Introduction
Neurodevelopmental disabilities (NDDs), such as
learning disabilities, developmental delay, cerebral palsy and
cognitive dysfunction, are one of the major diseases that affect
the quality of an individual's life from birth and harm the
physical and mental health of children and adolescents; they also
cause a heavy and far-reaching burden on families and society
(1). NDDs are not only related to
genetic factors (2,3) but also to medications taken during
pre-pregnancy and rubella infection in early pregnancy infections
(4,5). Environmental toxicants and industrial
chemicals, such as lead (6),
cadmium (7), arsenic (8), ethanol (9), methyl mercury (10), pesticides, organic solvents
(11) and endocrine disrupting
chemicals (EDCs) (12,13), which are abundant in the
environment, affect the developing brain, alter neurogenesis and
cause learning and memory deficits.
Neural stem cells (NSCs) exist in the nervous system
which can self-renew and differentiate into neurons, astrocytes and
oligodendrocytes (14). A moderate
number of NSCs are needed for the normal development of the nervous
system (15). The abnormal
proliferation or development of NSCs can cause severe neurological
developmental defects (15,16).
Various factors, such as bisphenol A (BPA) interfere with the
induction or occurrence of nerves by affecting the normal
proliferation or differentiation of NSCs, thereby producing the
neurodevelopmental toxicity expressed in severe NDDs (17).
NSCs change their response to external signals and
their developmental differentiation potential with different
developmental stages during neurogenesis (14). Some NSCs begin to express
neuron-specific genes such as glial fibrillary acidic protein
(GFAP) and microtubule-associated protein 2 (MAP2) to obtain
neuronal fate under the action of some regional differentiation
signals. However, Nestin and SRY-box 2 (Sox2) are expressed in the
population of undifferentiated NSCs that still maintain
pluripotency (18).
BPA is a synthetic phenolic compound widely used to
produce polycarbonate and epoxy resins for food containers, such as
cans and water dispensers (19).
BPA can leach from some of these polymers into water or food
products (19). Humans are exposed
to this compound through their diet and skin contact (20); BPA has been detected in >90% of
the human bodies surveyed in population-representative samples
(21,22). BPA exposure causes a wide range of
neurodevelopmental disorders, including cognitive impairment,
autism (23), neurodegeneration
(24-26)
and schizophrenia (27).
According to the development process of the
vertebrate nervous system, NSCs are the starting point for
differentiation into other nerve cells and are activated in
neurodegenerative diseases; they also play an important role in the
repair/replacement of nerve cells in lesion areas (28,29).
Both in vivo and in vitro experiments confirm that
BPA affects the proliferation and differentiation of NSCs (30), which triggers the development of
neurodegenerative diseases (31).
Previous studies have looked into whether the high
or low doses of BPA can influence NSCs, including the possible role
it may exert in NSCs or whether it can regulate NSC proliferation
and differentiation via the Wnt/β-Catenin, TGF-β or the estrogen
receptor signaling pathways (12,30,32).
Several previous studies on ovarian cancer cells confirm that BPA
can affect the TGF-β and the Wnt/β-Catenin signaling pathway
(33,34). Downstream Smads of TGF-β can
activate c-myc through zinc finger E-box-binding homeobox 1 (ZEB)1,
ZEB2, Snail zinc finger protein (SNAI)1, Snail2, Twist-related
protein 1 and PIM1, while c-myc can regulate the expression of
aurora kinases B (AURKB) to promote cell proliferation (35). The TGF-β signaling pathway can also
activate downstream factors to prevent cell differentiation by
forming myc-max heterodimers to inhibit Id2 expression
(36). In order to further
understand the mechanism of BPA-induced NDDs in infants, the
current study revealed that low-dose BPA (1 µM) enhanced the
stemness of human NSCs via the estrogen-related receptor α (ERRα)
(37) and the TGF-β signaling
pathways.
Materials and methods
Cell culture and treatment
Human NSCs (ReNcell® CX Immortalized
cells; cat. no. SCC007; MilliporeSigma) were provided by Dr Dan Lou
(Shanghai Municipal Center for Disease Control & Prevention,
Shanghai, China). Before cell resuscitation, Laminin (cat. no.
L-2020; MilliporeSigma) was thawed, diluted with DMEM (Gibco;
Thermo Fisher Scientific, Inc.) and plated on 35-mm petri dishes at
4˚C for 2 h. Then, 1x104 cells were seeded in the
laminin-coated 35-mm culture dishes filled with complete medium,
which was prepared by adding basic fibroblast growth factor (bFGF;
20 ng/ml) and EGF (20 ng/ml) to maintenance medium (cat. no.
SCM005; MilliporeSigma) for maintain cell stemnessand cultured in a
37˚C, 5% CO2 saturated humidity incubator. The fresh
cell culture medium was replaced every 1-2 days to form a stemness
maintenance model. According to the manufacturer's instructions of
ReNcell CX kit (cat. no. SCC009; MilliporeSigma), cell
differentiation was identified by double immunofluorescence
staining. The antibodies of β-tubulin III (1:500, ab215037; Abcam)
and GFAP (1:50, ab279290; Abcam) were used to form a neural
differentiation model after incubation overnight at 37 ˚C.
A total of 1x104 NSCs with 3 ml culture
medium were seeded into six-well plates in triplicate. The cells
were randomly divided into four subgroups as follows: Control group
(C), the medium contained EGF and bFGF without BPA; NSC stemness
maintenance and BPA stress group (SMB), the medium contained EGF
and bFGF, and BPA was added to the medium to a final concentration
of 1 µM; differentiation control group (DC), the medium had no EGF
and bFGF, and no BPA was added; and differentiation and BPA stress
group (DBS), the medium had no EGF and bFGF, and BPA was added to
the medium to a final concentration of 1 µM.
CRISPR-knockout of ERRα in hNSCs
Based on the CRISPR-DO website (Version 0.1,
cistrome.org/crispr/), a single guide
(sg)RNA target site (5'-GACAGAGACCGAGCCTCCTG-3') was identified in
the coding region of the ERRα gene. Subsequently, the
knockout all-in-one vector pCMV-Cas9-GFP-ERRα and negative vector
pCMV-Cas9-GFP (cat. no. CAS9GFPP) were constructed by Sigma-Aldrich
(Merck KGaA). NSCs with ERRα-knockout were obtained by
transfecting the pCMV-Cas9-GFP-ERRα all-in-one vector using a
lipidosome (Lipofectamine 3000, Thermo Fisher Scientific, Inc.) at
room temperature. The monoclonal cells were sorted 48 h
post-transfection by flow cytometry with blue (488 nm) lasers (BD
FACSAria; BD Biosciences) based on green fluorescent protein in
hNSC and expanded to extract the genome to screen cell lines with
ERRα gene frameshift mutation. The specific primers
(forward, 5'-GGTTGAAGGTTGCCTGGGGGCTAC-3'; reverse,
5'-GACTCTTTCAGAGCCACTGTAGAAG-3') were designed according to the
sgRNA targeting site for PCR to analyze the mutation of the sgRNA
targeting site. The PCR product was sent to Sangon Biotech
(Shanghai) Co., Ltd. for sequencing analysis.
Effects of BPA on NSC morphology
Wild-type hNSCs were inoculated into a six-well cell
culture plate at 10x105 cells/ml and cultured to 80%
confluence. Cells were treated with BPA (0, 0.1, 1 and 10 µM)
according to previous reports (12,30,32,38).
Each concentration of the experimental group was set up in
triplicate. After 24 h of exposure at 37˚C, the cells were observed
under a light microscope for morphology (scale bar, 20 µm).
Effects of BPA on NSC
proliferation
Cell proliferation was measured via the Cell
Counting Kit-8 (CCK-8) assay. The NSC stemness maintenance and BPA
stress group (SMB) and control group (C) were selected. A total of
10 µl of CCK-8 (CA1210; Beijing Solarbio Science & Technology
Co., Ltd.) was added to each well, and the cells were then
incubated for 2 h. The absorbance at 450 nm was measured using a
microplate reader (Thermo Fisher Scientific, Inc.). Proliferation
rates were evaluated at 0, 24, 48, 72 and 96 h after treatment with
BPA.
Effects of BPA on NSC apoptosis by
flow cytometry
Cells that reached 80% confluency were treated with
BPA at 0, 0.1, 1, 2.5, 5 and 10 µM for 24 h at 37˚C, and then
digested with Accutase™ (cat. no. SCR005; MilliporeSigma) for 3 min
at 37˚C to make cell suspension. The cell suspension was collected
in a centrifuge tube and centrifuged at 300 x g for 5 min at room
temperature. The supernatant was discarded and phosphate-buffered
saline (PBS) was added to pipette the cells evenly. Subsequently,
the cell suspension was centrifuged at 300 x g for 5 min at room
temperature, the supernatant was discarded again and the operation
was repeated once. NSCs were diluted with deionized water at a
ratio of 1:3, the diluent was pre-cooled at 4˚C, rinsed twice with
PBS and was centrifuged at 300 x g for 5 min at room temperature.
Finally, NSCs were resuspended using 250 µl of binding buffer
(E-CK-A151; Elabscience) in a liquid solution to adjust the
concentration to 1x106 cells/ml. A total of 100 µl cell
suspension was collected in a 5-ml flow tube, 5 µl Annexin V-FITC
and 10 µl PI staining solution were then used to stain the cells
for 15 min at room temperature in the dark. Finally, 200 µl of
diluted binding buffer was added to terminate the reaction and
immediately detected by flow cytometry (BD FACSAria; BD
Biosciences) and analyzed by FlowJo 10.8.0 (BD Biosciences).
Western blotting
The cell suspension was centrifuged at 300 x g for 5
min at room temperature, and the supernatant was discarded, total
protein was extracted from cells using lysis buffer (cat. no.
P0013; Beyotime Institute of Biotechnology) with protease
inhibitors PMSF(ST506; Beyotime) and kept on ice for 30 min.
Protein concentrations were determined by Bradford assay and then
20 µg each protein sample were boiled for 5 min, separated by 10%
SDS-PAGE, transferred onto a PVDF membrane, blocked with 5% skim
milk in TBST buffer (20% Tween) for 3 h at room temperature and
then incubated with primary antibodies at 4˚C overnight. Primary
antibodies were Nestin (1:1,000, ab6320; Abcam), Sox2 (1:1,000,
ab171380; Abcam), GFAP (1:1,000, ab279290; Abcam), Map2 (1:1,000,
ab281588; Abcam), ERRα (1:1,000, bs-6998R; Bioss), TGF-β1 (1:1,000,
ab215715; Abcam), α-tubulin (1:10,000, ab7291; Abcam), AURKB
(1:10,000, ab45145; Abcam) and Id2 (1:500, ab90055; Abcam). The
membranes were then washed three times with TBST and incubated with
a secondary antibody (1:30,000, Goat Anti-Mouse IgG H&L/HRP,
bs-40296G-HRP; 1:30,000, Goat Anti-Rabbit IgG H&L/HRP antibody,
bs-40295G-HRP; Bioss) at room temperature for 1 h. Protein bands
were visualized using ECL (MilliporeSigma).
Immunostaining
Cells were fixed with 4% paraformaldehyde for 15-30
min at room temperature. After washing with PBS for three times,
the cells were covered with 4% BSA solution containing 0.1% Triton
X-100 at room temperature for 1 h and then incubated overnight with
4% BSA-diluted primary antibodies β-tubulin III (1:500, ab215037;
Abcam) and GFAP (ab279290, 1:50; Abcam). The samples were removed
from the 4˚C condition on the second day, reheated at room
temperature for 30 min and then incubated with goat anti-rabbit IgG
Cy5 (1:500, bs-0295G-Cy5; Bioss) or goat anti-mouse IgG FITC
(1:500, bs-0296G-FITC; Bioss) at room temperature for 1 h. The
solution was then discarded, and the samples were washed with PBS
for three times. Nuclei were stained at room temperature with
Hoechst 33342 (C0030; Beijing Solarbio Science & Technology
Co., Ltd.). The NSCs differentiation were identified by detecting
the expression of β-tubulin III and GFAP under a fluorescence
microscope (scale bar, 20 µm; Zeiss AG).
RNA-seq and data analysis
Total RNA was isolated from hNSCs with TRIzol
(Takara Bio, Inc.) and assessed using a Qubit™ 3.0 Fluorometer
(Invitrogen; Thermo Fisher Scientific, Inc) for total RNA quantity
and integrity. RNA libraries were constructed using a QuantSeq 3'
mRNA-Seq library Prep kit FWD for Illumina (no. 016; Lexogen)
following the manufacturer's protocol. mRNA was purified using
Oligo(dT) magnetic beads and fragmented at 95˚C for 8 min with
fragmentation buffer. Using mRNA as a template, the first cDNA
strand was synthesized with random oligonucleotides and reverse
transcriptase under the following conditions: 5 min at 65˚C, 2 min
at 4˚C, 1 h at 42˚C and 10 min at 70˚C. The second cDNA strand was
synthesized by the addition of the buffer, dNTPs, RNase H and DNA
polymerase I under the following conditions: 2.5 h at 16˚C and 10
min at 70˚C. Following purification, end repair and ligation of the
sequencing adapter were performed according to the QuantSeq 3'
mRNA-Seq Library prep kit manufacturer's protocol.
The PCR products were analyzed on 2% agarose gel and
stained with ethidium bromide. The 150 nucleotide-long fragments
were isolated and purified by using QuantSeq 3' mRNA-Seq Library
prep kit and then analyzed by Beyotime Institute of Biotechnology
on a Illumina HiSeq 2000 (Illumina, Inc.). FASTQ files were
uploaded to the BaseSpace Suite (Illumina, Inc.) and aligned using
the RNA-Seq Alignment application (version 1.0.0), in which STAR
was selected to align the sequences and the maximum number of
mismatches was set to 14 following the recommendation by Lexogen
GmbH. Output files were analyzed using Cufflinks Assembly and DE
application (version 2.1.0) in the BaseSpace Suite to determine the
differentially expressed genes (DEGs), which were used to generate
an expression heatmap. GO and KEGG analysis were performed by
Beyotime Institute of Biotechnology using ClueGO (version 2.3.3)
and CluePedia (version 1.3.3), which are Cytoscape software
applications (version 3.5.1) (39).
Chromatin immunoprecipitation
(CHIP)-PCR
A total of 108 cells were used to isolate
genomic DNA. The cells were resuspended in DMEM culture medium and
fixed with 1.5% formaldehyde for 15 min at 4˚C. The cell pellets
were harvested and lysed by cell collection buffer [100 mM Tris-HCl
(pH 9.4); 10 mM DTT with complete protease inhibitor cocktail;
Roche Diagnostics] on ice for 20 min. The deposits were collected
and washed twice with PBS. Cell pellets were pretreated with a
nucleus/chromatin preparation buffer [buffer I: 10 mM EDTA, 0.5 mM
EGTA, 10 mM HEPES (pH 6.5), 0.25% Triton X-100; buffer II: 1 mM
EDTA, 0.5 mM EGTA, 10 mM HEPES (pH 6.5), 200 mM NaCl] and then
lysed with a nuclear lysis buffer [10 mM EDTA, 50 mM Tris-HCl (pH
8.1), 1% SDS, complete protease inhibitor cocktail] on ice for 15
min. Sonication on ice was performed to generate a DNA fragment of
400-800 bp with a no. 2 microtip at a power output of 25% (on for 6
sec and off for 30 sec for 10 times). Chromatin quantified to 100
µg of DNA was utilized for further immunoprecipitation and mixed
with immunoprecipitation (IP) buffer [2 mM EDTA, 150 mM NaCl, 20 mM
Tris-HCl (pH 8.1), 0.1% Triton X-100, complete protease inhibitor
cocktail) in a total volume of 1 ml. The chromatin was diluted at a
rate of 1:10 in IP buffer as the input. Protein A/G magnetic beads
(cat. no. B23202; Bimake) were washed twice with TE buffer [10 mM
Tris-HCl (pH 8.1), 1 mM EDTA). Then, 1 ml chromatin (containing 100
µg DNA) was mixed for IP with 60 µl of prepared beads (30 µl beads
A and 30 µl DNA beads G) in siliconized tubes and rotated at 4˚C
for 3 h. The supernatants were transferred to fresh microcentrifuge
tubes and 2 µg of ERRα antibody (1:200, GTX108166; GeneTex) or IgG
(1:500, GTX35035; GeneTex) was added to each sample and rotated
overnight at 4˚C. The following day, 40 µl of protein A/G magnetic
beads were added into each antibody-chromatin sample, and rotation
was performed for 2 h at 4˚C. The beads were harvested with a
magnetic holder and washed in the shaker sequentially for 5 min at
4˚C in 1 ml of wash buffer I [2 mM EDTA, 20 mM Tris-HCl (pH 8.1),
0.1% SDS, 1% Triton X-100, 150 mM NaCl] x 2, wash buffer II [2 mM
EDTA, 20 mM Tris-HCl (pH 8.1), 0.1% SDS, 1% Triton X-100, 500 mM
NaCl), and TE buffer 2X. The beads were resuspended in 250 µl of
extraction buffer (1% SDS and 0.1 M NaHCO3) and shook
(40 rpm) at room temperature for 30 min. The supernatants were
collected, and this step was repeated. The confluent supernatants
were chromatin eluted following the addition of 10 µg of RNase A
(cat. no. R5125; Sigma-Aldrich; Merck KGaA) for RNA digestion at
37˚C for 2 h. All samples, including IP and input samples, were
mixed with 120 µg of proteinase K to reverse formaldehyde
crosslinks at 65˚C overnight. DNA was purified with a PCR
purification kit (D0033; Beyotime) and the chromatin was dissolved
in 120 µl of TE buffer. Overall, 2 µl of DNA solution per well was
loaded for the PCR assay using TransTaq DNA Polymerase High
Fidelity (AP131-11; Transgen Biotech). The primers were as follows:
TGF-β1 promoter (target region), forward
5'-AAATTGGGGACAGTAAATGTATGGG-3', and reverse,
5'-TAGGAGAAGAGGGTCTGTCAACAT-3'; TGF-β1 ORF forward (control
region), 5'-CAACAATTCCTGGCG ATACC-3', and reverse
5'-GAACCCGTTGATGTCCACTT-3'. The thermocycling conditions were as
follows: 95˚C for 10 sec, followed by 30 cycles of 94˚C for 10 sec,
50˚C for 20 sec and 72˚C for 30 sec, and 72˚C for 1 min. PCR
products were detected by electrophoresis on a 12% agarose gel and
ethidium bromide staining.
Statistical analyses
All experiments were performed a minimum of three
times independently. Data are expressed as the mean of three
repeats ± standard deviation. Statistical comparisons of results
from multiple groups (data from NSC proliferation, GO analysis,
KEGG pathway analysis) were analyzed using one-way analysis of
variance. P<0.05 was considered to indicate a statistically
significant difference. All statistical analyses were performed
with the GraphPad Prism 6.0 program (GraphPad Software, Inc.).
Results
Low BPA concentration has no marked
effect on cell morphology
Under the ReNcell CX kit instructions, EGF and bFGF
were removed from the medium, which played roles in the maintenance
of NSCs. A differentiation model of human NSCs was constructed, and
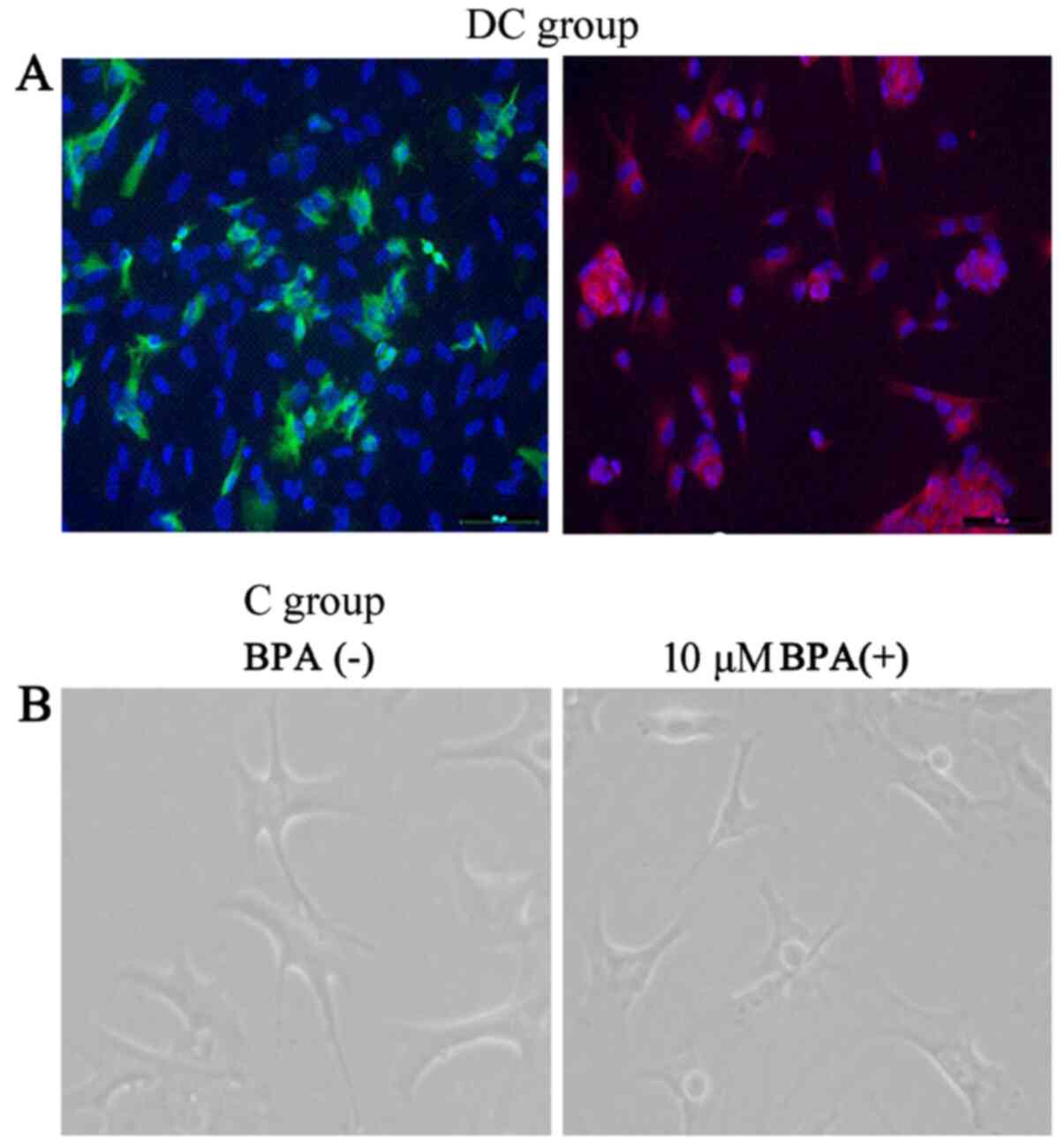
the expression profiles of GFAP and β-tubulin III were revealed in
cells, thereby indicating the formation of neurons (Fig. 1). The expression of GFAP and
β-tubulin III indicated that the NSCs have the ability promote
differentiate, and on this basis, the effects of different doses of
BPA on its morphology were investigated. The cells were added with
low doses (0.1, 1 and 10.0 µM) of BPA. As. a consequence of these
experiments, these gradient doses of BPA exhibited no remarkable
effect on cell morphology.
Low concentrations of BPA affects cell
proliferation and apoptosis
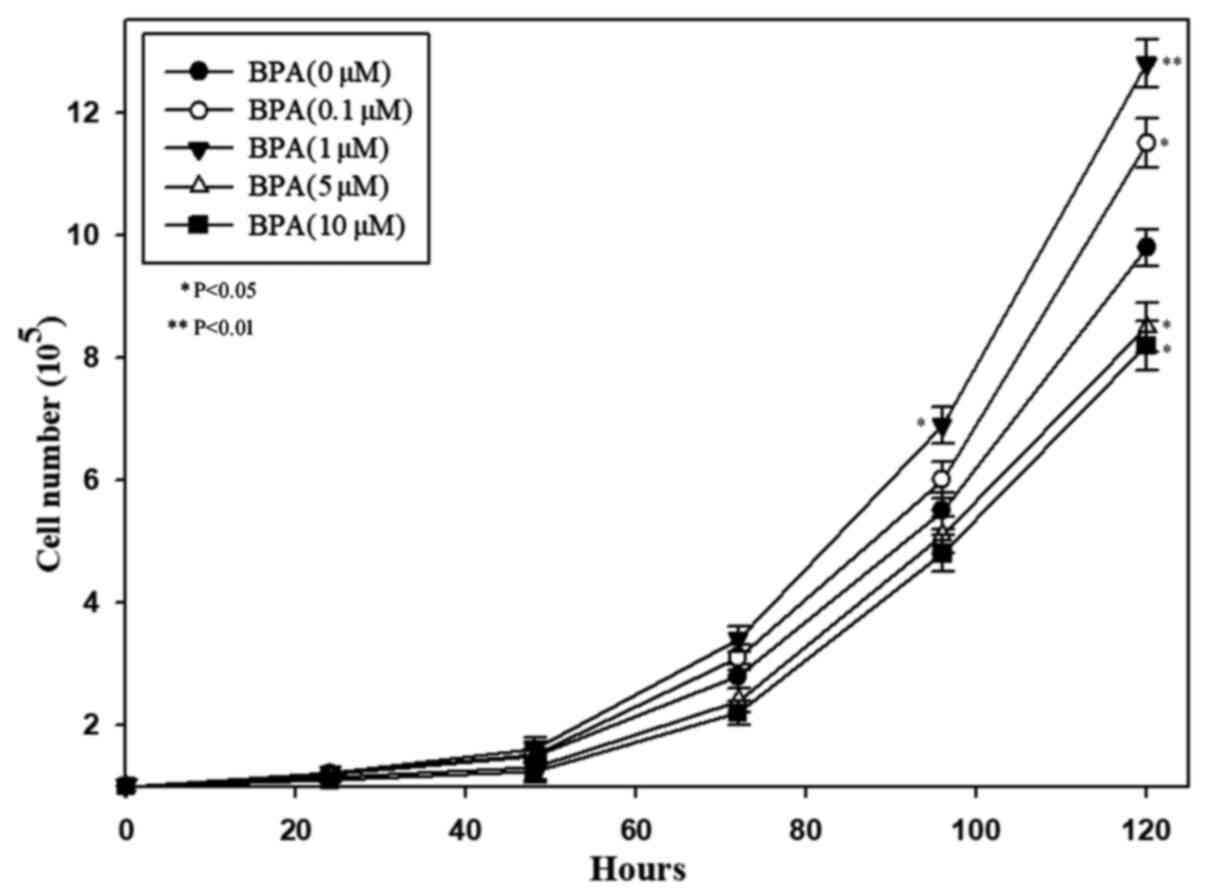
According to the effects of BPA on NSC morphology,
the effects of BPA on cell proliferation after cultivation at 0.1,
1, 5, and 10 µM BPA concentrations were analyzed using a CCK-8
assay. It revealed that 0.1 and 1 µM BPA markedly promoted cell
proliferation (Fig. 2) and that 1
µM BPA demonstrated a stronger promotion effect. However, it was
also observed that a concentration >5 µM could markedly inhibit
cell proliferation.
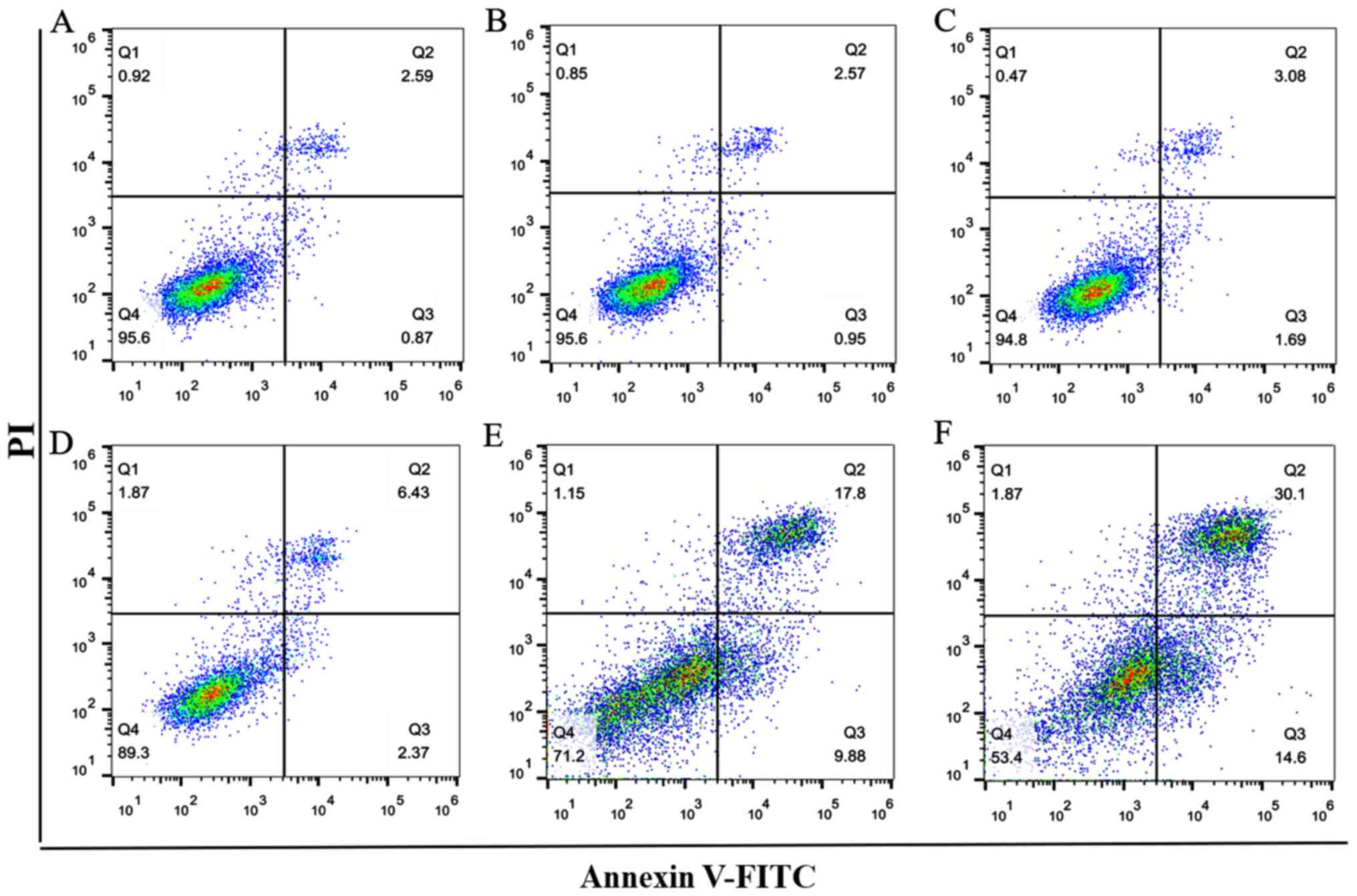
Flow cytometry was used to analyze the effects of
0.1, 1, 2.5, 5 and 10 µM BPA on NSC apoptosis (Fig. 3). Results revealed that the amount
of apoptosis induced in NSCs by <1 µM BPA (Fig. 3B and C) were similar compared with that of the
C group (Fig. 3A); whereas 2.5, 5
and 10 µM BPA caused a marked increase in NSC apoptosis compared
with the C (Fig. 3D-F). Thus, 1 µM
BPA was selected for subsequent molecular mechanism research after
considering the highest BPA concentration in serum in the present
population based on BPA characteristics (40).
BPA (1 µM) affects the key molecules
of cell differentiation and proliferation
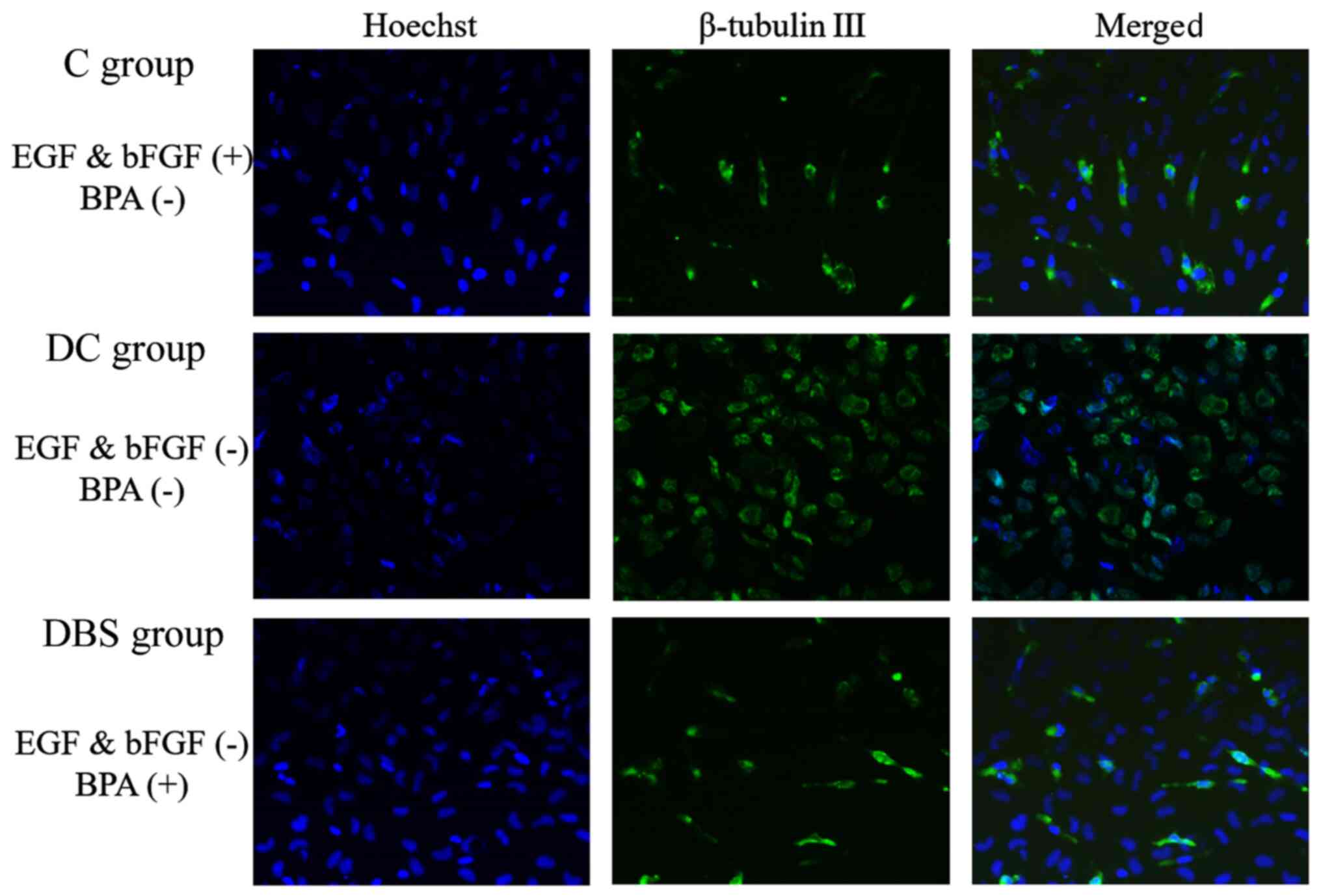
The NSC maintenance factors EGF and bFGF were
removed from medium. The increase in β-tubulin III expression in DC
group indicated the beginning of differentiation compared with C
group. When 1 µM BPA was added to the cell culture medium,
differentiation was inhibited, as seen by to the decrease in
β-tubulin III expression in DBS group compared with DC group
(Fig. 4).
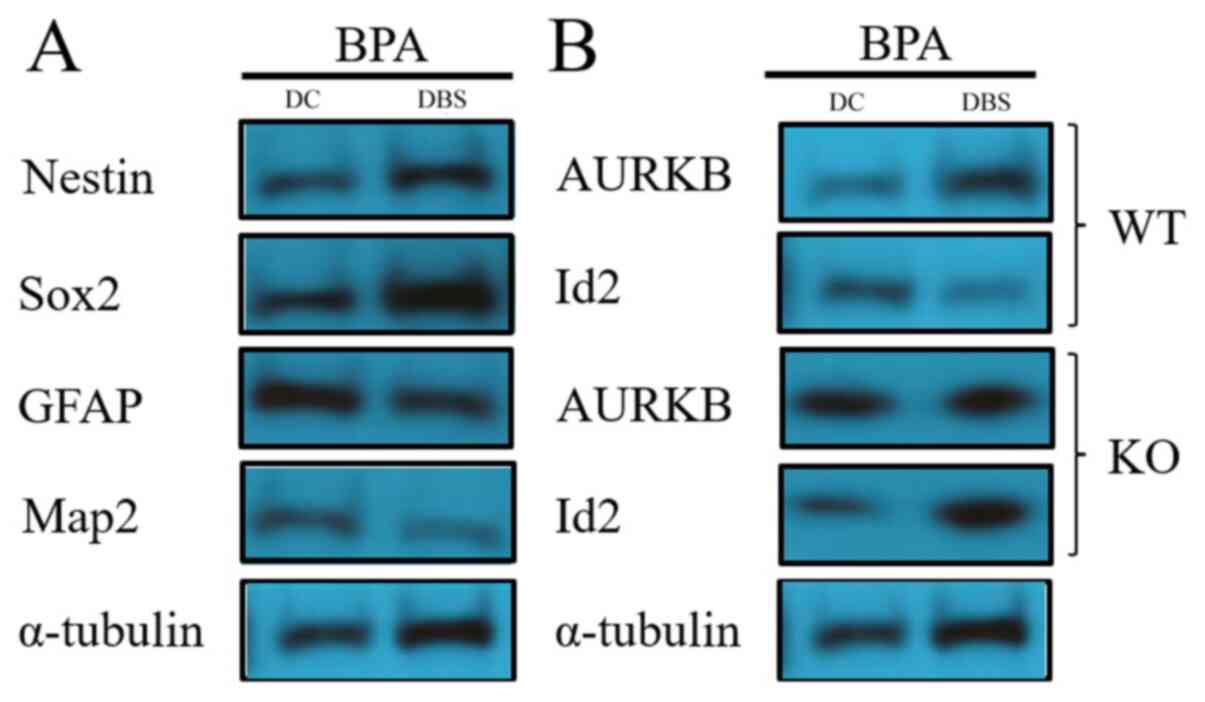
Western blotting was used to detect the expression
of partial genes in NSCs after exposure to 1 µM BPA. These genes
included maintenance marker genes, such as nestin and Sox2, and
differentiation marker genes, such as GFAP and MAP2. Western
blotting demonstrated that 1 µM BPA notably reduced GFAP and MAP2
expression levels in NSCs compared with the control; whereas 1 µM
BPA considerably increased the expression levels of nestin and Sox2
(Fig. 5). The results indicated
that the dose of 1 µM BPA affected the fate transfer mechanism of
human NSC maintenance and neural differentiation.
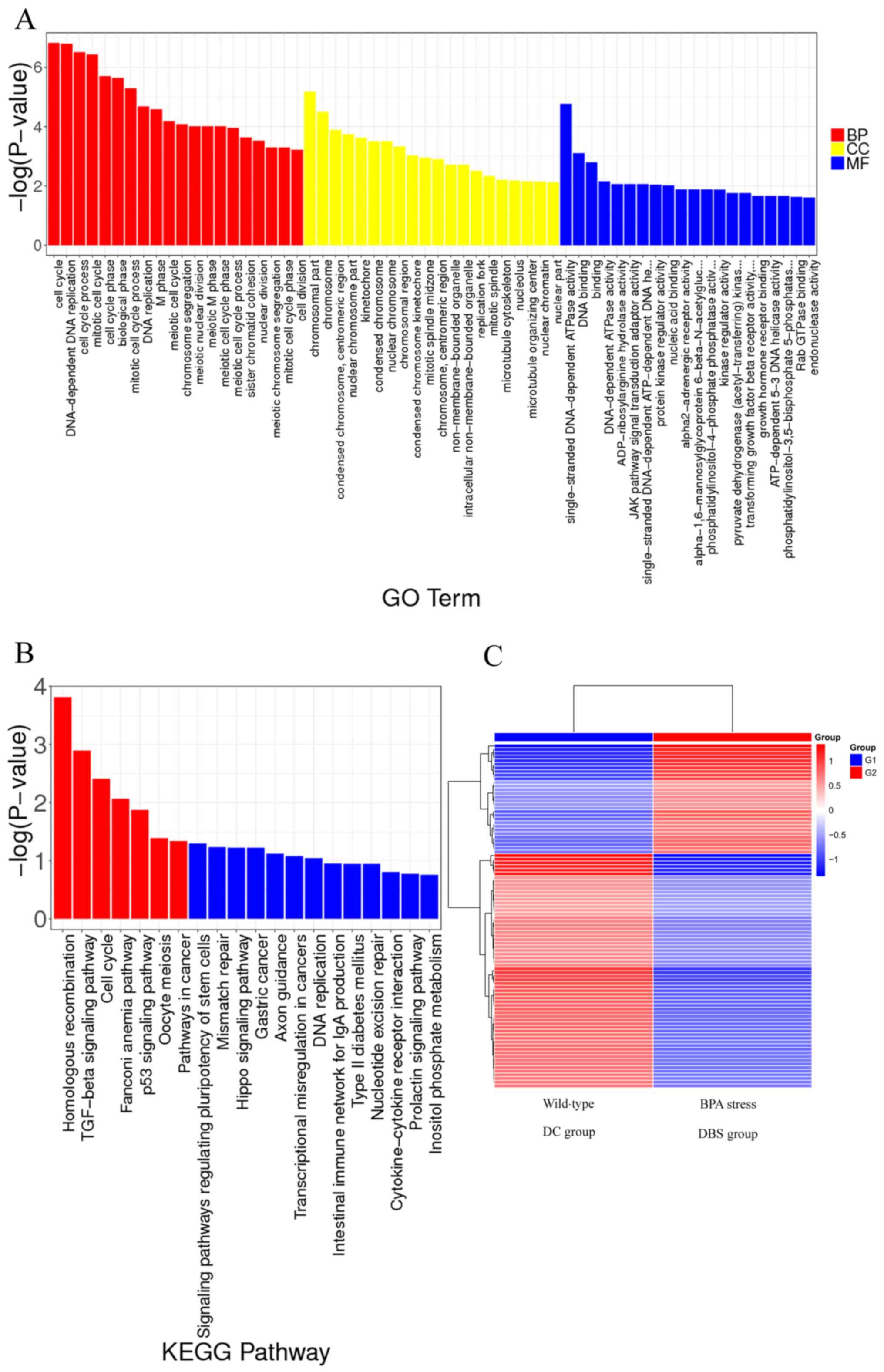
BPA promotes cell cycle
To classify and characterize the DEG functions and
pathways, a Gene Ontology classification was performed in addition
to a functional annotation of molecular functions (MFs), cellular
components (CCs) and biological processes (BPs). In contrast to the
control group, 2,124 gene expression (Processed data GSE185138)
levels were remarkably altered in the 1 µM BPA treated cells. Of
these gene expression levels, 259 were attributed to MFs, 1,397
were attributed to BPs and the remaining 468 transcripts belonged
to CCs. As presented in Fig. 6A,
the GO function analysis results indicated that treatment with 1 µM
BPA promoted ‘cell cycle’ (BPs).
BPA regulates the TGF-β pathway
The DEGs were used as input data, and KEGG pathway
analysis was performed to visualize the effects of 1 µM BPA on
specific cell signaling pathway-related gene expression changes
(Fig. 6C) and to screen for
biological pathways that may be potentially involved. The results,
including the significantly enriched KEGG signaling pathway
(P≤0.05), are presented in Fig.
6B. A total of seven signaling pathways were found with
remarkable activation of low-concentration (1 µM) BPA: ‘Homologous
recombination’, ‘TGF-β signaling pathway’, ‘cell cycle’, ‘Fanconi
anemia pathway’, ‘p53 signaling pathway’, ‘oocyte meiosis’ and
‘pathways in cancer’. Among these pathways, the TGF-β and p53
signaling pathways are closely related to cell differentiation fate
(12,30,32).
TGF-β gene can be activated by the estrogen signaling pathway
(41). DEG analysis demonstrated
that BPA had a significant influence on TGF-β and estrogen
signaling pathway, and the associated genes are summarized in
Table I.
 | Table IAssociated genes of the TGF-β and
estrogen signaling pathways. |
Table I
Associated genes of the TGF-β and
estrogen signaling pathways.
| Signaling
pathway | Number of
differentially expression gene | Number of
significantly differentially expressed genes (P < 0.05) | Gene name list | Classic Fisher | FDR |
|---|
| Estrogen | 98 | 16 | PRKACA, PLCB4,
GABBR1, GPER1, SHC1, ADCY3, ATF4,GNAS, ATF2, CALM2, HRAS,ERRA,
ADCY6, PLCB2, RAF1, CREB3L4 | 0.827 | 1 |
| TGF-β | 84 | 13 | SMAD4, SMAD5, E2F5,
SMAD1, TFDP1, RPS6KB2, AURKB, TGFB3, SKP1, TGFBR1, SMAD2, CREBBP,
ID2 | 0.956 | 1 |
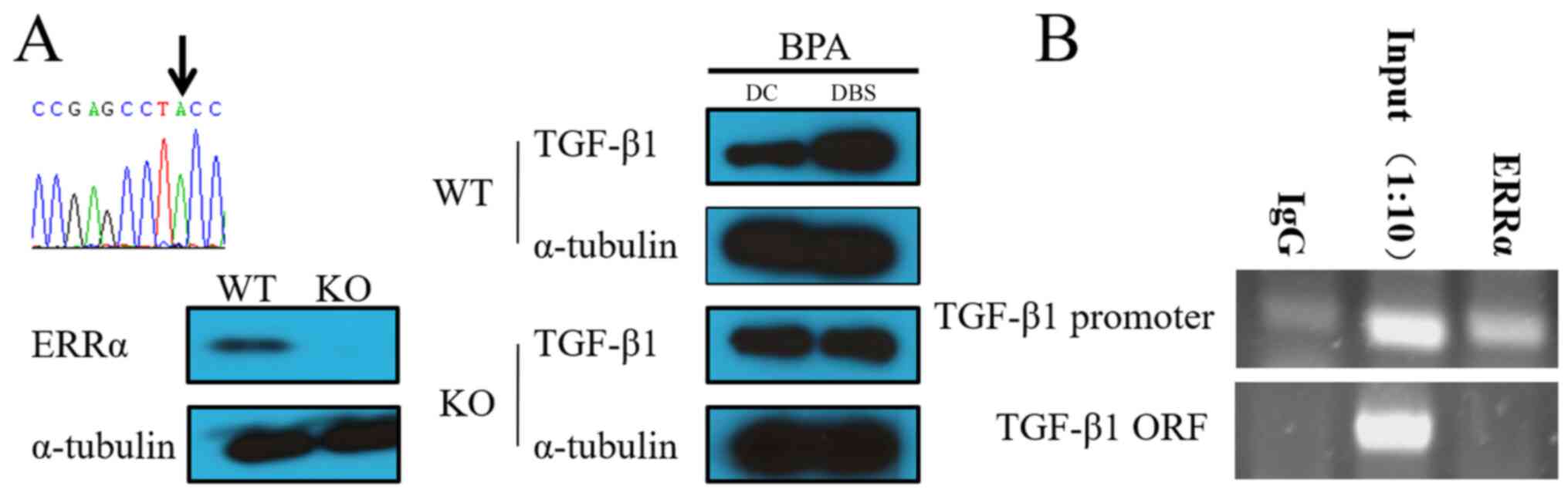
ERRα and TGF-β1 signaling pathways are
regulated by 1 µM BPA
BPA can bind to the ERR receptor to activate gene
expression (37). The present
study designed the sgRNA sequence (5'-GACAGAGACCGAGCCTCCTG-3') and
utilized CRISPR/Cas9 technology. In the current study, the
ERRα-knockout cells were obtained whose genomic coding
region was inserted into the ‘A’ base (Fig. 7A). The wild-type cell line (WT)
served as a control to analyze the TGF-β1 gene expression in
the ERRα gene-deficient cell line (knockout type cell line,
KO) under 1 µM BPA stress. In WT cell lines (Fig. 7A), TGF-β1 expression level
increased in the presence of BPA. In the ERRα-deficient cell
line, even in the presence of BPA, the expression of TGF-β1
was not revealed to be obviously different. These results indicated
that BPA might promote TGF-β1 gene expression by binding to
the ERRα receptor and the promoter of TGF-β1. CHIP-PCR
results also revealed that ERRα could bind to the promoter of
TGF-β1 under 1 µM BPA exposure (Fig. 7B).
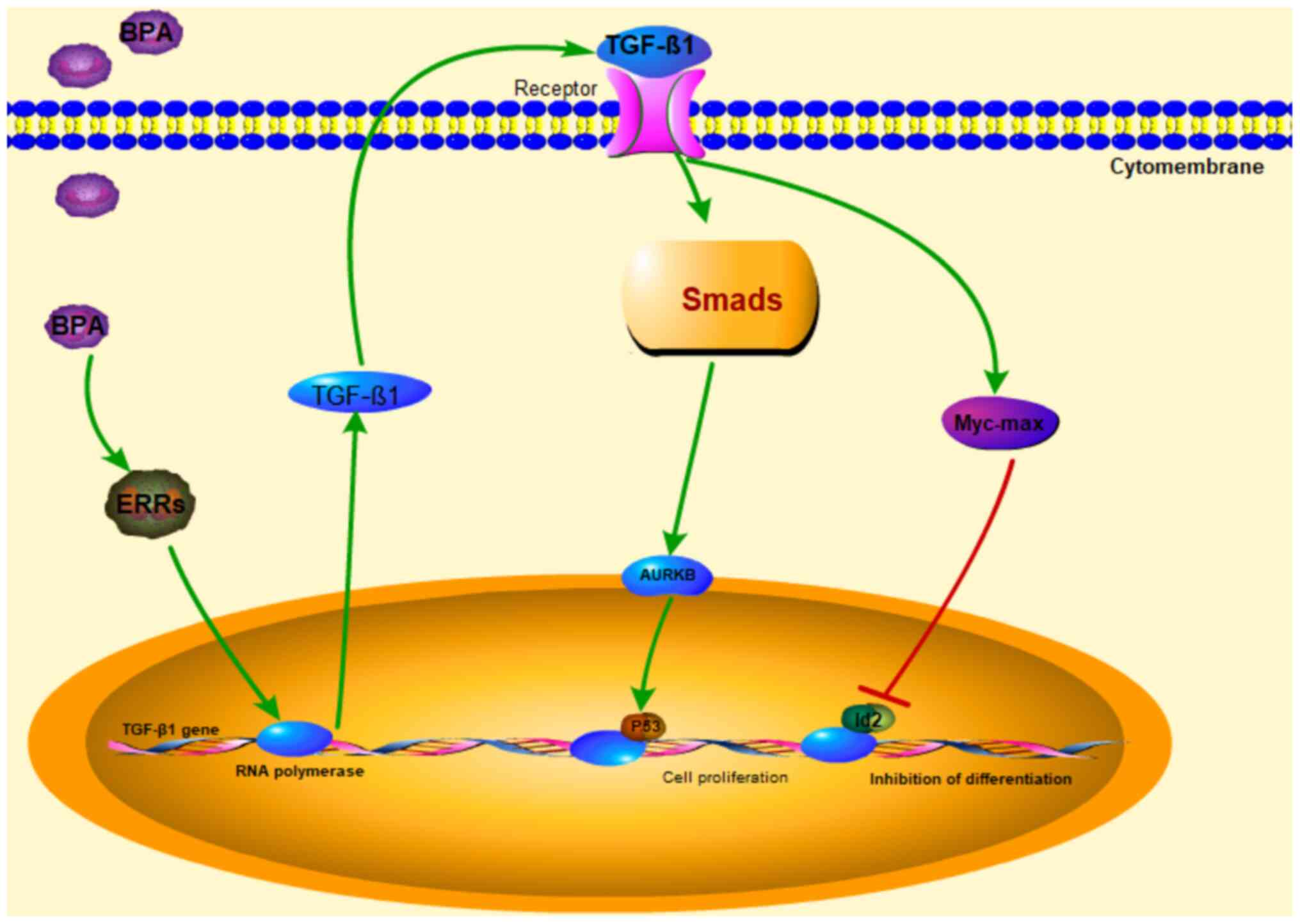
Changes in the TGF-β1 signaling
pathway downstream factors AURKB and Id2
AURKB and Id2 are downstream molecules of the TGF-β1
signaling pathway, which promote cell proliferation and inhibits
cell differentiation (35,36). Western blot analysis to detect the
protein expression levels of AURKB and Id2 in the NSCs exposed to 1
µM BPA (Fig. 5). The results
revealed of Fig. 5 revealed that 1
µM BPA could markedly decrease Id2 expression and increase
AURKB expression in WT NSCs; moreover, the same result was
obtained from the DEGs of RNA-seq analysis (Table I). In conclusion, the results in
the current study indicated that 1 µM BPA may activate the TGF-β1
signaling pathway through ERRα to regulate the fate transition of
the dry maintenance/neurological differentiation of human NSCs
(Fig. 8).
Discussion
BPA is found in breast milk and transferred from
pregnant women to their fetuses, affecting a developing embryo and
showing long-lasting deleterious effects during postnatal periods
(42,43). Children born from pregnant women
exposed to BPA have a much higher likelihood of attention deficit
hyperactivity disorder (11.2%) and social impairment compared with
children born to women without exposure to BPA (44).
BPA's deleterious effects on the nervous system are
well understood (31); however,
the effects of BPA on neurogenesis and the underlying cellular and
molecular mechanisms are not completely clear. The median
concentration of BPA in the urine of pregnant Chinese women is
4.8x10-3 µM (45), and
the concentration of free-BPA in serum is
4x10-6-9.1x10-2 µM (40). Based on BPA hydrophobicity and the
effects of low BPA concentration on morphology, proliferation and
apoptosis, the present study analyzed the effects of 1 µM BPA on
human NSCs. The results revealed that 1 µM BPA enhanced human NSC
proliferation but did not affect NSCs apoptosis. This implied that
1 µM BPA regulated NSC stemness via a molecular mechanism. However,
the present study did not further explore the effect of BPA on cell
proliferation changes via cell density, neutrosphere diameter and
cell cycle, as this study aimed to analyze the mechanism of 1 µM
BPA on NSCs stemness maintenance.
At 1 µM, BPA represses dopaminergic neuron
differentiation from human embryonic stem cells by downregulating
the expression of insulin-like growth factor 1(38). In addition, 0.01 and 0.1 µM BPA
promotes epithelial to mesenchymal transition via the canonical Wnt
pathway in ovarian cancer cells; the effect of 0.1 µM BPA on the
TGF-β pathway has also been confirmed by Chip-seq (33). These data suggest that the BPA
signaling pathway may differ in different cell types (38). The aforementioned results imply
that BPA can regulate different signaling pathways with different
concentration. In the present experiments, BPA at 1 µM
concentration was used to activate the TGF-β1 signaling
pathway.
Estrogen-related receptors are present in embryonic
stem cells and NSCs (46), and
their subtypes, including ERRα, ERRβ and ERRγ, can bind to BPA to
achieve BPA regulation (37). In
this process, ERRα can regulate cell proliferation and also
directly bind to the TGF-β1 gene promoter to induce its
expression (41). It is
hypothesized that BPA may bind to the ERR receptor and activate
TGF-β gene expression, thereby activating the TGF-β
signaling pathway.
BPA is involved in disrupting epigenetic
programming, thus altering brain development (47,48).
BPA causes developmental toxicity by inhibiting the proliferation
of neural progenitor cells and rat embryonic midbrain cells by
suppressing the ERK, JNK, CREB and p53 signaling pathways (32,49).
BPA induces reactive oxygen species (ROS) and oxidative stress in
rodent liver, sperm and brains (50,51).
Therefore, decreased NSC proliferation by BPA can be due to the ROS
generation, as enhanced ROS levels significantly reduce
neurogenesis (52).
The different signal pathway regulation regulated
the differentiation and proliferation of NSC. The differentiated
signal pathway activity is relatively low or inhibited when
proliferation is enhanced (53).
In the present study, 1 µM BPA could promote human NSC
proliferation and stemness. RNA-seq results revealed that 1 µM BPA
enhanced the cell cycle and activated the TGF-β signaling pathway.
Further experiments confirmed that BPA maintains cell stemness by
binding to the EERα receptor and activating the TGF-β1 signaling
pathway and downstream factors AURKB and Id2. Future
in vivo studies investigating BPA's toxic inhibition may
provide further insights into the mechanisms behind its
activity.
Acknowledgements
The authors would like to thank Dr Dan Lou at
Shanghai Municipal Center for Disease Control & Prevention
(Shanghai, China) for providing the hNSC.
Funding
Funding: The present work was supported by the Natural
Scientific Foundation of Shandong Province, China (grant no.
ZR2018MH038); and the Zibo City Integration Development Project
(grant no. 2019ZBXC320).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185138).
Authors' contributions
QW and XTi conceived and supervised the study. PD
and GY designed the study. XTu, YL, WC, YM and LW performed the
experiments and analyzed the data. PD and QW confirmed the
authenticity of the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of
interest.
References
|
1
|
Bellinger DC: Interpreting epidemiologic
studies of developmental neurotoxicity: Conceptual and analytic
issues. Neurotoxicol Teratol. 31:267–274. 2009.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Buxbaum JD and Hof PR: The emerging
neuroscience of autism spectrum disorders. Brain Res. 1380:1–2.
2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Maussion G, Diallo AB, Gigek CO, Chen ES,
Crapper L, Théroux JF, Chen GG, Vasuta C and Ernst C: Investigation
of genes important in neurodevelopment disorders in adult human
brain. Hum Genet. 134:1037–1053. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Arndt TL, Stodgell CJ and Rodier PM: The
teratology of autism. Int J Dev Neurosci. 23:189–199.
2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Daniels JL: Autism and the environment.
Environ Health Perspect. 114(A396)2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jaako-Movits K, Zharkovsky T, Romantchik
O, Jurgenson M, Merisalu E, Heidmets LT and Zharkovsky A:
Developmental lead exposure impairs contextual fear conditioning
and reduces adult hippocampal neurogenesis in the rat brain. Int J
Dev Neurosci. 23:627–635. 2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chow ES, Hui MN, Lin CC and Cheng SH:
Cadmium inhibits neurogenesis in zebrafish embryonic brain
development. Aquat Toxicol. 87:157–169. 2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tyler CR and Allan AM: Adult hippocampal
neurogenesis and mRNA expression are altered by perinatal arsenic
exposure in mice and restored by brief exposure to enrichment. PLoS
One. 8(e73720)2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ieraci A and Herrera DG: Single alcohol
exposure in early life damages hippocampal stem/progenitor cells
and reduces adult neurogenesis. Neurobiol Dis. 26:597–605.
2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Radonjic M, Cappaert NL, de Vries EF, de
Esch CE, Kuper FC, van Waarde A, Dierckx RA, Wadman WJ, Wolterbeek
AP, Stierum RH, et al: Delay and impairment in brain development
and function in rat offspring after maternal exposure to
methylmercury. Toxicol Sci. 133:112–124. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Landrigan PJ, Lambertini L and Birnbaum
LS: A research strategy to discover the environmental causes of
autism and neurodevelopmental disabilities. Environ Health
Perspect. 120:a258–a260. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kim K, Son TG, Park HR, Kim SJ, Kim HS,
Kim HS, Kim TS, Jung KK, Han SY and Lee J: Potencies of bisphenol A
on the neuronal differentiation and hippocampal neurogenesis. J
Toxicol Environ Health A. 72:1343–1351. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Itoh K, Yaoi T and Fushiki S: Bisphenol A,
an endocrine-disrupting chemical, and brain development.
Neuropathology. 32:447–457. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hirabayashi Y and Gotoh Y: Epigenetic
control of neural precursor cell fate during development. Nat Rev
Neurosci. 11:377–388. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Sanosaka T, Namihira M and Nakashima K:
Epigenetic mechanisms in sequential differentiation of neural stem
cells. Epigenetics. 4:89–92. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Temple S: The development of neural stem
cells. Nature. 414:112–117. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Negri-Cesi P: Bisphenol a interaction with
brain development and functions. Dose Response: Jun 17, 2015 (Epub
ahead of print). doi: 10.1177/1559325815590394.
|
|
18
|
Klajn A, Drakulic D, Tosic M, Pavkovic Z,
Schwirtlich M and Stevanovic M: SOX2 overexpression affects neural
differentiation of human pluripotent NT2/D1 cells. Biochemistry
(Mosc). 79:1172–1182. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kubwabo C, Kosarac I, Stewart B, Gauthier
BR, Lalonde K and Lalonde PJ: Migration of bisphenol A from plastic
baby bottles, baby bottle liners and reusable polycarbonate
drinking bottles. Food Addit Contam Part A Chem Anal Control Expo
Risk Assess. 26:928–937. 2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Vandenberg LN, Colborn T, Hayes TB,
Heindel JJ, Jacobs DR Jr, Lee DH, Shioda T, Soto AM, vom Saal FS,
Welshons WV, et al: Hormones and endocrine-disrupting chemicals:
Low-dose effects and nonmonotonic dose responses. Endocr Rev.
33:378–455. 2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Vandenberg LN, Hauser R, Marcus M, Olea N
and Welshons WV: Human exposure to bisphenol A (BPA). Reprod
Toxicol. 24:139–177. 2007.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chapin RE, Adams J, Boekelheide K, Gray LE
Jr, Hayward SW, Lees PS, McIntyre BS, Portier KM, Schnorr TM,
Selevan SG, et al: NTP-CERHR expert panel report on the
reproductive and developmental toxicity of bisphenol A. Birth
Defects Res B Dev Reprod Toxicol. 83:157–395. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
de Cock M, Maas YG and van de Bor M: Does
perinatal exposure to endocrine disruptors induce autism spectrum
and attention deficit hyperactivity disorders? Review. Acta
Paediatr. 101:811–818. 2012.PubMed/NCBI View Article : Google Scholar : Review.
|
|
24
|
Ooe H, Taira T, Iguchi-Ariga SM and Ariga
H: Induction of reactive oxygen species by bisphenol A and
abrogation of bisphenol A-induced cell injury by DJ-1. Toxicol Sci.
88:114–126. 2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Masuo Y and Ishido M: Neurotoxicity of
endocrine disruptors: Possible involvement in brain development and
neurodegeneration. J Toxicol Environ Health B Crit Rev. 14:346–369.
2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Huang B, Jiang C, Luo J, Cui Y, Qin L and
Liu J: Maternal exposure to bisphenol A may increase the risks of
Parkinson's disease through down-regulation of fetal IGF-1
expression. Med Hypotheses. 82:245–249. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Brown JS Jr: Effects of bisphenol-A and
other endocrine disruptors compared with abnormalities of
schizophrenia: An endocrine-disruption theory of schizophrenia.
Schizophr Bull. 35:256–278. 2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Schoenfeld TJ and Cameron HA: Adult
neurogenesis and mental illness. Neuropsychopharmacology.
40:113–128. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Reekmans K, Praet J, Daans J, Reumers V,
Pauwels P, Van der Linden A, Berneman ZN and Ponsaerts P: Current
challenges for the advancement of neural stem cell biology and
transplantation research. Stem Cell Rev Rep. 8:262–278.
2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tiwari SK, Agarwal S, Seth B, Yadav A, Ray
RS, Mishra VN and Chaturvedi RK: Inhibitory effects of bisphenol-A
on neural stem cells proliferation and differentiation in the rat
brain are dependent on Wnt/β-catenin pathway. Mol Neurobiol.
52:1735–1757. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Tiwari SK, Agarwal S, Tripathi A and
Chaturvedi RK: Bisphenol-A mediated inhibition of hippocampal
neurogenesis attenuated by curcumin via canonical Wnt pathway. Mol
Neurobiol. 53:3010–3029. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kim K, Son TG, Kim SJ, Kim HS, Kim TS, Han
SY and Lee J: Suppressive effects of bisphenol A on the
proliferation of neural progenitor cells. J Toxicol Environ Health
A. 70:1288–1295. 2007.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hui L, Li H, Lu G, Chen Z, Sun W, Shi Y,
Fu Z, Huang B, Zhu X, Lu W, et al: Low dose of bisphenol a
modulates ovarian cancer gene expression profile and promotes
epithelial to mesenchymal transition via canonical Wnt pathway.
Toxicol Sci. 164:527–538. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kim YS, Hwang KA, Hyun SH, Nam KH, Lee CK
and Choi KC: Bisphenol A and nonylphenol have the potential to
stimulate the migration of ovarian cancer cells by inducing
epithelial-mesenchymal transition via an estrogen receptor
dependent pathway. Chem Res Toxicol. 28:662–671. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
He J, Qi Z, Zhang X, Yang Y, Liu F, Zhao G
and Wang Z: Aurora kinase B inhibitor barasertib (AZD1152) inhibits
glucose metabolism in gastric cancer cells. Anticancer Drugs.
30:19–26. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Siegel PM, Shu W and Massagué J: Mad
upregulation and Id2 repression accompany transforming growth
factor (TGF)-beta-mediated epithelial cell growth suppression. J
Biol Chem. 278:35444–35450. 2003.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tohmé M, Prud'homme SM, Boulahtouf A,
Samarut E, Brunet F, Bernard L, Bourguet W, Gibert Y, Balaguer P
and Laudet V: Estrogen-related receptor γ is an in vivo receptor of
bisphenol A. FASEB J. 28:3124–3133. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Huang B, Ning S, Zhang Q, Chen A, Jiang C,
Cui Y, Hu J, Li H, Fan G, Qin L, et al: Bisphenol A represses
dopaminergic neuron differentiation from human embryonic stem cells
through downregulating the expression of insulin-like growth factor
1. Mol Neurobiol. 54:3798–3812. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen X, Bao HH, Wu WK, Yan SQ, Sheng J, Xu
YY, Gu CL, Huang K, Cao H, et al: Exposure to bisphenol A during
maternal pregnancy and the emotional and behavioral impact on their
preschool children. Chin J Epidemiol. 39:188–193. 2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
41
|
Huang X, Wang X, Shang J, Zhaang Z, Cui B,
Lin Y, Yang Y, Song Y, Yu S and Xia J: Estrogen related receptor
alpha triggers the migration and invasion of endometrial cancer
cells via up regulation of TGFB1. Cell Adhes Migr. 12:538–547.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Nishikawa M, Iwano H, Yanagisawa R, Koike
N, Inoue H and Yokota H: Placental transfer of conjugated bisphenol
A and subsequent reactivation in the rat fetus. Environ Health
Perspect. 118:1196–1203. 2010.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Schönfelder G, Wittfoht W, Hopp H,
Talsness CE, Paul M and Chahoud I: Parent bisphenol A accumulation
in the human maternal-fetal-placental unit. Environ Health
Perspect. 110:A703–A707. 2002.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Tewar S, Auinger P, Braun JM, Lanphear B,
Kimberly Yolton K, Epstein JN, Ehrlich S and Froehlich TE:
Association of bisphenol A exposure and
attention-deficit/hyperactivity disorder in a national sample of
U.S. children. Environ Res. 150:112–118. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhang Z, Alomirah H, Cho HS, Li YF, Liao
C, Minh TB, Mohd MA, Nakata H, Ren N and Kannan K: Urinary
bisphenol A concentrations and their implications for human
exposure in several Asian countries. Environ Sci Technol.
45:7044–7050. 2011.PubMed/NCBI View Article : Google Scholar
|
|
46
|
van der Laan S, Golfetto E, Vanacker JM
and Maiorano D: Cell cycle-dependent expression of Dub3, Nanog and
the p160 family of nuclear receptor coactivators (NCoAs) in mouse
embryonic stem cells. PLoS One. 9(e93663)2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Kundakovic M and Champagne FA: Epigenetic
perspective on the developmental effects of bisphenol A. Brain
Behav Immun. 25:1084–1093. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Dolinoy DC, Huang D and Jirtle RL:
Maternal nutrient supplementation counteracts bisphenol A-induced
DNA hypomethylation in early development. Proc Natl Acad Sci USA.
104:13056–13061. 2007.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Liu R, Xing L, Kong D, Jiang J, Shang L
and Hao W: Bisphenol A inhibits proliferation and induces apoptosis
in micromass cultures of rat embryonic midbrain cells through the
JNK, CREB and p53 signaling pathways. Food Chem Toxicol. 52:76–82.
2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Bindhumol V, Chitra KC and Mathur PP:
Bisphenol A induces reactive oxygen species generation in the liver
of male rats. Toxicology. 188:117–124. 2003.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Kabuto H, Hasuike S, Minagawa N and
Shishibori T: Effects of bisphenol A on the metabolisms of active
oxygen species in mouse tissues. Environ Res. 93:31–35.
2003.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Huang TT, Zou Y and Corniola R: Oxidative
stress and adult neurogenesis - effects of radiation and superoxide
dismutase deficiency. Semin Cell Dev Biol. 23:738–744.
2012.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhang L, Ye M, Zhu L, Cha J, Li C, Yao YG
and Mao B: Loss of ZC4H2 and RNF220 inhibits neural stem cell
proliferation and promotes neuronal differentiation. Cells.
9(E1600)2020.PubMed/NCBI View Article : Google Scholar
|