor, Inc.) was used to compute differential
expression.
RT-qPCR
RNA was extracted from the serum of all patients.
Total RNA was extracted with TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.), and its quality and quantity were
assessed using NanoDrop™ 2000c (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.). RNA was then reverse transcribed into
cDNA with a PrimeScript RT Reagent kit according to the
manufacturer's protocol (cat. no. RR047A; Takara Biotechnology Co.,
Ltd.) according to the manufacturer's protocol. RT-qPCR was
performed with SYBR™ Green PCR Master Mix kit (cat. no. 4368702;
Applied Biosystems; Thermo Fisher Scientific, Inc) using a
LightCycler® 480 Real-time PCR System [Roche Diagnostics
(Shanghai) Co., Ltd.]. GAPDH was used as an internal control. The
following primers were used: ANRIL forward,
5'-TGCTCTATCCGCCAATCAGG-3' and reverse, 5'-GGGCCTCAGTGGCACATACC-3';
HOXA11-AS forward, 5'-CGGCTAACAAGGAGATTTGG-3' and reverse,
5'-AGGCTCAGGGATGGTAGTCC-3' and GAPDH forward,
5'-AGCCACATCGCTCAGACAC-3' and reverse, 5'-GCCCAA TACGACCAAATCC-3'.
The thermocycling conditions for qPCR were as follows: 95˚C for 10
min; followed by 40 cycles of 95˚C for 15 sec and 60˚C for 60 sec
on an LightCycler® 480 Real-time PCR System. Relative
lncRNA expression levels were calculated using the
2-ΔΔCq method normalized to GAPDH (9). Each blood sample was analyzed in
duplicate, and all analyses were repeated three times.
Statistical analysis
Continuous variables are expressed as the mean ± SD,
whilst categorical variables are presented as counts or
percentages. Kolmogorov-Smirnov test was used to ensure that the
variables were normally distributed. The differences between
continuous variables were tested with unpaired Student's t-test,
and the differences between categorical variables were examined by
χ2 test. All statistical analyses were performed using
MedCalc (v2016; MedCalc Software Ltd). P<0.05 was considered to
indicate a statistically significant difference.
A logistic regression model performed on MedCalc was
used to test the association between the levels of ANRIL and/or
HOXA11-AS along with other known risk factors for ISR using
univariate and multivariate models. The multivariable model was
adjusted for SBP, DBP, sex, age, diabetes, acute coronary syndrome
(ACS), smoking, hs-CRP, low-density lipoprotein (LDL), ANRIL and
HOXA11-AS levels.
To assess differences in prognostic efficiency
involving ANRIL and HOXA11-AS, the area under the curve of a
receiver operating characteristic analysis (auROC) using MedCalc,
which is a measure of discrimination, was calculated. ISR was used
to evaluate the diagnostic performance of the scoring systems for
patients who underwent PCI. Furthermore, standard indices of
validity, such as the Youden index, specificity and sensitivity,
were calculated according to the ROC results.
Results
Study population
The smallest sample sizes were calculated using the
respective HOXA11-AS and ANRIL values. RT-qPCR quantification
indicated that, for HOXA11-AS, the mean and standard deviation for
the control group were 9.2850 and 3.020, whereas the mean and
standard deviation for the ISR group were 6.18 and 4.4. The α was
set as 0.05 and the β as 0.1, therefore the smallest sample size
for each group was 30. For ANRIL, the mean and standard deviation
for the control group were 1.535 and 0.8717, whereas the mean and
standard deviation for the ISR group were 2.69 and 1.0044. The α
was set as 0.05 and the β as 0.1, therefore the smallest sample
size for each group was 14. In conclusion, the sample size of the
present study was sufficient and did not affect the interpretation
or generalization of the data.
A total of 410 patients with single-vessel lesions
who received a DES at The Affiliated Hospital of Shaoxing
University and Shaoxing People's Hospital (China) between September
2015 and September 2017 were included in the present study. After
excluding those who did not meet the follow-up criteria, 296
patients remained. Baseline characteristics for each patient were
collected from the medical records. As presented in Table I, the mean age of the patients was
66.1±6.3 years, where 52.4% of all patients were male. A total of
17.6% patients (n=52) developed ISR during the follow-up coronary
angiography review. SBP, DBP and LDL levels in the ISR group were
significantly higher compared with those in the non-ISR group
(P<0.05). There were no differences in terms of body mass index,
age and hs-CRP between the two groups. In addition, the rate of
history of ACS and diabetes was significantly higher in the ISR
group (P<0.05).
Expression of ANRIL and HOXA11-AS
To identify potential lncRNAs involved in the
development of ISR, RNA sequencing was used to analyze and sequence
lncRNAs isolated from the serum of three patients with ISR and
three patients without ISR (accession no. GSE182225). As presented
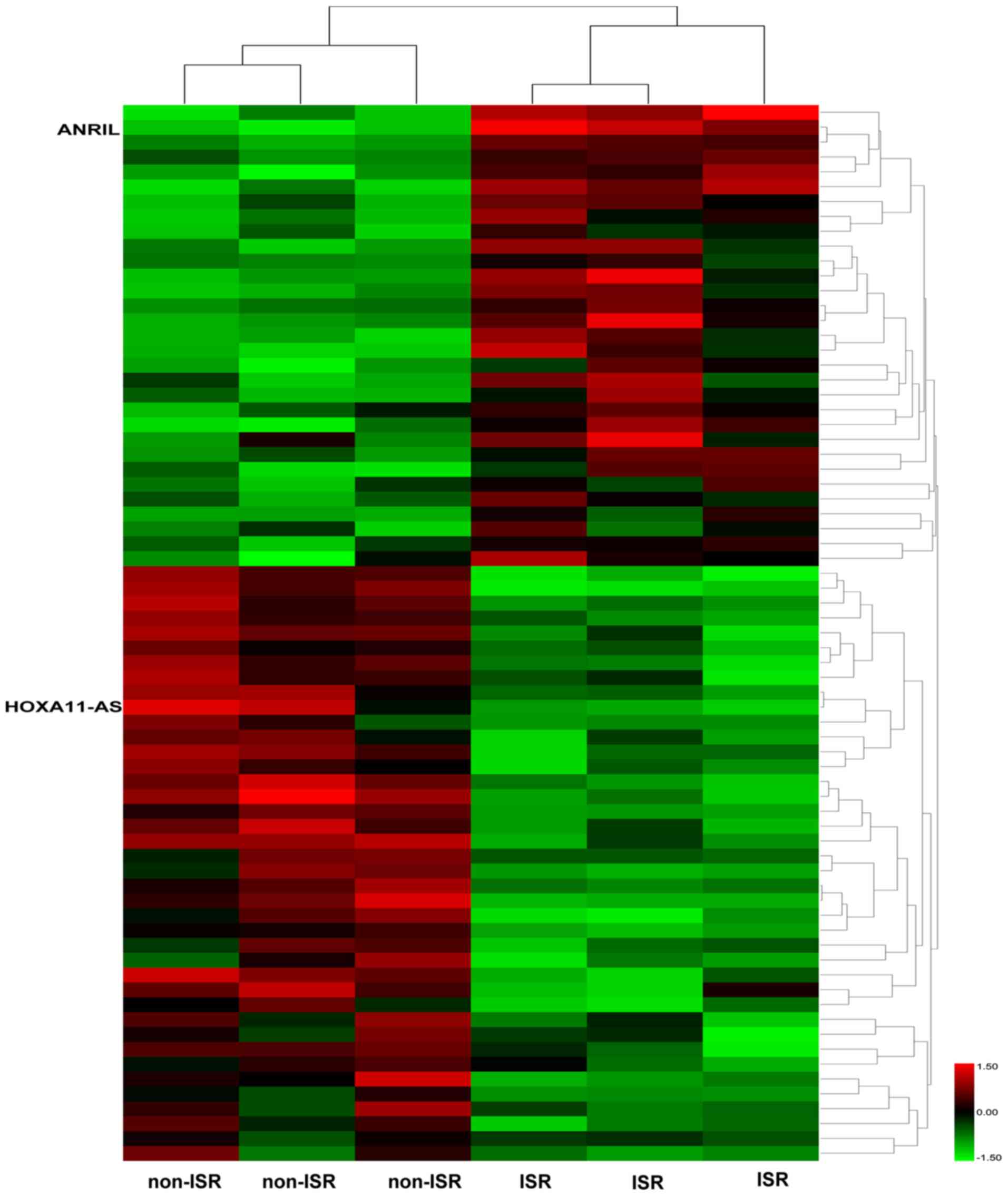
in Fig. 1, >30 lncRNAs
exhibited differences in expression between the two groups. As
ANRIL value for predicting ISR was reported by Wang et al
(8) and HOXA11-AS was identified
as potentially involved in ISR in our previous work (10), ANRIL and HOXA11-AS were selected
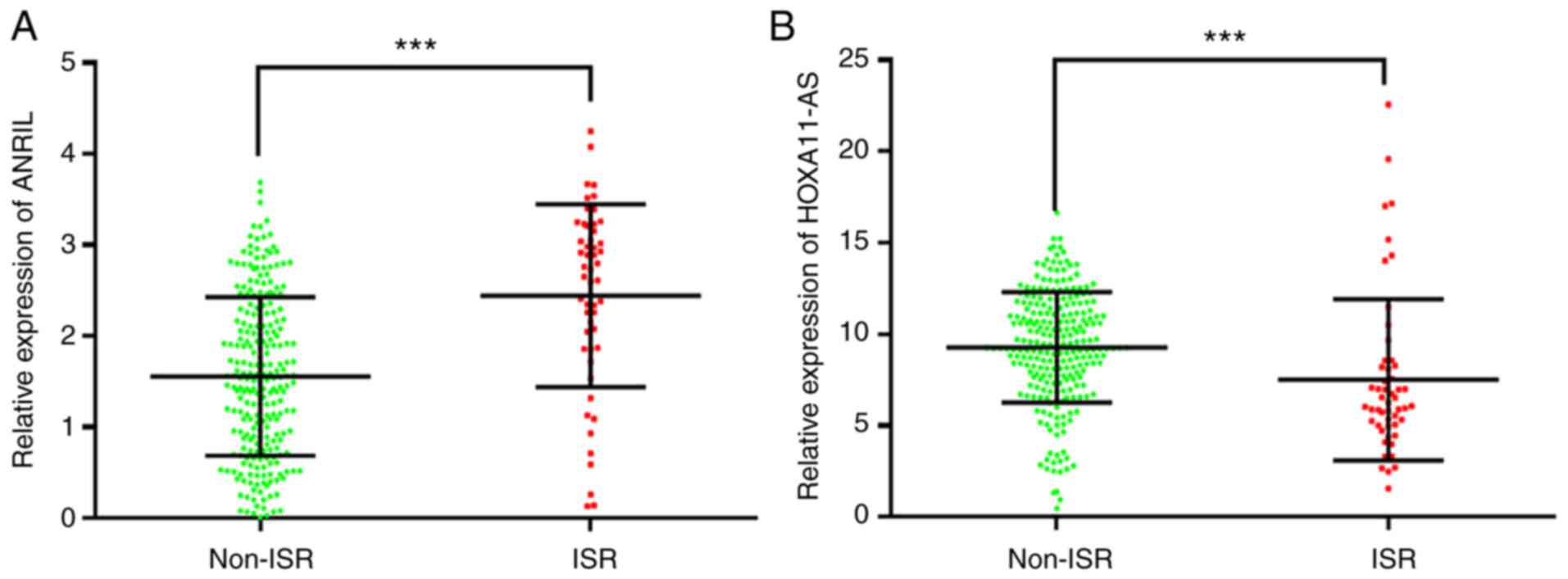
for further analysis. ANRIL and HOXA11-AS expression levels were
subsequently analyzed in all the patients (including 52 patients
with ISR and 244 patients without ISR) using RT-qPCR, which
revealed that ANRIL expression was significantly increased in the
ISR group compared with that in the non-ISR group, whilst HOXA11-AS
expression was significantly decreased (P<0.001), as shown in
Fig. 2.
Risk factor analysis for ISR
As shown in Table
II, univariate logistic regression revealed that SBP (odds
ratio (OR)=1.22; 95% confidence interval (CI)=1.03-1.98), DBP
(OR=1.09; 95% CI=1.03-1.88), ACS (OR=1.36; 95% CI=1.06-4.89),
diabetes (OR=1.92; 95% CI=1.61-4.19), LDL (OR=2.44; 95%
CI=1.46-8.04) and ANRIL expression (OR=2.95; 95% CI=1.68-8.08) were
significantly associated with ISR (P<0.05). In addition,
HOXA11-AS expression was negatively associated with ISR (OR=0.62;
95% CI=0.48-0.71; P<0.05). All variables were then entered into
the multivariate logistic regression analysis. As reported by
Table II, SBP (OR=1.31; 95%
CI=1.06-2.01), DBP (OR=1.05; 95% CI=0.99-1.96), ACS (OR=1.39; 95%
CI=1.14-3.27), diabetes (OR=1.88; 95% CI=1.42-6.21), LDL (OR=2.74;
95% CI=1.78-9.69) and ANRIL expression (OR=2.68; 95% CI=1.62-7.82)
were found to be independent risk factors for ISR (P<0.05),
whilst HOXA11-AS expression (OR=0.58; 95% CI=0.45-0.72; P<0.001)
was found to be an independent protective factor for ISR.
 | Table IILogistic regression analysis for
patients with drug-eluting stent implantation, with or without
in-stent restenosis. |
Table II
Logistic regression analysis for
patients with drug-eluting stent implantation, with or without
in-stent restenosis.
| | Univariate
analysis | Multivariate
analysis |
|---|
| Variables | OR | 95% CI | P-value | OR | 95% CI | P-value |
|---|
| Male | 0.99 | 0.49-1.94 | 0.722 | 0.96 | 0.41-2.07 | 0.603 |
| Age | 0.94 | 0.68-1.44 | 0.223 | 0.98 | 0.88-1.68 | 0.245 |
| Body mass
index | 1.09 | 0.29-1.99 | 0.485 | 1.09 | 0.30-1.85 | 0.502 |
| Systolic blood
pressure | 1.22 | 1.03-1.98 | 0.007 | 1.31 | 1.06-2.01 | 0.002 |
| Diastolic blood
pressure | 1.09 | 1.03-1.88 | 0.034 | 1.05 | 0.99-1.96 | 0.046 |
| Acute coronary
syndrome | 1.36 | 1.06-4.89 | 0.041 | 1.39 | 1.14-3.27 | 0.002 |
| Diabetes | 1.92 | 1.61-4.19 | 0.004 | 1.88 | 1.42-6.21 | 0.009 |
| Smoking | 1.05 | 0.53-1.60 | 0.692 | 1.02 | 0.24-1.38 | 0.416 |
| High-sensitivity
C-reactive protein | 1.14 | 0.78-1.56 | 0.265 | 1.09 | 0.42-1.95 | 0.301 |
| Low-density
lipoprotein | 2.44 | 1.46-8.04 | <0.001 | 2.74 | 1.78-9.69 | 0.001 |
| Lnc ANRIL relative
expression | 2.95 | 1.68-8.08 | 0.001 | 2.68 | 1.62-7.82 | 0.001 |
| Lnc HOXA11-AS
relative expression | 0.62 | 0.48-0.71 | 0.001 | 0.58 | 0.45-0.72 | 0.001 |
ROC analysis of ANRIL and HOXA11-AS
for ISR
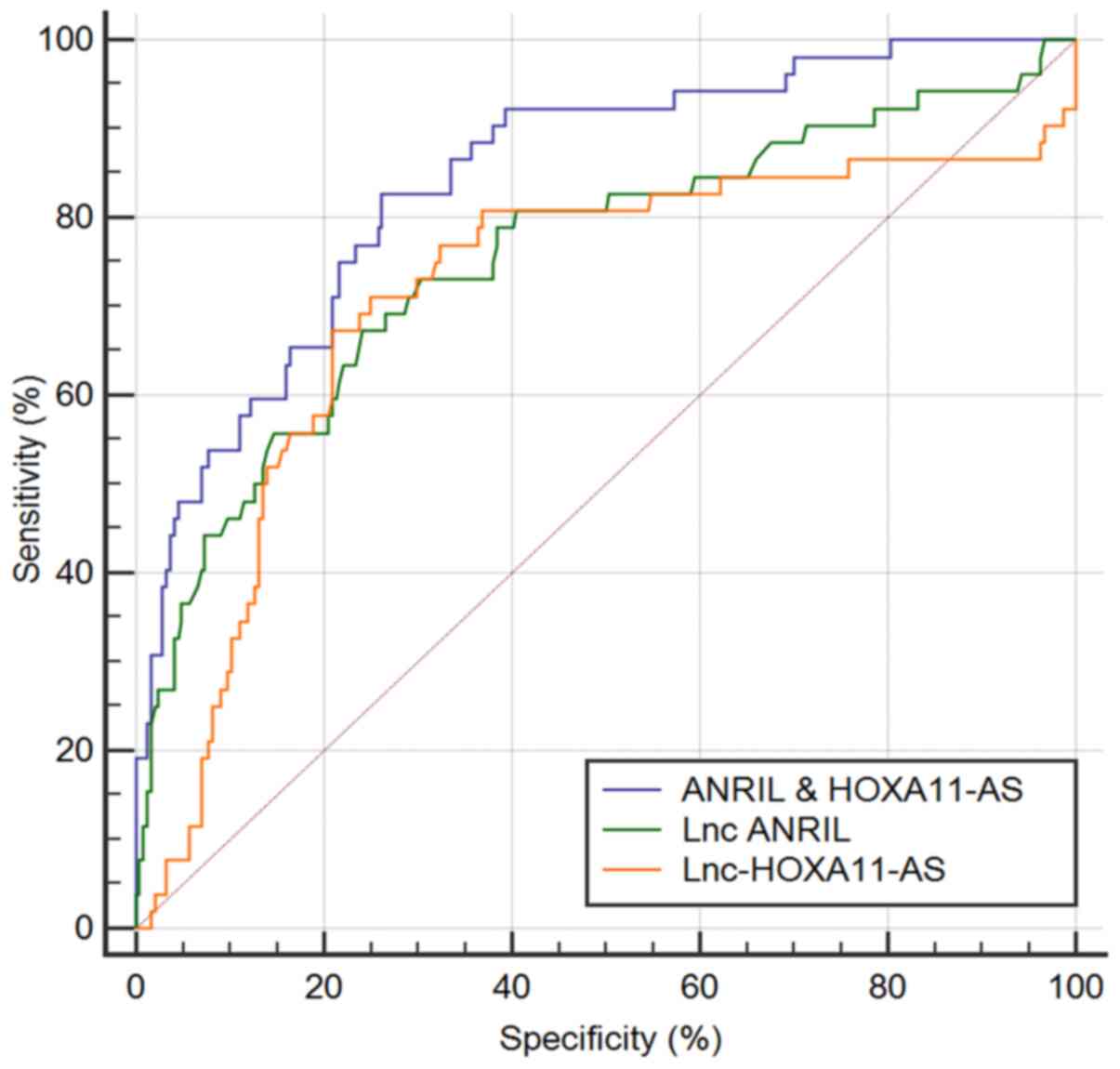
The ability of ANRIL and HOXA11-AS to predict ISR in
patients with single-vessel lesions who received a DES is presented
in Fig. 3 and Table III. The performance of ANRIL with
an auROC of 0.755 (95% CI=0.702-0.803) was higher than that of
HOXA11-AS with an auROC of 0.712 (95% CI=0.657-0.763), although the
difference was not statistically significant between the two
lncRNAs (data not shown). When using the best ANRIL cut-off value
of 2.25, the sensitivity was 67.3% and the specificity was 75.8%.
By contrast, when using a cut-off value for HOXA11-AS of 7.07, the
sensitivity was 67.31% and the specificity was 79.1%. ANRIL and
HOXA11-AS were then combined to obtain the ANRIL and HOXA11-AS
scores [ANRIL and HOXA11-AS score = -2.3517 + 1.12065 x ANRIL +
(-0.17024) x HOXA11-AS], for which coefficients were calculated by
multivariate logistic regression, with only ANRIL and HOXA11-AS
included. The score revealed an auROC of 0.844 (95%
CI=0.798-0.884), which was significantly higher than that of ANRIL
or HOXA11-AS (P<0.01). When a cut-off value of -1.67 was used
for this score, the sensitivity was 82.7% and the specificity was
73.8%. Therefore, the combined ANRIL and HOXA11-AS score exhibited
a better performance than either ANRIL or HOXA11-AS alone in
predicting ISR in patients with single-vessel lesions who received
a DES.
 | Table IIIROC analysis of using ANRIL and
HOXA11-AS for predicting ISR in patients with drug-eluting stent
implantation, with or without in-stent restenosis. |
Table III
ROC analysis of using ANRIL and
HOXA11-AS for predicting ISR in patients with drug-eluting stent
implantation, with or without in-stent restenosis.
| Parameter | auROC | 95% CI |
P-valuea |
P-valueb | Youden | Cut-off | Sensitivity
(%) | Specificity
(%) |
|---|
| ANRIL | 0.755 | 0.702-0.803 | <0.001 | 0.008 | 0.431 | 2.25 | 67.3 | 75.8 |
| HOXA11-AS | 0.712 | 0.657-0.763 | <0.001 | 0.002 | 0.464 | 7.07 | 67.31 | 79.10 |
| ANRIL and
HOXA11-AS | 0.844 | 0.798-0.884 | <0.001 | N/A | 0.564 | -1.67 | 82.7 | 73.8 |
Discussion
ISR occurrence is the result of a number of factors.
Several studies have shown that the type of coronary vascular
lesions, length of the lesions, diameter and length of the
implanted stent, types of stent, tandem stent and technical
proficiency of doctors can all affect the occurrence of ISR
(11,12). In addition, previous studies have
revealed that smoking, diabetes and hyperlipidemia are independent
risk factors for ISR (13). Sex
and age can also affect ISR occurrence (14). Early assessment of the risks of ISR
in patients after PCI and controlling the risk factors have been
considered to be one of the most effective means to prevent and
treat ISR (8,15).
lncRNAs are highly conserved molecular sequences
that are also relatively stable in the circulatory system,
rendering them potentially reliable molecular markers for disease
identification (16). Previous
studies have confirmed that some lncRNAs have diagnostic value for
CHD (15,17). For example, expression of the
lncRNA long intergenic noncoding RNA predicting cardiac remodeling
was found to be downregulated during the early phases after a
cardiac event, but remained elevated during latter stages after an
acute myocardial infarction (AMI) (18). In addition, myosin
heavy-chain-associated RNA transcript, a recently described lncRNA
that was found to protect the heart from hypertrophic remodeling,
demonstrated a positive correlation with the cardiac injury marker
cTnT in patients with AMI (5).
However, to the best of our knowledge, only one study has addressed
the association between plasma lncRNAs and ISR (8). In the present study, plasma samples
were collected from patients with and without ISR for analysis. RNA
sequencing results indicated that ANRIL expression was upregulated,
whilst HOXA11-AS was downregulated in plasma samples from patients
with ISR compared with patients without ISR. In addition, the
present results identified ANRIL to be an independent risk factor
for ISR, whereas HOXA11-AS was an independent protective factor for
ISR. Therefore, high ANRIL expression and low HOXA11-AS expression
are proposed to be predictors of high ISR risk. Furthermore, the
present data identified that the combined score of ANRIL and
HOXA11-AS was a superior predictor for ISR compared with either
ANRIL or HOXA11-AS alone.
Consistent with the present findings, the value of
ANRIL for predicting ISR was previously reported by Wang et
al (8). Accumulating evidence
has demonstrated that ANRIL is expressed in vascular endothelial
cells, VSMCs, mononuclear phagocytes and atherosclerotic plaques
(19-21).
ANRIL can not only regulate protein expression, but also
participate in post-translational modifications (20). Abnormal ANRIL expression has been
revealed to result in vascular endothelium injury, proliferation,
migration, senescence and apoptosis of VSMCs, in addition to
mononuclear cell adhesion and proliferation, glycolipid metabolism
disorders and DNA damage (21).
ANRIL accelerates atherosclerosis development and is a known risk
factor for CHD and ISR (8).
HOXA11-AS is a newly discovered lncRNA that has been
studied mainly in the context of tumor biology (22). Wang et al (23) reported that HOXA11-AS was
associated with cell cycle progression in glioma, associated with
tumor grade and lead to a poorer prognoses. In another study, Chen
et al (24) proposed that
HOXA11-AS may participate in the occurrence of cervical cancer by
regulating HOXA11. Furthermore, two previous studies revealed that
HOXA11-AS expression was associated with metastasis, invasion,
staging and prognosis in human ovarian cancer and lung
adenocarcinoma (25,26). These studies also proposed that
suppression of ovarian cancer by HOXA11-AS did not occur by
regulating the HOXA11 gene (25,26).
In 2017, a number of studies reported that HOXA11-AS could promote
the proliferation of tumor cells by regulating the expression of
microRNA (miR)-140-5p, large tumor suppressor kinase 1, miR-124 and
peptidyl arginine deiminase 2 (27-31).
However, to date, the role of HOXA11-AS in the cardiovascular field
has not been previously reported. The present study reported that
HOXA11-AS expression was decreased in patients with ISR and can
therefore function as a protective factor for ISR.
Multiplex detection demonstrated better predictive
performance than individual factors alone, which has also been
demonstrated in a number of previous studies (32,33).
Tesche et al (33)
demonstrated that a combination of coronary CT angiography-derived
non-calcified plaque volumes, lesion length and remodeling index
had a higher predictive value for ISR than each aforementioned
alone, which was clinically valuable for the prediction of ISR.
Consistent with this, the present results indicated that the
combination of ANRIL and HOXA11-AS significantly improved the
predictive accuracy for ISR compared with each lncRNA alone. With
the rapid development in technology and equipment, it is now
possible to simultaneously detect several different lncRNAs from a
single sample using multiplex microbead-based assays (34). Therefore, multiplex detection of
lncRNAs could represent a promising assay for predicting ISR.
In conclusion, the present results demonstrated that
HOXA11-AS expression was downregulated in the plasma of patients
with ISR, whereas ANRIL expression was upregulated. ANRIL was found
to be an independent risk factor for ISR, whilst HOXA11-AS was an
independent protective factor for ISR. In addition, high ANRIL
expression and low HOXA11-AS expression predicted a higher risks of
ISR. The present study subsequently revealed that the combined
score for ANRIL and HOXA11-AS proved to be superior at predicting
ISR compared with that for either ANRIL or HOXA11-AS alone.
Therefore, despite its limitations (lack of in vivo
investigations to confirm the role ANRIL or HOXA11-AS in ISR), the
present results support the use of the multiplex detection of
lncRNAs as a test for predicting ISR in the future.
Acknowledgements
Not applicable.
Funding
Funding: The present study is supported by the Basic Public
Welfare Research Project of Zhejiang Province (grant no.
LGF19H020002) and Science Project of Shaoxing City (grant no.
2018C30021).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The RNA sequencing data was uploaded to GEO database
(accession no. GSE182225; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182225).
Authors' contributions
All authors have read and approved the final
manuscript. ZJ and LL designed and performed the research. HS, WC
and HX helped collect and analyze the data. ZJ and LL confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The study protocol was approved by the Research
Ethics Committee of The Affiliated Hospital of Shaoxing University
(Shaoxing, China; approval no. 2015002). Written informed consent
was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hu SS, Gao RL, Liu LS, Zhu ML, Wang W and
Wang YJ: The abstract of 2018 report of Chinese cardiovascular
disease. Chin Circ Mag. 34:209–220. 2019.
|
|
2
|
Meier B: The first patient to undergo
coronary angioplasty - 23-year follow-up. N Engl J Med.
344:144–145. 2001.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lee MS and Banka G: In-stent restenosis.
Interv Cardiol Clin. 5:211–220. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Buccheri D, Piraino D, Andolina G and
Cortese B: Understanding and managing in-stent restenosis: A review
of clinical data, from pathogenesis to treatment. J Thorac Dis.
8:E1150–E1162. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Uchida S and Dimmeler S: Long noncoding
RNAs in cardiovascular diseases. Circ Res. 116:737–750.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ballantyne MD, Pinel K, Dakin R, Vesey AT,
Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, et
al: Smooth muscle enriched long noncoding RNA (SMILR) regulates
cell proliferation. Circulation. 133:2050–2065. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lai CF, Chen YT, Gu J, Nerbonne JM, Lin CH
and Yang KC: Circulating long noncoding RNA DKFZP434I0714 predicts
adverse cardiovascular outcomes in patients with end-stage renal
disease. Int J Cardiol. 277:212–219. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang F, Su X, Liu C, Wu M and Li B:
Prognostic value of plasma long noncoding RNA ANRIL for in-stent
restenosis. Med Sci Monit. 23:4733–4739. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang P, Meng L, Liu L and Peng F: The
value of circulating exosomes-mediated lncRNA HOXA11-AS in the
proliferation of vascular endothelial cell by up-regulating SOX4.
Journal of Wenzhou Medical Unversity. 51:639–645. 2021.(In
Chinese).
|
|
11
|
Shi KQ, Wu FL, Liu WY, Zhao CC, Chen CX,
Xie YY, Wu SJ, Lin XF, Chen YP, Wong DK, et al: Non-alcoholic fatty
liver disease and risk of in-stent restenosis after bare metal
stenting in native coronary arteries. Mol Biol Rep. 41:4713–4720.
2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bambagioni G, Di Mario C, Torguson R,
Demola P, Ali Z, Singh V, Skinner W, Artis A, Cate TT, Zhang C, et
al: Lipid-rich plaques detected by near-infrared spectroscopy
predict coronary events irrespective of age: A Lipid Rich Plaque
sub-study. Atherosclerosis. 334:17–22. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pleva L, Kukla P and Hlinomaz O: Treatment
of coronary in-stent restenosis: A systematic review. J Geriatr
Cardiol. 15:173–184. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li MK, Tsang AC, Tsang FC, Ho WS, Lee R,
Leung GK and Lui WM: Long-term risk of in-stent restenosis and
stent fracture for extracranial vertebral artery stenting. Clin
Neuroradiol. 29:701–706. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kumric M, Borovac JA, Martinovic D,
Ticinovic Kurir T and Bozic J: Circulating biomarkers reflecting
destabilization mechanisms of coronary artery plaques: are we
looking for the impossible? Biomolecules. 11(881)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Thum T and Condorelli G: Long noncoding
RNAs and microRNAs in cardiovascular pathophysiology. Circ Res.
116:751–762. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li M, Wang YF, Yang XC, Xu L, Li WM, Xia
K, Zhang DP, Wu RN and Gan T: Circulating long noncoding RNA LIPCAR
acts as a novel biomarker in patients with ST-segment elevation
myocardial infarction. Med Sci Monit. 24:5064–5070. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Holdt LM, Stahringer A, Sass K, Pichler G,
Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou
A, et al: Circular non-coding RNA ANRIL modulates ribosomal RNA
maturation and atherosclerosis in humans. Nat Commun.
7(12429)2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chi JS, Li JZ, Jia JJ, Zhang T, Liu XM and
Yi L: Long non-coding RNA ANRIL in gene regulation and its duality
in atherosclerosis. J Huazhong Univ Sci Technolog Med Sci.
37:816–822. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen L, Qu H, Guo M, Zhang Y, Cui Y, Yang
Q, Bai R and Shi D: ANRIL and atherosclerosis. J Clin Pharm Ther.
45:240–248. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wei C, Zhao L, Liang H, Zhen Y and Han L:
Recent advances in unraveling the molecular mechanisms and
functions of HOXA11 AS in human cancers and other diseases
(Review). Oncol Rep. 43:1737–1754. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang Q, Zhang J, Liu Y, Zhang W, Zhou J,
Duan R, Pu P, Kang C and Han L: A novel cell cycle-associated
lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA
transcript and is a biomarker of progression in glioma. Cancer
Lett. 373:251–259. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen J, Fu Z, Ji C, Gu P, Xu P, Yu N, Kan
Y, Wu X, Shen R and Shen Y: Systematic gene microarray analysis of
the lncRNA expression profiles in human uterine cervix carcinoma.
Biomed Pharmacother. 72:83–90. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Richards EJ, Permuth-Wey J, Li Y, Chen YA,
Coppola D, Reid BM, Lin HY, Teer JK, Berchuck A, Birrer MJ, et al:
A functional variant in HOXA11-AS, a novel long non-coding RNA,
inhibits the oncogenic phenotype of epithelial ovarian cancer.
Oncotarget. 6:34745–34757. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kim HJ, Eoh KJ, Kim LK, Nam EJ, Yoon SO,
Kim KH, Lee JK, Kim SW and Kim YT: The long noncoding RNA HOXA11
antisense induces tumor progression and stemness maintenance in
cervical cancer. Oncotarget. 7:83001–83016. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yu W, Peng W, Jiang H, Sha H and Li J:
LncRNA HOXA11-AS promotes proliferation and invasion by targeting
miR-124 in human non-small cell lung cancer cells. Tumour Biol: Oct
15, 2017 (Epub ahead of print). doi: 10.1177/1010428317721440.
|
|
28
|
Yu J, Hong JF, Kang J, Liao LH and Li CD:
Promotion of lncRNA HOXA11-AS on the proliferation of
hepatocellular carcinoma by regulating the expression of LATS1. Eur
Rev Med Pharmacol Sci. 21:3402–3411. 2017.PubMed/NCBI
|
|
29
|
Xu C, He T, Li Z, Liu H and Ding B:
Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth,
migration and invasion of glioma cells. Biomed Pharmacother.
95:1504–1513. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cui Y, Yi L, Zhao JZ and Jiang YG: Long
noncoding RNA HOXA11-AS functions as miRNA sponge to promote the
glioma tumorigenesis through targeting miR-140-5p. DNA Cell Biol.
36:822–828. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen D, Sun Q, Zhang L, Zhou X, Cheng X,
Zhou D, Ye F, Lin J and Wang W: The lncRNA HOXA11-AS functions as a
competing endogenous RNA to regulate PADI2 expression by sponging
miR-125a-5p in liver metastasis of colorectal cancer. Oncotarget.
8:70642–70652. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Japp NC, Souchek JJ, Sasson AR,
Hollingsworth MA, Batra SK and Junker WM: Tumor biomarker
in-solution quantification, standard production, and multiplex
detection. J Immunol Res. 2021(9942605)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tesche C, De Cecco CN, Vliegenthart R,
Duguay TM, Stubenrauch AC, Rosenberg RD, Varga-Szemes A, Bayer RR
II, Yang J, Ebersberger U, et al: Coronary CT angiography-derived
quantitative markers for predicting in-stent restenosis. J
Cardiovasc Comput Tomogr. 10:377–383. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Parsa SF, Vafajoo A, Rostami A, Salarian
R, Rabiee M, Rabiee N, Rabiee G, Tahriri M, Yadegari A, Vashaee D,
et al: Early diagnosis of disease using microbead array technology:
A review. Anal Chim Acta. 1032:1–17. 2018.PubMed/NCBI View Article : Google Scholar
|