Introduction
Breast cancer (BC) is one of the most common cancers
worldwide and is the main cause of mortality of female patients
with cancer (1,2). Surgery and systemic chemotherapy can
cure 70% of patients with BC, but the 5-year survival rate of those
with metastatic disease remains poor (3). Therefore, investigating the molecular
mechanism of BC metastasis may be helpful for the formulation of
treatment strategies for breast cancer.
Ring finger protein 6 (RNF6), a member
of E3 ubiquitin ligases, regulates a number of target genes that
are important for cell growth and survival (4,5).
Recent accumulating evidence indicated that RNF6 could promote the
degradation of its target protein by a ubiquitin proteasome
(6). A number of studies confirmed
that RNF6 is a carcinogenic gene in numerous types of cancer,
including prostate cancer, oesophageal squamous cell carcinoma and
lung adenocarcinoma (7-9).
For instance, the expression levels of RNF6 are elevated in
colorectal cancer, and high RNF6 levels is correlated with poor
prognosis (4,5). However, the effect of RNF6 on the
invasion and metastasis of BC and its potential mechanisms remain
to be elucidated.
The Hippo signalling pathway is an evolutionarily
conservative signalling pathway that plays a key role in
controlling the invasion and migration of tumor cells (10). At the core of the Hippo pathway is a
kinase cascade that activates mammalian STE20-like protein kinase
(MST) 1/2 phosphorylation and activates large tumour suppressor
kinase 1/2, which phosphorylates and inhibits the two major
downstream effectors of the Hippo pathway, YES-associated protein
(YAP) and transcriptional co-activator with PDZ-binding motif (TAZ)
(11,12). Dephosphorylated YAP and TAZ enter
the nucleus and induce target gene expression, which promotes cell
migration (13). Studies
demonstrated that YAP is overexpressed in some tumours, and
sustained expression of YAP promoted growth and tumour occurrence,
suggesting that the Hippo pathway plays a key role in tumorigenesis
(14,15). However, the mechanism of YAP
expression regulation in BC is still unclear.
To the best of our knowledge, the present study
first found that RNF6 was significantly upregulated in breast
cancer tissues compared with the adjacent normal tissues. The
expression and function of RNF6 in breast cancer cells was then
studied. RNF6 was also identified as a possible E3 ligase of MST1
(also known as S100A4), as it interacted with and promoted the
degradation of MST1. These data might highlight the role of RNF6 in
BC, which could serve as a valuable prognostic indicator and
potential therapeutic target for patients with BC.
Materials and methods
Patients and samples
BC samples were obtained from 146 patients (age,
42-72 years) with breast cancer at the Third Hospital of Nanchang
and the First Affiliated Hospital of Nanchang University (China)
from December 2007 to December 2015, immediately snap-frozen in
liquid nitrogen and stored at -80˚C for further analysis. All
patients provided written informed consent. The research procedure
was approved by the Ethics Committee of the Third Hospital of
Nanchang and the First Affiliated Hospital of Nanchang
University.
Cell lines
BC cell lines (BT549, MDA-MB-231, MDA-MB-453 and
MCF7) and normal human mammary gland cells (MCF10A) were purchased
from American Type Culture Collection. Cell lines were cultured in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with FBS
(Gibco; Thermo Fisher Scientific, Inc.) to a final concentration of
10% at 37˚C in a humidified incubator containing 5%
CO2.
Western blot assay
Protein from BT549, MDA-MB-231, MDA-MB-453, MCF7 and
normal human mammary gland cells (MCF10A) cells was extracted using
RIPA lysis buffer (MilliporeSigma) according to the manufacturer's
instructions. The total protein concentration was measured with a
BCA protein assay kit (Takara Biotechnology Co., Ltd.). The samples
(20 µg) were separated via 10% SDS-PAGE, then transferred onto PVDF
membranes. The membranes were blocked with 5% non-fat milk for 2 h
at room temperature, then treated with the following primary
antibodies overnight at 4˚C. The following primary antibodies were
used: Anti-RNF6 (1:1,000; cat. no. ab204506; Abcam), anti-YAP
(1:500; cat. no. 13584-1-AP; ProteinTech Group, Inc.), anti-MST1
(1:1,000; cat. no. 3682; Cell Signaling Technology, Inc.),
anti-ubiquitin (1:1,000; cat. no. 10201-2-AP; ProteinTech Group,
Inc.), anti-Cyr61 (1:500; cat. no. 26689-1-AP; ProteinTech Group,
Inc.), anti-connective tissue growth factor (1:500; cat. no.
23936-1-AP; ProteinTech Group, Inc.) and anti-cyclin E (1:500; cat.
no. 11554-1-AP; ProteinTech Group, Inc.). Tubulin expression
(1:2,000; cat. no.11224-1-AP; ProteinTech Group, Inc.) was used as
a loading control. After washing three times with Tris-buffered
saline-0.1% Tween-20, the membranes were incubated with
HRP-conjugated goat anti-rabbit IgG secondary anti-body (1:5,000;
cat. no. ab6728; Abcam) for 2 h at 4˚C. Finally, the protein bands
were visualized with ECL system (Thermo Fisher Scientific, Inc.)
and analyzed by densitometry using software ImageJ (National
Institutes of Health; version 1.48).
Immunohistochemistry (IHC)
staining
All BC tissue specimens were fixed with 10% neutral
formaldehyde solution for 4-6 h at room temperature, and were
dehydrated, waxed and wrapped. Then, the tissues were cut into 4-6
µm sections. The tissue sections were baked at 60˚C overnight.
Sections were blocked using 2% BSA (Sangon Biotech Co., Ltd.) for 1
h at room temperature. Anti-RNF6 (1:1,000; cat. no. ab204506;
Abcam) was applied to paraffin-embedded sections following
microwave antigen retrieval for 10 min in 0.01 mol/l citrate buffer
(pH, 6.0). Specimens were treated with 0.3% hydrogen peroxide in
methanol for 15 min at room temperature following incubation with
primary antibody to block endogenous peroxidase activity and
blocked with human serum (Gibco; Thermo Fisher Scientific, Inc.) to
minimize background reactivity at room temperature. Following the
primary antibody incubation, the sections were rinsed thrice with
PBS for 5 min and incubated with 50 µl HRP-conjugated secondary
goat anti-rabbit antibody (product code ab6721; 1:1,000; Epitomics;
Abcam) at room temperature for 20 min. Then, samples were washed
and incubated with ABC amplification system for 30 min at room
temperature (ready to use; cat. no. PK-7 200; Vector Laboratories,
Inc.). Finally, the chromogenic substrate 3,3'-diaminobenzidine
(DAB) was added. Furthermore, the nuclei were stained with
hematoxylin (ScyTek Laboratories, Inc.) for 1 min at room
temperature. Images were obtained using an image analyzer system
(Olympus BH-2 microscope; Olympus Corporation), digitized and the
DAB signal was quantified by ImageJ 1.51w software (National
Institutes of Health).
Constructs and plasmids
RNA duplexes for short hairpin (sh)RNA-mediated
RNF6 and YAP knockdown were synthesized by Shanghai
GenePharma Co., Ltd. shRNF6, pcDNA3.1-RNF6, shMST1,
pcDNA3.1-MST1 and pcDNA3.1-YAP plasmids were also purchased from
Shanghai GenePharma Co., Ltd. shRNA plasmid and overexpression
vector transfection in BC cells was performed using
Lipofectamine™ 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
concentration of shRNF6 and shYAP was 6x108
TU/ml, and 1x106 cells were transfected with RNF6
and YAP plasmids (10 µg). The Lipofectamine™
3000-plasmids complex was formed by mixing for 20 min at room
temperature. Then, complex was added to the cells and the cells
were cultured in serum-free Opti-MEM (Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C for 4 h, followed by incubation in
serum-containing medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) at 37˚C for 72 h. After 72 h, the cells were collected and
washed twice with PBS. Sequences were as follows: RNF6, shiRNA-1,
5'-UUUCUGAGUCUCCAUCACUUGCCGC-3', and shRNA-2,
5'-UUUCGCGAGUCUCUCUACUUCACGC-3'; YAP, shRNA-1,
5'-AAGGUGAUACUAUCAACCAAA-3', and shRNA-2,
5'-AAGACAUCUUCUGGUCAGAGA-3' and MST1, shRNA 1,
5'-GGGCACTGTCCGAGTAGCAGC-3', and shRNA 2,
5'-CCGTCTTTCCTTGAATACTTT-3'.
In vivo ubiquitination assay
For the in vivo ubiquitination assay,
RNF6-deficient or RNF6-overexpressing BC cells were treated with
MG132 (15 µg/ml; cat. no. A2585; APeXBIO Technology LLC) for 4-6 h
at 37˚C. Following further incubation for 2-3 h, cell lysates were
immunoprecipitated with anti-MST1 antibody (1:1,000; cat. no. 3682;
Cell Signaling Technology, Inc.). Ubiquitination levels of MST1
were measured with an anti-ubiquitin (Ub) antibody (1:1,000; cat.
no. 10201-2-AP; ProteinTech Group, Inc.).
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from BC and non-tumour
adjacent issues using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. RNA was reverse transcribed into cDNA using a Prime
Script RT Reagent kit with gDNA Eraser (Takara Biotechnology Co.,
Ltd.) according to the manufacturer's instructions. cDNA was used
as a template to perform qPCR on the ABI PRISM 7500 auto
fluorescence PCR instrument (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the SYBR Premix Ex Taq Kit (Takara
Biotechnology Co., Ltd.).
The primer sequences were as follows: RNF6 forward,
5'-AGAAGATGGCAGCAAGAGCG-3' and reverse,
5'-TCAAGTCAGGCTGAGATGCTAGT-3'; MST1 forward,
5'-GGGTCCCAGTAGCCAAGAT-3' and reverse, 5'-GAGGCACCACATACCATTCA-3';
YAP forward, 5'-GGATTTCTGCCTTCCCTGAA-3' and reverse,
5'-GATAGCAGGGCGTGAGGAAC-3'; GAPDH forward,
5'-CTTCGCTCTCTGCTCCTCCT-3' and reverse,
5'-GTTAAAAGCAGCCCTGGTGA-3'.
Migration and invasion assay
Transwell-based migration and invasion assays in BC
cell lines were conducted as previously described (6), with minor modifications. For the cell
invasion assay, the polycarbonate membranes of the upper
compartment of the chambers were precoated with a matrix gel.
In vivo metastasis assay
An experimental model of lung metastasis was used to
investigate the effects of RNF6 on BC metastasis. A total of 60
female BALB/c nude mice (age, 4-6 weeks; weight, 18-20 g; Shanghai
SLAC Laboratory Animal Co., Ltd.) were maintained in a 20˚C
environment with 40-60% humidity and a 12/12-h light/dark cycle,
andwith a standardized barrier system at the Experimental Animal
Center of Nanchang University. For each nude mouse,
1x106 MDA-MB-231 cells in 100 µl PBS were injected via
the tail vein. The mice were sacrificed 6 weeks after tumor
implantation. The nude mice were euthanized by cervical
dislocation; cessation of breathing, corneal reflex and heartbeat
were considered to indicate mortality. The lungs and major organs
were removed and fixed with 10% formalin. Subsequently, consecutive
tissue sections (4-6 µm) were made from each block of the lung. The
sections were stained with hematoxylin-eosin staining (H&E).
These slides were examined systematically using an image analyzer
system (Olympus BH-2 microscope; Olympus Corporation). The animal
study was approved by the Ethics Committee for Animal Experiments
of The First Affiliated Hospital of Nanchang University and The
Third Hospital of Nanchang (approval no. NDYFY2013D078).
Immunofluorescence assay
A total of 2x103 MDA-MB-231 cells were
placed on slides for 24 h, fixed with 4% polyformaldehyde for 30
min at room temperature, then treated with 0.3% Triton X-100/PBS
for 5 min at room temperature. The cells were then sealed with 5%
BSA (Beijing Solarbio Science & Technology Co., Ltd.) at room
temperature for 1 h and stained overnight with antibodies
(Anti-RNF6 1:500; cat. no. ab204506; Abcam and anti-MST1 (1:500;
cat. no. abab76822); Abcam.) at 4˚C. Following primary antibody
incubation, cells were incubated with fluorescent dye-bound
secondary antibodies (both 1:200; both Invitrogen; Thermo Fisher
Scientific, Inc.; cat. nos. A32723 and A-11035) for 1 h at room
temperature and stained with DAPI (5 mg/ml) for 15 sec at room
temperature.
Co-immunoprecipitation (Co-IP) and
glutathione S-transferase (GST) pull-down experiments
For the co-IP assay, RNF6 knockdown or overexpressed
BC cells were exposed to MG132 (15 µg/ml; cat. no. A2585; APeXBIO
Technology LLC) for 4-6 h at 37˚C before harvesting. Subsequently,
theBC cells were extracted using RIPA lysis buffer (MilliporeSigma)
according to the manufacturer's instructions, then the cell lysate
was immunoprecipitated with anti-MST1 and the ubiquitination levels
of MST1 were measured with anti-Ub antibody. For the GST pull-down
assay, BL-21 competent Escherichia coli (Beijing Solarbio
Science & Technology Co., Ltd.) was transformed with shRNF6 or
PcDNA-3.1-RNF6 plasmids and cultured overnight at room temperature.
GST fusion protein expression was induced with isopropyl
β-D-thiogalactoside (1 mg/ml) for 3-5 h at 20-30˚C. Cells were
harvested in RIPA lysis buffer(Beyotime Institute of Biotechnology)
and homogenized by sonication. Following centrifugation (14,000 g,
5 min, 4˚C), GST fusion proteins in supernatant were purified by
glutathione sepharose 4B beads according to the manufacturer's
instructions (GE Healthcare). An aliquot (5 µg) of glutathione or
GST-RNF6 protein was added to cell lysates, followed by overnight
incubation with gentle rotation at 4˚C and addition of
glutathione-sep harose 4B beads (GE Healthcare) for 3 h at room
temperature. The beads were collected by centrifugation (14,000 g,
5 min, 4˚C) and washed with ice-cold lysis buffer. MST1 protein was
detected by western blotting.
Cycloheximide (CHX) chase assay
To inhibit MST1 protein synthesis in BC cells, cells
were treated with 100 µg/ml CHX (Sigma-Aldrich; Merck KGaA) at 37˚C
for 4 h. A total of 5x105 MDA-MB-231 cells were
transfected with shRNA or shRNF6 plasmids (6x108 TU/ml,
Shanghai GenePharma Co., Ltd.) using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) The
Lipofectamine™ 3000-plasmids complex was formed by
mixing for 20 min at room temperature. Then, complex was added to
the cells and the cells were cultured in serum-free Opti-MEM
(Gibco; Thermo Fisher Scientific, Inc.) at 37˚C for 4 h, followed
by incubation in serum-containing medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc.) at 37˚C for 48 h. At 48 h
post-transfection, the cells were harvested at different time
points (0, 1, 3 and 5 h) following CHX treatment and the expression
of MST1 protein was detected by western blotting.
Statistical analysis
Data are presented as the mean ± SD of three
independent experiments. The unpaired Student's t-test method was
used to evaluate the significance between two groups while the
one-way analysis of variance method followed by Tukey's test was
used to estimate the difference among multiple groups. The
association between RNF6 expression and clinicopathological
characteristics of patients with BC was analysed using Fisher's
exact probabilities test. One-way ANOVA followed by Tukey's post
hoc test was used for multiple comparisons. All statistical
calculations were performed using SPSS 19.0 (IBM Corp.) and all
graphs were constructed with GraphPad Prism 6.0 (GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
RNF6 overexpression is correlated with
poor outcome in patients with BC
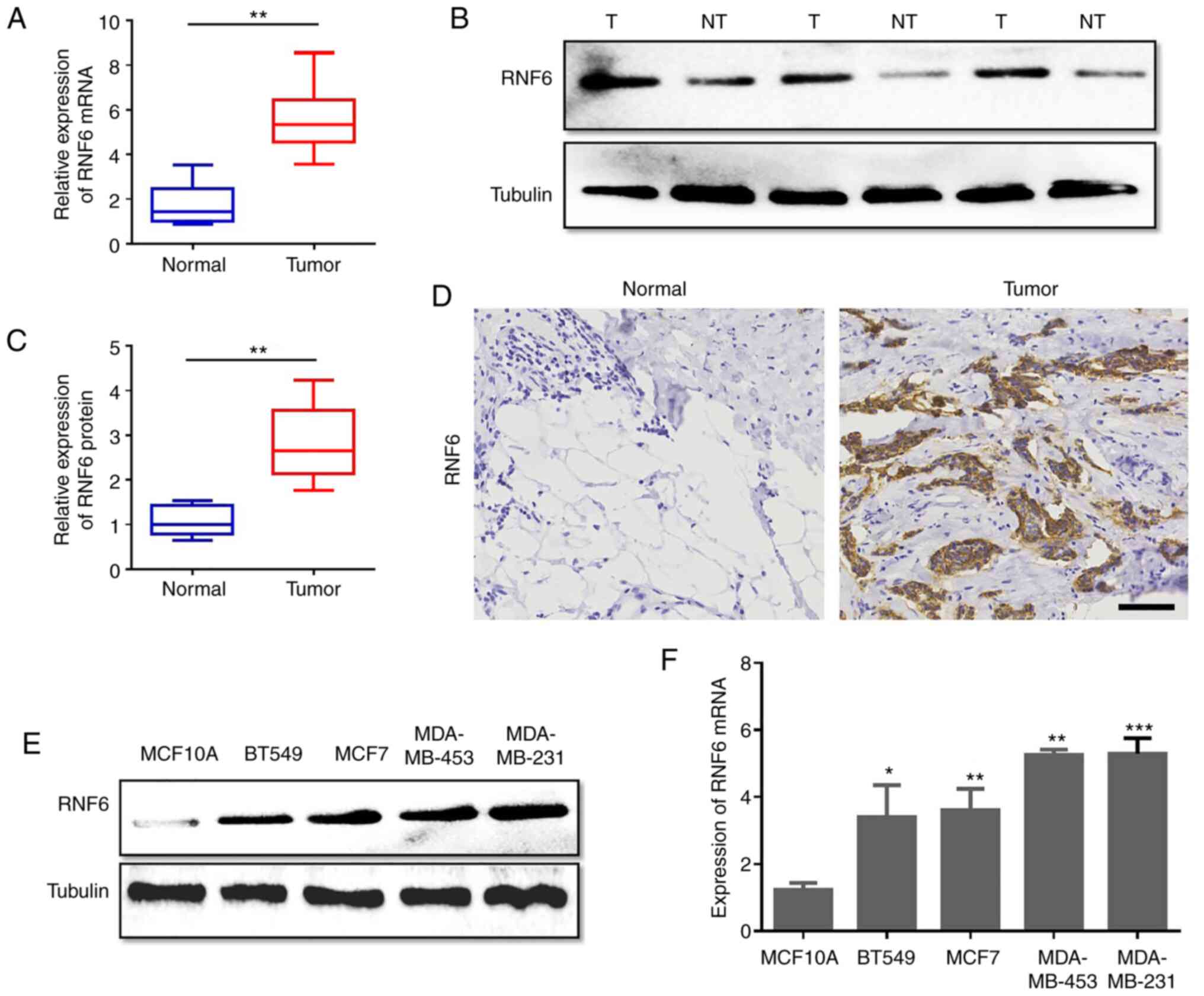
The potential role of RNF6 in the development and
progression of BC was investigated. Initially, the expression of
RNF6 in 146 BC and non-tumour adjacent tissue samples obtained from
patients with BC. was detected using RT-qPCR. The RT-qPCR data
revealed that RNF6 was highly expressed in primary breast cancer
tissues compared with adjacent control tissues (Fig. 1A). Furthermore, the protein levels
of 60 fresh BC tumour tissues and their paired non-tumour adjacent
tissues were quantitatively analysed. In accordance with increased
RNF6 mRNA levels, the levels of RNF6 protein in BC tissues were
significantly higher compared with adjacent normal tissues
(Fig. 1B and C). The levels of RNF6 protein in BC
paraffin-embedded tissue samples and corresponding normal paraffin
tissues was detected by IHC with an RNF6 antibody. RNF6 protein was
highly expressed in 65.75% (96 of 146) of the BC tissue samples
(Fig. 1D). To investigate the
expression of RNF6 in BC cells, the expression levels of RNF6 in BC
cell lines and normal human mammary gland cells (MCF10A) were
detected. RT-qPCR and western blot data showed that RNF6 expression
in BC cells was higher compared with MCF10A cells (Fig. 1E and F). These results demonstrated that high
RNF6 levels were observed in BC tissues.
To explore the correlation between RNF6
overexpression and BC clinicopathological parameters, the
expression of RNF6 in BC specimens and clinicopathological
characteristics were detected by IHC. RNF6 overexpression was
remarkably correlated with tumour stage and lymph node metastasis
stages (Table I). In addition, to
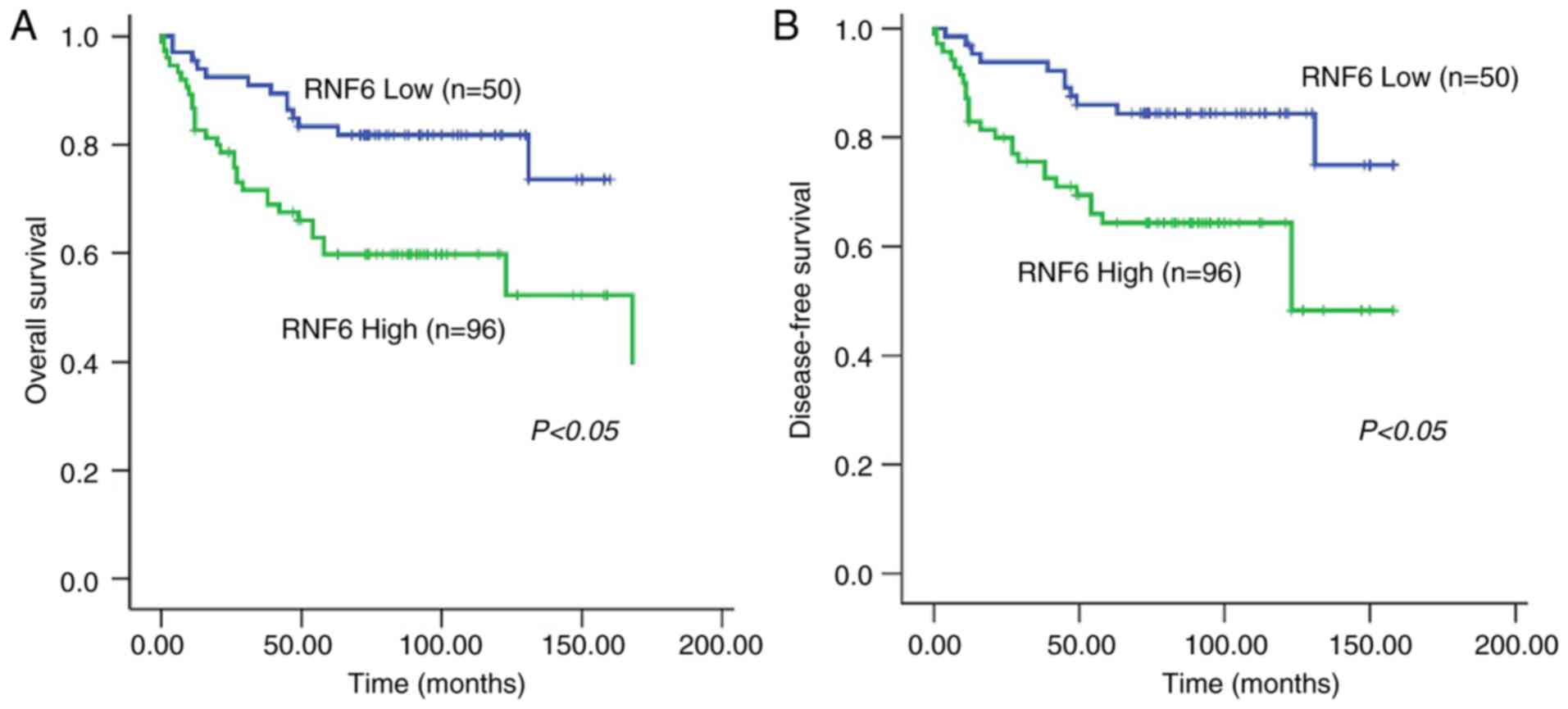
investigate the efficiency of RNF6 in the survival of patients with
BC, the association between RNF6 levels and the survival of
patients with BC was investigated. Patients with high RNF6 levels
were correlated with poor overall survival and poor disease-free
survival compared with patients with low levels of RNF6 (Fig. 2A and B). The results of multivariate Cox
regression analysis indicated that high RNF6 levels were an
independent predictor of poor survival in patients with BC
(Table II). In summary, these data
suggested that RNF6 was upregulated in BC tissues and associated
with unfavorable prognosis in patients with BC.
 | Table IAssociation between RNF6 expression
and clinicopathological features in 146 patients with breast
cancer. |
Table I
Association between RNF6 expression
and clinicopathological features in 146 patients with breast
cancer.
| | | RNF6
expression | |
|---|
| Parameters | Total cases
146 | High (n=96) | Low (n=50) | P-value |
| Age (years) | | | | P=0.302 |
|
<50 | 79 | 49 | 30 | |
|
≥50 | 67 | 47 | 20 | |
| Pathological
grade | | | | P=0.906 |
|
<2 | 116 | 76 | 40 | |
|
≥2 | 30 | 20 | 10 | |
| Clinical stage | | | | P=0.234 |
|
I-II | 97 | 67 | 30 | |
|
III-IV | 49 | 29 | 20 | |
| Tumor stage | | | | P<0.001 |
|
T1-T2 | 86 | 46 | 40 | |
|
T3-T4 | 60 | 50 | 10 | |
| Lymph node
metastasis stage | | | | P=0.004 |
|
N0-N1 | 97 | 56 | 41 | |
|
N2 | 49 | 40 | 9 | |
| Metastasis
stage | | | | P=0.067 |
|
M0 | 135 | 86 | 49 | |
|
M1 | 11 | 10 | 1 | |
 | Table IIUnivariate and multivariate analyses
of overall survival in patients with breast cancer. |
Table II
Univariate and multivariate analyses
of overall survival in patients with breast cancer.
| | Univariate
analysis | Multivariate
analysis |
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age
(≥50/<50) | 0.652 | 0.344-1.431 | 0.516 | - | - | - |
| Pathological grade
(<2/≥2) | 0.812 | 0.516-1.664 | 0.874 | - | - | - |
| Clinical stage
(I-II/III-IV) | 1.814 | 1.121-4.551 | 0.163 | - | - | - |
| Tumor stage
(T1-T2/T3-T4) | 1.757 | 1.163-3.146 | 0.012 | 1.642 | 0.925-3.426 | 0.074 |
| Lymph node
metastasis stage (N0-N1/N2) | 1.612 | 1.211-3.441 | 0.014 | 1.433 | 1.123-2.451 | 0.021 |
| Metastasis stage
(M0/M1) | 0.762 | 0.431-4.134 | 0.521 | - | - | - |
| RNF6 expression
(high/low) | 1.724 | 1.341-3.312 | 0.006 | 1.532 | 1.121-3.642 | 0.026 |
RNF6 promotes the migration and
invasion of BC in vivo and in vitro
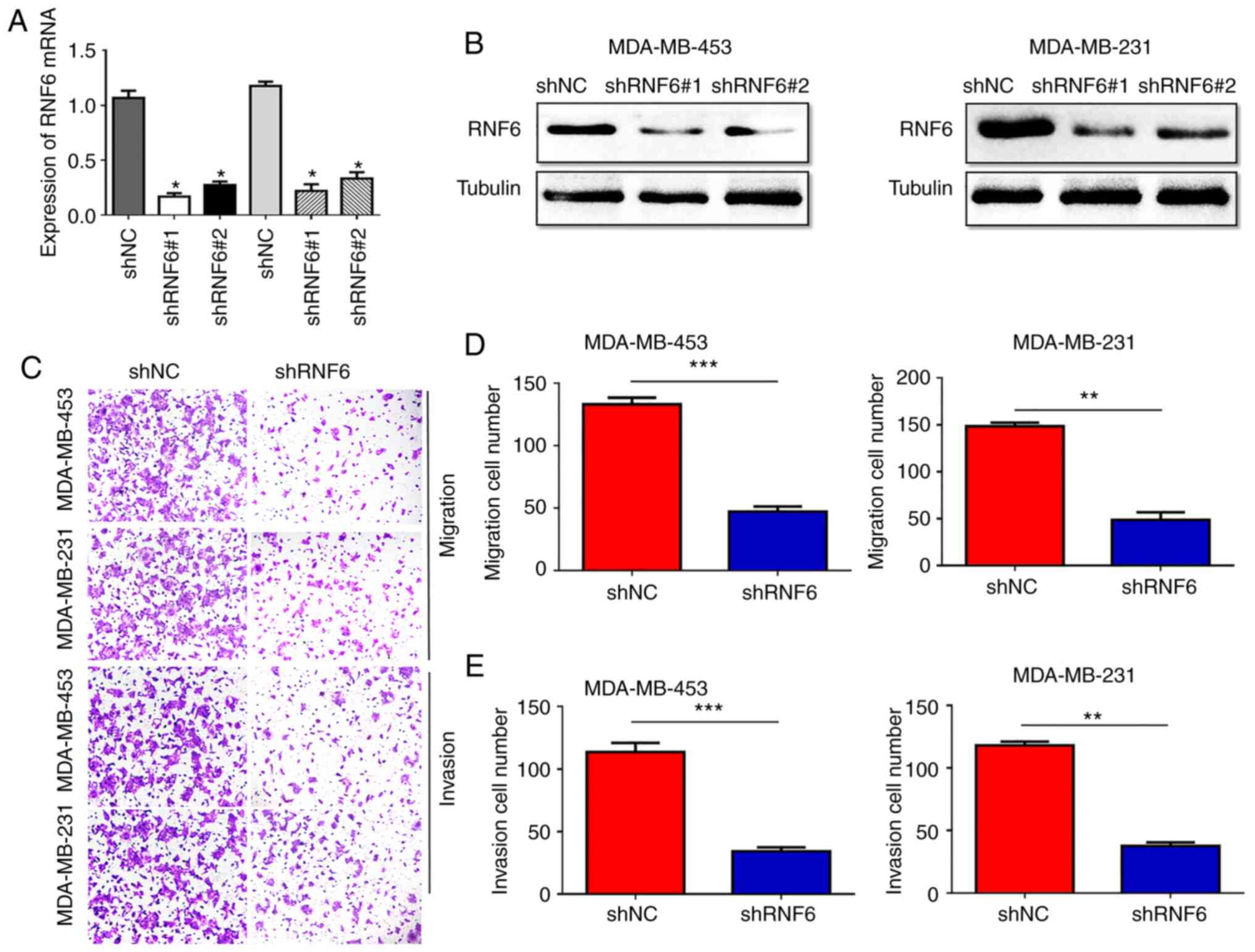
Since high RNF6 expression correlated with clinical
and lymph node metastasis stages, the role of RNF6 in the migration
and invasion of BC was investigated. RNF6 shRNA and corresponding
controls were used to silence RNF6 expression in MDA-MB-231 and
MDA-MB-453 cells with high endogenous RNF6 expression (Fig. 3A and B). A significant decrease in BC cell
migration was observed in the shRNF6 group compared with the shNC
group (Fig. 3C and D). Furthermore, stable RNF6 knockdown in
BC cells showed lower invasion rates compared with control cells
(Fig. 3E). Subsequently, the effect
of RNF6 on BC metastasis was investigated using an in vivo
tumour metastasis assay. The present results showed fewer lung
metastatic nodules in the shRNF6 group compared with the control
group (Fig. S1). In conclusion,
these data indicated that RNF6 promoted the migration and invasion
of BC in vivo and in vitro.
RNF6 facilitates the invasion and
migration of BC cells by YAP
To study the mechanism by which RNF6 regulates the
invasion and migration of BC cells, a luciferase reporter assay was
performed to screen the signalling pathway downstream of RNF6. As
shown in Fig. 4A and B, RNF6 knockdown inhibited YAP activity at
the basal level during YAP stimulation. Knockdown of RNF6
expression suppressed the expression of Cyr61, connective tissue
growth factor and cyclin E, which are target genes of YAP (Fig. 4C). RNF6 downregulation reduced YAP
protein levels compared with controls (Fig. 4D). These results indicated that RNF6
regulated the transcriptional activity of YAP by downregulating YAP
protein levels. To further investigate whether RNF6 mediates the
metastasis and invasion of BC by regulating YAP, YAP was
overexpressed in RNF6 knockdown BC cells. Protein expression levels
of RNF6 and YAP were measured by western blotting, and cell
migration and invasion were investigated by performing Transwell
assays. The western blotting image in Fig. 4E showed that RNF6 downregulation
decreased YAP expression and YAP upregulation attenuated the loss
of YAP expression in RNF6 knockdown BC cells. The migration and
invasion capabilities of MDA-MB-231 cells were significantly
reduced by RNF6 knockdown. However, YAP upregulation attenuated the
reduction in migration and invasion ability caused by RNF6
knockdown (Fig. 4F-H). YAP
expression was then knocked down in RNF6-overexpressing cells
(Fig. 4I), which reversed the
promoting effects of RNF6 on the migration of BC cells (Fig. 4J-L). Collectively, these findings
suggested that RNF6-promoted migration and invasion is
YAP-dependent in BC cells.
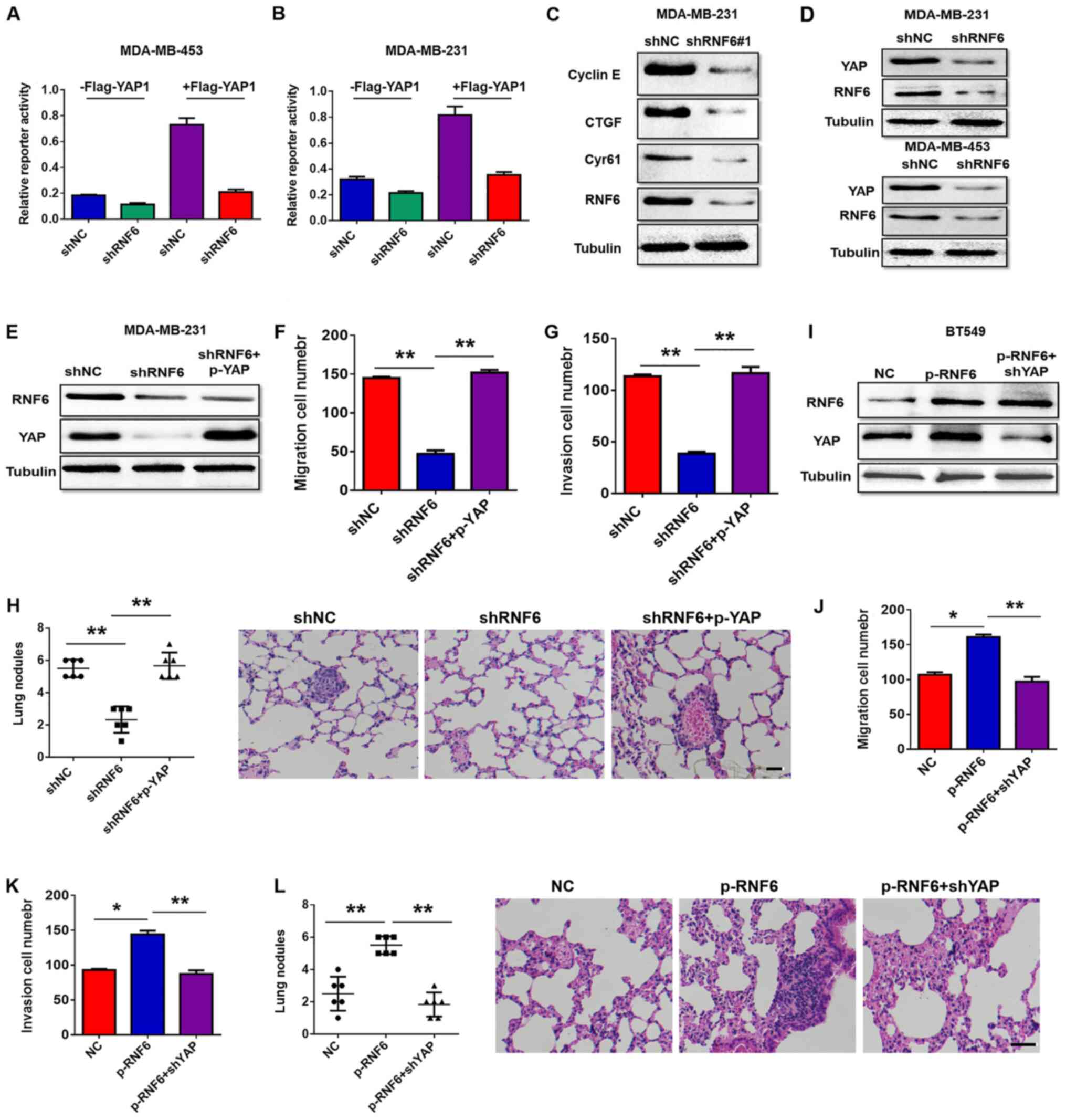 | Figure 4RNF6 promotes migration of breast
cancer cells via YAP in vitro and in vivo. Activity
of both endogenous and overexpressed YAP was measured using a YAP
reporter assay following RNAi-induced RNF6 knockdown in (A)
MDA-MB-453 and (B) MDA-MD-231 cells. (C) Levels of Cyr61, CTGF and
cyclin E, three target genes of YAP, were assessed by western
blotting in MDA-MB-231 cells stably transfected with shNC or
shRNF6. (D) Expression levels of YAP were assessed by western
blotting in BC cells stably transfected with shNC vector or shRNF6.
(E) Western blotting was performed to detect the expression of RNF6
and YAP. Transwell assay showed that upregulation of YAP
significantly rescued (F) cell migration and (G) invasion in
shRNF6-transfected MDA-MB-231 cells. (H) Statistical analysis of
lung metastatic nodules in RNF6 knockdown cells. Magnification,
x400. (I) Protein levels of RNF6 and YAP were detected by western
blotting. Transwell assay showed that YAP inhibition decreased
RNF6-enhanced (J) cell migration and (K) invasion. (L) Statistical
analysis of lung metastatic nodules in RNF6-overexpressing cells.
n=6/group. Scale bar, 50 µm. *P<0.01;
**P<0.01. RNF6, ring finger protein 6; YAP,
YES-associated protein; CTGF, connective tissue growth factor; sh,
short hairpin; NC, negative control. |
RNF6 regulates YAP expression via
MST1
Numerous studies have shown that MST1 is the main
component of YAP signalling (16,17),
and regulates the expression of YAP in pancreatic cancer (18,19).
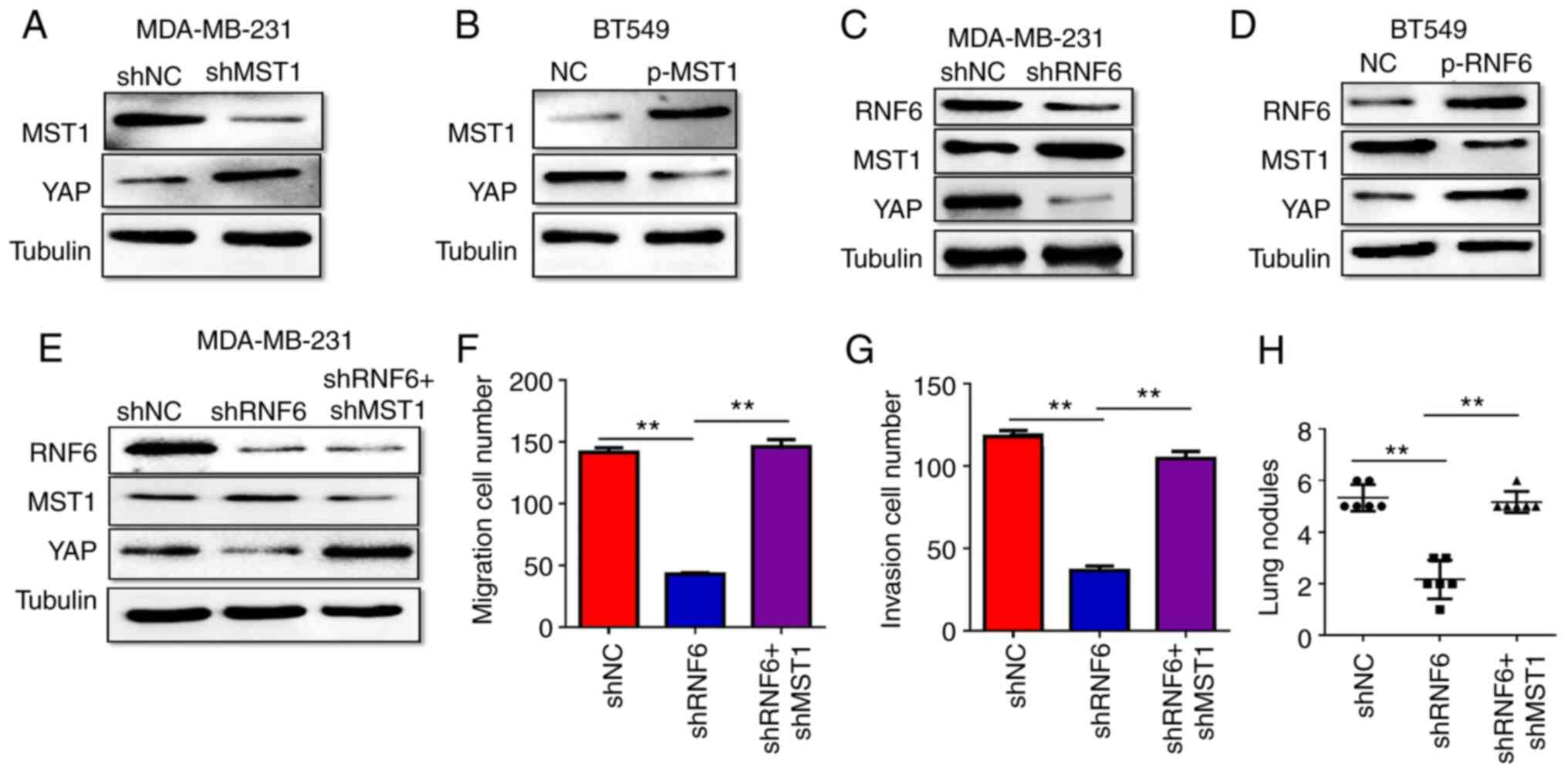
Therefore, the present study investigated whether MST1 is involved
in RNF6 regulation of YAP in BC. To confirm this hypothesis, the
change in YAP expression in shMST1-transfected MDA-MB-231 cells was
measured. Compared with their respective NC groups, MST1 knockdown
increased YAP expression in MDA-MB-231 cells (Fig. 5A), whereas MST1 upregulation had the
opposite effect in BC cells (Fig.
5B). These findings demonstrated that MST1 regulated YAP
expression in BC cells.
To determine whether RNF6 affects YAP expression via
MST1, the changes in MST1 and YAP expression in RNF6 knockdown
MDA-MB231 cells were further measured. RNF6 knockdown significantly
reduced YAP protein expression but increased MST1 protein
expression in MDA-MB231 cells compared with the shNC group
(Fig. 5C). By contrast, RNF6
overexpression significantly increased YAP protein expression but
decreased MST1 protein expression in BC cells compared with the NC
group (Fig. 5D). These findings
indicated that MST1 is involved in RNF6 regulation of YAP
expression. To further verify that RNF6 regulates YAP expression
via MST1 in BC cells, MST1 was knocked down in RNF6-deficient BC
cells. MST1 downregulation rescued decreased YAP expression in
RNF6-deficient MDA-MB231 cells (Fig.
5E), and the cell migration and invasion abilities were also
rescued in vitro and in vivo (Fig. 5F-H). These results demonstrated that
RNF6 regulated YAP-induced BC migration and invasion in an
MST1-dependent manner.
RNF6 promotes MST1 degradation
To further elucidate the mechanism of RNF6 in
regulating MST1 in BC cells, the interaction between RNF6 and MST1
was examined. Immunofluorescence analysis showed that RNF6 and MST1
protein were co-localised in MDA-MB-231 cells (Fig. 6A). In GST pull-down experiments, the
results showed that RNF6 is directly combined with MST1 in BC cells
(Fig. 6B). Co-IP results further
confirmed that there was a direct binding between RNF6 and MST1
(Fig. 6C). In general, these data
demonstrated that RNF6 protein interacts with MST1 protein.
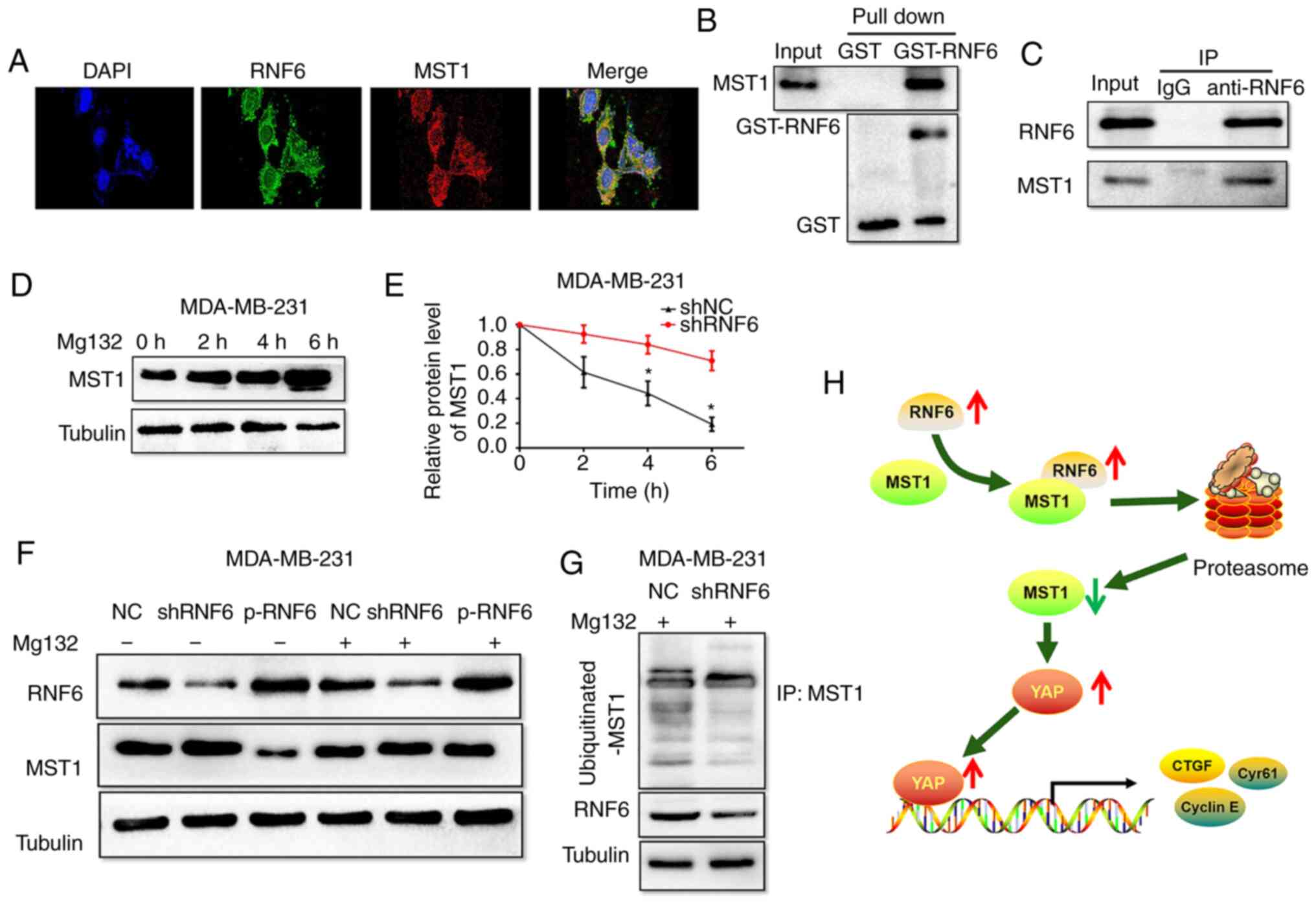 | Figure 6RNF6 accelerates ubiquitination and
degradation of MST1 in breast cancer cells. (A) Immunofluorescence
staining for MST1 and RNF6 in MDA-MB-231 cells. Magnification,
x400. Scale bar, 100 µm (B) MDA-MB-231 cell lysates were incubated
with GST-RNF6 or GST fusion protein and glutathione-sepharose 4B
beads for 3 h before washing. The elution was subsequently analyzed
by western blotting with anti-MST1 antibody. (C) MST1 was blotted
on immunoprecipitated endogenous RNF6 in MDA-MB-231 cell lysates.
(D) MDA-MB-231 cells were treated with MG132 (15 µmol/l) and the
protein levels of MST1 were detected. (E) RNF6 knockdown promoted
degeneration of MST1 protein in MDA-MB-231 cells.
*P<0.05 vs. shRNF6. (F) RNF6 had no effect on MST1
expression, as assessed by western blotting following transfection
with shRNF6 or p-RNF6 in BC cells. (G) Cells were treated with
proteasomal inhibitor MG132 (10 mmol/l). Cell lysates were prepared
and subjected to immunoprecipitation with anti-MST1 antibody. The
levels of Ub-attached MST were detected by western blotting
analysis with Ub antibody. (H) Proposed model by which RNF6
promotes migration and invasion of breast cancer by promoting
ubiquitination and degradation of MST1. YAP, YES-associated
protein; RNF6, ring finger protein 6; MST1, mammalian STE20-like
protein kinase 1; sh, short hairpin; Ub, ubiquitin; GST,
glutathione S-transferase; NC, negative control; p, plasmid. |
Since RNF6 is one of the most important E3 ubiquitin
ligases, the present study investigated whether RNF6 induces
ubiquitination and degradation of MST1 via the ubiquitin-proteasome
system (UPS). As shown in Fig. 6D,
significant time-dependent accumulation of MST1 protein was
observed in BC cells treated with the proteasome inhibitor MG132.
These results demonstrated that MST1 is degraded by the UPS in BC
cells. Subsequently, the degradation of MST1 protein in
RNF6-deficient cells following exposure to CHX was tested. The
degradation dynamics assay showed that the half-life of the
expressed MST1 was significantly decreased in shRNF6-transfected
MDA-MB-31 cells compared with shNC-transfected cells (Fig. 6E). The present results showed that
RNF6 silencing inhibited MST1 protein degradation in BC cells.
Moreover, MST1 protein levels remained unaltered after
dysregulation of RNF6 in BC cells following treatment with MG132
(Fig. 6F). Furthermore, RNF6
knockdown significantly reduced the ubiquitination levels of MST1
compared with the NC group (Fig.
6G). Collectively, these findings suggested that RNF6
accelerated the ubiquitination and degradation of MST1 in BC
cells.
Discussion
RNF6, a member of the E3 ligase ring finger family,
has been shown to play an important role in the progression of
malignant tumours (7,20). However, to the best of our
knowledge, the molecular mechanism of RNF6 in BC remains to be
elucidated. The present study showed that RNF6 expression levels in
the tumours of patients with BC significantly increased compared
with normal breast tissue. The present results showed that RNF6
overexpression was remarkably correlated with the clinical, tumour
and lymph node metastasis stages of BC. Furthermore, patients with
high RNF6 levels were correlated with poor overall survival and
disease-free survival compared with patients with low RNF6 levels.
The results of multivariate Cox regression analysis indicated that
high RNF6 levels were an independent predictor of poor survival in
patients with BC. In summary, these data suggested that RNF6 acts
as an oncogene in BC tissues and associated with unfavorable
prognosis in patients with BC.
Metastasis is a crucial biological feature in cancer
and is closely associated with cancer progression (21-23).
The high invasiveness and distant metastasis of BC are considered
as the main obstacles of patients with BC to obtain satisfactory
prognosis (21-23).
Therefore, it is of significance to understand the mechanism behind
BC metastasis. The present study showed a significant decrease in
BC cell migration in the shRNF6 group. Stable RNF6 knockdown in BC
cells resulted in lower migration compared with control cells.
Furthermore, the present results showed that RNF6 downregulation
reduced the expression of YAP, and YAP upregulation reduced the
loss of YAP expression in RNF6-deficient BC cells. The present
findings demonstrated that RNF6 knockdown significantly reduced the
migration and invasion ability of cancer cells, and YAP
upregulation rescued the decreased migration and invasion ability
induced by RNF6 knockdown. These results demonstrated that RNF6
regulates YAP-induced BC migration and invasion.
Subsequently, the molecular mechanism of RNF6
regulating YAP expression was investigated. Studies have shown that
the carcinogenic Hippo-YAP pathway plays a key role in regulating
BC progression, in which MST1 is activated by oxidative stress and
plays a central role in the Hippo pathway, controlling cell
proliferation, differentiation and apoptosis during development
(24-29).
The present results showed that MST1 knockdown significantly
increased YAP expression in BC cells, whereas MST1 upregulation had
the opposite effect in BC cells. These findings suggested that MST1
regulated YAP expression in BC cells. Furthermore, the present data
showed that MST1 downregulation rescued decreased YAP expression in
RNF6-deficient MDA-MB231 cells, and the cell migration and invasion
abilities were also rescued. These results demonstrated that RNF6
regulates YAP-induced BC migration and invasion in an
MST1-dependent manner.
Finally, the mechanism by which RNF6 regulated MST1
expression in BC was further explored. Previous studies have
reported that tumor necrosis factor receptor-associated factor 6
regulates YAP signaling by promoting the ubiquitination and
degradation of MST1 in pancreatic cancer (30). The present results showed that RNF6
is a novel E3 ubiquitin ligase of MST1, and RNF6 induced
ubiquitination and degradation of MST1 through the UPS. This
conclusion is mainly based on the following reasons. First, the
present results showed that RNF6 protein interacted with MST1
protein. Second, significant accumulation of MST1 protein was
observed in BC cells treated with MG132. These results indicated
that MST1 is degraded by the UPS in BC cells. Third, the present
results showed that RNF6 silencing promoted MST1 protein
degradation in BC cells. Moreover, MST1 protein levels remained
unchanged after dysregulation of RNF6 in BC cells following
treatment with MG132. In addition, RNF6 knockdown significantly
reduced the ubiquitination levels of MST1.
In conclusion, the present study provided evidence
that RNF6 plays an oncogenic role in BC. We not only clarified the
role of RNF6 in breast cancer metastasis, but also provided
insights into the mechanism of RNF6 via mediating the MST1/YAP axis
(Fig. 6H). These results
highlighted the important role of RNF6 expression, which may be
used as a prognostic indicator and therapeutic target for future
breast cancer treatment.
Supplementary Material
Quantification of the number of
lesions in the lungs of animals injected in the tail vein with (A)
MDA-MB-231 and (B) MDA-MB-453 cells. Data are presented as the mean
± SD (n=10). RNF6, ring finger protein 6; sh, short hairpin; NC,
negative control. Scale bar, 50 μm. **P<0.01;
***P<0.001 vs. NC group.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by a grant from the National
Natural Science Foundation of China (grant no. 81960503).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YH and YZ performed the in vitro experiments
and analyzed the data. YH performed the animal experiments. YZ
collected the clinical tissue samples. ZJ and YH designed the
study, wrote the manuscript and revised the manuscript. All authors
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of the First Affiliated Hospital of Nanchang University
and the Third Hospital of Nanchang, and written informed consent
was obtained from the parents or guardians of all patients. The
animal studies were approved by the Ethical Committee of the First
Affiliated Hospital of Nanchang University and the Third Hospital
of Nanchang.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chan BK, Wiseberg-Firtell JA, Jois RH,
Jensen K and Audisio RA: Localization techniques for guided
surgical excision of non-palpable breast lesions. Cochrane Database
Syst Rev. (CD009206)2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sharma R, Sharma R, Khaket TP, Dutta C,
Chakraborty B and Mukherjee TK: Breast cancer metastasis: Putative
therapeutic role of vascular cell adhesion molecule-1. Cell Oncol
(Dordr). 40:199–208. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Liu L, Zhang Y, Wong CC, Zhang J, Dong Y,
Li X, Kang W, Chan FKL, Sung JJY and Yu J: RNF6 promotes colorectal
cancer by activating the Wnt/β-catenin pathway via ubiquitination
of TLE3. Cancer Res. 78:1958–1971. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liang Q, Ma D, Zhu X, Wang Z, Sun TT, Shen
C, Yan T, Tian X, Yu T, Guo F, et al: RING-finger protein 6
amplification activates JAK/STAT3 pathway by modifying SHP-1
ubiquitylation and associates with poor outcome in colorectal
cancer. Clin Cancer Res. 24:1473–1485. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cai J, Xiong Q, Jiang X, Zhou S and Liu T:
RNF6 facilitates metastasis and radioresistance in hepatocellular
carcinoma through ubiquitination of FoxA1. Exp Cell Res.
374:152–161. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xu K, Shimelis H, Linn DE, Jiang R, Yang
X, Sun F, Guo Z, Chen H, Li W, Chen H, et al: Regulation of
androgen receptor transcriptional activity and specificity by
RNF6-induced ubiquitination. Cancer Cell. 15:270–282.
2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lo HS, Hu N, Gere S, Lu N, Su H, Goldstein
AM, Taylor PR and Lee MP: Identification of somatic mutations of
the RNF6 gene in human esophageal squamous cell carcinoma. Cancer
Res. 62:4191–4193. 2002.PubMed/NCBI
|
|
9
|
Qin X, Chen S, Qiu Z, Zhang Y and Qiu F:
Proteomic analysis of ubiquitination-associated proteins in a
cisplatin-resistant human lung adenocarcinoma cell line. Int J Mol
Med. 29:791–800. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hippo signaling mediates ferroptosis in
malignant mesothelioma. Cancer Discov. 9(1155)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Turunen SP, von Nandelstadh P, Öhman T,
Gucciardo E, Seashore-Ludlow B, Martins B, Rantanen V, Li H,
Höpfner K, Östling P, et al: FGFR4 phosphorylates MST1 to confer
breast cancer cells resistance to MST1/2-dependent apoptosis. Cell
Death Differ. 26:2577–2593. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gomez M, Kulaberoglu Y and Hergovich A:
MST1/2 kinase assays using recombinant proteins. Methods Mol Biol.
1893:319–331. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ji XY, Zhong G and Zhao B: Molecular
mechanisms of the mammalian Hippo signaling pathway. Yi Chuan.
39:546–567. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
He L, Yuan L, Sun Y, Wang P, Zhang H, Feng
X, Wang Z, Zhang W, Yang C, Zeng YA, et al: Glucocorticoid receptor
signaling activates TEAD4 to promote breast cancer progression.
Cancer Res. 79:4399–4411. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tang DE, Dai Y, Lin LW, Xu Y, Liu DZ, Hong
XP, Jiang HW and Xu SH: STUB1 suppresseses tumorigenesis and
chemoresistance through antagonizing YAP1 signaling. Cancer Sci.
110:3145–3156. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chen M, Zhang H, Shi Z, Li Y, Zhang X, Gao
Z, Zhou L, Ma J, Xu Q, Guan J, et al: The MST4-MOB4 complex
disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a
pro-oncogenic role in pancreatic cancer. J Biol Chem.
293:14455–14469. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Dent P, Booth L, Roberts JL, Liu J,
Poklepovic A, Lalani AS, Tuveson D, Martinez J and Hancock JF:
Neratinib inhibits Hippo/YAP signaling, reduces mutant K-RAS
expression, and kills pancreatic and blood cancer cells. Oncogene.
38:5890–5904. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lipkowitz S and Weissman AM: RINGs of good
and evil: Ring finger ubiquitin ligases at the crossroads of tumour
suppression and oncogenesis. Nat Rev Cancer. 11:629–643.
2011.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Fang S, Lorick KL, Jensen JP and Weissman
AM: Ring finger ubiquitin protein ligases: Implications for
tumorigenesis, metastasis and for molecular targets in cancer.
Semin Cancer Biol. 13:5–14. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhu L, Tong G, Chen J, Wang Y, Wang S,
Zhao M, Li J and Ma J: Cloning and identification of a novel RNF6
transcriptional splice variant Spg2 in human development. Sci China
C Life Sci. 51:302–307. 2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Viale G, Hanlon Newell AE, Walker E,
Harlow G, Bai I, Russo L, Dell'Orto P and Maisonneuve P: Ki-67
(30-9) scoring and differentiation of Luminal A- and Luminal B-like
breast cancer subtypes. Breast Cancer Res Treat. 178:451–458.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen JY, Lai YS, Chu PY, Chan SH, Wang LH
and Hung WC: Cancer-derived VEGF-C increases chemokine production
in lymphatic endothelial cells to promote CXCR2-dependent cancer
invasion and MDSC recruitment. Cancers (Basel).
11(1120)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Paoletti C, Miao J, Dolce EM, Darga EP,
Repollet MI, Doyle GV, Gralow JR, Hortobagyi GN, Smerage JB, Barlow
WE and Hayes DF: Circulating tumor cell clusters in metastatic
breast cancer patients: A SWOG S0500 translational medicine study.
Clin Cancer Res. 25(0208.2019)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
D'Agostino L, Nie Y, Goswami S, Tong K, Yu
S, Bandyopadhyay S, Flores J, Zhang X, Balasubramanian I, Joseph I,
et al: Recycling endosomes in mature epithelia restrain tumorigenic
signaling. Cancer Res. 79:4099–4112. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Salah Z, Melino G and Aqeilan RI:
Correction: Negative regulation of the Hippo pathway by E3
ubiquitin ligase ITCH is sufficient to promote tumorigenicity.
Cancer Res. 79(3007)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Pan Z, Tian Y, Cao C and Niu G: The
emerging role of YAP/TAZ in tumor immunity. Mol Cancer Res.
17:1777–1786. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu J, Minikes AM, Gao M, Bian H, Li Y,
Stockwell BR, Chen ZN and Jiang X: Intercellular interaction
dictates cancer cell ferroptosis via NF2-YAP signalling. Nature.
572:402–406. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Gobbi G, Donati B, Do Valle IF, Reggiani
F, Torricelli F, Remondini D, Castellani G, Ambrosetti DC,
Ciarrocchi A and Sancisi V: The Hippo pathway modulates resistance
to BET proteins inhibitors in lung cancer cells. Oncogene.
38:6801–6817. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Huang W, Hu H, Zhang Q, Wu X, Wei F, Yang
F, Gan L, Wang N, Yang X and Guo AY: Regulatory networks in
mechanotransduction reveal key genes in promoting cancer cell
stemness and proliferation. Oncogene. 38:6818–6834. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li JA, Kuang T, Pu N, Fang Y, Han X, Zhang
L, Xu X, Wu W, Wang D, Lou W and Rong Y: TRAF6 regulates YAP
signaling by promoting the ubiquitination and degradation of MST1
in pancreatic cancer. Clin Exp Med. 19:211–218. 2019.PubMed/NCBI View Article : Google Scholar
|