Introduction
Inflammation is an immune response to infections and
tissue injury. An appropriate inflammatory response is
indispensable in protecting the body from internal and external
factors (1). However, excessive and
irreversible inflammation causes destruction of normal tissues,
which may result in diseases such as cancer, sepsis,
atherosclerosis and autoimmunity (2).
Lipopolysaccharide (LPS) is a naturally-derived
glycolipid compound that is commonly used to evaluate the
anti-inflammatory effects of macrophages in vitro. As one of
the cell membrane components of gram-negative bacteria (3), LPS is recognized by toll-like receptor
4 and stimulates mitogen-activated protein kinase (MAPK) and
nuclear factor-kappa B (NF-κB) signaling, key pathways of the
inflammatory response (4). As a
result, LPS signaling promotes the secretion of inflammatory
mediators such as inducible nitric oxide synthase (iNOS),
cyclooxygenase-2 (COX-2) and nitric oxide (NO) and induces the
expression of inflammatory cytokines such as interleukin (IL)-6,
IL-1β and tumor necrosis factor-alpha (TNF-α) (5). Additionally, the antioxidant effects
of upregulating nuclear factor erythroid 2-related factor 2 (Nrf2)
and heme oxygenase-1 (HO-1) are known to suppress the expression of
various LPS-induced inflammatory factors. HO-1 is a protein
associated with stress, ischemia and inflammatory conditions, and
primarily exhibits protective, anti-inflammatory, antioxidant and
anti-proliferative effects (6).
Therefore, the anti-inflammatory mechanisms of MAPKs and NF-κB, and
the antioxidant effects of HO-1 expression, are common research
targets for the treatment of inflammation.
A variety of diseases share the same underlying
pathophysiological mechanisms of inflammation, and research into
natural medicines to treat inflammation has gained considerable
popularity. Fritillaria thunbergii Miquel (FTM) is located
in the stem of Fritillaria thunbergii, a herbal medicine
that is commonly used in East Asian regions including Korea, China
and Japan. FTM (or ‘Jeol-pae-mo’ in Korean) has traditionally been
used to effectively reduce heat, remove phlegm and prevent coughing
(7). In a recent study, FTM was
shown to inhibit the expression of cytokines following PMA/A23187
stimulation of HMC-1 cells, and to inhibit cytokine expression in
an acute inflammatory BALB/c mouse model (8). In addition, peimine and peiminie, the
main active ingredients of FTM, were found to improve DNCB-induced
atopy (9,10). However, to the best of our
knowledge, there are no studies of the LPS-induced inflammatory
factors and antioxidant effects of the extract and fractions of FTM
in macrophages.
In the present study, the potential
anti-inflammatory effects of FTM extract and fractions were
investigated through the verification of NO and prostaglandin
E2 (PGE2) expression. To improve our
understanding of the anti-inflammatory properties of FTM, its
effects on the expression of MAPKs, NF-κB, pro-inflammatory
cytokines and HO-1 were also investigated.
Materials and methods
Reagents
Dulbecco's modified Eagle's medium (DMEM) was
obtained from Welgene, Inc., Gyeongsan, Korea. LPS,
dimethylsulfoxide (DMSO), fetal bovine serum (FBS) and
penicillin/streptomycin (P/S) were procured from Gibco; Thermo
Thermo Fisher Scientific, Inc.
C27H45NO3:
(3β,5α,6α,22β)-Cevane-3,6,20-triol (peimine) and
C27H43NO3 (peiminine) were
supplied by Abcam. The Cell Counting Kit-8 (CCK-8) was purchased
from Dojindo Molecular Technologies, Inc. Griess reagent,
2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) assay
kit (cat. no. CS0790), the bicinchoninic acid assay (BCA) kit,
protease inhibitor, and phosphatase inhibitor 2 and 3 cocktails
were supplied by Sigma-Aldrich (Merck KGaA). IL-1β, IL-6 and TNF-α
enzyme-linked immunosorbent assay (ELISA) kits were obtained from
BD Biosciences and the PGE2 ELISA kit was supplied by
R&D Systems, Inc. The reverse transcriptase and
SYBR® Green reagents were purchased from Invitrogen
(Thermo Fisher Scientific, Inc.) and Taq polymerase was purchased
from Kapa Biosystems (Roche Diagnostics). PCR primers were designed
by GenoTech Corp. The anti-iNOS (cat. no. sc-651), anti-COX-2 (cat.
no. sc-1746), anti-Nrf2 (cat. no. sc-722), anti-actin (cat. no.
sc-8432) and anti-lamin B (cat. no. sc6216) primary antibodies, and
the enhanced chemiluminescence (ECL) reagent were supplied by Santa
Cruz Biotechnology, Inc. The anti-HO-1 (cat. no. GTX61906) antibody
was obtained from GeneTex, Inc. Anti-NF-κB (cat. no. 8242S),
anti-extracellular signal-regulated kinase 1/2 (ERK; cat. no.
4695S), anti-c-Jun N-terminal kinase (JNK; cat. no. 9258S),
anti-p38 (cat. no. 9212L), anti-phosphorylated (p)-NF-κB (cat. no.
3033S), anti-p-ERK (cat. no. 4370S), anti-p-JNK (cat. no. 4668S)
and anti-p-p38 (cat. no. 4511S) antibodies were supplied by Cell
Signaling Technology, Inc. Horseradish peroxidase (HRP)-conjugated
secondary antibodies (cat. nos. 115-035-062 and 111-035-045) were
obtained from Jackson ImmunoResearch Laboratories, Inc. All
chemicals and reagents were of cell culture grade.
FTM preparation
FTM was obtained from Omniherb and verified by
Professor Yungmin Bu of the Herbology Laboratory, College of Korean
Medicine, Kyung Hee University (Seoul, Korea); FTM voucher
specimens are stored in the medicine storage room of the Anatomy
Laboratory, College of Korean Medicine, Kyung Hee University. For
extraction with methanol (MtOH), 100 g FTM was stored in 1 l MtOH
(80%) for 1 week. The extract was concentrated and lyophilized to
obtain 10.1 g of powder (yield ratio, 10.1%). For fractionation,
800 g FTM was immersed in 3 l MtOH (80%) for another week. The
extract was filtered using filter paper and subsequently
concentrated. The concentrate was sequentially fractionated using
hexane, chloroform and ethyl acetate according to the polarity of
the solvent. Briefly, 250 ml hexane was added to 500 ml of the
water fraction and sufficiently mixed; the water and hexane
fractions were separated, and 250 ml hexane was added to the water
fraction once again; this sequence was repeated three times.
Chloroform and ethyl acetate were then sequentially added to
separate and collect each fraction, which was then concentrated,
dried and used to assess activity. The hexane fraction was not
extracted. Finally, 2.23 g chloroform (yield ratio, 0.28%) and 0.41
g ethyl acetate (yield ratio, 0.05%) were extracted, resulting in a
final aqueous solution fraction of 11 g (yield). To extract FTM
with ethanol, 100 g FTM was extracted by immersion in 1 L Et-OH
(80%) for 1 week. The extract was concentrated and lyophilized to
obtain 5.3 g powder (yield ratio, 5.3%). All extracts were stored
at -4˚C and dissolved in DMSO prior to use.
High-performance liquid chromatography
(HPLC) analysis
To qualitatively evaluate the FTM, HPLC
(Waters® 2695/UV detector 2487 system) was performed
using peimine and peiminine, constituent compounds of FTM. The
XBridge™ C18 column (250x6 mm, 5 µm) was used for analysis, with a
flow rate of 1 ml/min and a sample injection volume of 10 µl.
H2O and acetonitrile (pH 10) were used as the binary
mobile phase, and the detection wavelength of the chromatogram was
set to 220 nm.
Cell culture and viability assay
The RAW 264.7 murine macrophage cell line was
purchased from the Korean Cell Line Bank (Seoul, Korea). Cells were
cultured in DMEM containing 10% FBS and 1% P/S at 37˚C (5%
CO2, 95% humidity). To determine the effects of FTM on
cellular viability, RAW 264.7 cells (5x104 cells/well)
were seeded into 96-well culture plates, and cultured in serum-free
medium for 24 h in the presence or absence of FTM (12.5, 25, 50 and
100 µg/ml). Then, 10 µl CCK-8 reagent was added to each well and
the plates were incubated for an additional 2 h. Cell viability was
determined by absorbance measurement at a wavelength of 450 nm
(VersaMax microplate reader; Molecular Devices, LLC). The
absorbance values are expressed as a percentage of the non-treated
cells, and a value of <90% was considered to indicate
cytotoxicity. Serum-free medium was used to eliminate
serum-associated interference.
Nitric oxide (NO) production
To determine the effects of FTM on the production of
NO, RAW 264.7 the cells (5x104 cells/well) were seeded
into 96-well culture plates and cultured for 24 h in the presence
or absence of LPS (1 µg/ml) and/or FTM (12.5, 25 and 50 µg/ml). To
evaluate the expression of NO, 100 µl Griess reagent was added to
50 µl cell culture supernatant and allowed to react at room
temperature for 30 min. The absorbance was measured using an ELISA
reader at a wavelength of 540 nm, and NO levels were calculated
using a sodium nitrite standard curve.
ELISA
To investigate the effects of FTM on the release of
inflammatory cytokines and PGE2, RAW 264.7 cells
(2x105 cells/well) were seeded into 24-well culture
plates and incubated with or without LPS (1 µg/ml) and/or FTM
(12.5, 25 and 50 µg/ml) for 24 h. Inflammatory cytokine (IL-6; cat
no. 555240; IL-1β, cat. no. 559603; and TNF-α, cat. no. 555268) and
PGE2 (cat. no. 555268; KGE004B) expression levels were
measured using the corresponding ELISA kit per the manufacturers'
instructions.
Western blot analysis
To investigate the effects of ethyl acetate fraction
of FTM (EAFM) on the expression of NF-κB and MAPKs, RAW 264.7 cells
(2x106 cells/well) were seeded into 60π dishes and
incubated with or without LPS (1 µg/ml) and/or EAFM (12.5, 25 and
50 µg/ml) for 30 min. To confirm the effect of EAFM on the
expression of iNOS and COX-2, RAW 264.7 cells (2x106
cells/well) were seeded into 6-well culture plates and incubated
with or without LPS (1 µg/ml) and/or EAFM (12.5, 25 and 50 µg/ml)
for 24 h. The cytoplasmic and nuclear proteins of RAW 264.7 cells
were extracted using NE-PER™ Nuclear and Cytoplasmic Extraction
Reagents (Invitrogen; Thermo Fisher Scientific, Inc.). The cells
were lysed in RIPA buffer (composition: 50 mM Tris-Cl, 150 mM NaCl,
1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS), and protease
inhibitor and phosphatase inhibitor 2 and 3 cocktails were added to
the extraction solutions. The protein concentration was quantified
(30 µg) using a BCA assay kit with a bovine serum albumin standard.
Equal amounts of protein was separated by 10% SDS-PAGE and
electrotransferred onto nitrocellulose membranes. The membranes
were blocked with 5% skim milk and incubated with primary
antibodies at 4˚C for 24 h (iNOS, dilution ratio: 1:1,000, 130 kDa;
COX-2, dilution ratio: 1:200, 70 kDa; β-actin, dilution ratio:
1:1,000, 42 kDa; p-ERK, dilution ratio: 1:1,000, 44, 42 kDa; ERK,
dilution ratio: 1:1,000, 44, 42 kDa; p-JNK, dilution ratio:
1:1,000, 54, 46 kDa; JNK, dilution ratio: 1:1,000, 54, 46 kDa;
p-p38, dilution ratio: 1:1,000, 40 kDa; p38, dilution ratio:
1:1,000, 40 kDa; NF-κB, dilution ratio: 1:500, 65 kDa; p-NF-κB,
dilution ratio: 1:500, 65 kDa; Lamin B, dilution ratio: 1:1,000, 67
kDa; Nrf2, dilution ratio: 1:400, 120 kDa; HO-1, dilution ratio:
1:2,000, 33 kDa). HRP-conjugated secondary antibodies were then
added and the membranes were incubated for 1 h at room temperature.
Protein expression was visualized with an ECL reagent. The density
of the bands was evaluated using the ImageJ software (version
1.51j8; National Institutes of Health) and the expression level of
each protein was normalized to that of β-actin.
Reverse transcription PCR
To investigate the effects of EAFM on the expression
of iNOS, COX-2 and pro-inflammatory cytokines, RAW 264.7 cells
(2x106 cells/well) were seeded into 6-well culture
plates and incubated with or without LPS (1 µg/ml) and/or EAFM
(12.5, 25, 50 µg/ml) for 6 h. The expression of HO-1 was treated
with EAFM (12.5, 25 and 50 µg/ml) for 24 h. Total cellular RNA was
extracted using RNAiso (Total RNA extraction reagent; Takara Bio,
Inc.) according to the manufacturers' protocol, and cDNA was
synthesized using reverse transcriptase. PCR was conducted using a
C1000 Touch™ Thermal Cycler (Bio-Rad Laboratories, Inc.). The PCR
cycles were as follows: 35-40 cycles of 1 min at 94˚C
(denaturation), 30 sec at 55-58˚C (annealing) and a 1 min 72˚C
(extension), using Taq polymerase. The sequences of the primers are
displayed in Table I. PCR products
were electrophoresed on a SYBR® Green stained agarose
gel; the band density was evaluated using ImageJ software and mRNA
expression was normalized to that of β-actin.
 | Table IPrimer sequences used for reverse
transcription PCR. |
Table I
Primer sequences used for reverse
transcription PCR.
| Gene name | Sequence
(5'-3') | Accession no. | Annealing
temperature, ˚C | Cycles | Base pairs |
|---|
| Nos2 (iNOS) | F:
CACCTTGGAGTTCACCCAGT | NM_010927.4 | 58 | 30 | 170 |
| | R:
ACCACTCGTACTTGGGATGC | NM_001313921.1 | | | |
| | | NM_001313922.1 | | | |
| Ptgs2 (COX-2) | F:
AGAAGGAAATGGCTGCAGAA | NM_011198.4 | 55 | 35 | 194 |
| | R:
GCTCGGCTTCCAGTATTGAG | | | | |
| Il6 (IL-6) | F:
AGTTGCCTTCTTGGGACTGA | NM_001314054.1 | 58 | 30 | 223 |
| | R:
TTCTGCAAGTGCATCATCGT | | | | |
| Tnf (TNF-α) | F:
GCAGAAGAGGCACTCCCCCA | NM_001278601.1 | 58 | 30 | 280 |
| | R:
GATCCATGCCGTTGGCCAGG | | | | |
| Il1b (IL-1β) | F:
GGCTGTGGAGAAGCTGTGGC | NM_008361.4 | 55 | 30 | 386 |
| | R:
GGGTGGGTGTGCCGTCTTTC | | | | |
| Hmox1 (HO-1) | F:
GAGAATGCTGAGTTCAT | NM_010442.2 | 58 | 25 | 548 |
| | R:
ATGTTGAGCAGGAAGGC | | | | |
| Actb (β-actin) | F:
TTCTACAATGAGCTGCGTGT | NM_007393 | 58 | 30 | 456 |
| | R:
CTCATAGCTCTTCTCCAGGG | | | | |
ABTS radical scavenging activity
To investigate the effects of EAFM on ABTS radical
scavenging activity, RAW 264.7 cells (2x106 cells/well)
were seeded into 6-well culture plates and incubated with or
without FTM (12.5, 25 and 50 µg/ml) for 24 h. The experiment was
carried out according to the manufacturer's protocol. The ABTS
latical scavenging ability was analyzed compared to RAW 264.7 cells
without treatment.
Statistical analysis
All experiments were independently conducted at
least three times. All data are expressed as the mean ±
standard error of the mean (SEM) and were analyzed using GraphPad
prism software (version 5.01; GraphPad Software, Inc.). The
significance of the experimental results was analyzed using one-way
ANOVA test, followed by Tukeys's multiple comparison post hoc
analysis.
Results
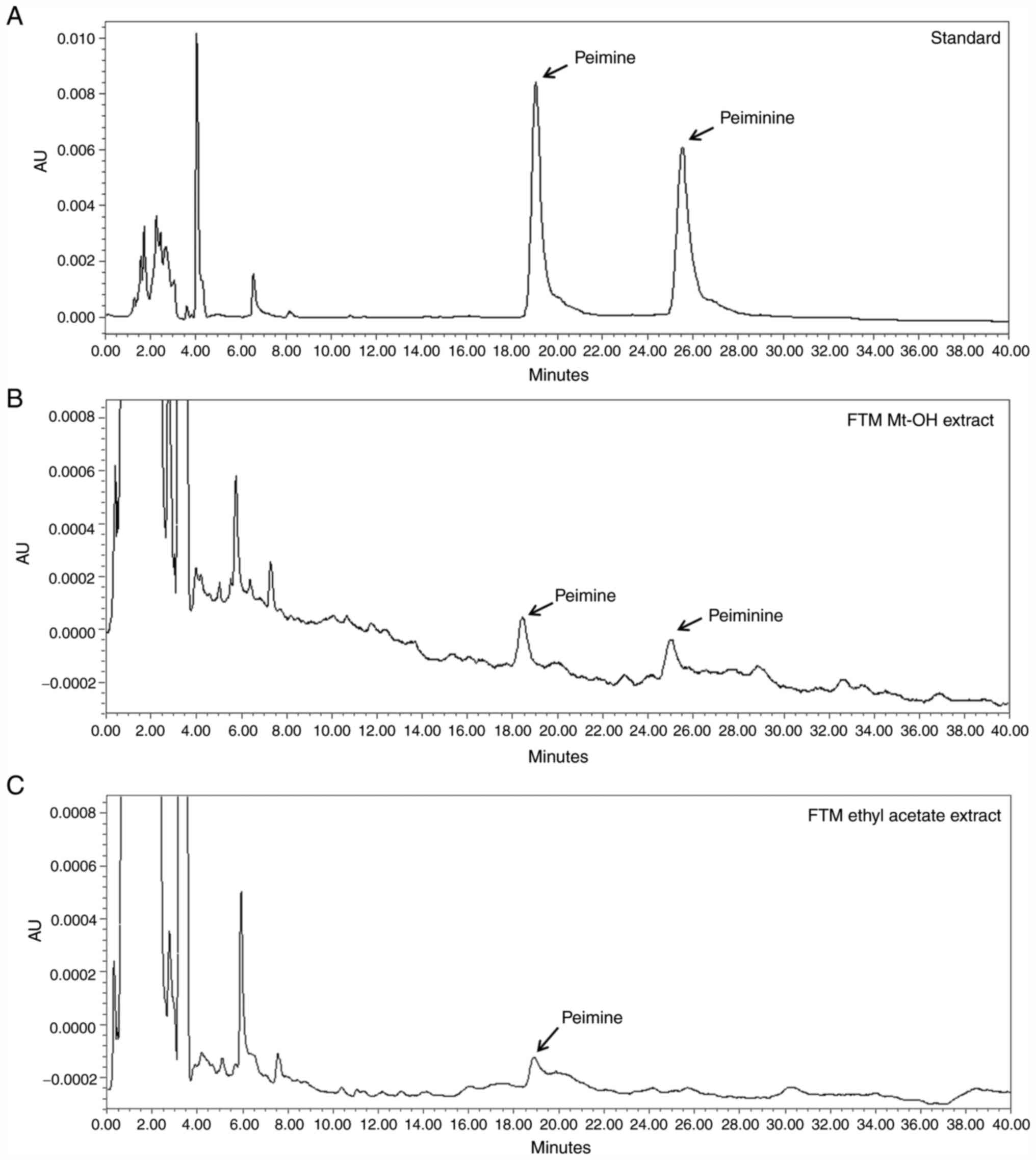
Qualitative analysis of FTM
Peimine and peiminine are bioactive marker compounds
of FTM (11,12). As shown in Fig. 1A and B, FTM Mt-OH extract revealed a number of
chromatogram peaks for 0-30 min, and peimine and peiminie were
identified at the same time points as for the standard; 19.607 and
26.316 min, respectively. As a result of examining the chromatogram
peak of EAFM, peimine was detected at 19.353 min, but peiminine was
not. (Fig. 1C).
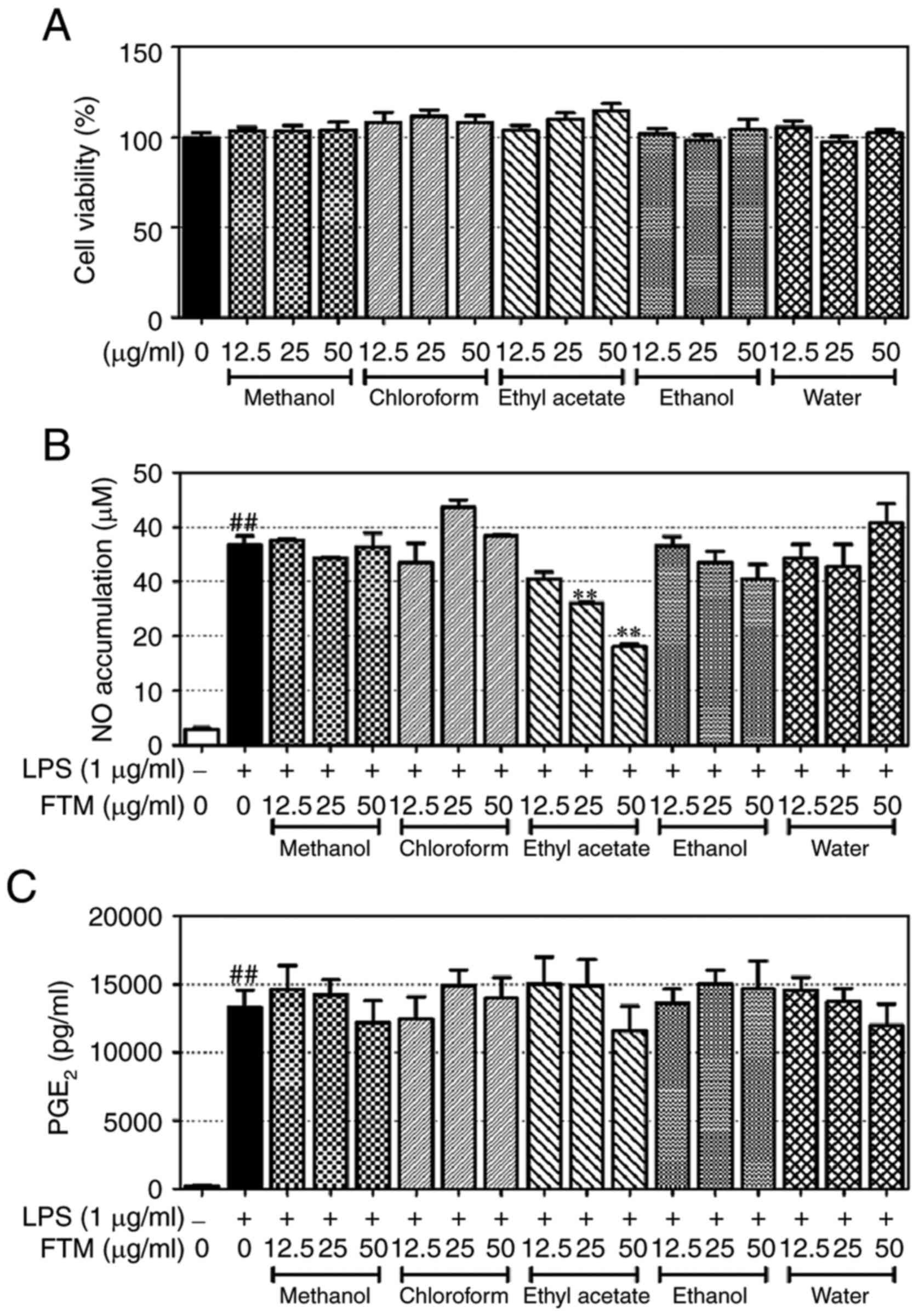
EAFM significantly inhibits NO
production
Prior to experimentation, the cytotoxicity of FTM
was verified using a CCK-8 kit. FTM extract and all fractions
showed no significant cytotoxicity at concentrations of 12.5, 25
and 50 µg/ml (Fig. 2A). As a
result, concerns regarding FTM cytotoxicity were eliminated. The
inhibitory effects of the FTM extract and fractions on NO
expression were then assessed using Griess reagent and ELISA. EAFM
was found to inhibit the generation of NO in a
concentration-dependent manner (Fig.
2B); none of the other extracts and fractions had a significant
effect on NO expression. Furthermore, none of the extracts and
fractions (including those of FTM) significantly altered the
expression of PGE2 (Fig.
2C).
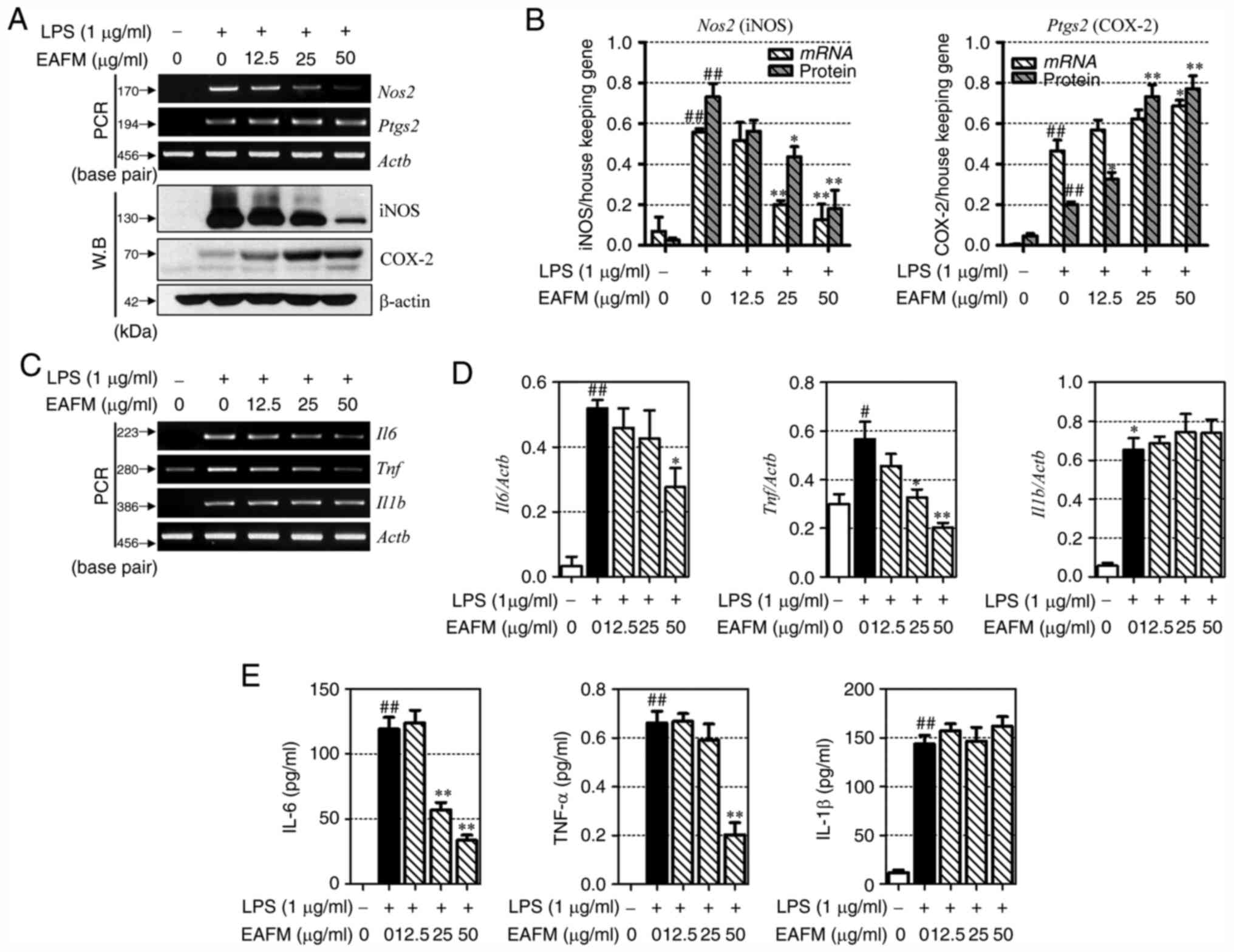
EAFM inhibits the expression of iNOS
and inflammatory cytokines
To investigate the anti-inflammatory effects of
EAFM, iNOS and COX-2 expression were verified at the mRNA and
protein levels. LPS stimulation of RAW 264.7 cells induced the
expression of iNOS and COX-2. EAFM suppressed iNOS expression in a
concentration-dependent manner, but increased the expression of
COX-2 (Fig. 3A and B). IL-6, TNF-α and IL-1β are
representative pro-inflammatory cytokines expressed in macrophages.
To confirm the inflammatory potential of EAFM, the protein levels
of these cytokines in the RAW 264.7 cell culture medium, as well as
the mRNA expression levels, were investigated. As shown in Fig. 3C-E, LPS stimulation upregulated the
expression of IL-6, TNF-α and IL-1β in RAW 264.7 cells. IL-6 and
TNF-α mRNA expression was subsequently suppressed by EAFM in a
concentration-dependent manner (Fig.
3C and D). However, EAFM did
not significantly affect the mRNA expression levels of IL-1β.
Furthermore, LPS induced the expression of pro-inflammatory
cytokines in the culture medium of RAW 264.7 cells, of which IL-6
and TNF-α, but not IL-1β, were significantly inhibited by EAFM
administration (Fig. 3E).
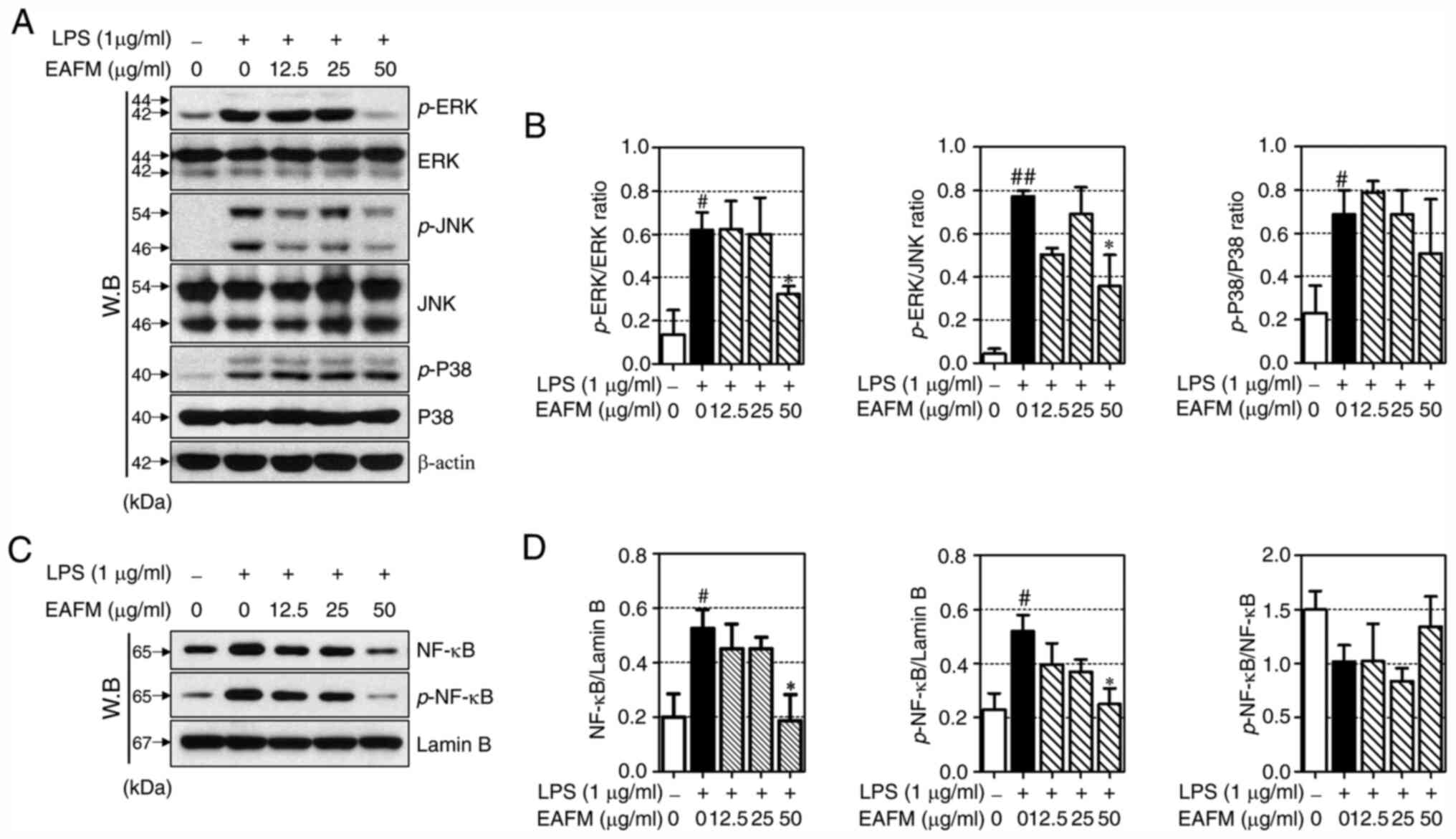
EAFM inhibits the phosphorylation of
ERK and JNK, and inhibits the nuclear translocation of NF-κB
To investigate the anti-inflammatory signaling
mechanisms of EAFM, the expression of MAPKs (ERK, JNK and p38) and
NF-κB was investigated by western blotting. LPS stimulation induced
the phosphorylation of ERK, JNK and p38; furthermore, treatment
with EAFM significantly inhibited ERK and JNK phosphorylation at a
concentration of 50 µg/ml (Fig. 4A
and B). LPS stimulation initiates
various inflammatory responses by promoting the nuclear
translocation of NF-κB. In the present study, LPS induced the
expression and phosphorylation of NF-κB in nuclear proteins.
Furthermore, EAFM was confirmed to markedly inhibit the expression
and phosphorylation of NF-κB at a concentration of 50 µg/ml
(Fig. 4C and D).
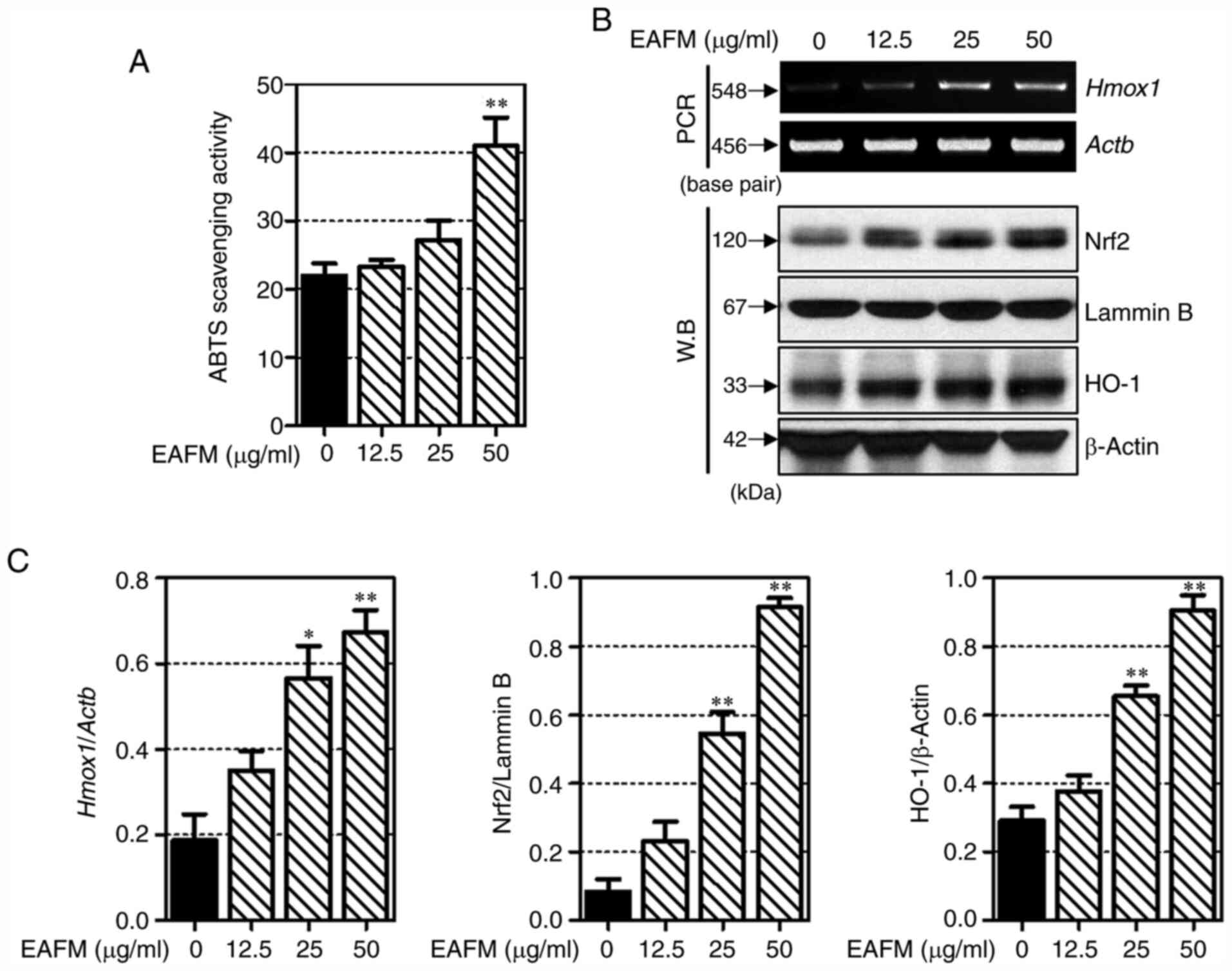
EAFM exerts antioxidant effects by
activating ABTS radical scavenging and inducing Nrf2 and HO-1
expression
Next, The ABTS radical scavenging ability of EAFM
was confirmed. As shown in Fig. 5A,
EAFM increased the ABTS radical scavenging ability in a
concentration-dependent manner. In particular, the EAFM of 100
µg/ml was significantly increased compared to the non-treated
cells. the mRNA expression levels of HO-1 were determined to
confirm that the anti-inflammatory properties of EAFM resulted from
an antioxidant effect. As shown in Fig.
5B and C, EAFM induces HO-1
expression at the mRNA level. The effect of EAFM on the expression
of Nrf2 (a transcription factor of HO-1) was verified by western
blotting. EAFM was also found to increase the expression of Nrf2
and HO-1 at the protein level. These results indicate that EAFM
prevents oxidative damage by promoting HO-1 expression in
macrophages.
Discussion
The aim of the present study was to reveal the
mechanisms by which EAFM regulates inflammation and oxidation. The
RAW 264.7 murine macrophage cell line is commonly used to verify
the anti-inflammatory effects of drugs, and to evaluate the
associated signaling pathways. Inflammatory mechanisms are complex
processes regulated by cytokine interactions and the induction of
various pro-inflammatory genes (13). Macrophages are distributed
throughout the human body and serve an important role in these
inflammatory process. Macrophages provide immediate defense against
pathogens prior to leukocyte migration, and LPS stimulation induces
the secretion of macrophage inflammatory mediators such as
interleukins, TNF-α, iNOS and COX-2(3). The pharmacological reduction of
LPS-induced inflammatory mediators is considered to effectively
alleviate various symptoms, including macrophage-associated
inflammatory reactions. Therefore, an LPS-induced RAW 264.7 cell
inflammation model was used in the present study.
NO is a well-known modulator of the inflammatory
response, and is important for the defense against infectious
organisms. However, it can also detrimentally effect host tissues.
NO reacts with molecular oxygen to generate reactive nitrogen
species, which can result in various modifications to cellular
function (14). As a result, NO is
considered to be an important factor of the inflammatory response.
The expression of NO is regulated by iNOS, which is not expressed
under normal conditions, but generated following the release of
inflammatory cytokines such as TNF-α and IL-1β. In the present
study, EAFM suppressed the expression of NO as well as iNOS mRNA
and protein (15,16). This indicates that the
NO-suppressive effect of EAFM is the result of iNOS regulation.
PGE2 is produced in macrophages and is
involved in vasodilation, pain and fever in the early stages of the
inflammatory response and COX-2 plays a major role in the
expression of PGE2 (17). In previous studies, COX-2 inhibitors
ablated PGE2 synthesis, and consequently suppressed
inflammation. In addition, LPS-induced hepatotoxicity was
alleviated in COX-2 gene-positive mice. Therefore, PGE2
and COX-2 have been used as anti-inflammatory indicators in
numerous studies. However, in the present study, in contrast to the
NO and iNOS results, EAFM had no effect on the expression of
PGE2 and increased COX-2 expression. The reason for the
existence of the difference in expression between COX-2 and PGE2 is
assumed to be as follows. PGE2 metabolism is first
catalyzed by hydroxyprostaglandin dehydrogenase (15-PGDH) (18). Previous studies have shown that
limited degradation of PGE2 through decreased expression of 15-PGDH
can lead to tumor growth (19),
whereas elevation of 15-PGDH in lung cancer cells inhibited the
expression of PGE2 (20). Probably, EAFM seems to control the
expression of PGE2 through upregulation of 15-PGDH
without affecting COX-2. However, additional research will have to
be conducted to test this hypothesis.
In addition, the reason why EAFM suppresses iNOS,
but not COX-2 expression is presumed to be as follows: The
intracellular expression pathways of iNOS and COX-2 are complex,
and the expression of these indicators depends on the degree of
promoter dependence between them. The iNOS promoter contains
cis-acting elements such as NF-κB, activator protein 1 (AP-1),
CCAAT/enhancer-binding protein beta (C/EBPβ) and signal transducer
and activator of transcription (Stat), whereas the COX-2 promoter
includes cis-acting components such as NF-κB, C/EBPβ and cis-acting
replication elements (CRE) elements (21,22).
The activities of different promoters vary depending on the cell
type and stimulant. In the present study, the COX-2 promoter was
confirmed to be less dependent on NF-κB than the iNOS promoter. As
the results of pro-inflammatory cytokine expression indicate, EAFM
inhibited IL-6 and TNF-α by suppressing NF-κB. These results
suggest that EAFM may not inhibit COX-2 expression by complementing
other promoters of COX-2, highlighting the need for further
investigation.
Cytokines are small secretory proteins that
facilitate intercellular communication. Pro-inflammatory cytokines
are produced by activated macrophages, and representative cytokines
such as IL-6, TNF-α and IL-1β, are involved in both acute and
chronic inflammatory signaling (13,23).
IL-6 is rapidly produced in response to infection and tissue
damage. In previous studies, IL-6-knockout animals displayed
reduced susceptibility to disease symptoms in a variety of
conditions, including Castleman's disease, rheumatoid arthritis and
inflammatory myopathies (24).
TNF-α is considered to be a master regulator of inflammatory
cytokine expression and the abnormal production of TNF-α is
associated with the development of various diseases, including
rheumatoid arthritis, Crohn's disease and atherosclerosis.
Therefore, TNF-α is a proposed target for the treatment of numerous
inflammatory diseases (25). IL-1β
is induced at the site of inflammation and generally promotes the
expression of PGE2 and COX-2(26). IL-1β injection induces fever,
headache, muscle and joint pain in humans by initiating
inflammatory reactions (27). In
the present study, EAFM significantly inhibited the expression of
IL-6 and TNF-α, indicating its ability to regulate acute
LPS-induced inflammatory responses. However, EAFM did not
significantly affect IL-1β expression, which supports previous
results suggesting that EAFM does not inhibit the subsequent
expression of COX-2 and PGE2.
The NF-κB pathway is one of the primary mechanisms
by which macrophages express pro-inflammatory factors (28). In its inactive state, NF-κB is bound
to inhibitor of NF-κB subunit α (IκB) in the cytoplasm; following
stimulation, IκB is degraded and NF-κB translocates to the nucleus
to initiate various inflammatory responses. MAPKs serve regulatory
roles in cellular proliferation and differentiation, and the
regulation of cellular responses to cytokines and stress. There are
three MAPK signaling pathways that involve ERK, JNK or p38, which
transmit information from the extracellular environment to the
nucleus. In addition, published studies indicate that MAPKs are
involved in the regulation of COX-2 and iNOS expression (29). In the present study, EAFM inhibited
both the nuclear transfer and phosphorylation of NF-κB, as well as
ERK and JNK phosphorylation. This indicates that the potential for
EAFM to inhibit NO and inflammatory cytokine production is mediated
by NF-κB, ERK and JNK. However, although EAFM inhibits the nuclear
potential and phosphorylation of NF-κB, there is a limitation that
it is not clear why it does not affect IL-1β.
There are several ways to measure the antioxidant
effect, and the results are slightly different. ABTS is a
relatively stable free radical and is widely used for measuring
antioxidant activity along with DPPH. This method can measure both
lipophilic or hypophilic substances (30). In this study, EAFM improved the ABTS
radical scavenging ability. These findings mean that EAFM improves
the ability to remove free radicals, which means that it has
antioxidant effects. The Nrf2/HO-1 axis is a major defense
mechanism against oxidative stress, where HO-1 expression converts
heme to biliverdin/bilirubin, carbon monoxide (CO) and free iron
(6). The generation of
biliverdin/bilirubin has a strong antioxidant effect, and CO also
has anti-inflammatory properties. In activated macrophages, HO-1
induction reduces the production of IL-6, TNF-α and IL-1β and HO-1
overexpression decreases the expression of iNOS, which is produced
following COX-2 initiation. NO and PGE2 production is
also suppressed. HO-1 is dependently regulated by Nrf2; following
nuclear translocation, Nrf2 binds to an antioxidant response
element, regulating antioxidant mechanisms and serving an important
role in the defense against oxidative stress. In the present study,
EAFM notably increased the expression of both Nrf2 and HO-1. These
results indicate that the antioxidant effect of EAFM, at least in
part, influences preceding anti-inflammatory mechanisms.
In conclusion, the results if the present study
confirm that EAFM suppresses the expression of NO and
pro-inflammatory cytokines by inhibiting MAPK/NF-κB signaling
pathways. Additionally, EAFM also exerts its antioxidant effects by
inducing HO-1 expression. The results of these experiments
highlight the potential of EAFM as a future treatment for
inflammatory diseases. The following limitations exist in this
study. i) Peimine and peiminine, known as the main components of
FTM, have been demonstrated in previous studies to have
anti-inflammatory effects (9,31).
However, referring to the results of the HPLC experiment, it was
confirmed that peiminie was not present in the constituents of
EAFM, and a very small amount of peimine was included. Therefore,
further studies are needed to determine which chemicals of EAFM
exhibit anti-inflammatory effects. ii) In this study, the
expression of COX-2 increased with EAFM treatment. Referring to the
anti-inflammatory effect of Xanthii fructus before (32), it showed anti-inflammatory effect
similar to the result of this study, but the expression of COX-2
increased as the drug was treated. It can be inferred that there is
some similarity between EAFM and Xanthii fructus, and it is
suggested that further research is needed in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YS conceptualized the study. JHK, MK and EYK
performed the experiments. SH, BK and HSJ statistically analyzed
the data. HSJ, KS and MWS interpreted the results. JHK drafted the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng
J, Li Y, Wang X and Zhao L: Inflammatory responses and
inflammation-associated diseases in organs. Oncotarget.
9:7204–7218. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Okin D and Medzhitov R: Evolution of
inflammatory diseases. Curr Biol. 22:R733–R740. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Alexander C and Rietschel ET: Bacterial
lipopolysaccharides and innate immunity. J Endotoxin Res.
7:167–202. 2001.PubMed/NCBI
|
|
4
|
Küper C, Beck FX and Neuhofer W: Toll-like
receptor 4 activates NF-κB and MAP kinase pathways to regulate
expression of proinflammatory COX-2 in renal medullary collecting
duct cells. Am J Physiol Renal Physiol. 302:F38–F46.
2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Aderem A and Ulevitch RJ: Toll-like
receptors in the induction of the innate immune response. Nature.
406:782–787. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Loboda A, Damulewicz M, Pyza E, Jozkowicz
A and Dulak J: Role of Nrf2/HO-1 system in development, oxidative
stress response and diseases: An evolutionarily conserved
mechanism. Cell Mol Life Sci. 73:3221–3247. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Herbology Editorial Committee of Korean
Medicine: Herbology. Younglimsa, Seoul, 2004 (In Korean).
|
|
8
|
Cho IH, Lee MJ, Kim JH, Han NY, Shin KW,
Sohn Y and Jung HS: Fritillaria ussuriensis extract inhibits the
production of inflammatory cytokine and MAPKs in mast cells. Biosci
Biotechnol Biochem. 75:1440–1445. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lim JM, Lee B, Min JH, Kim EY, Kim JH,
Hong S, Kim JJ, Sohn Y and Jung HS: Effect of peiminine on
DNCB-induced atopic dermatitis by inhibiting inflammatory cytokine
expression in vivo and in vitro. Int Immunopharmacol. 56:135–142.
2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Park JH, Lee B, Kim HK, Kim EY, Kim JH,
Min JH, Kim S, Sohn Y and Jung HS: Peimine inhibits the production
of proinflammatory cytokines through regulation of the
phosphorylation of NF-κB and MAPKs in HMC-1 cells. Pharmacogn Mag.
13 (Suppl 2):S359–S364. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
He J, He Y and Zhang AC: Determination and
visualization of peimine and peiminine content in Fritillaria
thunbergii bulbi treated by sulfur fumigation using
hyperspectral imaging with chemometrics. Molecules.
22(1402)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wu X, Chan SW, Ma J, Li P, Shaw PC and Lin
G: Investigation of association of chemical profiles with the
tracheobronchial relaxant activity of Chinese medicinal herb beimu
derived from various fritillaria species. J Ethnopharmacol.
210:39–46. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang JM and An J: Cytokines,
inflammation, and pain. Int Anesthesiol Clin. 45:27–37.
2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xue Q, Yan Y, Zhang R and Xiong H:
Regulation of iNOS on immune cells and its role in diseases. Int J
Mol Sci. 19(3805)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Coleman JW: Nitric oxide in immunity and
inflammation. Int Immunopharmacol. 1:1397–1406. 2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Korhonen R, Lahti A, Kankaanranta H and
Moilanen E: Nitric oxide production and signaling in inflammation.
Curr Drug Targets Inflamm Allergy. 4:471–479. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ricciotti E and FitzGerald GA:
Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol.
31:986–1000. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cho H and Tai HH: Inhibition of
NAD+-dependent 15-hydroxyprostaglandin dehydrogenase (15-PGDH) by
cyclooxygenase inhibitors and chemopreventive agents.
Prostaglandins Leukot Essent Fatty Acids. 67:461–465.
2002.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ding Y, Tong M, Liu S, Moscow JA and Tai
HH: NAD+-linked 15-hydroxyprostaglandin dehydrogenase (15-PGDH)
behaves as a tumor suppressor in lung cancer. Carcinogenesis.
26:65–72. 2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hazra S, Batra RK, Tai HH, Sharma S, Cui X
and Dubinett SM: Pioglitazone and rosiglitazone decrease
prostaglandin E2 in non-small-cell lung cancer cells by
up-regulating 15-hydroxyprostaglandin dehydrogenase. Mol Pharmacol.
71:1715–1720. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kang YJ, Wingerd BA, Arakawa T and Smith
WL: Cyclooxygenase-2 gene transcription in a macrophage model of
inflammation. J Immunol. 177:8111–8122. 2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lee AK, Sung SH, Kim YC and Kim SG:
Inhibition of lipopolysaccharide-inducible nitric oxide synthase,
TNF-alpha and COX-2 expression by sauchinone effects on
I-kappaBalpha phosphorylatio, C/EBP and AP-1 activation. Br J
Pharmacol. 139:11–20. 2003.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Feghali CA and Wright TM: Cytokines in
acute and chronic inflammation. Front Biosci. 2:d12–d26.
1997.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Tanaka T, Narazaki M and Kishimoto T: IL-6
in inflammation, immunity, and disease. Cold Spring Harb Perspect
Biol. 6(a016295)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Parameswaran N and Patial S: Tumor
necrosis factor-alpha signaling in macrophages. Crit Rev Eukaryot
Gene Expr. 20:87–103. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Dinarello CA: Proinflammatory cytokines.
Chest. 118:503–508. 2000.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Janik JE, Miller LL, Longo DL, Powers GC,
Urba WJ, Kopp WC, Gause BL, Curti BD, Fenton RG, Oppenheim JJ, et
al: Phase II trial of interleukin 1 alpha and indomethacin in
treatment of metastatic melanoma. J Natl Cancer Inst. 88:44–49.
1996.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lawrence T: The nuclear factor NF-kappaB
pathway in inflammation. Cold Spring Harb Perspect Biol.
1(a001651)2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kim EK and Choi EJ: Pathological roles of
MAPK signaling pathways in human diseases. Biochim Biophys Acta.
1802:396–405. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
More GK and Makola RT: In-vitro analysis
of free radical scavenging activities and suppression of
LPS-induced ROS production in macrophage cells by Solanum
sisymbriifolium extracts. Sci Rep. 10(6493)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yi PF, Wu YC, Dong HB, Guo Y, Wei Q, Zhang
C, Song Z, Qin QQ, Lv S, Wu SC and Fu BD: Peimine impairs
pro-inflammatory cytokine secretion through the inhibition of the
activation of NF-κB and MAPK in LPS-induced RAW264.7 macrophages.
Immunopharmacol Immunotoxicol. 35:567–572. 2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yeom M, Kim JH, Min JH, Hwang MK, Jung HS
and Sohn Y: Xanthii fructus inhibits inflammatory responses
in LPS-stimulated RAW 264.7 macrophages through suppressing NF-κB
and JNK/p38 MAPK. J Ethnopharmacol. 176:394–401. 2015.PubMed/NCBI View Article : Google Scholar
|