Introduction
Serratia spp., particularly the species
Serratia marcescens (S. marcescens), are ubiquitous
environmental gram-negative bacteria. They are members of the
Enterobacteriaceae family that flourish in all kinds of
environments and have become important pathogens and were included
in those accounting for hospital-acquired infections in recent
years. In general, Serratia spp. does not pose a threat to
individuals with normal immunity. However, patients who are
hospitalized in the neonatal department, (pediatric) intensive care
unit (ICU) or bone marrow transplant and oncology units are
immunocompromised or immunosuppressed and the severity of infection
is markedly increased (1-3).
Serratia spp. infections are also associated with other
complications, such as infections in the lower respiratory tract,
urinary tract, wounds and the bloodstream (4). Studies have also indicated that
Serratia spp. may give rise to corneal ring infiltrates in
patients with human immunodeficiency virus through contaminated
contact lenses (5,6). Among the infections caused by
Serratia spp., bloodstream infection (BSI) is the most
prevalent and fatal (7). Hence, a
rapid and sensitive characterization method is urgently needed to
guide anti-infection therapies (8).
Carbapenem-resistant Enterobacteriaceae (CRE)
have become a serious problem worldwide. Resistance to carbapenems
is rising year on year due to the spread of carbapenemases, the
mutation of penicillin-binding proteins, decreased membrane
permeability and overexpression of efflux pumps (9,10). For
CRE, the predominant mechanism contributing to such resistance
involves carbapenemases, particularly β lactamase Klebsiella
pneumoniae carbapenemase (blaKPC), and type B and type D
carbapenemases. Genes encoding these carbapenemases have also been
detected in S. marcescens (11,12).
In addition, other types of carbapenemases, including S.
marcescens enzymes, Serratia metallo-β-lactamase,
Guiana extended-spectrum β-lactamase and Germany
imipenemase, were detected in S. marcescens isolated in
different regions (13-18).
The high hydrolytic efficiency of these carbapenemases when acting
on carbapenem substrates combined with the natural resistance of
S. marcescens to polymyxin B and polymyxin E (two restricted
antibiotics used for multidrug-resistant bacteria infections),
makes the treatment of S. marcescens infections challenging
in the clinical setting (19).
Loop-mediated isothermal amplification (LAMP) is a
high-specificity, high-efficiency and rapid amplification
technology based on PCR. It uses a DNA polymerase with high
displacement strand activity and three pairs of specially designed
primers to amplify the target sequence (20). LAMP is frequently used to detect
infectious diseases among humans, livestock and plants. There are
various detection methods for the reaction endpoint, including the
turbidity method, agarose gel electrophoresis and UV light
detection method. The turbidity method is widely used due to its
simple operation and real-time mode. This method detects the
by-product of the LAMP reaction
(Mg2P2O7, magnesium pyrophosphate,
a white precipitate) at an optical density of 650 nm every 6 sec. A
variety of microorganisms have been successfully detected by using
this method, such as Mycoplasma pneumoniae, Salmonella spp.
and Clostridium difficile (21-23).
Due to the prevalence of carbapenem-resistant
Serratia spp. in China (24-28),
the present study aimed to develop a LAMP method for the rapid
characterization of blaKPC producing Serratia spp.
(from pure cultures and clinical specimens) with the
calcein/Mn2+ complex and a constant turbidity recording
system and the reaction characteristics were assessed.
Materials and methods
Bacterial strains and preparation of
templates
In the present study, 31 bacterial strains isolated
from patients were used, including three different strains of
Serratia spp. (S. marcescens DY12303, S.
liquefaciens DY12165 and S. rubidaea DY12122). The
characteristics of these 31 bacterial strains are listed in
Table I. S. marcescens
DY12303 with blaKPC-2 was obtained from a patient who was
admitted to the ICU at Henan Province Hospital of Traditional
Chinese Medicine (TCM; Second Affiliated Hospital of Henan
University of TCM, Zhengzhou, China) and validated by a Vitek-2
Compact automated system for ID/AST (Bio Merieux). This strain was
used as the positive control for Serratia spp.
identification and blaKPC detection. Sequencing of 16s
ribosomal (r)RNA and blaKPC-2 were performed using the PCR
primers PCR F27, R1492, KPC-forward (F) and KPC-reverse (R) (see
Table III). The PCR mixture
contained 10 µl PCR 2X master mix reagent (Tiangen Biotech Co.,
Ltd.), 1 µl 2 µM forward and reverse primers, 1 µl (2 ng) of
template and 7 µl double-distilled (dd)H2O. The
procedure was as follows: Initial denaturation at 94˚C for 5 min,
followed by 35 cycles of 94˚C for 30 sec as the denaturation step,
58˚C for 30 sec as the annealing step and 72˚C for 30 sec as the
extension step. For the final extension step, the reaction was held
at 72˚C for 10 min. The product was sequenced by Sangon
Biotechnology Co., Ltd. and the sequencing results of 16s rRNA and
blaKPC-2 were 100% identical to the S. marcescens and
blaKPC-2 sequences in GenBank. Genomic DNA from bacteria was
extracted using a QIAamp DNA Mini kit (cat. no. 51304; Qiagen GmbH)
following the manufacturer's protocol. Purified DNA was diluted
100-fold and saved as a template for the experiments.
 | Table IBacterial strains used in the present
study. |
Table I
Bacterial strains used in the present
study.
| Species | Resistance
mechanism | Source |
|---|
| S.
marcescens DY12303 | KPC-2 | Clinical
isolate |
| S.
liquefaciens DY12165 | - | Clinical
isolate |
| S. rubidaea
DY12122 | - | Clinical
isolate |
| E. coli
DY12123 | - | Clinical
laboratorya |
| K.
pneumoniae DY12326 | - | Clinical
isolate |
| A. baumannii
DY12522 | - | Clinical
isolate |
| P. vulgaris
DY12184 | - | Clinical
laboratory |
| P.
aeruginosa DY12122 | VIM-2 | Clinical
laboratory |
| S. sonnel
DY12531 | - | Clinical
laboratory |
| Y. pestis
DY12189 | - | Clinical
laboratory |
| C. freundii
DY12218 | - | Clinical
laboratory |
| S. typhi
DY12161 | - | Clinical
laboratory |
| M. morganii
DY12439 | - | Clinical
laboratory |
| P. rettgeri
DY12210 | - | Clinical
laboratory |
| A. baumannii
FY12252 | OXA-23 | Clinical
isolate |
| P.
aeruginosa DY12476 | - | Clinical
laboratory |
| K. oxytoca
DY12151 | - | Clinical
isolate |
| E. cloacae
DY12105 | - | Clinical
isolate |
| E. coli
DY12228 | NDM-1 | Clinical
laboratory |
| K.
pneumoniae DY12274 | - | Clinical
isolate |
| P. mirabilis
DY12407 | - | Clinical
laboratory |
| S.
maltophilia DY12143 | - | Clinical
laboratory |
| E. coli
DY12295 | MCR-1 | Clinical
laboratory |
| B. cepacia
DY12521 | VIM-1 | Clinical
isolate |
| K.
pneumoniae DY12384 | MCR-1 | Clinical
isolate |
| A. baumannii
DY12223 | NDM-1 | Clinical
isolate |
| S.
maltophilia DY12143 | blaL1 | Clinical
isolate |
| S. typhi
DY12161 | TEM-1 | Clinical
laboratory |
| E. aerogenes
DY12121 | CTX-M-3 | Clinical
isolate |
| E. cloacae
DY12315 | NDM-1 | Clinical
isolate |
| P.
aeruginosa DY12476 | IMP-9 | Clinical
isolate |
 | Table IIISequences of primers used for
amplification of LuxS and blaKPC. |
Table III
Sequences of primers used for
amplification of LuxS and blaKPC.
| Primer |
Type/definition | Sequence
(5'-3') |
|---|
| U5-FIP | Forward inner |
GCTTTCCAGGCATCGGCAACGCACCGGTTTCTACATGAG |
| U5-BIP | Reverse inner |
ATGGCCGACGTGCTGAAAGTACTGGTACTCGTTCAGCTCA |
| U5-F3 | Forward outer |
GGCGTGGAGATTATCGACAT |
| U5-B3 | Reverse outer |
CGAGTGCATGTGGTAGGTAC |
| U5-LF | Loop forward |
TTCCGGCACGCCGATCA |
| U5-LB | Loop reverse |
GACCGACCAGCGCAAGA |
| K17-FIP | Forward inner |
CAGCACAGCGGCAGCAAGACTGAGGAGCGCTTCCCA |
| K17-BIP | Reverse inner |
TCCGTTACGGCAAAAATGCGCTCGTCATGCCTGTTGTCAGAT |
| K17-F3 | Forward outer |
GGCGCAACTGTAAGTTACCG |
| K17-B3 | Reverse outer |
TCACTGTATTGCACGGCG |
| K17-LF | Loop forward |
GCCCTTGAATGAGCTGCAC |
| K17-LB | Loop reverse |
GGTTCCGTGGTCACCCA |
| LuxS-F | PCR forward |
TTGATCTGCGCTTTTGCCGC |
| LuxS-R | PCR reverse |
TTCACCACCACGCCGTTATC |
| KPC-F | PCR forward |
CGGTCAGTCCGTTTGTTC |
| KPC-R | PCR reverse |
CTTGGTCGGTCTGTAGGG |
| F27 | 16S rRNA PCR |
AGAGTTTGATCCTGGCTCAG |
| R1492 | 16S rRNA PCR |
ACGGCTACCTTGTTACGACTT |
Primer design
As the LuxS gene of Serratia spp. diverged
from other luxS genes of bacterial species, such as Escherichia
coli, Salmonella enterica, Pantoea ananatis,
Listeria monocytogenes, Yersinia pestis, and
Erwenia amylovora the luxS gene, which is associated with
the quorum sensing system of Serratia spp., was selected as
the target sequence. The complete luxS gene sequences of S.
marcescens ATCC274 (GenBank accession no. AJ628150), S.
odorifera (GenBank accession no. GG753567), S. lique
faciens ATCC27592 (GenBank accession no. CP006252) and S.
rubidaea (GenBank accession no. CP014474) were obtained from
the National Center for Biotechnology Information (NCBI) GenBank
database (https://www.ncbi.nlm.nih.gov/genbank/) and compared
using CLUSTAL W2.1. The sequence alignment results (Fig. S1) were further analyzed using
Primer Explorer software V5 (http://primerexplorer.jp/lampv5e/index.html), and
forward outer (F3), backward outer (B3), forward internal (FIP),
backward internal (BIP), forward loop (LF) and backward loop (LB)
primers were designed to optimize the amplification reaction.
To design universal LAMP primers for blaKPC,
KPC-1 to KPC-32 (the GenBank accession no. of each blaKPC
subtype is listed in Table II)
were compared using CLUSTAL W2.1(29). The sequence alignment results were
tackled with the same procedure for Serratia spp. To obtain
the most appropriate primers, three sets of primers were designed
for Serratia spp. and blaKPC. The sensitivity and
specificity of the LAMP assay were then compared with those of
conventional PCR. To complete this comparison experiment, luxS-F
and luxS-R primers for S. marcescens identification and
KPC-F and KPC-R for blaKPC identification were designed. The
primers were synthesized by Sangon Biotechnology Co., Ltd. and the
primer sequences are listed in Table
III.
 | Table IISubtypes of blaKPC used in
blaKPC primer design. |
Table II
Subtypes of blaKPC used in
blaKPC primer design.
| Subtype of
blaKPC | GenBank accession
no. |
|---|
| KPC-1 | AF297554 |
| KPC-2 | AY034847 |
| KPC-3 | AB557734 |
| KPC-4 | FJ473382 |
| KPC-5 | EU400222 |
| KPC-6 | EU555334 |
| KPC-7 | EU729727 |
| KPC-8 | FJ234412 |
| KPC-9 | FJ624872 |
| KPC-10 | GQ140348 |
| KPC-11 | HM066995 |
| KPC-12 | HQ641422 |
| KPC-13 | HQ342889 |
| KPC-14 | JX524191 |
| KPC-15 | KC433553 |
| KPC-16 | KC465199 |
| KPC-17 | KC465200 |
| KPC-18 | NG049251 |
| KPC-19 | KJ775801 |
| KPC-21 | NG049254 |
| KPC-22 | NG049255 |
| KPC-24 | NG049256 |
| KPC-25 | KU216748 |
| KPC-26 | KX619622 |
| KPC-27 | KX828722 |
| KPC-28 | NG052581 |
| KPC-30 | KY646302 |
| KPC-31 | NG055494 |
| KPC-32 | NG055495 |
Specificity and sensitivity evaluation
of the LAMP assay
To evaluate the sensitivity of the LAMP assay, pure
genomic DNA of S. marcescens DY12303 was extracted by using
a QIAamp DNA Mini Kit (Qiagen GmbH). DNA quantity and quality were
determined with a NanoDrop-1000 Spectrometer (Thermo Fisher
Scientific Inc.). The pure genomic DNA was further subjected to
serial 10-fold dilutions over a log7 scale with ddH2O.
Template concentrations ranging from 392.0 ng/µl to 0.392 pg/µl
were measured to compare the detection limit difference between the
LAMP assay and conventional PCR.
To determine the specificity, 18 common
non-Serratia clinical isolates were evaluated using the
Serratia spp. identification LAMP assay. In addition, 12
clinical isolates carrying prevalent resistance genes were used for
the specificity evaluation of blaKPC LAMP assays (details of
the bacterial strains are presented in Table I).
LAMP assay protocol and product
detection
A 25-µl volume of the reaction system of the DNA
amplification kit (Eiken Chemical Co., Ltd.) was applied to the
LAMP assay. The main components of the reaction system were
recorded in the instruction book for the d kit. The final
concentrations of primers required for one reaction were as
follows: For FIP and BIP 40 pmol, for LF and LB 20 pmol, and for F3
and B3 5 pmol. Finally, 1 µl genomic DNA template was added to the
reaction tube (Eiken Chemical Co., Ltd.) containing the materials
required for the LAMP reaction. The reaction proceeded at 65˚C for
a duration of 60 min. For inactivated products, a reaction
temperature of 80˚C for 5 min in a constant-temperature dry bath
was used. To detect LAMP products, two different methods were
adopted. The first was direct visual inspection; 1 µl of
calcein/Mn2+ complex (Eiken Chemical Co., Ltd.) as the
indicator was added to 24 µl of LAMP reactants prior to the LAMP
reaction. In positive reactions, the color changed from orange to
green, whereas in negative reactions, there was no color change.
The color change was easily observed under natural light or with
the aid of ultraviolet light (365 nm wavelength). An analytical
method was also used for reaction monitoring. This method used an
LA-230 Loopamp real-time turbidimeter (Eiken Chemical Co., Ltd.) to
monitor the turbidity (the white precipitation caused by the
presence of magnesium pyrophosphate,
Mg2P2O7). The LAMP assay was
assessed in real-time by recording the optical density at 650 nm
every 6 sec and the turbidity was recorded with the turbidimeter
software.
Conventional PCR protocol and product
detection
For conventional PCR, 50-µl reaction mixtures were
employed. The main components were as follows: 25 µl PCR 2X master
mix reagent (Tiangen Biotech Co., Ltd.), 1 µM forward and reverse
primers (luxS and KPC; see Table
II) and 1 µl of the DNA template. The procedure for
conventional PCR was an initial incubation at 94˚C for 2 min, 35
cycles at 94˚C for 30 sec as a denaturation step, 58˚C for 30 sec
as the annealing step, and 72˚C for 30 sec as the extension step.
For the final extension step, the reaction was held at 72˚C for 10
min. The PCR products were separated by electrophoresis using 1%
agarose gels (Amresco) and GelRed (Biotium) as a nucleic acid dye
at the recommended concentrations. A Gel Doc XR+ imaging system
(Bio-Rad Laboratories, Inc.) was used to image the PCR
products.
Evaluation of inhibition in clinical
specimens
To evaluate inhibition in clinical specimens, pure
DNA extracted by QIAamp DNA Mini Kit (Qiagen GmbH) from the sputum
and blood of healthy donors were mixed 1:1 with serially diluted
genomic DNA extracted from the clinical isolate S.
marcescens DY12303 with blaKPC as templates for the LAMP
assay. Thus, the final concentration of templates ranged from 19.6
to 0.0196 pg/µl. Subsequently, 1 µl of DNA template was added to 24
µl of the reaction mixture; the concentrations in the mixture for
LAMP were described above. Finally, 25 µl of the LAMP assay mixture
was used to evaluate the inhibitory characteristics of the pure DNA
extracted from sputum and blood.
LAMP assay evaluation of clinical
specimens
Clinical specimens required for the present study
were collected at the Henan Province Hospital of TCM (Zhengzhou,
China) from January to December 2018. The age range of patients was
4-82 years old, with a mean age of ± SD is 43.67±23.44. The sex
ratio was 208 (male): 151 (female). Sputum specimens obtained from
the ICU, Neurosurgery department and Oncology department were
collected to investigate the efficacy of the LAMP assay for
detecting Serratia spp. with blaKPC. First, a
conventional culture method was adopted-all clinical specimens were
inoculated, cultured, separated and identified in accordance with
the national clinical inspection operating procedures of China and
the results were used as a gold standard. In short, the clinical
samples were inoculated into 5% blood agar medium and cultured at
37˚C for 18-24 h. For Gram-negative bacilli, the isolated strains
were biochemically identified according to Bergey's Manual
Systematic Bacteriology (30) and
compared with positive control strains. In short, the difference
between Serratia marcescens and other bacteria in the same
genus was arabinose-negative, raffinose-negative and
xylose-negative; it may ferment sugars including maltose, glucose,
mannose and sucrose. Each specimen was then stored at -20˚C for DNA
extraction. To reduce DNA loss during the extraction process,
sputazyme (Kyokuto) was used to reduce the viscosity. In brief, the
proper volume of sputazyme was added to 500 µl sputum, then boiled
for 5 min and subsequently centrifuged at 6,600 x g for 10 min at
room temperature. The DNA in the supernatants was used as
templates. A QIAamp DNA Microbiome Kit (Qiagen GmbH) was used to
extract bacterial DNA from the sputum specimens according to the
manufacturer's instructions. Both unpurified and purified DNA were
used as templates for LAMP assays and real-time PCR assay
(Serratia marcescens genesig Standard kit, Primerdesign Co.,
Ltd.) to compare the two methods.
Real-time PCR protocol and product
detection
A Serratia marcescens genesig Standard kit
(Primerdesign Co., Ltd.) was used for real-time PCR in accordance
with the manufacturer's protocol. The final volume of the S.
marcescens Probe-based Fluorescent quantitative PCR was 20 µl,
which contained 10 µl Precision PLUS 2X qPCR Master Mix, 1 µl S.
marcescens primer/probe mix, 5 µl (2 ng) of templates and 4 µl
RNase-free water. PCR cycling parameters were as follows: 95˚C for
2 min, followed by 50 cycles at 95˚C for 10 sec and 60˚C for 1
min.
Data analysis
Sensitivity, specificity, positive predictive value
(PPV), negative predicted value (NPV) and positive likelihood ratio
(PLR) were calculated by using Clinical Calculator 1 (http://vassarstats.net/clin1.html#return). The
calculations were as follows: Sensitivity=A/(A+C) x100%,
Specificity=D/(D+B) x100%, PPV=A/(A+B) x100%, NPV=D/(D+C) x100%,
PLR= Sensitivity/(1-Specificity) x100%, where A indicates
positivity according to LAMP and conventional culture, B positivity
according to LAMP and negativity on conventional culture, C
negativity on LAMP and positivity on conventional culture and D
negativity on LAMP and conventional culture.
Results
Optimal primers for the LAMP assay of
Serratia spp. with blaKPC
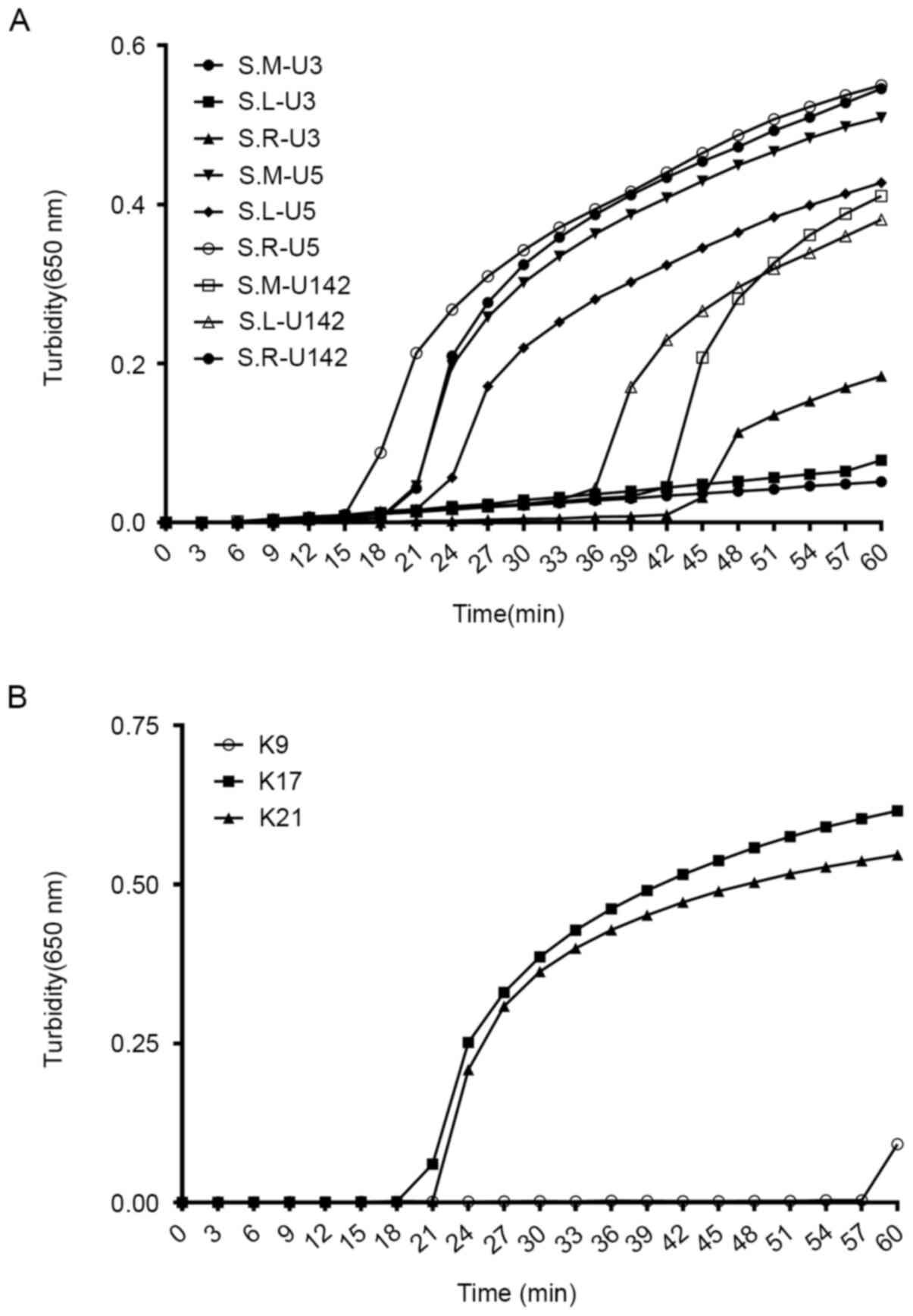
A total of three sets of candidate primers were
designed for the detection of Serratia spp. and
blaKPC. Under the same reaction conditions, it was observed
that the primer U5 set amplified the target sequence at 15, 18 and
21 min for S. marcescens DY12303, S. liquefaciens
DY12165 and S. rubidaea DY12122, respectively. For the
remaining two sets, either low efficiency or amplification failure
was observed. Therefore, the U5 sets were chosen, which were able
to amplify the target gene in the shortest time, as the optimal
reaction primers for Serratia spp. identification (Fig. 1A). For blaKPC detection,
primer K17 was confirmed as the fastest and most appropriate primer
(Fig. 1B). Accordingly, the U5 and
K17 primer sets (listed in Table
III) were chosen as the optimal primers for the detection of
Serratia spp. with blaKPC in the LAMP assay.
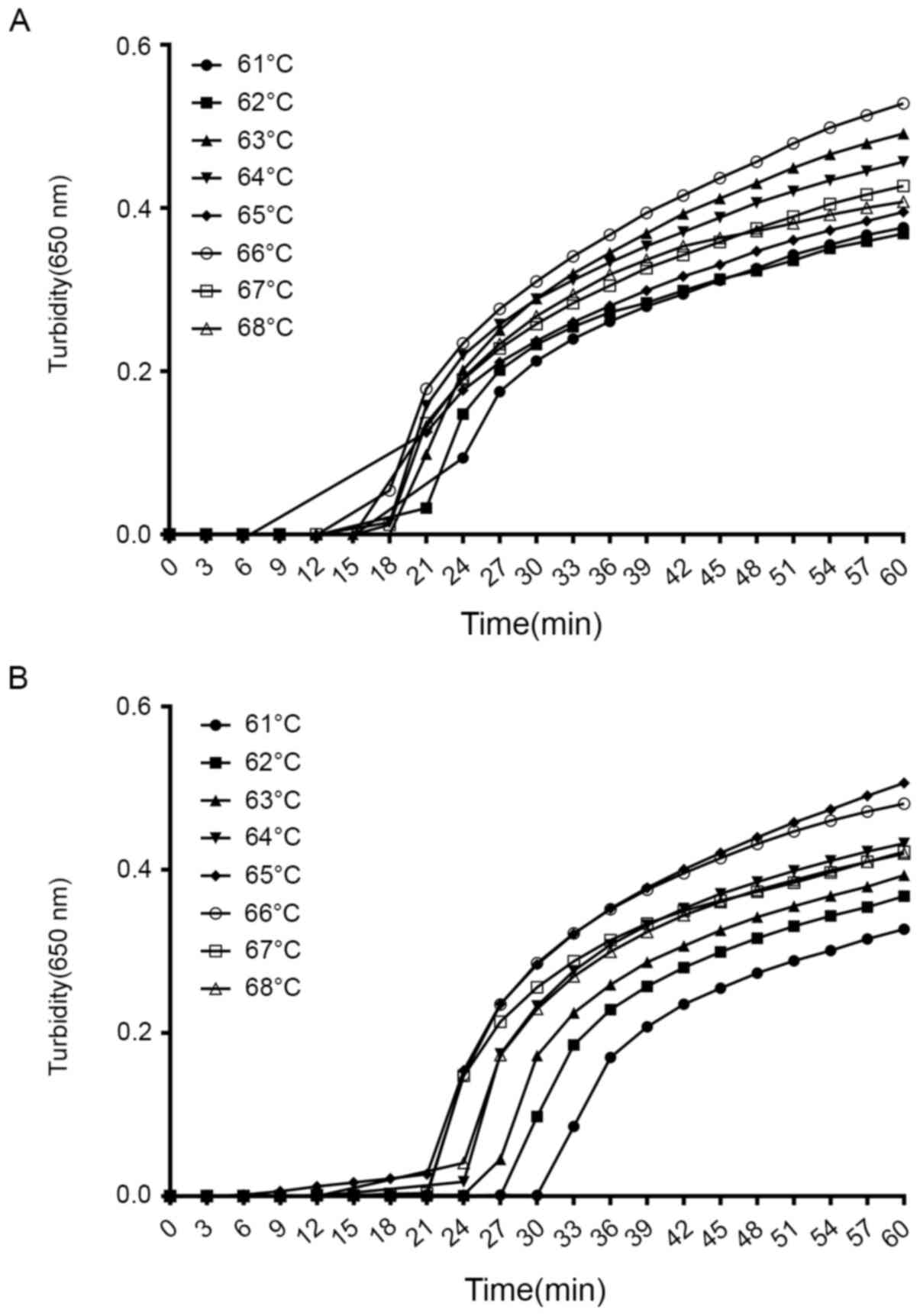
Optimal temperatures for the LAMP
assay of Serratia spp. with blaKPC
To determine the most suitable reaction conditions
for the selected primers in the LAMP reaction for the detection of
Serratia spp. with blaKPC, several temperatures from
61 to 68˚C at 1˚ intervals were evaluated. As indicated in Fig. 2A and B, 64-67˚C was the most suitable
temperature range. Based on these results, 65˚C was chosen as the
reaction temperature.
Specificity of the LAMP assay for
Serratia spp. with blaKPC
To estimate the specificity of the LAMP assay, S.
marcescens DY12303 was used as the positive control and
ddH2O as the negative control. Furthermore, 18
non-Serratia spp. bacterial strains and 12 clinical isolates
without blaKPC were tested in the specificity experiments.
Fig. 3A and B indicate that an increase in the
turbidity curve only occurred when DNA from S. marcescens
DY12303 was used as the template. For the negative control
(ddH2O) and the other reference strains, no significant
amplification was detected. These results suggested that the
primers selected had excellent specificity.
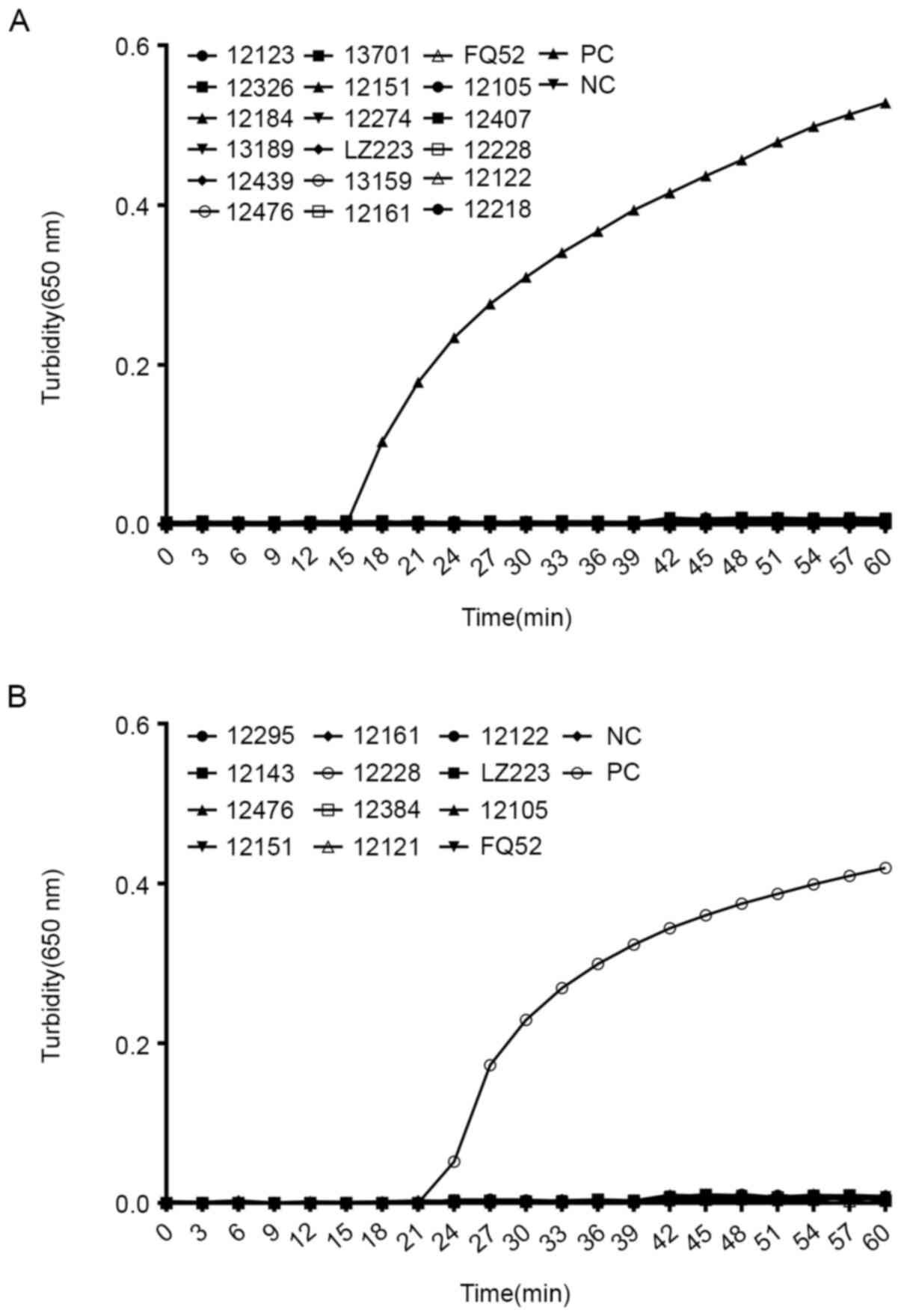 | Figure 3Specificity of the LAMP assay for
Serratia spp. harboring blaKPC. (A)
Specificity of the LAMP reaction for S. marcescens
identification. Samples: PC, positive control (S. marcescens
DY 12303); NC, negative control (double-distilled water); 12123,
E. coli DY12123; 12326, K. pneumoniae DY12326; 12223,
A. baumannii DY12223; 12184, P. vulgaris DY12184;
12122, P. aeruginosa DY12122; 12531, S. sonnel
DY12531; 12189, Y. pestis DY12189; 12218, C. freundii
DY12218; 12161, S. typhi DY12161; 12439, M. morganii
DY12439; 12210, P. rettgeri DY12210; 12252, A.
baumannii FY12252; 12476, P. aeruginosa DY12476; 12151,
K. oxytoca DY12151; 12105, E. cloacae DY12105; 12228,
E. coli DY12228; 12274, K. pneumoniae DY12274; 12407,
P. mirabilis DY12407. (B) Specificity of the LAMP reaction
for blaKPC detection. Samples: PC, positive control (S.
marcescens DY 12303); NC, negative control (double-distilled
water); 12295, E. coli DY12295; 12521, B. cepacia
DY12521; 12384, K. pneumoniae DY12384; 12223, A.
baumannii DY12223; 12143, S. maltophilia DY12143; 12161,
S. typhi DY12161; 12121, E. aerogenes DY12121; 12315,
E. cloacae DY12315; 12476, P. aeruginosa DY12476;
12228, E. coli DY12228; 12122, P. aeruginosa DY12122;
12252, A. baumannii FY12252. Turbidity was monitored by a
Loopamp real-time turbidimeter at 650 nm every 6 sec. Amplification
was performed at 65˚C for 60 min. LAMP, loop-mediated isothermal
amplification; blaKPC, β lactamase Klebsiella
pneumoniae carbapenemase. |
Sensitivity of the LAMP assay for
Serratia spp. with blaKPC
As indicated in Fig.
4A-1 and B-1, the detection
limit of the LAMP assay for Serratia spp. and blaKPC
was 3.92 pg/µl when monitored by a real-time turbidimeter recording
system. In addition, a direct visual method was adopted to monitor
the results of the LAMP assay for Serratia spp. and
blaKPC. Prior to the LAMP reaction, 1 µl of
calcein/Mn2+ complex was added to 24 µl of the prepared
LAMP reaction mixture. All positive reactions turned green, whereas
the negative reactions still remained orange (Fig. 4A-2 and B-2). It was concluded that these two
detection methods had similar sensitivities. To compare
sensitivities, conventional PCR was also performed by using
luxS and blaKPC primers. The amount of template added
in the conventional PCR was the same as that for the LAMP assay. It
was observed that the detection limit of conventional PCR for luxS
and KPC was 39.2 pg/µl (Fig. 4A-3
and B-3). Taken together, these
results suggested that the LAMP assay was 10 times more sensitive
than conventional PCR.
 | Figure 4Sensitivity comparison between the
LAMP assay and conventional PCR for Serratia spp. harboring
blaKPC. The pure genomic DNA extracted from S.
marcescens DY12303 was diluted in a serial 10-fold dilution.
Both LAMP reactions and PCRs were carried out in duplicate for each
dilution point. Tubes and lanes: 1, 392.0 ng/µl; 2, 39.2 ng/µl; 3,
3.92 ng/µl; 4, 392.0 pg/µl; 5, 39.2 pg/µl; 6, 3.92 pg/µl; 7, 0.392
pg/µl; 8, negative control (double-distilled H2O). (A-1
and B-1) The turbidity of the LAMP assays for S. marcescens
(A-1) and blaKPC (B-1) were monitored by a Loopamp real-time
turbidimeter at 650 nm every 6 sec. (A-2 and B-2) 1 µl of
fluorescent detection reagent was added to 25 µl of LAMP reaction
mixture prior to the LAMP reaction. In positive reactions, the
color changed from orange to green. Otherwise, there was no color
change. (A-3 and B-3) The PCR products were analyzed by 1% agarose
gel electrophoresis and stained with GelRed. LAMP, loop-mediated
isothermal amplification; blaKPC, β lactamase Klebsiella
pneumoniae carbapenemase; M, marker. |
Inhibition analysis of clinical
specimens
Pure genomic DNA extracted from the clinical isolate
S. marcescens DY12303, which has blaKPC-2, was used
as a positive control and seeded into DNA extracted from the blood
and sputum of healthy donors and reaction curves were compared with
those of samples from the pure culture. As indicated in Fig. 4A-1 and B-1, compared with the pure culture
samples, the sensitivity of the luxS and KPC LAMP assay for the
spiked samples was unchanged, at 1.96 pg/µl (Fig. 5A and B). Thus, it was concluded that the DNA
extracted from the sputum and blood did not interfere with the LAMP
assay.
Performance comparison of the LAMP
assay, conventional culture and real-time PCR in detecting clinical
specimens of Serratia spp. with blaKPC
In the present study, sputum specimens collected at
the Henan Province Hospital of TCM (Zhengzhou, China) were used to
examine the feasibility, sensitivity and specificity of the LAMP
assay. The LAMP assay was compared with conventional culture and
the results of real-time PCR kits. From January to December 2018, a
total of 359 sputum specimens were collected. For conventional
culturing combined with Taqman probe real-time PCR for
blaKPC, 70 positive specimens of blaKPC producing
Serratia spp. were detected. For the LAMP assay, with
non-purified DNA (boiling method) from sputum specimens as
templates, 62 positive specimens of blaKPC producing
Serratia spp. were detected. The sensitivity and specificity
of the LAMP assay were 94.3 and 94.8%, respectively. However, when
the same templates were used for real-time PCR directly, no
amplification products were detected due to the existence of
interfering substances in the sputum. When purified DNA extracted
from the sputum specimens was used as a template for the LAMP
assay, the sensitivity and specificity was 94.3 and 94.5%,
respectively. When the same templates were used in a real-time PCR
assay, the sensitivity was 94.3% and the specificity was 96.0%,
which was similar to those of the LAMP assay. The results of the
LAMP assay are presented in Table
IV. The present results revealed that the LAMP reaction had
excellent sensitivity and specificity, and was less susceptible to
interfering substances in clinical specimens than the real-time PCR
method.
 | Table IVSensitivity and specificity of
Serratia spp. with β lactamase Klebsiella pneumoniae
carbapenemase in the loop-mediated isothermal amplification
assay. |
Table IV
Sensitivity and specificity of
Serratia spp. with β lactamase Klebsiella pneumoniae
carbapenemase in the loop-mediated isothermal amplification
assay.
| | Culture (n) | |
|---|
| Sample/result | Positive | Negative | Sensitivity
(%) | Specificity
(%) | PPV (%) | NPV (%) | PLR (%) |
|---|
| Sputum
directly | | | | | | | |
|
Positive | 66 | 15 | 94.3
(85.3-98.2) | 94.8
(91.4-97.0) | 81.5
(71.0-88.9) | 98.6
(96.1-99.5) | 18.26
(11.1-29.8) |
|
Negative | 4 | 274 | | | | | |
| Extracted DNA | | | | | | | |
|
Positive | 66 | 16 | 94.3
(85.3-98.2) | 94.5
(91.0-96.7) | 80.5
(70.0-88.1) | 98.6
(96.1-99.5) | 17.0
(10.5-27.5) |
|
Negative | 4 | 273 | | | | | |
Discussion
Serratia spp. are the smallest
gram-negative bacilli in the environment and belong to the
Enterobacteriaceae family
In the clinical setting, the most commonly
encountered species are S. marcescens and S.
liquefaciens. S. marcescens is generally considered as an
innocuous, non-pathogenic, environmental organism. As its red
colonies are easily recognized by the naked eye, it is frequently
used as a popular biological marker (31). However, this organism has recently
been thought to be an important opportunistic pathogen that may
lead to various types of infectious disease in vulnerable
populations, such as patients at ICUs and hematopathy departments.
Nosocomial infections caused by S. marcescens have reached
1-2%. Infections may take place in any part of the body, with the
respiratory tract and the blood stream being the most common
(32). Furthermore, in pediatric
patients, S. marcescens may give rise to meningitis
according to case reports (33).
S. marcescens infection may also cause endocarditis and
osteomyelitis in patients with heroin addiction (34).
Serratia spp.-associated BSI usually occurs
due to contaminated intravenous solutions and is a life-threatening
condition that must be diagnosed and treated as soon as possible
(35). In recent decades, routine
BSI diagnosis has included continuous monitoring of the blood
culture by automated systems, subcultivating on an appropriate
solid medium and bacterial identification using commercial kits,
requiring ~72 h. Although the detection time has been shortened by
the introduction of mass spectrometry as an alternative method for
bacterial identification directly from positive blood cultures, the
specialized equipment and high cost of this method restrict its use
in general hospitals and health care units (36). Furthermore, this method cannot be
used to detect resistance markers, such as blaKPC, in
Serratia spp. Therefore, the development of a convenient,
rapid, sensitive detection method for Serratia spp. with
blaKPC is required for the diagnosis and treatment of
Serratia spp.-associated BSI.
In our experience, lassical culturing used to detect
Serratia spp requires >48 h to identify the target
pathogen and antibiotic spectrum. Furthermore, the sensitivity of
the culture method is insufficient for the detection of
Serratia spp. with blaKPC. To the best of our
knowledge, a method based on LAMP assays for detecting
blaKPC harboring Serratia spp. has not been
previously reported. Compared with conventional bacterial culture
and identification methods, LAMP assays may proceed at a constant
temperature and the time required is only 1 h, saving time and
effort. Furthermore, heparin, a commonly used anticoagulant in
serum and plasma, which is well-known as an inhibitory substance
for conventional PCR, does not affect LAMP assays (37). It's also possible for LAMP assays to
detect a pathogen directly from biological fluids, such as blood,
pleuroperitoneal fluids and cerebrospinal fluid (38).
After consideration of the epidemiology of
Serratia spp. and the advantages of the LAMP assay, the
present study aimed to design primers specifically for
Serratia spp. with the most prevalent carbapenemase
resistance gene, blaKPC, for use in a LAMP assay. The LAMP
assay allows clinical specimens to be screened more easily and
rapidly than existing techniques.
The sensitivity analysis suggested that the
detection limit of the LAMP method reached 3.92 pg/µl for
Serratia spp. with blaKPC. A sensitivity
comparison with conventional PCR revealed that the LAMP was 10
times more sensitive than conventional PCR. In the specificity
analysis, the luxS gene from Serratia spp. (~516 bp)
was selected as the target sequence for Serratia spp.
identification. It was indicated that Serratia spp. was able
to be detected within 30 min and 19 non-Serratia spp. family
bacterial strains tested negative, demonstrating good specificity.
Furthermore, 12 bacterial strains without blaKPC also
produced negative results. In summary, the blaKPC primers
designed in the present study were able to detect all subtypes of
blaKPC.
It was previously demonstrated that for the LAMP
assay, it is unnecessary to purify the DNA from clinical specimens
(39). However, exogenous DNA and
inhibitors may markedly reduce the sensitivity of PCR. To assess
the anti-interference performance of the LAMP method, DNA was
extracted from the sputum and blood of healthy donors and spiked
specimens were prepared using genomic DNA extracted from the
DY12303 bacterial strain and extracts from sputum and blood. The
results indicated that the limit of bacterial detection in the LAMP
assay with spiked specimens was similar to that for the pure
culture.
To investigate the adaptability of the LAMP assay
for clinical specimens, 359 sputum specimens were collected and the
DNA was extracted with a commercial DNA extraction kit and the
boiling method. The detection results for the LAMP assay and
real-time PCR were compared with conventional culture methods. The
results indicated that the detection efficiency of the LAMP assay
was almost the same as that of the conventional culture method and
was not affected by the DNA extraction method. For the real-time
PCR, the detection efficiency was the same as for the conventional
culture method when purified DNA from sputum was used as templates,
but no amplification products were detected when non-purified DNA
extracted by the boiling method was used as the template. From
these results, it was concluded that the LAMP assay had good
sensitivity, specificity and efficiency compared with conventional
culture and real-time PCR methods. Furthermore, in the present
study, the fluorescent reagent (calcein/Mn2+ complex)
that was added to the reaction mixture made the detection results
more visual and easier to determine. The advantages of the LAMP
assay suggested that this method was more suitable than
conventional molecular methods, such as regular PCR and real-time
PCR, for the rapid detection of blaKPC-producing
Serratia spp. in clinical specimens.
In the LAMP reaction, it is not necessary for all of
the primers to link with the target sequence. FIP and BIP primers
are of great importance and if these primers bind to the target
sequence, the signal is positive. However, further studies are
required to confirm this assumption (40). Although the amplification principle
of the LAMP method is complex, the performance of this method is
more feasible and efficient than that of other methods. The
operation of the assay may be accomplished under isothermal
conditions, which may be achieved in a primary laboratory. Due to
its favorable characteristics, this LAMP assay may be adopted by
inspection and quarantine departments to perform site inspections
for blaKPC-producing Serratia spp. In ICUs of
hospitals, the LAMP assay may be used as a means of point-of-care
testing to detect resistant bacteria and guide antibiotic
usage.
In summary, a detection method based on a LAMP assay
for blaKPC-producing Serratia spp. was successfully
established. The sensitivity, specificity and feasibility were
indicated to be superior to conventional culture methods and
molecular biotechnology methods. The most significant feature of
the LAMP assay presented in the present study is that it is easy
for the staff of a primary laboratory to master. Thus, this assay
may have a positive impact in primary hospitals, disease control
and prevention departments, as well as inspection and quarantine
departments. Furthermore, the present study offers a novel
perspective on the infectious characteristics of Serratia
spp. and their resistance mechanisms, leading to a greater
understanding of antibiotic screening for the treatment of critical
care patients.
Supplementary Material
Serratia spp. luxS gene
alignment using CLUSTAL W2.1. Rows: 1-, S. marcescens
ATCC274; 2-, S. liquefaciens ATCC27592; 3-, S.
odorifera; 4-, S. rubidaea.
Acknowledgements
Not applicable.
Funding
This work was supported by the TCM Administration
of Henan Province (grant no. 2018ZY0104).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
YL and WL conceived the study. XL, DZ, and CW
performed the experiments. XZ and DP collected the clinical strains
and performed the data analysis. DP and WL wrote the manuscript. YL
and XL confirmed the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The procedures in the present study were approved
and documented by the Ethics Committee of Henan Province Hospital
of TCM (Zhengzhou, China). All research complied with the
declaration of Helsinki and the relevant rules of the Ethics
Committee. Informed consent for participation in the study was
obtained from all participants (or their parent or legal
guardian).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ogtrop ML, van Zoeren-Grobben D,
Verbakel-Salomons EM and van Boven CP: Serratia marcescens
infections in neonatal departments: Description of an outbreak and
review of the literature. J Hosp Infect. 36:95–103. 1997.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Manning ML, Archibald LK, Bell LM,
Banerjee SN and Jarvis WR: Serratia marcescens transmission
in a pediatric intensive care unit: A multifactorial occurrence. Am
J Infect Control. 29:115–119. 2001.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dessì A, Puddu M, Testa M, Marcialis MA,
Pintus MC and Fanos V: Serratia marcescens infections and
outbreaks in neonatal intensive care units. J Chemother.
21:493–499. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hsieh S and Babl FE: Serratia
marcescens cellulitis following an iguana bite. Clin Infet Dis.
28:1181–1182. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Chaidaroon W and Supalaset S: Corneal ring
infiltrates caused by Serratia marcescens in a patient with
human immunodeficiency virus. Case Rep Ophthalmol. 7:359–363.
2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gastmeier P: Serratia marcescens:
An outbreak experience. Front Microbiol. 5(81)2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Knowles S, Herra C, Devitt E, O'Brien A,
Mulvihill E, McCann SR, Browne P, Kennedy MJ and Keane CT: An
outbreak of multiply resistant Serratia marcescens: The
importance of persistent carriage. Bone Marrow Transplant.
25:873–877. 2000.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gupta N, Hocevar SN, Moulton-Meissner HA,
Stevens KM, McIntyre MG, Jensen B, Kuhar DT, Noble-Wang JA, Schnatz
RG, Becker SC, et al: Outbreak of Serratia marcescens
bloodstream infections in patients receiving parenteral nutrition
prepared by a compounding pharmacy. Clin Infect Dis. 59:1–8.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sullivan KV, Deburger B, Roundtree SS,
Ventrola CA, Blecker-Shelly DL and Mortensen JE: Pediatric
multicenter evaluation of the Verigene gram-negative blood culture
test for rapid detection of inpatient bacteremia involving
gram-negative organisms, extended-spectrum beta-lactamases, and
carbapenemases. J Clin Microbiol. 52:2416–2421. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tojo M, Fujita T, Ainoda Y, Nagamatsu M,
Hayakawa K, Mezaki K, Sakurai A, Masui Y, Yazaki H, Takahashi H, et
al: Evaluation of an automated rapid diagnostic assay for detection
of Gram-negative bacteria and their drug-resistance genes in
positive blood cultures. PLoS One. 9(e94064)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nasri E, Subirats J, Sànchez-Melsió A,
Mansour HB, Borrego CM and Balcázar JL: Abundance of carbapenemase
genes (blaKPC, blaNDM and
blaOXA-48) in wastewater effluents from Tunisian
hospitals. Environ Pollut. 229:371–374. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Srisrattakarn A, Lulitanond A,
Wilailuckana C, Charoensri N, Wonglakorn L, Saeniamla P, Daduang J
and Chanawong A: Rapid and simple identification of carbapenemase
genes, bla NDM, bla OXA-48, bla VIM, bla
IMP-14 and bla KPC groups, in Gram-negative
bacilli by in-house loop-mediated isothermal amplification with
hydroxynaphthol blue dye. World J Microbiol Biotechnol.
33(130)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hopkins KL, Findlay J, Meunier D, Cummins
M, Curtis S, Kustos I, Mustafa N, Perry C, Pike R and Woodford N:
Serratia marcescens producing SME carbapenemases: An
emerging resistance problem in the UK? J Antimicrob Chemother.
72:1535–1537. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Pollett S, Miller S, Hindler J, Uslan D,
Carvalho M and Humphries RM: Phenotypic and molecular
characteristics of carbapenem-resistant Enterobacteriaceae
in a health care system in Los Angeles, California, from 2011 to
2013. J Clin Microbiol. 52:4003–4009. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wachino J, Yoshida H, Yamane K, Suzuki S,
Matsui M, Yamaqishi T, Tsutsui A, Konda T, Shibayama K and Arakawa
Y: SMB-1, a novel subclass B3 metallo-beta-lactamase, associated
with ISCR1 and a class 1 integron, from a carbapenem-resistant
Serratia marcescens clinical isolate. Antimicrob Agents
Chemother. 55:5143–5149. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
de Vries JJ, Baas WH, van der Ploeg K,
Heesink A, Degener JE and Arends JP: Outbreak of Serratia
marcescens colonization and infection traced to a healthcare
worker with long-term carriage on the hands. Infect Control Hosp
Epidemiol. 27:1153–1158. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Nodari CS, Siebert M, Matte UDS and Barth
AL: Draft genome sequence of a GES-5-producing Serratia
marcescens isolated in southern Brazil. Braz J Microbiol.
48:191–192. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wendel AF, Kaase M, Autenrieth IB, Peter
S, Oberhettinger P, Rieber H, Pfeffer K, Mackenzie CR and Willmann
M: Protracted regional dissemination of GIM-1-producing Serratia
marcescens in western germany. Antimicrob Agents Chemother.
61:e01880–16. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Mahlen SD: Serratia infections: From
military experiments to current practice. Clin Microbiol Rev.
24:755–791. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wong YP, Othman S, Lau YL, Son R and Chee
HY: Loop mediated isothermal amplification (LAMP): A versatile
technique for detection of microorganisms. J Appl Microbiol.
124:626–643. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sakai J, Maeda T, Tarumoto N, Misawa K,
Tamura S, Imai K, Yamaguchi T, Iwata S, Murakami T and Maesaki S: A
novel detection procedure for mutations in the 23S rRNA gene of
Mycoplasma pneumoniae with peptide nucleic acid-mediated
loop-mediated isothermal amplification assay. J Microbiol Methods.
141:90–96. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mashooq M, Kumar D, Niranjan AK, Agarwal
RK and Rathore R: Development and evaluation of probe based real
time loop mediated isothermal amplification for Salmonella: A new
tool for DNA quantification. J Microbiol Methods. 126:24–29.
2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lin M, Liu W, Wang P, Tan J, Zhou Y, Wu P,
Zhang T, Yuan J and Chen Y: Rapid detection of ermB gene in
Clostridium difficile by loop-mediated isothermal
amplification. J Med Microbiol. 64:854–861. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cai JC, Zhou HW, Zhang R and Chen GX:
Emergence of Serratia marcescens, Klebsiella
pneumoniae, and Escherichia coli Isolates possessing the
plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in
intensive care units of a Chinese hospital. Antimicrob Agents
Chemother. 52:2014–2018. 2008.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lin X, Hu Q, Zhang R, Hu Y, Xu X and Lv H:
Emergence of Serratia marcescens isolates possessing
Carbapenem-hydrolysing β-lactamase KPC-2 from China. J Hosp Infect.
94:65–67. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou T, Zhang X, Guo M, Ye J, Lu Y, Bao Q
and Chi W: Phenotypic and molecular characteristics of
carbapenem-non-susceptible Enterobacteriaceae from a teaching
hospital in Wenzhou, southern China. Jpn J Infect Dis. 66:96–102.
2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang R, Liu L, Zhou H, Chan EW, Li J,
Fang Y, Li Y, Liao K and Chen S: Nationwide surveillance of
clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in
China. EBioMedicine. 19:98–106. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li B, Feng J, Zhan Z, Yin Z, Jiang Q, Wei
P, Chen X, Gao B, Hou J, Mao P, et al: Dissemination of
KPC-2-encoding IncX6 plasmids among multiple
Enterobacteriaceae Species in a Single Chinese Hospital.
Front. Microbiol. 9(478)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Thompson JD, Higgins DG and Gibson TJ:
CLUSTAL W: Improving the sensitivity of progressive multiple
sequence alignment through sequence weighting, position-specific
gap penalties and weight matrix choice. Nucleic Acids Res.
22:4673–4680. 1994.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Garrity G, Staley JT, Boone DR, De Vos P,
Goodfellow M and Rainey FA: Bergey's manual® of
systematic bacteriology. In: Vol 2 The Proteobacteria, Part B: The
Gammaproteobacteria. Springer, New York, NY, 2006.
|
|
31
|
Joyner J, Wanless D, Sinigalliano CD and
Lipp EK: Use of quantitative real-time PCR for direct detection of
Serratia marcescens in marine and other aquatic
environments. Appl Environ Microbiol. 80:1679–1683. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ferrer R, Martin-Loeches I, Phillips G,
Osborn TM, Townsend S, Dellinger RP, Artiqas A, Schorr C and Levy
MM: Empiric antibiotic treatment reduces mortality in severe sepsis
and septic shock from the first hour: Results from a
guideline-based performance improvement program. Crit Care Med.
42:1749–1755. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Khanna A, Khanna M and Aggarwal A:
Serratia marcescens-a rare opportunistic nosocomial pathogen
and measures to limit its spread in hospitalized patients. J Clin
Diagn Res. 7:243–246. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Farhoudi HO, Banks T and Ali N: A case
report of serratia endocarditis with review of the literature. Med
Ann Dist Columbia. 43:401–406. 1974.PubMed/NCBI
|
|
35
|
Su JR, Blossom DB, Chung W, Gullion JS,
Pascoe N, Heseltine G and Srinivasan A: Epidemiologic investigation
of a 2007 outbreak of Serratia marcescens bloodstream
infection in Texas caused by contamination of syringes prefilled
with heparin and saline. Infect Control Hosp Epidemiol. 30:593–595.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
36
|
Chemaly RF, Rathod DB, Sikka MK, Hayden
MK, Hutchins M, Horn T, Tarrand J, Adachi J, Nguyen K, Trenholme G
and Raad I: Serratia marcescens bacteremia because of
contaminated prefilled heparin and saline syringes: a multi-state
report. Am J Infect Control. 39:521–524. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Enosawa M, Kageyama S, Sawai K, Watanabe
K, Notomi T, Onoe S, Mori Y and Yokomizo Y: Use of loop-mediated
isothermal amplification of the IS900 sequence for rapid detection
of cultured Mycobacterium avium subsp. paratuberculosis. J Clin
Microbiol. 41:4359–4365. 2003.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Dhama K, Karthik K, Chakraborty S, Tiwari
R, Kapoor S, Kuamr A and Thomas P: Loop-mediated isothermal
amplification of DNA (LAMP): A new diagnostic tool lights the world
of diagnosis of animal and human pathogens: A review. Pak J Biol
Sci. 17:151–166. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Notomi T, Mori Y, Tomtia N and Kanda H:
Loop-mediated isothermal amplification (LAMP): Principle, features,
and future prospects. J Microbiol. 53:1–5. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lin WH, Zou BJ, Song QX and Zhou GH:
Progress in multiplex loop-mediated isothermal amplification
technology. Yi Chuan. 37:899–910. 2015.PubMed/NCBI View Article : Google Scholar
|