Introduction
Perineural invasion was traditionally considered as
the primary type of interaction between tumor cells and nerve cells
in the tumor microenvironment, and is considered critical in the
invasion and metastasis of pancreatic, colorectal and gastric
cancer, as well as other malignant tumors (1,2).
Previous studies showed that there is a new way of communication
between tumor cells and nerve cells, which is called tumor
cell-mediated neurogenesis (3-6).
In addition, newborn nerves stimulate the proliferation and
metastasis of tumor cells. In 2008, Ayala et al (7) described the phenomenon of neurogenesis
in prostate cancer, and defined cancer-associated neurogenesis and
cancer-related axonogenesis. The newborn nerves were revealed to
act as ‘channels’, leading to the migration of tumor cells, which
is closely associated with metastasis and degeneration of tumors
(5,7). A similar phenomenon has recently been
reported in which the co-culture of dorsal root ganglion (DRG)
cells with pancreatic cancer cells led to a significant increase in
the density of DRG neurons (8).
Under these conditions, tumor cells could not only proliferate
rapidly, but also showed a tendency to grow close to the DGR cells,
which suggested that tumor cells can stimulate nerve growth
(8,9). In 2013, Magnon et al (5) confirmed that the sympathetic nerves
could release norepinephrine to activate the β2 and β3 adrenergic
receptors, leading to the onset of prostate cancer; whereas
parasympathetic nerve-released acetylcholine could bind to type І
muscarinic receptor to promote the proliferation and metastasis of
prostate cancer. Since then, numerous pathological studies have
observed the existence of newly developed nerves in a variety of
malignant tumors (4-6,10,11).
Although the associated mechanisms have not been elucidated yet,
the understanding of cancer-related neurogenesis may provide a
novel approach for cancer treatment.
Neurogenesis is a complex process involving numerous
factors, and the growth of axons serves a crucial role in it
(12). New axons need neurotrophins
to promote their growth, and axon guidance cues guide them to
extend to specific areas (13,14).
It is worth noting that nerve growth factor (NGF) is one of the
most important members of the neurotrophic factor family. It
possesses dual biological functions of neuron nutrition and axon
growth promotion, and plays important regulatory roles in the
development, differentiation, growth and regeneration of neurons
(15). Meanwhile, protein gene
product 9.5 (PGP9.5) is a ubiquitin hydrolase, which regulates
cellular processes in the cell hydrolysis pathway, including cell
cycle division and cell death (16). Previous studies have detected a high
expression of PGP9.5 in numerous tumors, including colorectal,
pancreatic, esophageal and bladder cancer (16-18).
In a previous study, in tumor tissues, PGP9.5 induced an increase
in cyclin ubiquitination resulting in uncontrolled growth of
undifferentiated cells, which is one of the key factors leading to
oncogene activation (19).
An increasing number of studies have confirmed that
tumor cells can obtain NGF through autocrine and paracrine routes
to assist their survival, proliferation and metastasis. For
example, several types of cancer cells, including breast cancer,
have been shown to produce autocrine NGF, and its high mRNA and
protein expression levels in these cells have been confirmed
(20). Of note, NGF binding
receptors in cancer cells are highly consistent with the receptors
in nerve cells, which are able to bind to tropomyosin receptor
kinase A and p75 neurotrophin receptor, to mediate the survival and
proliferation of tumor cells (20-22).
In addition, paracrine NGF from breast cancer cells could affect
the growth and development of adjacent neurons in addition to their
own (20). It has been reported
that NGF can effectively induce the differentiation of embryonic
neural precursor cells (20), and
the overexpression of proNGF in prostate cancer tissue could also
effectively induce neurogenesis (21).
In certain tumor cells, including breast cancer
cells, autocrine NGF has been shown to not only be involved in
their development, but may also affect the growth and development
of neighboring neurons through the paracrine system. Thus, NGF may
be a key molecule involved in regulating cancer-related
neurogenesis, which might play a crucial role in the signal
transmission system that controls the related nerve tumor growth,
and enhances the migration, invasion and metastasis of tumor cells.
In the present study, PGP9.5 was used as a neuronal marker, and its
positive expression level was scored. Furthermore, recombinant NGF
lentiviral overexpression, knockout and silencing plasmids were
constructed, and whether NGF affects neuron growth was investigated
and preliminarily confirmed. Furthermore, the successfully
constructed plasmids could be used to verify the hypothetical roles
of NGF in cancer-associated neurogenesis and cancer-related
axonogenesis.
Materials and methods
Expression of PGP9.5 in breast
cancer
In total, 92 immunohistochemically identified breast
cancer tissues were collected from patients (age, 24-79 years; mean
age, 51.5±12 years) at the First College of Clinical Medical
Science, China Three Gorges University (Yichang, China) between
October 2014 and November 2015; patients provided written informed
consent and the study was approved by the Ethics Committee of the
First Affiliated Hospital of Three Gorges University. Patients
could be any age and pathological grade, and with or without
metastasis (Table SI). Samples
were used to perform immunohistochemistry using recombinant
anti-PGP9.5 antibody (cat. no. ab109261; Abcam). The clinical data
of the specimens were classified by patient age, tumor size and
degree of differentiation, which in turn was classified according
to the Nottingham system (Table I)
(23). Immunohistochemistry was
carried out according to the EnVision two-step method as previously
described (24), and PBS was used
as a negative control. PGP9.5 was used as a neuronal marker, and
its positive expression level was scored according to the following
positive criteria: Cells that contain brown and yellow granules in
the cytoplasm were considered positive, and the total number of
positive cells in 100 cells in every five fields of view was
counted. The average value was calculated and further scored
according to the staining intensity and the number of positive
cells. The staining intensity criteria were as follows: 0 was
considered negative, where there were no brown and yellow granules
in the cytoplasm; 1 was considered weakly positive, with light
yellow granules; 2 was considered positive, with yellow granules;
and 3 was considered strongly positive, with brown and yellow
granules. The number of positive cells was scored according to the
following criteria: 0 was considered absence of positive cells; 1
was assigned when the percentage of positive cells was <25%; 2
corresponded to 25-50% positive cells; and 3 was assigned if the
percentage of positive cells was >50%.
 | Table IRelationship between PGP9.5
expression and the clinicopathological features of breast
cancer. |
Table I
Relationship between PGP9.5
expression and the clinicopathological features of breast
cancer.
|
Characteristics | Total cases | Positive rate
(%) | P-value |
|---|
| Age (years) | | | |
|
≤50 | 44 | 39 (88.6) | 0.867 |
|
>50 | 48 | 42 (87.5) | |
| Tumor size
(cm) | | | |
|
≤2 | 58 | 50 (86.2) | 0.707 |
|
>2 | 34 | 31 (91.2) |
| Pathological
grade | | | |
|
I | 29 | 21 (72.4) | 0.005 |
|
II-III | 63 | 60 (95.2) | |
Construction of recombinant NGF
lentiviral overexpression plasmids
MDA-MB231 and T-47D cell lines (The Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences) were
cultured in a 5% CO2 incubator at 37˚C with DMEM
(high-glucose) (cat. no. 31053028; Gibco; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (cat. no. 10099141; Gibco;
Thermo Fisher Scientific, Inc.) and 1% pen/strep. Genomic DNA of
the breast cancer cell lines MDA-MB231 and T-47D was obtained using
Ezup Column Animal Genomic DNA Purification kit (cat. no. B518251;
Sangon Biotech Co., Ltd.), which was subsequently used to construct
recombinant pMD18-T-NGF plasmids using the pMD™ 18-T Vector Cloning
kit (cat. no. 6011; Takara Bio, Inc.) with the primers listed in
Table II. Recombinant pMD18-T-NGF
was sequenced and used as a template to amplify NGF and NGF-FLAG.
The PCR cycling conditions were as follows: Initial denaturation at
95˚C for 5 min; followed by 34 cycles at 95˚C for 40 sec, 56.2˚C
for 50 sec and 72˚C for 30 sec; and a final extension at 72˚C for 5
min. The FLAG-labeled NGF (PCR product of pMD18-T-NGF-FLAG) was
further linked to pCDH-CMV-MCS-EF1-puro (#vt1480; YouBio) to
construct the recombinant FLAG-labeled NGF overexpression plasmid
pCDH-CMV-MCS-EF1-puro-NGF-FLAG (pCDH-NGF-FLAG) with
XbaI/BamHI enzyme loci. NGF (PCR product of
pMD18-T-NGF) was also directly linked to pCDH-CMV-MCS-EF1-puro to
construct the NGF overexpression plasmid pCDH-CMV-MCS-EF1-puro-NGF
(pCDH-NGF). Furthermore, 3 µg proteolipid protein (pLP)1, 3 µg pLP2
and 3 µg pLP/VSVG from ViraPower™ Lentiviral Packaging Mix (cat.
no. K4975-00; Invitrogen; Thermo Fisher Scientific, Inc.), 3 µg
pCDN-NGF, 3 µg pCDN-NGF-FLAG, 3 µg pCDH-puro and 3 µg pCHD-EGFP
were transfected into 5 ml 1.2x106 293FT cells (cat. no.
R70007; Invitrogen; Thermo Fisher Scientific, Inc.) using 36 µl
Lipofectamine® 2000 reagent (Thermo Fisher Scientific,
Inc.), and incubated overnight at 37˚C in a humidified 5%
CO2 incubator, according to the manufacturer's protocol,
and the vector (pCDH-puro) was used as a negative control. After 72
h, the supernatants were collected, sterilized by filtration (0.45
µm filter; cat. no. SLHV033RB; EMD Millipore) and were used to
infect the breast cancer cell lines MDA-MB231 and T-47D
(2.5x106 cells; twice, every 24 h; MOI, 8) at 37˚C, and
bright field and fluorescence microscopy were used to detect the
infection efficiency. After 48 h, the infected cells were used to
screen target cells by puromycin selection; 2 and 6 µg/ml puromycin
were used for MDA-MB-231 and T-47D, respectively. The
puromycin-screening time was determined according to the results of
a preliminary puromycin-resistance experiment.
 | Table IIRecombinant overexpression NGF primer
pairs. |
Table II
Recombinant overexpression NGF primer
pairs.
| Primer name | Sequences
(5'-3') | Restriction
enzyme |
|---|
| pCDH-NGF-F |
TGCTCTAGAATGTCCATGTTGTTCTACACTCTG | BamHI |
| pCDH-NGF-R |
CGCGGATCCTCAGGCTCTTCTCACAGCCTT | XbaI |
|
pCDH-NGF-FLAG-F |
TGCTCTAGAATGTCCATGTTGTTCTACACTCTG | |
|
pCDH-NGF-FLAG-R |
CGCGGATCCTCACTTATCGTCGTCATCCTTGTAATCGGCCTTCTCACAGCCTT | |
The puromycin-screened breast cancer cells,
including the overexpression (pCDH-NGF and pCDH-NGF-FLAG), vector
(pCDH-puro) and untransfected groups (MB231 and T-47D cells), were
seeded onto 100-mm culture plates (2.5x106 cells in DMEM
in triplicate. After 72-h incubation at 37˚C, the cells were
collected, and the RNA was extracted by RNAsimple kit (cat. no.
DP419; Tiangen Biotech Co., Ltd.), according to the manufacturer's
instructions, and reverse transcribed into cDNA using the Takara
Reverse Transcriptase kit (cat. no. RR036Q; Takara Bio, Inc.). The
reverse transcription (RT) conditions were 65˚C for 5 min, 42˚C for
40 min and 70˚C for 15 min. GAPDH was used as the internal
reference (GAPDH primer sequence: forward, 5'-AGGTGAAGGTCGGAGTCA-3'
and reverse, 5'-GGTCATTGATGGCAACAA-3') to normalize NGF expression.
Fluorescence quantitative PCR (qPCR) [TB Green Premix Ex Taq II
(Tli RNase H Plus); cat. no. RR820A; Takara Bio, Inc.] was
performed to detect the expression levels of NGF with the following
primers: forward, 5'-GGCAGACCCGCAACATTACT-3'and reverse,
5'-CACCACCGACCTCGAAGTC-3' using a CFX96 Touch™ Real-Time
PCR Detection system (Bio-Rad Laboratories, Inc.) as follows:
Initial denaturation at 95˚C for 5 min; followed by 34 cycles of
95˚C for 30 sec, 56˚C for 60 sec and 72˚C for 60 sec; and a final
extension at 95˚C for 5 sec. The results were quantified using the
2-ΔΔCq method (25).
Measurement of NGF expression levels
by ELISA and western blotting
The overexpression (pCDH-NGF-FLAG), vector
(pCDH-puro) and untransfected (MDA-MB231 or T-47D cells) groups
were seeded (2.5x106 cells in DMEM) onto 100-mm culture
plates in triplicate. After 72-h incubation at 37˚C, the cell
supernatants from the vector and normal groups were collected (~8.5
ml per dish) and analyzed with a human NGF-β ELISA kit PicoKine
(cat. no. EK0469; Boster Biological Technology) using their
original concentrations, whereas the supernatants obtained from the
overexpression group were diluted 1:200 and further detected in
parallel with the other groups using the same ELISA kit. ELISA was
conducted according to the manufacturer's protocol.
In addition, proteins from cell supernatants were
concentrated using 15-ml 3-kDa ultrafiltration centrifuge tubes
(cat. no. UFC900396; EMD Millipore) at 5,000 x g for 20 min at 4˚C.
The protein concentrations were determined with a BCA Protein Assay
kit (cat. no. P0010; Beyotime Institute of Biotechnology). A total
of 25 µg proteins from each group were incubated with SDS loading
buffer at 95˚C for 10 min and then separated by SDS-PAGE on 15%
gels. Proteins were then transferred to nitrocellulose membranes,
which were blocked with 5% skim milk for 1 h at room temperature.
The protein expression levels of NGF-β in the pCDH-NGF-FLAG group
were detected with rabbit anti-FLAG antibody (1:1,000; cat. no.
F7425; Sigma-Aldrich; Merck KGaA) in TBS-0.1% Tween-20 overnight at
4˚C, followed by detection with IRDye® 680LT goat
anti-rabbit (1:20,000; cat. no. 926-68021; LI-COR Biosciences) at
room temperature for 2 h. The protein expression of β-actin (rabbit
polyclonal antibody; 1:1,000; cat. no. D110001; Sangon Biotech Co.,
Ltd.) was used as the internal reference, and the protein bands
were visualized on the CLx Dual-color Infrared Laser Imaging system
(Odyssey®; LI-COR Biosciences). RealBand 3-color High
Range Protein Marker (cat. no. C620014-0250; Sangon Biotech Co.,
Ltd.) was used as a marker.
Construction of recombinant NGF
lentiviral CRISPR/Cas9 knockout plasmids
NGF-guide (g)RNA1, -gRNA2 and -gRNA3 were designed
and synthesized by Sangon Biotech Co., Ltd., and used for the
construction of recombinant NGF-CRISPR/Cas9 plasmids. The
non-sequencing NGF-gRNA was used as a negative control. Exon 3 of
NGF was the targeted exon, which affected the signal peptide
domain. The designed NGF-gRNAs (Table
III) were annealed and linked to the cas9 vector lentiCRISPRv2
(cat. no. 52961; Addgene) to construct LentiCRISPRv2-NGF-gRNAs
(-1/2/3, non-sequencing). The constructed recombinant plasmids were
validated by ampicillin resistance, appearing at the correct
molecular weight on a 1% agarose gel and sequenced by a commercial
company (Sangon Biotech Co., Ltd.).
 | Table IIINGF-gRNA primer pairs. |
Table III
NGF-gRNA primer pairs.
| Primer name | Sequences
(5'-3') |
|---|
| NGF-gRNA1-F |
CACCGCGATAGCTGCACGCGTGGCG |
| NGF-gRNA1-R |
AAACCGCCACGCGTGCAGCTATCGC |
| NGF-gRNA2-F |
CACCGAGCTTTTCTGATCGGCATAC |
| NGF-gRNA2-R |
AAACGTATGCCGATCAGAAAAGCTC |
| NGF-gRNA3-F |
CACCGTCGCCGCTTTTTAAACAGCC |
| NGF-gRNA3-R |
AAACGGCTGTTTAAAAAGCGGCGAC |
|
NGF-gRNA-Fa |
CACCGGCGTCGGAGCGGCAGAACTC |
|
NGF-gRNA-Ra |
AAACGAGGCGGTACGGACGGCCGCC |
|
Lenti-CRISPR-NGF-F |
TTCGGGTTTATTACAGGGACA |
|
Lenti-CRISPR-NGF-R |
GACTGTGGGCGATGTGC |
|
MV-CRISPR-NGF-F |
GTGCATAGCGTAATGTCCAT |
|
MV-CRISPR-NGF-R |
CTATAAATTACCATGCAGTCCTT |
| GAPDH-F |
AGGTGAAGGTCGGAGTCA |
| GAPDH-R |
GGTCATTGATGGCAACAA |
The verified 3 µg LentiCRISPRv2-NGF-gRNA2/3
(sequencing proved the gRNA1 was incorrect), 3 µg pCMV-VSV-G (cat.
no. 8454; Addgene), 3 µg pLJM1-EGFP (cat. no. 19319; Addgene), 3 µg
psPAX2 (cat. no. 12260; Addgene) and 3 µg negative control
(non-sequencing gRNA) were transfected into 5 ml 1.2x106
293FT cells by 36 µl Lipofectamine 2000. After a 7-h incubation at
37˚C, the cell supernatants were collected, filter sterilized and
used to infect the breast cancer cell lines MDA-MB231 and T-47D
(2.0x105 cells/ml; MOI, 8; twice, every 24 h), 48 h
post-infection, the infected cells were further used to screen
target cells by puromycin selection as described in NGF
overexpression section. After puromycin selection the infected
single clone was further sorted using flow cytometry (MoFlo Astrios
EQ Cell Sorter; Beckman Coulter, Inc.).
To select the target cells that had been correctly
CRISPR/Cas9-edited by NGF-gRNA2/3, the genomic DNA of sorted cells
was obtained by Ezup Column Animal Genomic DNA Purification kit
(cat. no. B518251; Sangon Biotech Co., Ltd.) and used as template
to perform standard PCR (EXTaq; cat. no. RR001A; Takara
Biotechnology Co., Ltd.). The PCR cycling conditions were as
follows: Initial denaturation at 95˚C for 5 min; followed by 34
cycles of 95˚C for 40 sec, 56˚C for 50 sec and 72˚C for 54 sec; and
a final extension at 72˚C for 5 min. The standard PCR NGF primers
used are presented in Table II,
and the NGF was detected by 2% agarose gel electrophoresis and
visualized by ethidium bromide (cat. no. A600195; Sangon Biotech
Co., Ltd.). Furthermore, 1.2x102 PCR-verified cells were
seeded onto 96-microwell plates in triplicate. Then, the cells were
collected from different wells to verify NGF (726 bp) by standard
PCR, and the open-reading frame (ORF) of NGF was further sequenced
(Sangon Biotech, Co., Ltd.) to verify the CRISPR/Cas9-edited cells.
After three rounds of selection by sorting, PCR and sequencing, as
aforementioned, the CRISPR/Cas9-edited mutants (MDA-MB231-E3-A and
MDA-MB231-E3-B) cells were verified.
CRISPR/Cas9-edited MDA-MB231-E3-A and MDA-MB231-E3-B
cells were seeded onto 60-mm culture plates (2.5x105
cells in 5 ml DMEM) in triplicate. After 72-h incubation at 37˚C,
total cell RNA was collected for NGF qPCR as aforementioned, and
GAPDH was used as the internal reference. The cycling conditions
were as follows: Initial denaturation at 95˚C for 5 min; followed
by 35 cycles of 95˚C for 30 sec, 56˚C for 60 sec and 72˚C for 60
sec; and a final extension at 95˚C for 5 sec.
Construction of recombinant lentiviral
NGF-silencing plasmids
The NGF-short hairpin (sh)RNA1/2/3 and non-targeting
negative control shRNA were designed (Table IV) and synthesized by Sangon
Biotech, Co., Ltd.; these sequences were annealed (95˚C for 4 min
and 70˚C for 10 min) and linked to the lentiviral vector pLKO.1-TRC
(cat. no. 10878 Addgene). pLKO.1-NGF-shRNA1, -shRNA2, -shRNA3 and
negative shRNA were further verified by ampicillin-resistance,
double enzyme digestion assays by EcoRI and NcoI, and
sequenced (Sangon Biotech, Co., Ltd.). The non-targeting shRNA was
used as a negative control.
 | Table IVNGF-shRNA primer pairs. |
Table IV
NGF-shRNA primer pairs.
| Primer name | Sequences
(5'-3') |
|---|
| NGF-shRNA1-F |
CCGGGCAGACCCGCAACATTACTGTCTCGAGACAGTAATGTTGCGGGTCTGCTTTTTG |
| NGF-shRNA1-R |
AATTCAAAAAGCAGACCCGCAACATTACTGTCTCGAGACAGTAATGTTGCGGGTCTGC |
| NGF-shRNA2-F |
CCGGGAGAGGTGAACATTAACAACACTCGAGTGTTGTTAATGTTCACCTCTCTTTTTG |
| NGF-shRNA2-R |
AATTCAAAAAGAGAGGTGAACATTAACAACACTCGAGTGTTGTTAATGTTCACCTCTC |
| NGF-shRNA3-F |
CCGGGGGATATGGTACAACCCTTGTCTCGAGACAAGGGTTGTACCATATCCCTTTTTG |
| NGF-shRNA3-R |
AATTCAAAAAGCATTGACTCAAAGCACTGGACTCGAGTCCAGTGCTTTGAGTCAATGC |
|
NGF-shRNAs-Fa |
CCGGGGGATATGGTACACCGCGGTTTATCCGGATAGATACCTCGAGGTATCTATCCGG |
| NGF-shRNAs-R |
AATTCAAAAAGCATTAAAAGCGGTTTATCCGGATAGATACCTCGAGGTATCTATCCGG |
| q-NGF-F |
GGCAGACCCGCAACATTACT |
| q-NGF-R |
CACCACCGACCTCGAAGTC |
| GAPDH-F |
AGGTGAAGGTCGGAGTCA |
| GAPDH-R |
GGTCATTGATGGCAACAA |
Verified 3 µg pLKO.1-NGF-shRNA1/3 (sequencing proved
that shRNA2 was incorrect), 3 µg -shRNA (non-targeting shRNA as the
negative control), 3 µg pLP1, 3 µg pLP2, 3 µg pLP/VSVG and 3 µg
pLKO.1 were transfected into 5 ml 1.2x106 293FT cells
using 36 µl Lipofectamine 2000, along with empty lentiviral vectors
as a control. After a 72-h incubation at 37˚C, the supernatants
were collected, filter sterilized and further used to infect the
breast cancer cell lines MDA-MB231 and T-47D (twice, every 24 h;
MOI, 8). A total of 48 h post-infection, the infected cells were
used to select target cells by puromycin selection as
aforementioned.
MDA-MB231-pLKO-NGF-shRNA1/3,
T-47D-pLKO-NGF-shRNA1/3, MDA-MB231-pLKO-shRNA (negative control),
T-47D-pLKO-shRNA (negative control) and MDA-MB231 or T-47D cells
were seeded onto 60-mm culture plates (2.5x105 cells in
5 ml DMEM) in triplicate. After 72 h, total cell RNA was collected
for NGF qPCR, as aforementioned, using the following cycling
conditions: Initial denaturation at 95˚C for 5 min; followed by 35
cycles of 95˚C for 30 sec, 50˚C for 60 sec and 72˚C for 60 sec; and
a final extension at 95˚C for 5 sec. The GAPDH was used as the
internal reference.
Detection of cell viability in breast
cancer cells containing different recombinant plasmids
Cell viability of MDA-MB231-pCDH-NGF-FLAG
(overexpression group), MDA-MB231 (untreated group),
MDA-MB231-pLKO-NGF-shRNA1 (silent group) and MDA-MB231-E3-A
(knockout group) was determined by MTT assays. Cells were seeded
(2.5x104 cells/ml; 4,000 cells/well) onto 96-microwell
plates in triplicate. After 24, 48, 72 and 96 h at 37˚C, cells were
collected for MTT assay. Briefly, MTT (200 µg/ml; prepared in
serum-free DMEM) was added to the wells, and the cells were
incubated at 37˚C for 4 h. The supernatants were removed, and 150
µl dimethyl sulfoxide was added to each well to dissolve the purple
formazan crystals. After shaking for 15 min at room temperature,
the optical density was detected at 490 nm using a Multiskan
Spectrum microplate spectrophotometer (Thermo Fisher Scientific,
Inc.). Experiments were repeated three times.
Influence of NGF on PC12 cell neuronal
differentiation activity
PC-12 is a classic neuronal cell line. Studies have
suggested that NGF can induce PC-12 cell differentiation (26-28).
Thus, undifferentiated PC12 cells (The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences) were cultured in
RPMI-1640 medium (cat. no. SH30809.018; Hyclone; Cytiva)
supplemented with 10% horse serum (cat. no. 26050088; Gibco; Thermo
Fisher Scientific, Inc.) and 5% FBS, and were treated with 10, 50
and 100 ng/ml NGF cell supernatants from NGF-overexpressing
MDA-MB231 cells at 37˚C. NGF was obtained from the supernatants of
NGF-overexpressing cells and the concentrations were confirmed by
ELISA (Human NGF/NGF-β ELISA kit; cat. no. EK0469; Boster
Biological Technology). NGF-β standards was used to establish the
standard curve. The cell number and axon length were calculated by
fluorescence microscopy (NIKON Ti-s inverted fluorescence
microscope; Nikon Corporation); 250 cells from five fields of
vision (n=50) were analyzed.
Statistical analysis
All data are presented as the mean ± standard
deviation. Statistical analysis was assessed by Kruskal-Wallis
followed by Dunn's post hoc test for multiple comparison test;
Student's t-test was used to compare two groups.
Clinicopathological characteristics were compared by χ2.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Association between PGP9.5 expression
level and clinicopathological features of breast cancer
The 92 clinical specimens of breast cancer were
collected and classified according to the age of the patients,
tumor size and histological grade (Table I; Table
SI); the tissue samples were used to examine PGP9.5 expression
by immunohistochemical analysis (Fig.
S1); PGP9.5 was used as a neuronal marker. The results showed
that the positive expression rate of PGP9.5 was not associated with
the age or tumor size of patients with breast cancer (both
P>0.05), whereas it was positively related with the pathological
grade of breast cancer (P<0.05).
Construction of recombinant NGF
lentiviral overexpression plasmids
Genomic DNA from MDA-MB231 and T-47D breast cancer
cells was extracted and used to construct recombinant pMD18-T-NGF
plasmids, which were verified by PCR and sequencing, and used as a
template to construct NGF-FLAG using primers containing FLAG
sequences and XbaI/BamHI enzyme loci. The
double-enzyme digested product was further linked to the vector
pCDH-CMV-MCS-EF1-puro to construct a recombinant NGF lentiviral
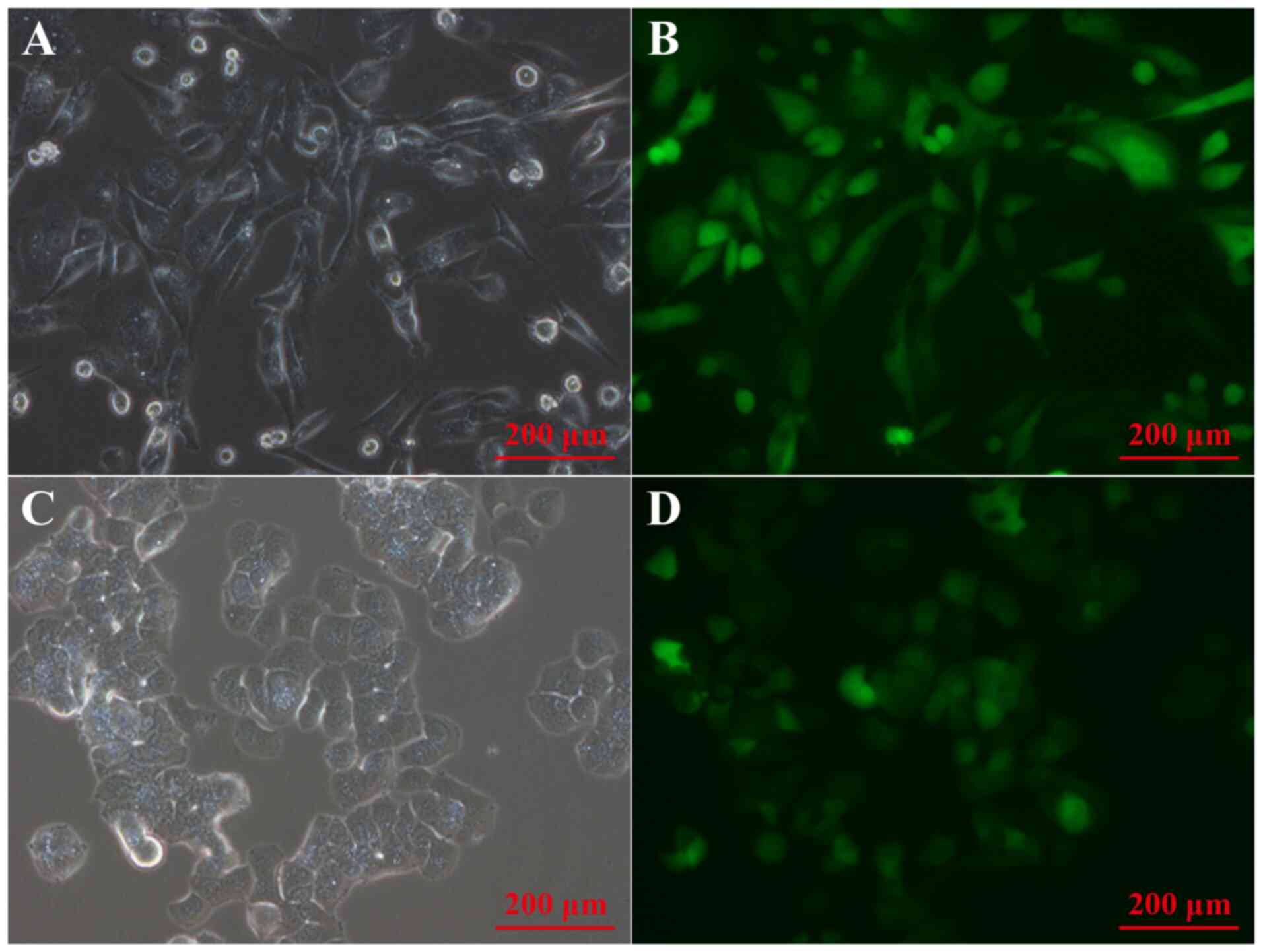
overexpression plasmid; the infection efficiency was verified by
fluorescence microscopy. Bright field and fluorescence microscopy
were used to observe the infection efficiency of NGF lentiviral
overexpression vector in breast cancer cells, and the results
showed that NGF lentiviral overexpression plasmids could
efficiently infect breast cancer cells (Fig. 1).
The puromycin-screened breast cancer cells,
including the overexpression (pCDH-NGF and pCDH-NGF-FLAG), vector
(pCDH-puro) and untransfected (MDA-MB231 and T-47D cells) groups,
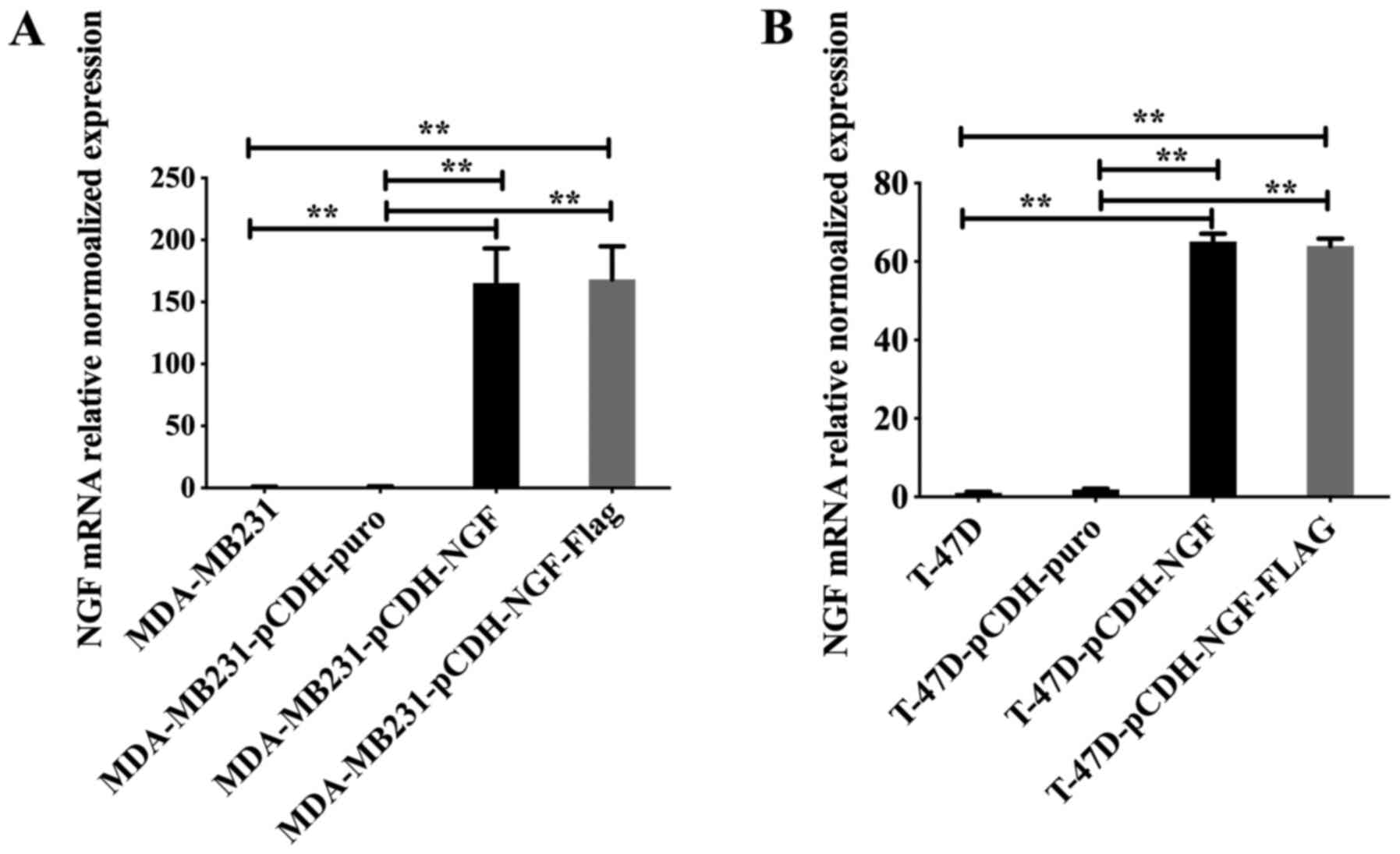
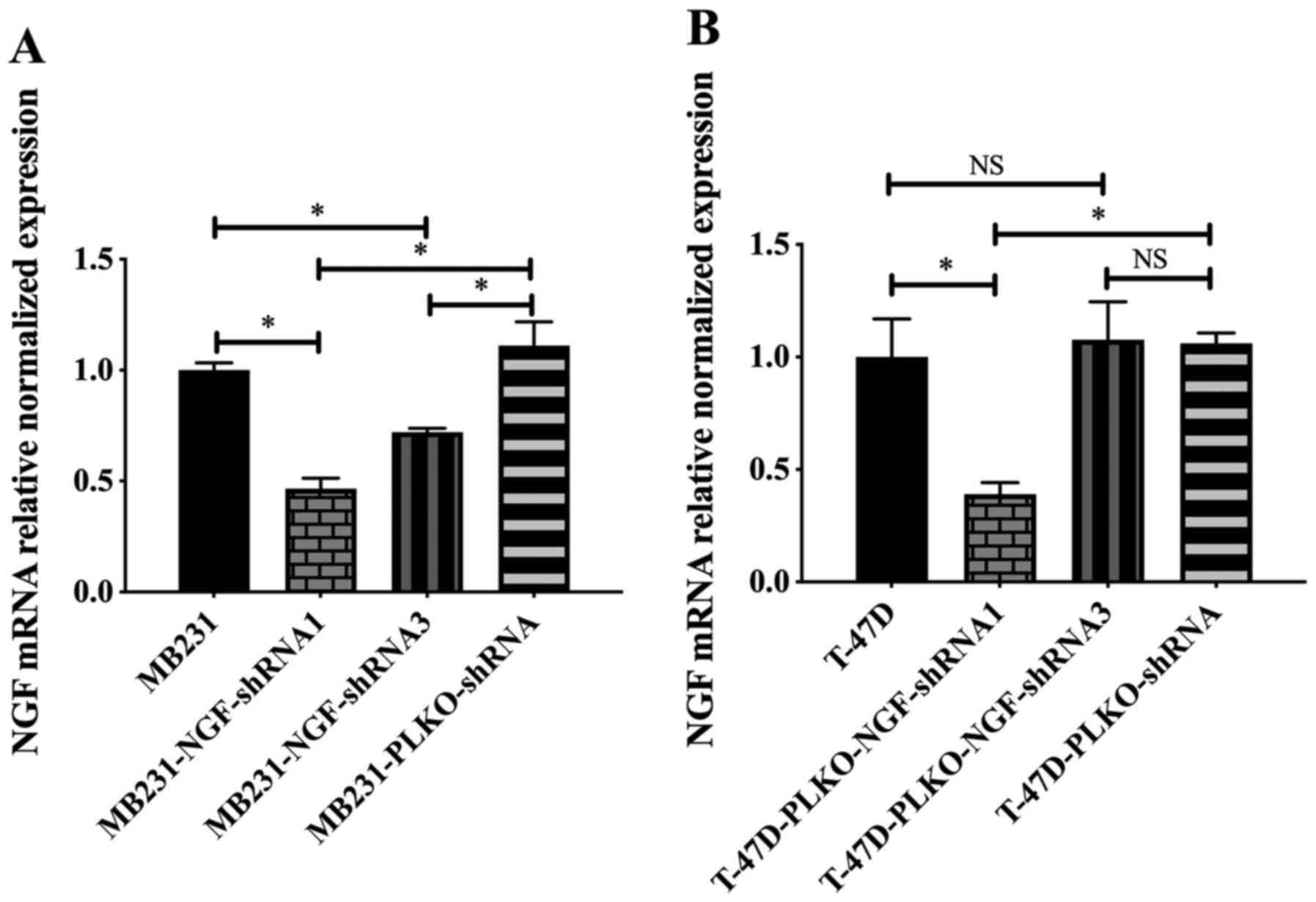
were collected and NGF mRNA expression levels were examined by qPCR
(Fig. 2). The results showed that
NGF mRNA expression levels in the overexpression groups were
significantly higher compared with those in the untransfected and
pCDH-puro groups (P<0.01), whereas there was no statistically
significant difference between the untransfected and pCDH-puro
groups (P>0.05) (Fig. 2),
suggesting the successful construction and infection of recombinant
NGF lentiviral overexpression plasmids.
Levels of NGF in breast cancer cells
transfected with overexpression plasmid
The cell supernatants from the untransfected
(MDA-MB-231 and T-47D), vector (MDA-MB231-pCHD-NGF-puro and
T-47D-pCDH-NGF-puro) and overexpression (MDA-MB231-pCHD-NGF-FLAG
and T-47D-pCDH-NGF-FLAG) groups were collected and analyzed by
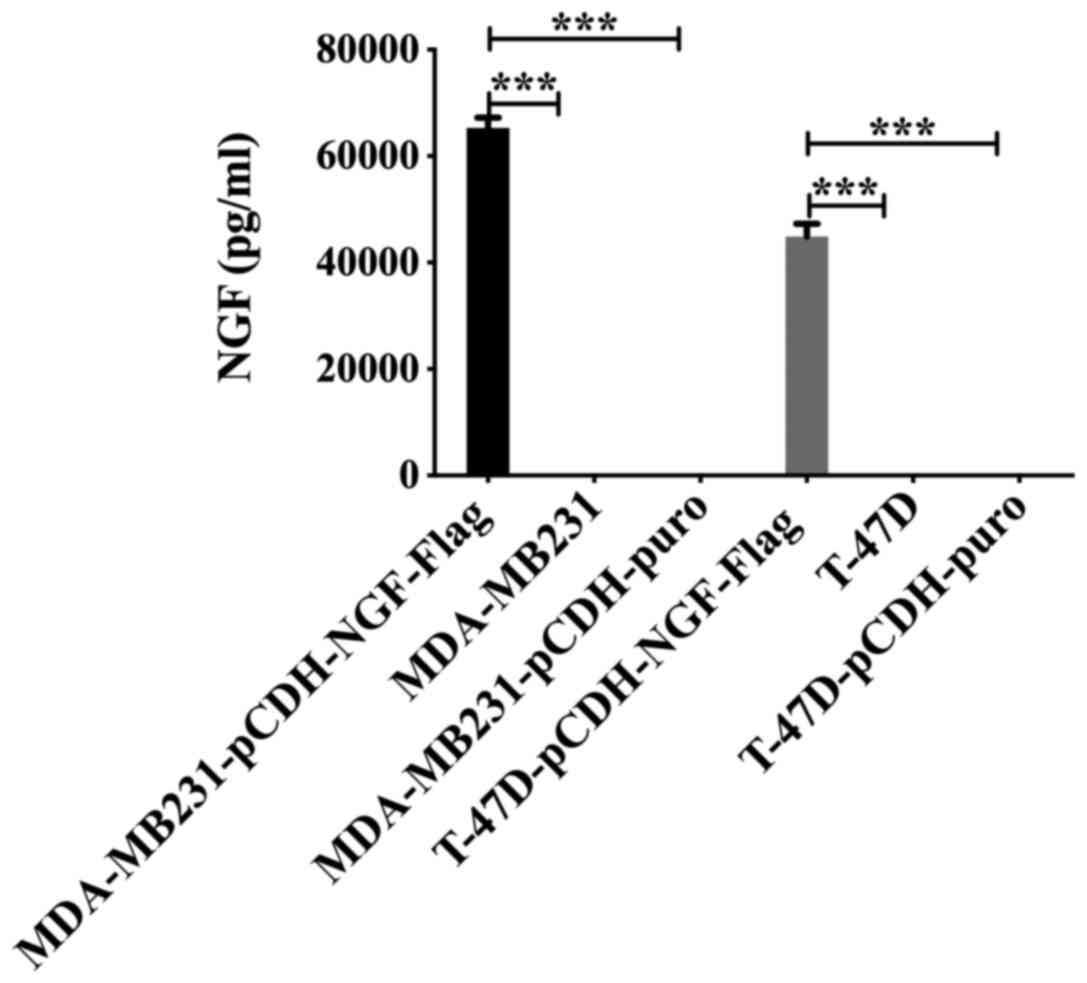
NGF-β ELISA. The results showed that the levels of NGF-β in the
overexpression groups were significantly higher than those in the
untransfected and pCDH-puro groups (P<0.001), whereas the levels
of NGF-β showed no statistically significant difference between the
untransfected and pCDH-puro groups (P>0.05) (Fig. 3).
The cell supernatants from the untransfected
(MDA-MB231 and T-47D), overexpression (MDA-MB231-pCHD-NGF-FLAG and
T-47D-pCDH-NGF-FLAG) and vector (MDA-MB231-pCHD-NGF-puro and
T-47D-pCDH-NGF-puro) groups were concentrated by ultrafiltration
centrifuge tubes, and β-actin (42.0 kDa; lower row) was used as the
internal reference. The results showed that NGF overexpression
breast cancer cell lines could secrete mature NGF protein (NGF,
13.5 kDa; FLAG, 1 kDa; FLAG binds to NGF, the total size is 14.5
kDa; upper row), whose expression levels were markedly higher than
those of the vector and untransfected groups (Fig. 4).
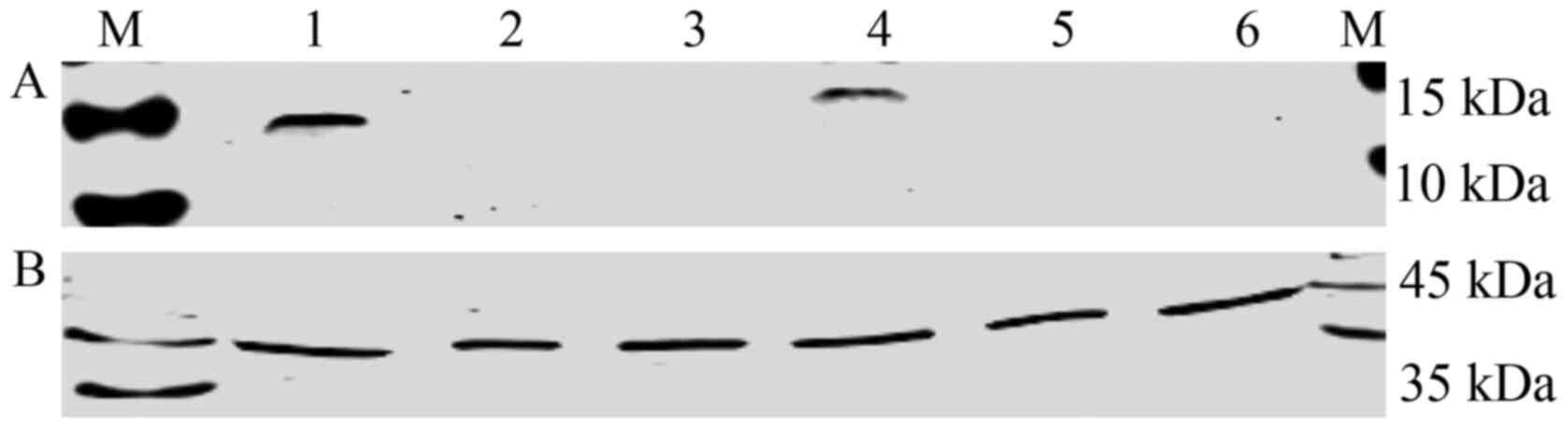 | Figure 4NGF protein expression levels in NGF
lentiviral overexpression vector-infected breast cancer cells using
anti-FLAG antibody. (A) Upper row, 1, MDA-MB231-pCDH-NGF-FLAG; 2,
MDA-MB231-pCDH-puro; 3, MDA-MB231; 4, T-47D-pCDH-NGF-FLAG; 5,
T-47D-pCDH-puro; 6, T-47D. Overexpression groups,
MDA-MB231-pCHD-NGF-FLAG and T-47D-pCDH-NGF-FLAG; untransfected
cells groups, MDA-MB231 and T-47D; pCDH-puro groups,
MDA-MB-231-pCHD-NGF-puro and T-47D-pCDH-NGF-puro. (B) Lower row,
β-actin was used as internal reference. M, marker; NGF, nerve
growth factor; puro, puromycin. |
Together, these results demonstrated that
MDA-MB231-pCHD-NGF-FLAG and T-47D-pCDH-NGF-FLAG-infected breast
cancer cells contained NGF overexpression plasmids.
Construction of recombinant NGF
lentiviral knockout plasmids
The results showed that NGF lentiviral knockout
plasmids could efficiently infect MDA-MB231 and T-47D breast cancer
cells (Fig. 5).
Flow cytometry was used to sort individual
CRISPR/Cas9-edited cells, which were subsequently verified by PCR
and agarose gel electrophoresis (Fig.
S2; 726 bp), and the PCR identified-cells were used for further
sequencing (Fig. 6). After
repetition of the described analysis three times, the genotype of
the breast cancer cell could be confirmed. As shown in Fig. 7, MDA-MB231-E3-A was a homozygous
(biallelic) knockout cell line, whereas MDA-MB231-E3-B was
heterozygous (monoallelic) knockout cell line.
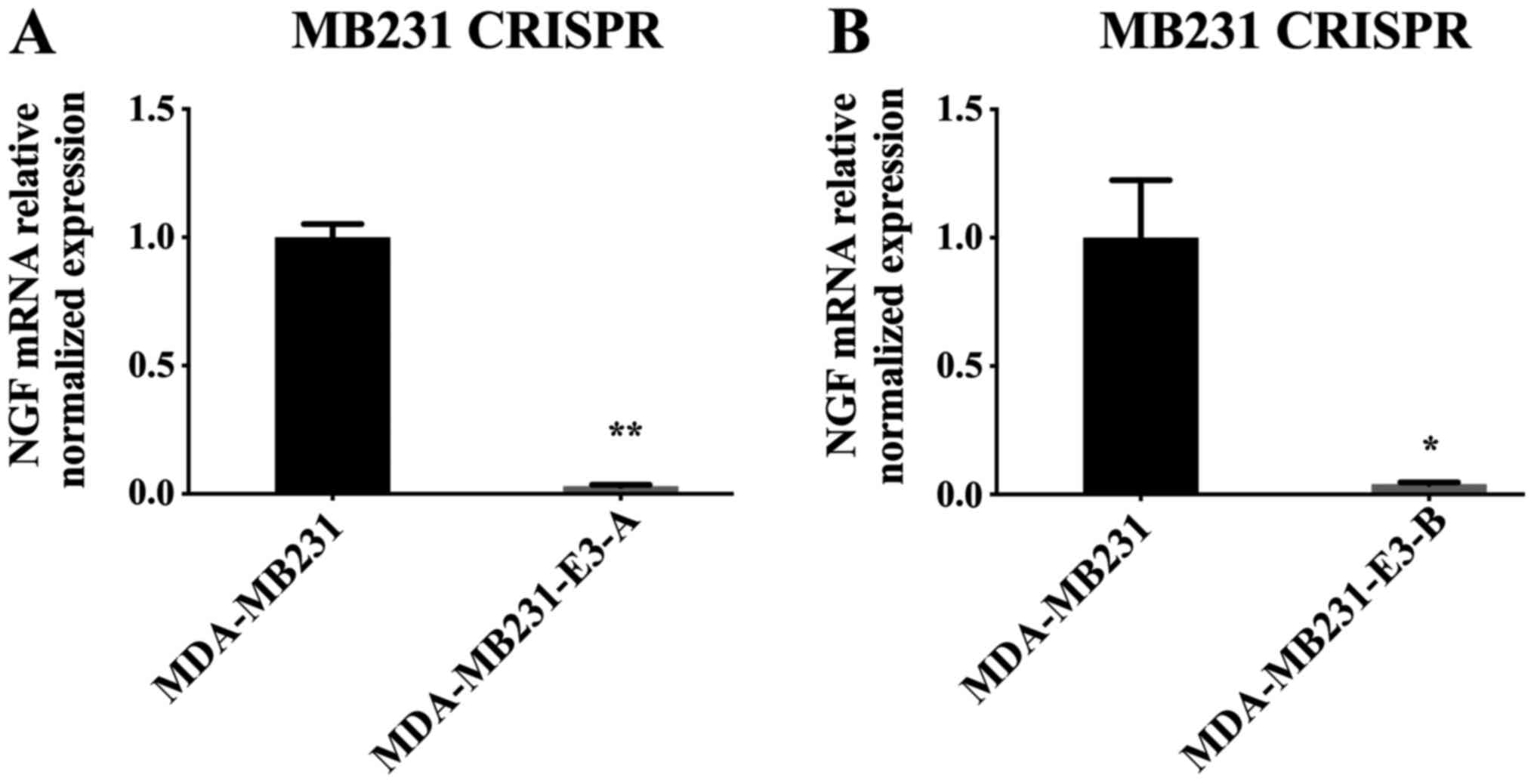
Puromycin-screened breast cancer cells were
collected, including the knockout (MDA-MB231-E3-A and -E3-B) and
untreated cells (MDA-MB231) groups, and their RNA was extracted and
used for RT-qPCR analysis. The results showed that the NGF mRNA
expression levels in the knockout groups were significantly lower
compared with those in the normal group (P<0.01; Fig. 8), suggesting the successful
construction and infection of recombinant NGF lentiviral knockout
plasmids.
Construction of recombinant shRNA-NGF
lentiviral plasmids
The designed NGF-shRNA1/2/3s and negative control
shRNA were designed and inserted into the lentiviral vector pLKO.1
using the primers listed in Table
III. pLKO-NGF-shRNA1/2/3 and control shRNA were verified by
ampicillin-resistant assay, double enzyme digestion assay and
sequenced. The results showed that shRNA2 failed to knockdown NGF,
thus only shRNA1 and 3 were used in subsequent experiments.
Puromycin-screened breast cancer cells, including
the silenced (MDA-MB-231-NGF-shRNA1 and -shRNA3, and
T-47D-NGF-shRNA1 and -shRNA3), negative control
(MDA-MB231-pLKO-shRNA and T-47D-pLKO-shRNA) and untreated
(MDA-MB231 and T-47D) groups, were examined by qPCR. The results
showed that NGF mRNA expression levels in the MDA-MB231-NGF-shRNA1,
T-47D-NGF-shRNA1 and MDA-MB231-NGF-shRNA3 groups were significantly
lower compared with those in the untreated cells and negative
control groups (P<0.05); whereas no statistically significant
differences were identified when T-47D-NGF-shRNA3 was compared with
untreated and negative control groups (P>0.05) (Fig. 9), which suggested the successful
construction and infection of the recombinant shRNA-NGF lentiviral
plasmids MDA-MB231-NGF-shRNA1 and T-47D-NGF-shRNA1.
Levels of NGF in knockout and
knockdown MDA-MB231 breast cancer cells
Culture supernatants from the knockout
(MDA-MB231-E3-A and -E3-B), silenced (MDA-MB231-pLKO-NGF-shRNA1),
negative control (MDA-MB231-pLKO-shRNA) and untreated (MDA-MB231)
groups were collected and analyzed using an NGF-β ELISA kit. The
results showed that the levels of NGF-β in the knockout and shRNA
groups were significantly lower than those in the negative control
groups (P<0.05), whereas the levels of NGF-β in the knockout
groups were lower than those in the shRNA silenced group
(P<0.001) (Fig. 10).
Detection of cell viability in breast
cancer cells transfected with the different recombinant
plasmids
The overexpression (MDA-MB231-pCDH-NGF-FLAG),
knockout (MDA-MB231-E3-A), silenced (MDA-MB231-pLKO-NGF-shRNA1),
negative control (MDA-MB231-pCDH-NGF-puro, lentiCRISPRv-NGF-gRNA
and MDA-MB231-pLKO-NGF-shRNA) and untreated (MDA-MB231) groups were
used to examine the effects on the viability of breast cancer cells
transfected with different recombinant plasmids. The results showed
that the viability of cells in the NGF overexpression group was
significantly higher compared with the negative control group at
24, 48, 72 and 96 h (P<0.05), whereas the viability of the NGF
knockout and silenced groups was lower than that of the negative
control groups at 24 or 48 h (P<0.05) and showed no
statistically significant difference at 72 or 96 h (Fig. 11).
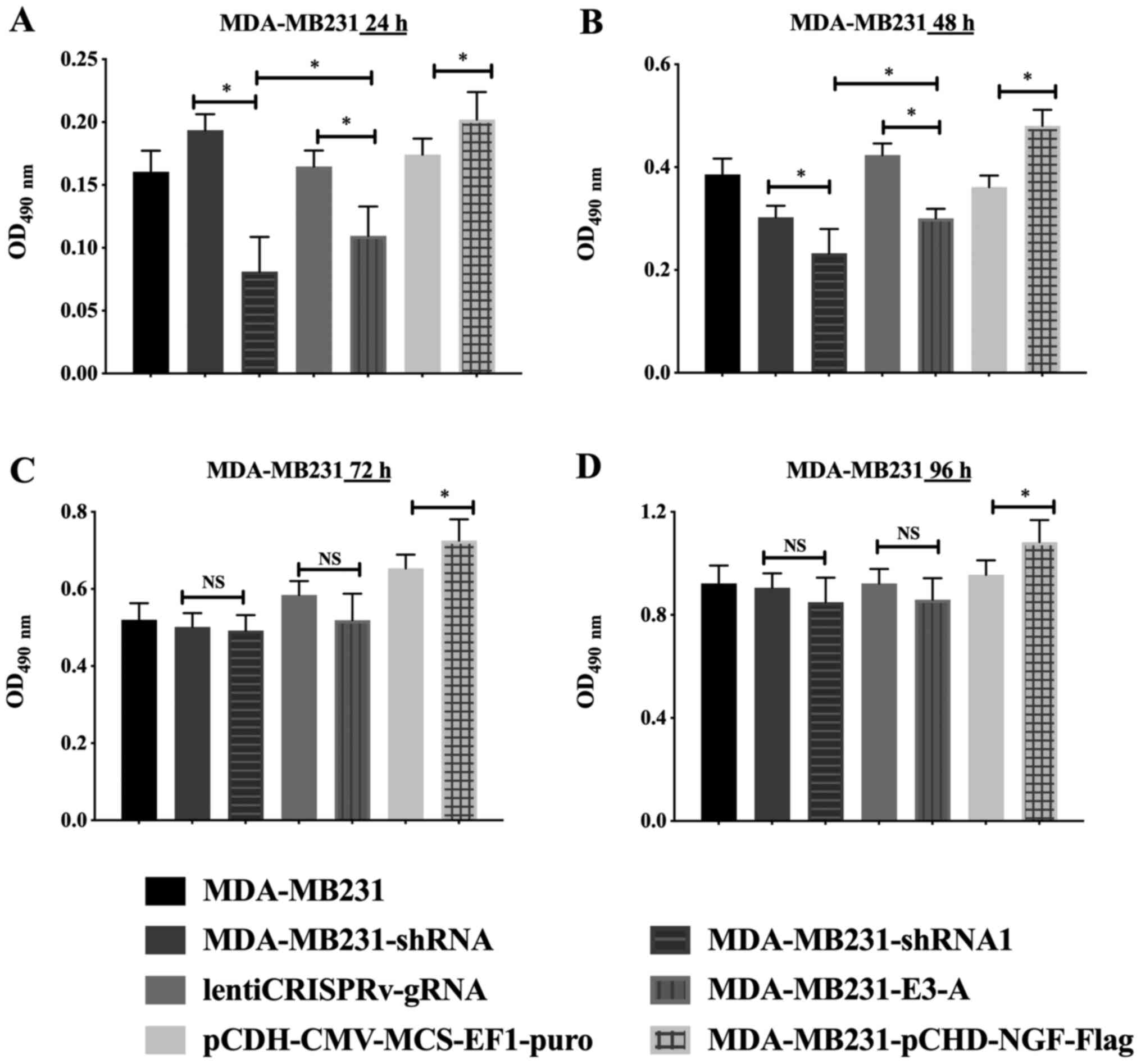 | Figure 11Detection of breast cancer cell
viability in cells with different recombinant plasmids.
Overexpression group, MDA-MB231-pCDH-NGF-FLAG; overexpression
negative group, MDA-MB231-pCDH-NGF-puro; knockout group,
MDA-MB231-E3-A; knockout negative group, lentiCRISPRv-NGF-gRNA;
silent group, MDA-MB231-pLKO-NGF-shRNA1; silent negative group,
MDA-MB231-pLKO-NGF-shRNA; normal group, MDA-MB231. The effects on
the viability of breast cancer cells transfected with different
recombinant plasmids at (A) 24, (B) 48, (C) 72 and (D) 96 h were
detected. Data are presented as the mean ± SD of three independent
experiments. *P<0.05. NGF, nerve growth factor;
shRNA, short hairpin RNA; gRNA, guide RNA; puro, puromycin; OD,
optical density. |
Influence of NGF on neuronal
differentiation activity of PC12 cells
The classic neuronal cell line, PC12, was cultured
and treated with cell supernatants containing 10, 50 and 100 ng/ml
NGF from NGF-overexpressing MDA-MB231 cells. The results showed
that the maximum protrusion length, and the number of protrusions
and the maximum diameter of cells were increased with the
increasing NGF concentrations (Fig.
12; Table V), which suggested
that PC12 cells could be altered by NGF in a
concentration-dependent manner.
 | Table VInfluence of NGF on the formation of
PC12 cell protrusion (n=50, cell numbers). |
Table V
Influence of NGF on the formation of
PC12 cell protrusion (n=50, cell numbers).
| Group | Number of
protrusions | Maximum diameter
(µm) | Longest protrusion
length (µm) |
|---|
| Control | 0.6 | 6.5 | 2.5 |
| NGF (10 ng/ml) | 3.0a | 7.9 | 5.1a |
| NGF (50 ng/ml) | 3.52a | 9.3a | 8.16a |
| NGF (100
ng/ml) | 3.8a | 11.2a | 13.54a |
Discussion
Nerve fibers, like blood vessels and lymphatics, are
spread throughout the body. Numerous studies have reported that
cancer angiogenesis and lymphangiogenesis occur in the cancer
microenvironment (29-32),
but the potential biological effects of cancer-related neurogenesis
have been rarely reported. Tumor cells can use a large number of
factors secreted by nerve fibers to create microenvironments to
help them survive and proliferate, such as epinephrine,
catecholamines and adrenaline (7,33-38).
At the same time, tumor cells can stimulate the production of
neurons by secreting neurotrophic factors and axonal guiding
molecules (11). The interaction
between tumor cells and their microenvironment promotes tumor
development (39). In general, if
the tumor is diagnosed early, surgical resection of the primary
lesion can be used for an effective cure. Tumor cells can invade
surrounding tissues and develop into invasive diseases, leading to
severe consequences. The majority of cancer-related deaths are
closely associated with the formation of invasive tumor tissues.
Numerous cytokines and soluble components, such as IL-1β, IL-6,
IL-17 and IL-32 in the tumor microenvironment affect tumorigenesis,
and the microenvironment is continually changing according to the
evolution of the tumor. Recruitment of a variety of cytokines to
the surrounding environment can help to form the secretory
environment of soluble factors, and also assist in the regeneration
of blood vessels and the lymphatic system (40-47).
In the early 1970s, Folkman et al (48) first discovered the tumor
angiogenesis ability. Since then, numerous studies have attempted
to determine the mechanism of angiogenesis and develop
corresponding treatments (49,50).
Angiogenesis markedly affects the development of the tumor and is
involved in the development of in situ invasive tumors.
Vascular endothelial growth factor (VEGF) A and its receptor 2
(VEGFR2; also known as FLK1) play an important role in it (51). Currently, treatments based on VEGF
can effectively improve the survival time of patients with a
malignant tumor, but only for a few months (52). Lymphangiogenesis is an important
cause of tumor cell invasion (53).
Previous studies have shown that lymphangiogenic factors affect the
proliferation of tumor cells, and the specific marker of lymphatic
angiogenesis is LYVE-1 (54-58).
In addition, VEGF-c and VEGF-d play a role in lymph node metastasis
in multiple human tumor cells, which demonstrates the importance of
this factor in tumor metastasis (54,58,59).
Similar to angiogenesis and lymphangiogenesis,
neurogenesis also occurs in tumor cells. Although previous studies
have demonstrated that bladder (59,60),
prostate (61), breast (62) and pancreas (63) cancers contain nerve endings, only in
prostate cancer have newborn axons been identified (64). Numerous studies have shown that
neuropeptides and neurotransmitters in the tumor microenvironment
affect neurogenesis (61-64).
Early studies suggested that nerve fibers guide the
migration of the invading nerve cells (63). Other studies have found that the
nervous system regulates numerous secreted molecules associated
with the development of tumors. For example, neurotransmitters are
associated with the immune suppression of tumors and induce changes
in angiogenesis and vascular density (65-67).
The presence of neurons in the tumor tends to lead to a poor
prognosis (68). Besides, the
nervous system and the tumor are mutually beneficial, since tumor
cells can secrete nerve molecules (69-71)
and axonal guide molecules (72) to
stimulate the nerve endings to penetrate the tumor.
Previous data showed that NGF serves crucial
regulatory roles in the development, differentiation, growth and
regeneration of neurons (73). In
the present study, 92 clinical cases of breast cancer were
collected, and the presence of neurons in the breast cancer
microenvironment was verified by immunohistochemistry. To explore
the role of NGF in cancer-related neurogenesis, recombinant NGF
lentiviral overexpression, knockout and silencing plasmids were
constructed. Furthermore, whether NGF affected neuron growth was
preliminary confirmed, and the results showed that the maximum
length, and number of protrusions and maximum diameter were
increased with the increase in NGF concentration.
For further direction, since previous studies
reported that as a classical tumor angiogenesis regulatory factor,
VEGF also plays a role similar to neurotrophic factor and axon
guidance factor, and is involved in the processes of axon growth,
neuronal cell migration, axon guidance and neural connection
(51,74). VEGF not only affects the growth of
primary cortical neurons as a neurotrophic factor, but also
stimulates the growth of axons and neurons in dorsal root ganglia
and regulates the migration of neural crest cells and astrocytoma
cells (51,75,76).
However, whether it is involved in cancer-related neurogenesis
remains unknown. Multiple differential expression plasmids of VEGF
will be constructed to detect if it is a downstream target of NGF.
Since there are few studies regarding the role of NGF in
cancer-associated neurogenesis and cancer-related axonogenesis
(2,7,9,11), the
differential expression plasmids constructed in the present study
may provide an important basis for further studies to elucidate its
role in this phenomenon.
Supplementary Material
Tissue samples were used to examine
PGP9.5 expression by immunohistochemistry. PGP9.5 was used as a
neuronal marker. (A) Normal breast tissue. (B and C) Ductal
carcinoma in situ. (D) Invasive ductal carcinoma.
Magnification, x40.
Preliminary selection of
CRISPR/Cas9-edited cell by PCR and gel electrophoresis. Flow
cytometry-sorted cells were preliminary selected by PCR and 2%
agarose gel electrophoresis (visualized by ethidium bromide), and
CRISPR/Cas9-edited cells were verified by sequencing. M, marker
(DL2000 DNA ladder); 1-16, PCR production (726 bp).
Characteristics of patients.
Acknowledgements
The authors would like to thank Mr. Ding Zhang (The
First College of Clinical Medical Science, China Three Gorges
University; Yichang, China) for providing the patients' breast
cancer samples.
Funding
Funding: Financial support for this project was provided by the
Hubei Office of Education Foundation (grant no. Q20151204)
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL, PW, JZ and CY performed the experiments. JW
conceived the project and analyzed the results. LZ drafted the
manuscript and analyzed the results. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Approval was received from the Ethics Committee of
The First College of Clinical Medical Science, China Three Gorges
University (Yichang, China), and written consent from patients were
obtained prior to using the breast tissues for research related
activities, including paper publication.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interest.
References
|
1
|
Hutchings C, Phillips JA and Djamgoz MBA:
Nerve input to tumours: Pathophysiological consequences of a
dynamic relationship. Biochim Biophys Acta Rev Cancer.
1874(188411)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cervantes-Villagrana RD, Albores-Garcia D,
Cervantes-Villagrana AR and García-Acevez SJ: Tumor-induced
neurogenesis and immune evasion as targets of innovative
anti-cancer therapies. Signal Transduct Target Ther.
5(99)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jiang SH, Hu LP, Wang X, Li J and Zhang
ZG: Neurotransmitters: Emerging targets in cancer. Oncogene.
39:503–515. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhu Y, Zhang GN, Shi Y, Cui L, Leng XF and
Huang JM: Perineural invasion in cervical cancer: Pay attention to
the indications of nerve-sparing radical hysterectomy. Ann Transl
Med. 7(203)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Magnon C, Hall SJ, Lin J, Xue X, Gerber L,
Freedland SJ and Frenette PS: Autonomic nerve development
contributes to prostate cancer progression. Science.
341(1236361)2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Deborde S, Omelchenko T, Lyubchik A, Zhou
Y, He S, McNamara WF, Chernichenko N, Lee SY, Barajas F, Chen CH,
et al: Schwann cells induce cancer cell dispersion and invasion. J
Clin Invest. 126:1538–1554. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Ayala GE, Dai H, Powell M, Li R, Ding Y,
Wheeler TM, Shine D, Kadmon D, Thompson T, Miles BJ, et al:
Cancer-related axonogenesis and neurogenesis in prostate cancer.
Clin Cancer Res. 14:7593–7603. 2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Dai H, Li R, Wheeler T, Ozen M, Ittmann M,
Anderson M, Wang Y, Rowley D, Younes M and Ayala GE: Enhanced
survival in perineural invasion of pancreatic cancer: An in vitro
approach. Hum Pathol. 38:299–307. 2007.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Albo D, Akay CL, Marshall CL, Wilks JA,
Verstovsek G, Liu H, Agarwal N, Berger DH and Ayala GE:
Neurogenesis in colorectal cancer is a marker of aggressive tumor
behavior and poor outcomes. Cancer. 117:4834–4845. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Huang D, Su S, Cui X, Shen X, Zeng Y, Wu
W, Chen J, Chen F, He C, Liu J, et al: Nerve fibers in breast
cancer tissues indicate aggressive tumor progression. Medicine
(Baltimore). 93(e172)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhao Q, Yang Y, Liang X, Du G, Liu L, Lu
L, Dong J, Han H and Zhang G: The clinicopathological significance
of neurogenesis in breast cancer. BMC Cancer.
14(484)2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chowdary PD, Che DL and Cui B:
Neurotrophin signaling via long-distance axonal transport. Annu Rev
Phys Chem. 63:571–594. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vanderhaeghen P and Cheng HJ: Guidance
molecules in axon pruning and cell death. Cold Spring Harb Perspect
Biol. 2(a001859)2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mancino M, Ametller E, Gascon P and
Almendro V: The neuronal influence on tumor progression. Biochim
Biophys Acta. 1816:105–118. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Adriaenssens E, Vanhecke E, Saule P,
Mougel A, Page A, Romon R, Nurcombe V, Le Bourhis X and Hondermarck
H: Nerve growth factor is a potential therapeutic target in breast
cancer. Cancer Res. 68:346–351. 2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mandelker DL, Yamashita K, Tokumaru Y,
Mimori K, Howard DL, Tanaka Y, Carvalho AL, Jiang WW, Park HL, Kim
MS, et al: PGP9.5 promoter methylation is an independent prognostic
factor for esophageal squamous cell carcinoma. Cancer Res.
65:4963–4968. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lee YM, Lee JY, Kim MJ, Bae HI, Park JY,
Kim SG and Kim DS: Hypomethylation of the protein gene product 9.5
promoter region in gallbladder cancer and its relationship with
clinicopathological features. Cancer Sci. 97:1205–1210.
2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wen W, Liu G, Jin K and Hu X: TGF-β1
induces PGP9.5 expression in CAFs to promote the growth of
colorectal cancer cells. Oncol Rep. 37:115–122. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ohta T and Fukuda M: Ubiquitin and breast
cancer. Oncogene. 23:2079–2088. 2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Dollé L, Adriaenssens E, El
Yazidi-Belkoura I, Le Bourhis X, Nurcombe V and Hondermarck H:
Nerve growth factor receptors and signaling in breast cancer. Curr
Cancer Drug Targets. 4:463–470. 2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bradshaw RA, Pundavela J, Biarc J,
Chalkley RJ, Burlingame AL and Hondermarck H: NGF and ProNGF:
Regulation of neuronal and neoplastic responses through receptor
signaling. Adv Biol Regul. 58:16–27. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liss MA, Gordon A, Morales B, Osann K,
Skarecky D, Lusch A, Zaldivar F and Ahlering TE: Urinary nerve
growth factor as an oncologic biomarker for prostate cancer
aggressiveness. Urol Oncol. 32:714–719. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mei F, Liu JY and Xue WC: Histological
grading of invasive breast carcinoma: Nottingham histological
grading system. Zhonghua Bing Li Xue Za Zhi. 48:659–664.
2019.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
24
|
Rao SJ, Rao JBM and Rao PJ:
Immunohistochemical analysis of stromal fibrocytes and
myofibroblasts to envision the invasion and lymph node metastasis
in oral squamous cell carcinoma. J Oral Maxillofac Pathol.
21:218–223. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Dikmen M: Comparison of the effects of
curcumin and RG108 on NGF-induced PC-12 Adh cell differentiation
and neurite outgrowth. J Med Food. 20:376–384. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Schimmelpfeng J, Weibezahn KF and
Dertinger H: Quantification of NGF-dependent neuronal
differentiation of PC-12 cells by means of neurofilament-L mRNA
expression and neuronal outgrowth. J Neurosci Methods. 139:299–306.
2004.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chan GKL, Hu WWH, Zheng ZX, Huang M, Lin
YXY, Wang CY, Gong AGW, Yang XY, Tsim KWK and Dong TTX: Quercetin
potentiates the NGF-induced effects in cultured PC 12 cells:
Identification by HerboChips showing a binding with NGF. Evid Based
Complement Alternat Med. 1502457:2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cai S, Cheng X, Liu Y, Lin Z, Zeng W, Yang
C, Liu L, Chukwuebuka OA and Li W: EYA1 promotes tumor angiogenesis
by activating the PI3K pathway in colorectal cancer. Exp Cell Res.
367:37–46. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ernst BP, Mikstas C, Stover T, Stauber R
and Strieth S: Association of eIF4E and SPARC expression with
lymphangiogenesis and lymph node metastasis in hypopharyngeal
cancer. Anticancer Res. 38:699–706. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wang J, Huang Y, Zhang J, Xing B, Xuan W,
Wang H, Huang H, Yang J and Tang J: NRP-2 in tumor
lymphangiogenesis and lymphatic metastasis. Cancer Lett.
418:176–184. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zanotto-Filho A, Rajamanickam S, Loranc E,
Masamsetti VP, Gorthi A, Romero JC, Tonapi S, Gonçalves RM, Reddick
RL, Benavides R, et al: Sorafenib improves alkylating therapy by
blocking induced inflammation, invasion and angiogenesis in breast
cancer cells. Cancer Lett. 425:101–115. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xia Y, Wei Y, Li ZY, Cai XY, Zhang LL,
Dong XR, Zhang S, Zhang RG, Meng R, Zhu F and Wu G: Catecholamines
contribute to the neovascularization of lung cancer via
tumor-associated macrophages. Brain Behav Immun. 81:111–121.
2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kim-Fuchs C, Le CP, Pimentel MA,
Shackleford D, Ferrari D, Angst E, Hollande F and Sloan EK: Chronic
stress accelerates pancreatic cancer growth and invasion: A
critical role for beta-adrenergic signaling in the pancreatic
microenvironment. Brain Behav Immun. 40:40–47. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lamkin DM, Sloan EK, Patel AJ, Chiang BS,
Pimentel MA, Ma JC, Arevalo JM, Morizono K and Cole SW: Chronic
stress enhances progression of acute lymphoblastic leukemia via
β-adrenergic signaling. Brain Behav Immun. 26:635–641.
2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu Y, Yu X and Zhuang J: Epinephrine
stimulates cell proliferation and induces chemoresistance in
myeloma cells through the β-adrenoreceptor in vitro. Acta Haematol.
138:103–110. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang
D, Li T, Wang CZ, Tan YX, Ding J, et al: ADRB2 signaling promotes
HCC progression and sorafenib resistance by inhibiting autophagic
degradation of HIF1alpha. J Hepatol. 65:314–324. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Lu R, Fan C, Shangguan W, Liu Y, Li Y,
Shang Y, Yin D, Zhang S, Huang Q, Li X, et al: Neurons generated
from carcinoma stem cells support cancer progression. Signal
Transduct Target Ther. 2(16036)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Marusyk A and Polyak K: Tumor
heterogeneity: Causes and consequences. Biochim Biophys Acta.
1805:105–117. 2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
West NR, McCuaig S, Franchini F and Powrie
F: Emerging cytokine networks in colorectal cancer. Nat Rev
Immunol. 15:615–629. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Mantovani A, Barajon I and Garlanda C:
IL-1 and IL-1 regulatory pathways in cancer progression and
therapy. Immunol Rev. 281:57–61. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Berraondo P, Sanmamed MF, Ochoa MC,
Etxeberria I, Aznar MA, Pérez-Gracia JL, Rodríguez-Ruiz ME,
Ponz-Sarvise M, Castañón E and Melero I: Cytokines in clinical
cancer immunotherapy. Br J Cancer. 120:6–15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Galdiero MR, Marone G and Mantovani A:
Cancer inflammation and cytokines. Cold Spring Harb Perspect Biol.
10(a028662)2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Najafi M, Goradel NH, Farhood B, Salehi E,
Solhjoo S, Toolee H, Kharazinejad E and Mortezaee K: Tumor
microenvironment: Interactions and therapy. J Cell Physiol.
234:5700–5721. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yao M, Brummer G, Acevedo D and Cheng N:
Cytokine regulation of metastasis and tumorigenicity. Adv Cancer
Res. 132:265–367. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Hong JT, Son DJ, Lee CK, Yoon DY, Lee DH
and Park MH: Interleukin 32, inflammation and cancer. Pharmacol
Ther. 174:127–137. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Setrerrahmane S and Xu H: Tumor-related
interleukins: Old validated targets for new anti-cancer drug
development. Mol Cancer. 16(153)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Folkman J, Merler E, Abernathy C and
Williams G: Isolation of a tumor factor responsible for
angiogenesis. J Exp Med. 133:275–288. 1971.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Fischer C, Jonckx B, Mazzone M, Zacchigna
S, Loges S, Pattarini L, Chorianopoulos E, Liesenborghs L, Koch M,
De Mol M, et al: Anti-PlGF inhibits growth of
VEGF(R)-inhibitor-resistant tumors without affecting healthy
vessels. Cell. 131:463–475. 2007.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Fischer C, Mazzone M, Jonckx B and
Carmeliet P: FLT1 and its ligands VEGFB and PlGF: Drug targets for
anti-angiogenic therapy? Nat Rev Cancer. 8:942–956. 2008.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ellis LM and Hicklin DJ: VEGF-targeted
therapy: Mechanisms of anti-tumour activity. Nat Rev Cancer.
8:579–591. 2008.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Alitalo K, Tammela T and Petrova TV:
Lymphangiogenesis in development and human disease. Nature.
438:946–953. 2005.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Stacker SA, Baldwin ME and Achen MG: The
role of tumor lymphangiogenesis in metastatic spread. FASEB J.
16:922–934. 2002.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lala PK, Nandi P and Majumder M: Roles of
prostaglandins in tumor-associated lymphangiogenesis with special
reference to breast cancer. Cancer Metastasis Rev. 37:369–384.
2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Dieterich LC and Detmar M: Tumor
lymphangiogenesis and new drug development. Adv Drug Deliv Rev.
99:148–160. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Garnier L, Gkountidi AO and Hugues S:
Tumor-associated lymphatic vessel features and immunomodulatory
functions. Front Immunol. 10(720)2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Paduch R: The role of lymphangiogenesis
and angiogenesis in tumor metastasis. Cell Oncol (Dordr).
39:397–410. 2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Achen MG and Stacker SA: Tumor
lymphangiogenesis and metastatic spread-new players begin to
emerge. Int J Cancer. 119:1755–1760. 2006.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Seifert P, Benedic M and Effert P: Nerve
fibers in tumors of the human urinary bladder. Virchows Arch.
440:291–297. 2002.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Seifert P and Spitznas M: Axons in human
choroidal melanoma suggest the participation of nerves in the
control of these tumors. Am J Ophthalmol. 133:711–713.
2002.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ventura S, Pennefather J and Mitchelson F:
Cholinergic innervation and function in the prostate gland.
Pharmacol Ther. 94:93–112. 2002.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Mitchell BS, Schumacher U, Stauber VV and
Kaiserling E: Are breast tumours innervated? Immunohistological
investigations using antibodies against the neuronal marker protein
gene product 9.5 (PGP 9.5) in benign and malignant breast lesions.
Eur J Cancer. 30A:1100–1103. 1994.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Kayahara M, Nakagawara H, Kitagawa H and
Ohta T: The nature of neural invasion by pancreatic cancer.
Pancreas. 35:218–223. 2007.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Ayala GE, Wheeler TM, Shine HD, Schmelz M,
Frolov A, Chakraborty S and Rowley D: In vitro dorsal root ganglia
and human prostate cell line interaction: Redefining perineural
invasion in prostate cancer. Prostate. 49:213–223. 2001.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Godbout JP and Glaser R: Stress-induced
immune dysregulation: Implications for wound healing, infectious
disease and cancer. J Neuroimmune Pharmacol. 1:421–427.
2006.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lee JW, Shahzad MM, Lin YG, Armaiz-Pena G,
Mangala LS, Han HD, Kim HS, Nam EJ, Jennings NB, Halder J, et al:
Surgical stress promotes tumor growth in ovarian carcinoma. Clin
Cancer Res. 15:2695–2702. 2009.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Thaker PH, Han LY, Kamat AA, Arevalo JM,
Takahashi R, Lu C, Jennings NB, Armaiz-Pena G, Bankson JA, Ravoori
M, et al: Chronic stress promotes tumor growth and angiogenesis in
a mouse model of ovarian carcinoma. Nat Med. 12:939–944.
2006.PubMed/NCBI View
Article : Google Scholar
|
|
68
|
Entschladen F, Drell TL IV, Lang K, Joseph
J and Zaenker KS: Tumour-cell migration, invasion, and metastasis:
Navigation by neurotransmitters. Lancet Oncol. 5:254–258.
2004.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Geldof AA, Van Haarst EP and Newling DW:
Neurotrophic factors in prostate and prostatic cancer. Prostate
Cancer Prostatic Dis. 1:236–241. 1998.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Ricci A, Greco S, Mariotta S, Felici L,
Bronzetti E, Cavazzana A, Cardillo G, Amenta F, Bisetti A and
Barbolini G: Neurotrophins and neurotrophin receptors in human lung
cancer. Am J Respir Cell Mol Biol. 25:439–446. 2001.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Dollé L, El Yazidi-Belkoura I,
Adriaenssens E, Nurcombe V and Hondermarck H: Nerve growth factor
overexpression and autocrine loop in breast cancer cells. Oncogene.
22:5592–5601. 2003.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Chédotal A, Kerjan G and Moreau-Fauvarque
C: The brain within the tumor: New roles for axon guidance
molecules in cancers. Cell Death Differ. 12:1044–1056.
2005.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Sofroniew MV, Howe CL and Mobley WC: Nerve
growth factor signaling, neuroprotection, and neural repair. Annu
Rev Neurosci. 24:1217–1281. 2001.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Melincovici CS, Boşca AB, Şuşman S,
Mărginean M, Mihu C, Istrate M, Moldovan IM, Roman AL and Mihu CM:
Vascular endothelial growth factor (VEGF)-key factor in normal and
pathological angiogenesis. Rom J Morphol Embryol. 59:455–467.
2018.PubMed/NCBI
|
|
75
|
Deyama S, Bang E, Kato T, Li XY and Duman
RS: Neurotrophic and antidepressant actions of brain-derived
neurotrophic factor require vascular endothelial growth factor.
Biol Psychiatry. 86:143–152. 2019.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Kutcher ME, Klagsbrun M and Mamluk R: VEGF
is required for the maintenance of dorsal root ganglia blood
vessels but not neurons during development. FASEB J. 18:1952–1954.
2004.PubMed/NCBI View Article : Google Scholar
|