Introduction
Gestational diabetes mellitus (GDM), the most common
metabolic disorder in pregnancy, refers to glucose intolerance
occurring in the second and third trimesters of pregnancy, leading
to hyperglycemia of varying severity (1). The main drivers of the increased
prevalence of GDM include maternal obesity, physical inactivity and
increased maternal age (2). GDM
increases the risk of severe pregnancy complications for mother and
child, including birth trauma, shoulder dystocia, macrosomia,
neonatal hypoglycemia and hyperbilirubinemia, and stillbirth
(3). The patients with GDM have a
significantly increased lifetime risk for type 2 diabetes, while
their offspring are more likely to develop type 2 diabetes,
obesity, metabolic and cardiovascular disease later in life
(4). Therefore, effective
prevention and early diagnosis of patients with GDM are very
important.
A previous study has reported that maternal diabetes
may lead to alterations in placental structure and function, as
well as fetal malformation, which has principally been attributed
to hyperglycemia (5). The placenta
is a highly specific organ for the exchange of gases, nutrients and
metabolites between the mother and fetus during pregnancy and is
essential for the maintenance of pregnancy (6). Trophoblasts are the first cell lineage
to differentiate, invade and migrate to the vascular tissue of the
placenta and fetal membrane during pregnancy (7). The inhibition of trophoblast cell
viability, invasion and migration of trophoblast cells may
contribute toward maldevelopment of placental tissues, which has
been reported in certain studies (8,9).
Therefore, trophoblast cells with normal biological functions serve
crucial roles in placental development and are widely used in the
construction of GDM cell models.
MicroRNAs (miRNAs/miRs) are a novel class of small
non-coding RNAs, which regulate gene expression via binding to the
3'-untranslated region (3'-UTR) of the target gene (10). miRNAs are important metabolic and
developmental regulators during pregnancy and are involved in the
development of GDM (11,12). Certain studies have reported that
miRNAs may serve as potential biomarkers of GDM (13,14),
suggesting that miRNAs may be therapeutic targets of GDM. A
previous study reported that the expression of miR-1323 was
significantly upregulated in patients with GDM (15). In addition, miR-1323 has been
demonstrated to be associated with the placenta and, therefore, may
be involved in the regulation of trophoblast function (16). The role of TP53INP1 in metabolic
regulation has been reported (17),
suggesting that TP53INP1 may regulate GDM. Notably, Zhang et
al (18) reported that TP53INP1
was a direct target of miR-1323 and may reverse the effect of
miR-1323 on the proliferation of hepatocellular carcinoma cells
(18). However, the clinical
significance and biological function of miR-1323 in patients with
GDM remain unclear. To the best of our knowledge, the interaction
between miR-1323 and TP53INP1 has not been revealed in the
pathogenesis of GDM to understand the molecular mechanisms.
The present study aimed to assess the expression of
miR-1323 in the serum of patients with GDM and trophoblastic cells.
Additionally, the present study investigated the diagnostic value
of serum miR-1323 in GDM. Furthermore, the effect of miR-1323 on
trophoblast viability was analyzed and the correlation between
miR-1323 and TP53INP1 was assessed to further understand the
underlying molecular mechanisms involved in the biological function
of miR-1323.
Materials and methods
Patients and blood sample
collection
The experimental protocols were approved by the
Ethics Committee of Weifang Maternal and Child Health Hospital
(Weifang, China), and all participants provided written informed
consent. The sample size in the present study was determined based
on the margin of error (set at 10%) and confidence level (set at
95%); therefore, the minimum sample size should be 96. The present
study recruited 110 patients with GDM from Weifang Maternal and
Child Health Hospital between March 2014 and May 2018. The patients
were diagnosed according to the guidelines of American Diabetes
Association (19), and 78 healthy
pregnant females were recruited as the control group. The
demographic and clinical characteristics of all recruits are
summarized in Table I, and there
were no significant differences in age, body mass index, gestation
and placental weight between the two groups (all P>0.05). The
average age of patients with GDM was 33.118±3.402 years (range,
27-42 years) and the average age of the healthy pregnant females
was 32.385±3.579 years (range, 26-41 years). The patients with
pre-gestational diabetes, multiple gestations accompanied with
further complications and those taking medications were all
excluded from the present study. Blood samples were collected from
participants at 24-28 weeks gestational following an overnight
fast, and fasting blood glucose (FBG) was measured and recorded by
using the glucose oxidase method (20). Additionally, the cut-off value for
FBG to diagnose GDM was 5.1 mM/l. Meanwhile, serum was isolated
from blood by centrifugation at 1,500 x g for 10 min at 4˚C and
stored at -80˚C for further use.
 | Table IClinical characteristics of the
females included in the present study. |
Table I
Clinical characteristics of the
females included in the present study.
| Parameter | Control (n=78) | GDM (n=110) | P-value |
|---|
| Age, years | 32.385±3.579 | 33.118±3.402 | 0.156 |
| BMI,
kg/m2 | 22.587±3.614 | 23.466±3.326 | 0.087 |
| Pregnancy weeks,
weeks | 25.218±2.196 | 25.591±1.757 | 0.198 |
| Placental weight,
kg | 0.586±0.088 | 0.616±0.155 | 0.128 |
Cell culture and treatment
Human trophoblast HTR-8/SVneo and BeWo cell lines
were purchased from Cell Bank of Type Culture Collection of Chinese
Academy of Sciences. HTR-8/SVneo cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; including 1,000 mg/l glucose;
Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
and BeWo cells were cultured in F-12 medium (Gibco; Thermo Fisher
Scientific, Inc.), supplemented with 10% FBS. Both HTR-8/SVneo and
BeWo cells were incubated at 37˚C in a humidified atmosphere
containing 5% CO2. The cells were divided into two
groups: The high glucose (HG)-treated group and the normal cell
group. Cells in the HG group were cultured in HG medium with 25 mM
glucose, while cells in the normal group were incubated in normal
medium with 5 mM glucose at 37˚C for 72 h.
Transfection of HTR-8/SVneo and BeWo
cells
HTR-8/SVneo and BeWo cells were cultured at 37˚C for
24 h. Subsequently, the cell transfection vectors, including 50 nM
miR-1323 mimic (5'-UCAAAACUGAGGGGCAUUUUCU-3'), 100 nM miR-1323
inhibitor (5'-AGAAAAUGCCCCUCAGUUUUGA-3'), 50 nM miR-1323 mimic
negative control (mimic NC; 5'-UUCUCCGAACGUGUCACGU-3') and 100 nM
miR-1323 inhibitor NC (5'-CAGUACUUUUGUGUAGUACAA-3'), which were
obtained from Shanghai GenePharma Co., Ltd., were used for cell
transfection to regulate the expression of miR-1323 in trophoblast
cells. The TP53INP1 overexpression vector pcDNA3.1-TP53INP1 (50 nM)
was constructed to promote the expression of TP53INP1 in
trophoblast cells, and the empty vector was used as a control for
the overexpression vector pcDNA3.1-TP53INP1. The aforementioned
vectors were respectively transfected into trophoblast cells using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols. Cell
transfection was performed at 37˚C for 6 h, and subsequently the
transfection reagent was removed. After 48 h of transfection, the
cells were used for subsequent experiments.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA, including miRNA, was extracted from serum
samples and cells using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). The purity and concentration of
the RNA was evaluated using a NanoDrop 2000 (Thermo Fisher,
Scientific, Inc.). Single-strand cDNA was synthesized using the
obtained RNA and RT-primer
(5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGAAAATG-3') using a
PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.) and
stored at -20˚C. The conditions for cDNA synthesis were as follows:
42˚C for 30 min and 85˚C for 5 sec.
The miR-1323 expression in serum and cells, and mRNA
expression of the TP53INP1 gene in cells were examined using
RT-qPCR, which was performed using a SYBR Green I Master Mix kit
(Invitrogen; Thermo Fisher Scientific, Inc.) and a 7300 Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
All the procedures were performed according to the manufacturer's
protocols. The following thermocycling conditions were used for the
qPCR: 95˚C for 10 min, 40 cycles of 95˚C for 30 sec, 60˚C for 15
sec and 72˚C for 15 sec. The miR-1323 and TP53INP1 expression
levels were normalized to U6 and β-actin, respectively. The primer
sequences were as follows: miR-1323 forward,
5'-GCCGAGUCAAAACUGAGG-3' and reverse, 5'-CTCAACTGGTGTCGTGGA-3'; U6
forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse,
5'-AACGCTTCACGAATTTGCGT-3'; TP53INP1 forward,
5'-GCACCCTTCAGTCTTTTCCTGTT-3' and reverse,
5'-GGAGAAAGCAGGAATCACTTGTATC-3'; β-actin forward,
5'-CTGGGACGACATGGAGAAAA-3' and reverse, 5'-AAGGAAGGCTGGAAGAGTGC-3'.
Each value was calculated using the 2-ΔΔCq method
(21).
MTT assay
The viability of HTR-8/SVneo and BeWo cells was
determined by MTT assay. Cells were seeded onto 96-well microplates
at a density of 3x104 cells/well and incubated at 37˚C
for 24 h. At the time points of 0, 24, 48 and 72 h, MTT (0.5 mg/ml)
was added into the wells and was further incubated for 4 h. Next,
the supernatant was removed and 200 µl dimethyl sulfoxide was added
into the wells. The cell viability was examined by reading the
absorbance at 490 nm.
Western blot analysis
The cells were lysed using RIPA lysis buffer (Thermo
Fisher Scientific, Inc.) to obtain total proteins. The BCA method
was used to determine the concentration of proteins. Following
quantitation with a WBC Protein Quantitation kit (Thermo Fisher
Scientific, Inc.), total proteins were separated via SDS-PAGE (12%
gel) and transferred onto PVDF membranes (EMD Millipore). After 4 h
of blocking with 5% skimmed milk at 4˚C, the primary antibodies,
including anti-TP53INP1 (1:500; cat. no. A04229; Wuhan Boster
Biological Technology, Ltd.) and anti-β-actin (dilution, 1:500;
cat. no. BA2305; B Wuhan Boster Biological Technology, Ltd.), were
incubated with the membranes at 4˚C overnight. Next, the membranes
were incubated with an HRP-conjugated secondary antibody (1:5,000;
cat. no. BA1054; Wuhan Boster Biological Technology, Ltd.) at 37˚C
for 2 h. In this analysis, β-actin was used as an internal control.
Subsequently, an enhanced chemiluminescence BeyoECL Plus kit
(Beyotime Institute of Biotechnology) was applied for protein band
visualization. The protein bands were quantified using ImageJ
Software v1.46 (National Institutes of Health).
Luciferase reporter assay
According to the TargetScan's bioinformatics
prediction (version 7.2; www.targetscan.org/vert_72), complementary sequences
of miR-1323 were searched at the 3'-UTR of TP53INP1. To confirm the
interaction between miR-1323 and TP53INP1, a luciferase reporter
assay was performed. The wild-type (WT) 3'-UTR containing the
binding site of miR-1323 or mutant-type (MT) 3'-UTR were inserted
into the pGL-control vector (Promega). According to the
manufacturer's protocols, the combined vectors were co-transfected
into HTR-8/SVneo and BeWo cells with miR-1323 mimic, miR-1323
inhibitor or the NCs using Lipofectamine 3000 reagent. After 48 h
of transfection, relative luciferase activity was measured using a
Dual-Luciferase Reporter assay system (Promega Corporation) and
normalized to Renilla luciferase activity.
Statistical analysis
All statistical analyses were performed using SPSS
21.0 software (IBM Corp.) and GraphPad Prism 7.0 software (GraphPad
Software, Inc.). All data are presented as the mean ± standard
deviation. Data between the two groups were compared using unpaired
Student's t-test. One-way analysis of variance followed by Tukey's
test was used to analyze the differences between multiple groups.
Pearson's correlation analysis was performed to assess the
correlation between indicators. Receiver operating characteristic
(ROC) curves were plotted to assess the diagnostic value of
miR-1323 in patients with GDM. P<0.05 was considered to indicate
a statistically significant difference.
Results
Diagnostic value of upregulated serum
miR-1323 and its correlation with FBG in patients with GDM
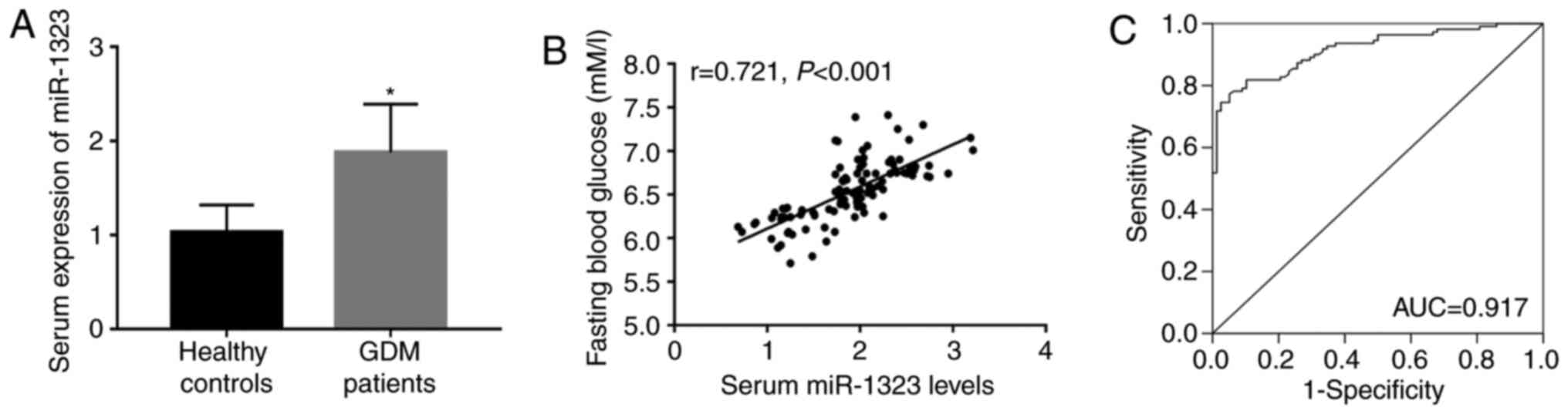
As shown in Fig. 1A,
a significant increase in serum expression of miR-1323 was observed
in patients with GDM, compared with that in the healthy controls
(P<0.05). Increased blood glucose is a characteristic of GDM,
and Fig. 1B shows a positive
correlation between miR-1323 and FBG (r=0.721; P<0.001). In
addition, in patients with GDM the FBG level was 6.532±0.344 mM/l
and the coefficient of variation was 5.267%. The ROC curve
indicated that miR-1323 had high diagnostic accuracy with an AUC of
0.917. At an optimal cut-off value of 1.489, the sensitivity was
77.27% and the specificity was 94.87% (Fig. 1C).
Effect of HG on miR-1323 expression
and trophoblast viability in HTR-8/SVneo and BeWo cells
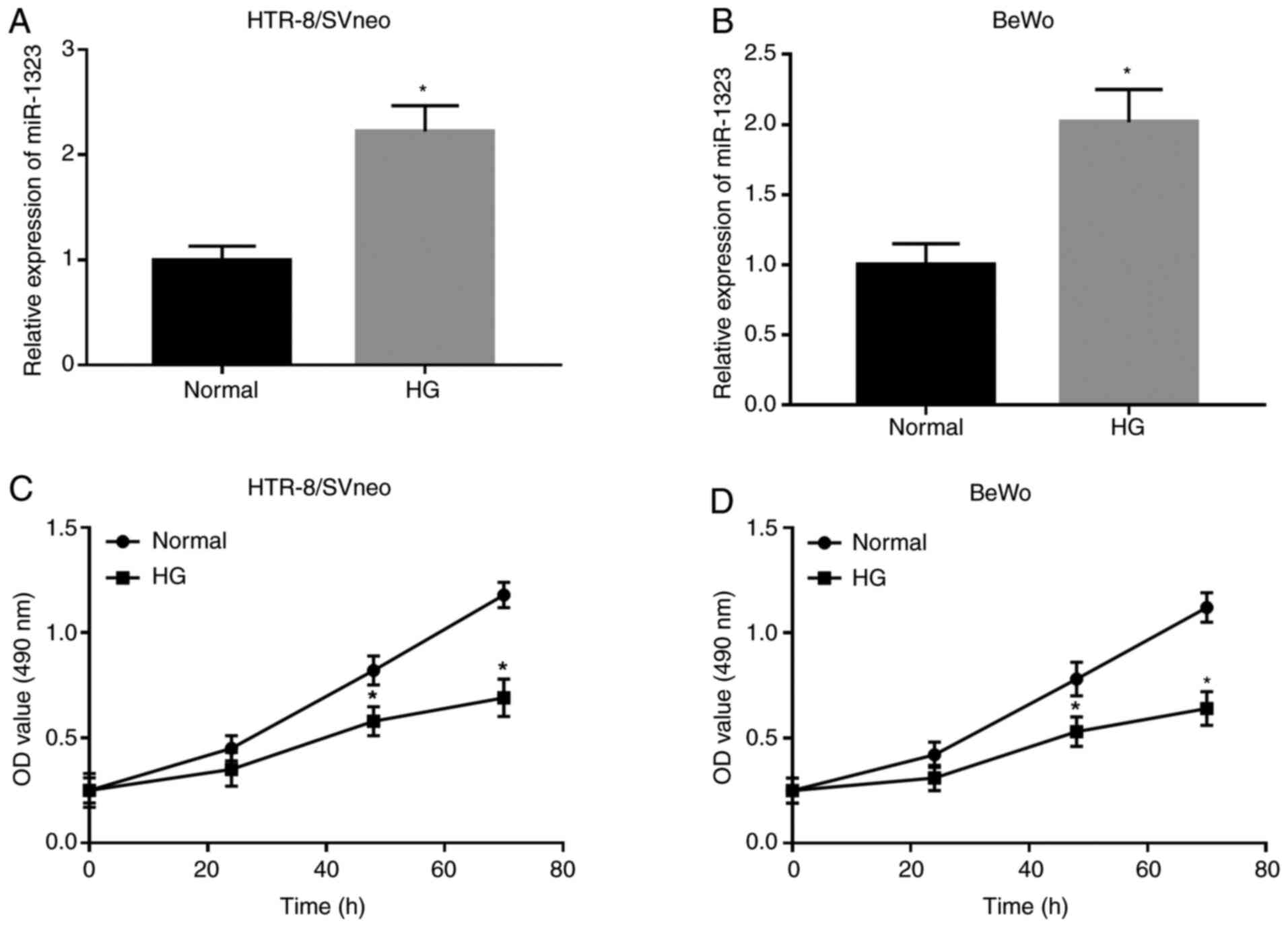
Following HG treatment, the expression of miR-1323
was upregulated in HG-treated cells, compared with that in normal
cells in HTR-8/SVneo and BeWo cell lines (all P<0.05; Fig. 2A and B). In HTR-8/SVneo and BeWo cells, the
trophoblast viability was significantly downregulated in HG-treated
cells compared with that in normal cells at 48 and 72 h (all
P<0.05; Fig. 2C and D).
Effect of miR-1323 on trophoblast
viability in HG-treated HTR-8/SVneo and BeWo cells
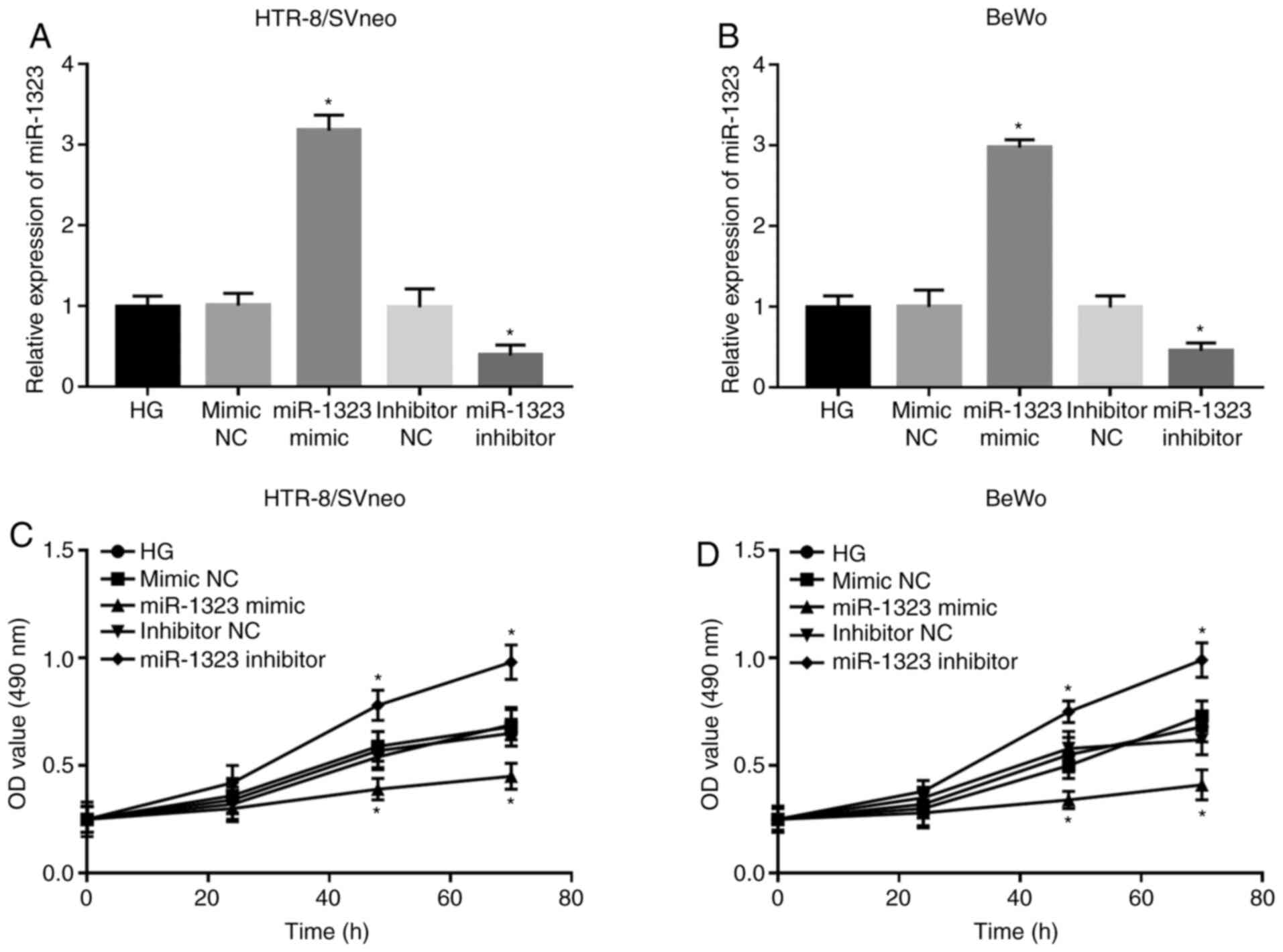
Following cell transfection, the expression of
miR-1323 was significantly upregulated by the miR-1323 mimic, but
was significantly downregulated by the miR-1323 inhibitor in
HG-treated HTR-8/SVneo and BeWo cells (all P<0.05; Fig. 3A and B). The analyses of trophoblast viability
suggested that the cell viability was markedly suppressed by the
miR-1323 overexpression, but was significantly promoted by
miR-1323-knockdown at 48 and 72 h in both HTR-8/SVneo and BeWo
cells (all P<0.05; Fig. 3C and
D).
miR-1323 directly regulates TP53INP1
expression in HTR-8/SVneo and BeWo cells
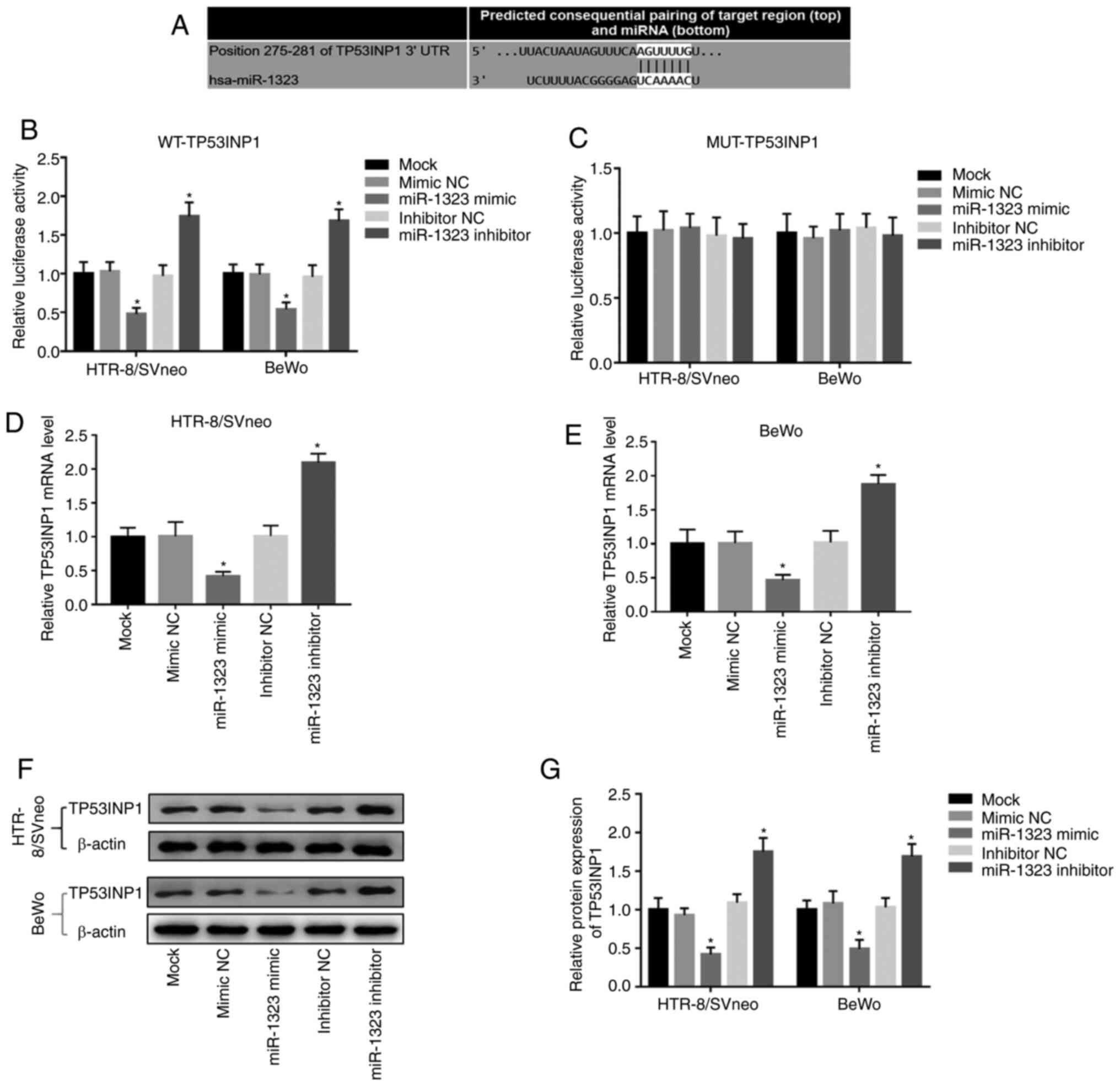
In order to further confirm the association of
miR-1323 with TP53INP1, a luciferase reporter assay was performed.
The putative binding site of miR-1323 at the 3'-UTR of TP53INP1 was
identified (Fig. 4A). In the WT
group, the relative luciferase activity was inhibited by the
overexpression of miR-1323, but was promoted by the knockdown of
miR-1323 in HTR-8/SVneo and BeWo cells (Fig. 4B). However, there were no changes in
the MUT group in HTR-8/SVneo and BeWo cells (Fig. 4C). Additionally, Fig. 4D and E demonstrate that the overexpression of
miR-1323 inhibited, while the knockdown of miR-1323 promoted, the
TP53INP1 mRNA expression in HTR-8/SVneo and BeWo cells (all
P<0.05). Furthermore, the relative protein expression of
TP53INP1 in two cell lines was detected, and it was revealed that
the protein expression of TP53INP1 was inhibited by overexpression
of miR-1323 and was promoted by the knockdown of miR-1323 (all
P<0.05; Fig. 4F and G).
Effect of miR-1323/TP53INP1 on
trophoblast viability in HG-treated HTR-8/SVneo and BeWo cells
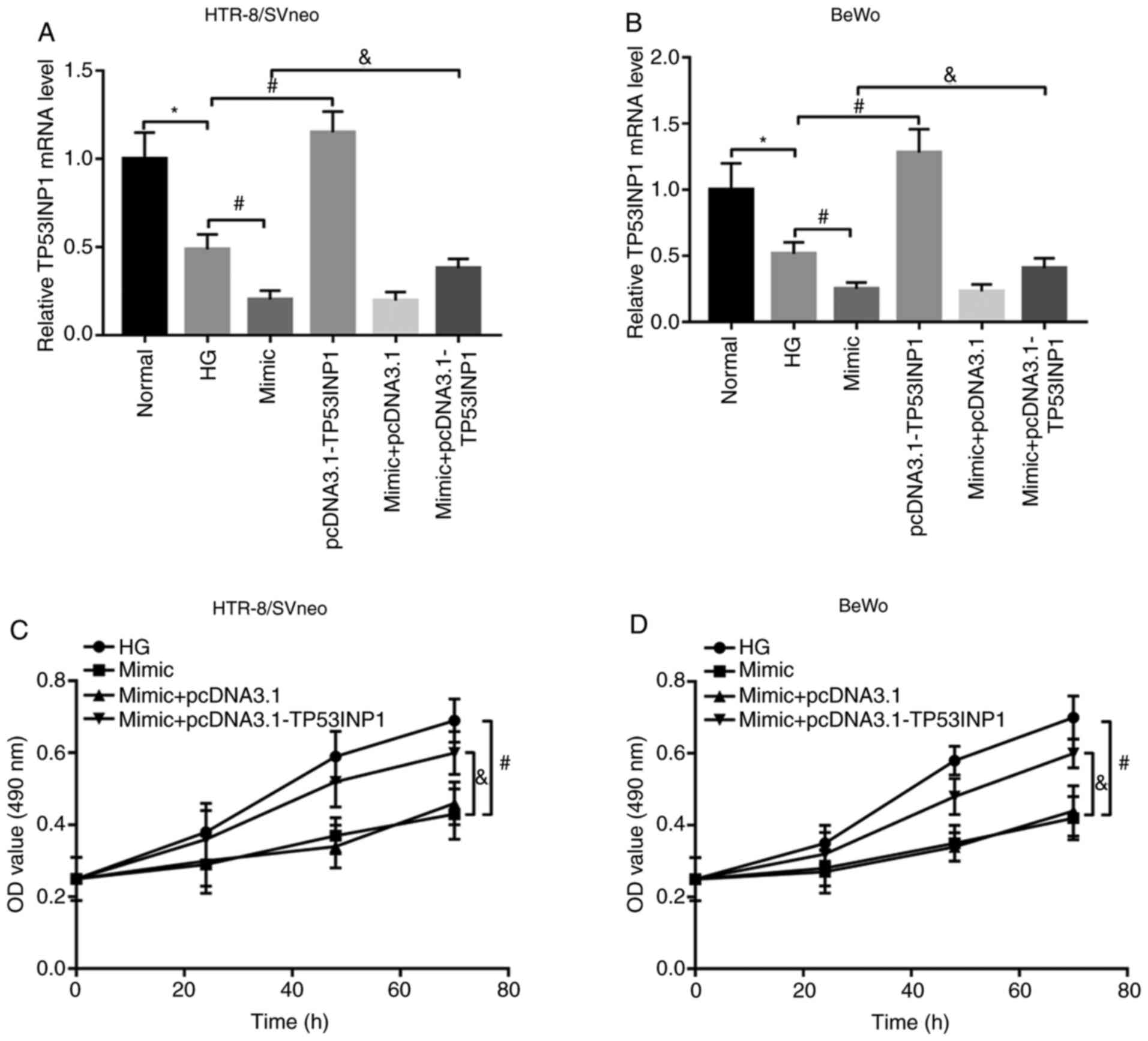
The expression of TP53INP1 was suppressed by HG
treatment, and was promoted by the pcDNA3.1-TP53INP1 in HG-treated
trophoblast cells. Additionally, the expression of TP53INP1, which
was suppressed by miR-1323 overexpression, was promoted by
pcDNA3.1-TP53INP1 in HG-treated HTR-8/SVneo and BeWo cells (all
P<0.05; Fig. 5A and B). The inhibition of trophoblast viability
because of miR-1323 overexpression was reversed by the
overexpression of TP53INP1 (all P<0.05; Fig. 5C and D). These data suggested that miR-1323 may
inhibit the trophoblast cell viability by suppressing TP53INP1 in
HTR-8/SVneo and BeWo cells.
Discussion
In recent years, microRNAs (miRNAs) have been
recognized as important molecules in biological processes and are
associated with various human diseases (22). For example, Fang et al
(23) reported that miR-185
promoted the progression of atherosclerosis by targeting STIM1. A
study by Liu et al (24)
demonstrated that miR-34a promoted renal fibrosis by downregulating
Klotho in tubular epithelial cells. Certain miRNAs with abnormal
expression have been reported in GDM, including miR-132(25) and miR-185(26). In addition, it has been reported
that miR-1323 is upregulated in patients with GDM (15) and may be involved in the regulation
of trophoblast function (16).
These studies highlighted the roles of miRNAs in GDM. The present
study also identified that serum miR-1323 expression was
upregulated in patients with GDM and was positively correlated with
FBG. Therefore, we hypothesized that miR-1323 may be involved in
GDM.
miRNAs have been considered as ideal diagnostic
tools for various human diseases due to their abnormal expression
and stability in blood samples (27). For example, decreased levels of
serum miR-103 may be used as non-invasive diagnostic biomarkers in
patients with sepsis (28) and
decreased levels of serum miR-375 may serve as biomarkers for the
diagnosis of osteosarcoma (29).
Considering the increased expression of miR-1323 in patients with
GDM, ROC curves were plotted based on the miR-1323 expression and
revealed that miR-1323 may be a potential diagnostic biomarker for
patients with GDM. Abnormal serum miR-1323 expression has been
reported to be a candidate biomarker for numerous other diseases,
including lung adenocarcinoma (30)
and breast cancer (31). Therefore,
we hypothesized that serum miR-1323 may be a novel and effective
diagnostic biomarker for patients with GDM.
It is well known that the normal growth and
development of the placenta is one of the important steps of a
healthy pregnancy. However, patient with GDM often have placental
dysplasia (5). HG is a
characteristic of GDM, resulting in impaired trophoblast function,
thereby inhibiting normal development of the placenta (32). Therefore, HG-induced trophoblast
cells are widely used to study the molecules involved in the
biological function of trophoblast cells during the pathological
process of GDM. In the present study, HG markedly inhibited the
viability of HTR-8/SVneo and BeWo cells, which is consistent with
the results of a previous study (25).
Previous studies have demonstrated that miRNAs may
regulate the viability or proliferation of trophoblasts in
diseases, including GDM. For example, upregulation of miR-211
repressed the proliferation of JEG-3 and BeWo cells in
choriocarcinoma (33). Peng et
al (34) reported that miR-137
suppressed the viability of HTR-8/SVneo cells in patients with GDM.
In the present study, miR-1323 was upregulated in HG-induced
trophoblast cells, compared with the normal trophoblast cells.
Additionally, miR-1323 overexpression inhibited, and miR-1323
knockdown promoted the viability of HTR-8/SVneo and BeWo cells in
the HG-treated cells. Additionally, previous studies have
demonstrated that miR-1323 may affect disease by regulating cell
viability or proliferation. For example, suppression of miR-1323
has been reported to promote breast cancer cell viability (31). In addition, a study by Gillet et
al (15) reviewed the
literature and reported that miR-1323 was involved in trophoblast
proliferation in pregnant females. Therefore, we hypothesized that
miR-1323 may exert a cytotoxic role in GDM by inhibiting
trophoblast cell viability.
TP53INP1, a key stress protein with
antioxidant-associated tumor suppressor function, is involved in
cell cycle arrest and apoptosis (35). Numerous studies (18,36,37)
have demonstrated that TP53INP1 may serve as a target of miRNA and
was regulated by miRNA in diseases. For example, Du et al
(36) reported that TP53INP1 was a
target of miR-142-5p and was inhibited by miR-142-5p in heart
failure caused by myocardial infarction. Li et al (37) found that TP53INP1 was a target of
miR-155-5p and regulated by miR-155-5p in cervical cancer. Notably,
a study by Zhang et al (18)
suggested that TP53INP1 was a direct target of miR-1323 and was
inhibited by miR-1323 in hepatocellular carcinoma. Therefore, the
present study focused on the effect of miR-1323 on the levels of
TP53INP1 in patients with GDM. The result suggested that TP53INP1
was the target of miR-1323, the mRNA and protein levels of TP53INP1
were downregulated by miR-1323-overexpression. In addition, it has
been reported that miRNAs regulate cell viability by regulating
TP53INP1 in other diseases. For example, Ye et al (38) reported that miR-3934-5p may inhibit
the cell viability of neuroblastoma by targeting TP53INP1. TP53INP1
expression was negatively regulated by miR-301a, and
TP53INP1/miR-301a was involved in gastric cancer cell viability
(39). Zhang et al (18) reported that miR-1323 promoted the
proliferation of hepatocellular carcinoma cells by targeting the
TP53INP1. In the present study, in trophoblasts co-transfected with
miR-1323 and TP53INP1 overexpression vectors, in was revealed that
the cell viability inhibited by miR-1323 overexpression was
reversed by TP53INP1 overexpression. These results suggested that
the inhibition of trophoblast viability by miR-1323 may be achieved
by inhibiting TP53INP1. miR-1323 may be a potential therapeutic
target for GDM treatment.
In conclusion, the results of the present study
suggested that the upregulated level of serum miR-1323 may serve as
a diagnostic biomarker for GDM. In addition, knockdown of miR-1323
in trophoblast cells may promote the trophoblast cell viability
under HG conditions by targeting TP53INP1, suggesting that miR-1323
may be a potential therapeutic target for GDM treatment. However,
only in vitro experiments were conducted in the present
study, and further in vivo experiments are required to
confirm the role of the miR-1323/TP53INP1 axis in the pathogenesis
of GDM. Another limitation was identified in the present study. A
recent study has reported that the HTR-8/SVneo cell line contains
two populations, one of epithelial and one of mesenchymal origin
(40). However, this was not taken
into consideration in the current study, which is a limitation.
Therefore, further studies need to focus on the different
populations in the HTR-8/SVneo cells, and further conclusions may
emerge.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL and YL designed and conceived the study,
conducted the clinical studies, analyzed the clinical data, wrote
the manuscript and confirmed the authenticity of the raw data. JZ
performed the cell experiments and analyzed the corresponding data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from each
patient and the experimental procedures were approved by the Ethics
Committee of Weifang Maternal and Child Health Hospital (Weifang,
China).
Patient consent for publication
Written informed consent for publication was
obtained from each participant.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chiefari E, Arcidiacono B, Foti D and
Brunetti A: Gestational diabetes mellitus: An updated overview. J
Endocrinol Invest. 40:899–909. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lavery JA, Friedman AM, Keyes KM, Wright
JD and Ananth CV: Gestational diabetes in the United States:
Temporal changes in prevalence rates between 1979 and 2010. BJOG.
124:804–813. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Committee on Practice
Bulletins-Obstetrics. ACOG Practice Bulletin No. 190: Gestational
diabetes mellitus. Obstet Gynecol. 131:e49–e64. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yung HW, Alnaes-Katjavivi P, Jones CJ,
El-Bacha T, Golic M, Staff AC and Burton GJ: Placental endoplasmic
reticulum stress in gestational diabetes: The potential for
therapeutic intervention with chemical chaperones and antioxidants.
Diabetologia. 59:2240–2250. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Edu A, Teodorescu C, Dobjanschi CG, Socol
ZZ, Teodorescu V, Matei A, Albu DF and Radulian G: Placenta changes
in pregnancy with gestational diabetes. Rom J Morphol Embryol.
57:507–512. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang Y, Ji L, Peng Z, Lai R, Zhang X, Xu
Y, Chen Z, Liu R, Zhong Y, Hu H and Wang L: Silencing DAPK3 blocks
the autophagosome-lysosome fusion by mediating SNAP29 in
trophoblast cells under high glucose treatment. Mol Cell
Endocrinol. 502(110674)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chang SC and Vivian Yang WC: Hyperglycemia
induces altered expressions of angiogenesis associated molecules in
the trophoblast. Evid Based Complement Alternat Med.
2013(457971)2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zong S, Li C, Luo C, Zhao X, Liu C, Wang
K, Jia W, Bai M, Yin M, Bao S, et al: Dysregulated expression of
IDO may cause unexplained recurrent spontaneous abortion through
suppression of trophoblast cell proliferation and migration. Sci
Rep. 6(19916)2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tian FJ, Qin CM, Li XC, Wu F, Liu XR, Xu
WM and Lin Y: Decreased stathmin-1 expression inhibits trophoblast
proliferation and invasion and is associated with recurrent
miscarriage. Am J Pathol. 185:2709–2721. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gao Y, She R, Wang Q, Li Y and Zhang H:
Up-regulation of miR-299 suppressed the invasion and migration of
HTR-8/SVneo trophoblast cells partly via targeting HDAC2 in
pre-eclampsia. Biomed Pharmacother. 97:1222–1228. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dias S, Pheiffer C, Abrahams Y, Rheeder P
and Adam S: Molecular biomarkers for gestational diabetes mellitus.
Int J Mol Sci. 19(2926)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tryggestad JB, Vishwanath A, Jiang S,
Mallappa A, Teague AM, Takahashi Y, Thompson DM and Chernausek SD:
Influence of gestational diabetes mellitus on human umbilical vein
endothelial cell miRNA. Clin Sci (Lond). 130:1955–1967.
2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pheiffer C, Dias S, Rheeder P and Adam S:
Decreased expression of circulating miR-20a-5p in South African
women with gestational diabetes mellitus. Mol Diagn Ther.
22:345–352. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yoffe L, Polsky A, Gilam A, Raff C,
Mecacci F, Ognibene A, Crispi F, Gratacós E, Kanety H, Mazaki-Tovi
S, et al: Early diagnosis of gestational diabetes mellitus using
circulating microRNAs. Eur J Endocrinol. 181:565–577.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gillet V, Ouellet A, Stepanov Y,
Rodosthenous RS, Croft EK, Brennan K, Abdelouahab N, Baccarelli A
and Takser L: miRNA profiles in extracellular vesicles from serum
early in pregnancies complicated by gestational diabetes mellitus.
J Clin Endocrinol Metab. 104:5157–5169. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Na Q, Wang D and Song W: Underexpression
of 4 placenta-associated microRNAs in complete hydatidiform moles.
Int J Gynecol Cancer. 22:1075–1080. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Seillier M, Pouyet L, N'Guessan P, Nollet
M, Capo F, Guillaumond F, Peyta L, Dumas JF, Varrault A, Bertrand
G, et al: Defects in mitophagy promote redox-driven metabolic
syndrome in the absence of TP53INP1. EMBO Mol Med. 7:802–818.
2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang F, Yang C, Xing Z, Liu P, Zhang B,
Ma X, Huang L and Zhuang L: LncRNA GAS5-mediated miR-1323 promotes
tumor progression by targeting TP53INP1 in hepatocellular
carcinoma. Onco Targets Ther. 12:4013–4023. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
American Diabetes Association. Diagnosis
and classification of diabetes mellitus. Diabet Care. 35 (Suppl
1):S64–S71. 2012.
|
|
20
|
Tiongco RE, Bituin A, Arceo E, Rivera N
and Singian E: Salivary glucose as a non-invasive biomarker of type
2 diabetes mellitus. J Clin Exp Dent. 10:e902–e907. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen X, Gong Y, Zhang DH, You ZH and Li
ZW: DRMDA: Deep representations-based miRNA-disease association
prediction. J Cell Mol Med. 22:472–485. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fang M, Li Y, Wu Y, Ning Z, Wang X and Li
X: miR-185 silencing promotes the progression of atherosclerosis
via targeting stromal interaction molecule 1. Cell Cycle.
18:682–695. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liu Y, Bi X, Xiong J, Han W, Xiao T, Xu X,
Yang K, Liu C, Jiang W, He T, et al: MicroRNA-34a promotes renal
fibrosis by downregulation of klotho in tubular epithelial cells.
Mol Ther. 27:1051–1065. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhou X, Xiang C and Zheng X: miR-132
serves as a diagnostic biomarker in gestational diabetes mellitus
and its regulatory effect on trophoblast cell viability. Diagn
Pathol. 14(119)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Qi S and Wang X: Decreased expression of
miR-185 in serum and placenta of patients with gestational diabetes
mellitus. Clin Lab. 65:2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bertoli G, Cava C and Castiglioni I: The
potential of miRNAs for diagnosis, treatment and monitoring of
breast cancer. Scand J Clin Lab Invest Suppl. 245 (Suppl
1):S34–S39. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yang M, Zhao L and Sun M: Diagnostic value
of miR-103 in patients with sepsis and noninfectious SIRS and its
regulatory role in LPS-induced inflammatory response by targeting
TLR4. Int J Genomics. 2020(2198308)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu W, Zhao X, Zhang YJ, Fang GW and Xue
Y: MicroRNA-375 as a potential serum biomarker for the diagnosis,
prognosis, and chemosensitivity prediction of osteosarcoma. J Int
Med Res. 46:975–983. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhao H, Zheng C, Wang Y, Hou K, Yang X,
Cheng Y, Che X, Xie S, Wang S, Zhang T, et al: miR-1323 promotes
cell migration in lung adenocarcinoma by targeting Cbl-b and is an
early prognostic biomarker. Front Oncol. 10(181)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Xu Y and Liu M: MicroRNA-1323
downregulation promotes migration and invasion of breast cancer
cells by targeting tumor protein D52. J Biochem. 168:83–91.
2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Peng HY, Li MQ and Li HP: High glucose
suppresses the viability and proliferation of HTR8/SVneo cells
through regulation of the miR137/PRKAA1/IL6 axis. Int J Mol Med.
42:799–810. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ji L and Li X: Long noncoding RNA MEG3 is
a tumor suppressor in choriocarcinoma by upregulation of
microRNA-211. J Cell Physiol. 234:22911–22920. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Peng HY, Li MQ and Li HP: miR-137
restricts the viability and migration of HTR-8/SVneo cells by
downregulating FNDC5 in gestational diabetes mellitus. Curr Mol
Med. 19:494–505. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Nishimoto M, Nishikawa S, Kondo N,
Wanifuchi-Endo Y, Hato Y, Hisada T, Dong Y, Okuda K, Sugiura H,
Kato H, et al: Prognostic impact of TP53INP1 gene expression in
estrogen receptor α-positive breast cancer patients. Jpn J Clin
Oncol. 49:567–575. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Du J, Yang ST, Liu J, Zhang KX and Leng
JY: Silence of LncRNA GAS5 protects cardiomyocytes H9c2 against
hypoxic injury via sponging miR-142-5p. Mol Cells. 42:397–405.
2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li N, Cui T, Guo W, Wang D and Mao L:
miR-155-5p accelerates the metastasis of cervical cancer cell via
targeting TP53INP1. Onco Targets Ther. 12:3181–3196.
2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ye W, Liang F, Ying C, Zhang M, Feng D and
Jiang X: Downregulation of microRNA-3934-5p induces apoptosis and
inhibits the proliferation of neuroblastoma cells by targeting
TP53INP1. Exp Ther Med. 18:3729–3736. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Dou J, Tu D, Zhao H and Zhang X: LncRNA
PCAT18/miR-301a/TP53INP1 axis is involved in gastric cancer cell
viability, migration, and invasion. J Biochem. 168:547–555.
2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Abou-Kheir W, Barrak J, Hadadeh O and
Daoud G: HTR-8/SVneo cell line contains a mixed population of
cells. Placenta. 50:1–7. 2017.PubMed/NCBI View Article : Google Scholar
|